Abstract
We previously identified WRAP53 as an antisense transcript that regulates the p53 tumor suppressor. The WRAP53 gene also encodes a protein essential for Cajal body formation and involved in cellular trafficking of the survival of motor neuron complex, the telomerase enzyme and small Cajal body-specific RNAs to Cajal bodies. Here, we show that the WRAP53 protein is overexpressed in a variety of cancer cell lines of different origin and that WRAP53 overexpression promotes cellular transformation. Knockdown of the WRAP53 protein triggers massive apoptosis through the mitochondrial pathway, as demonstrated by Bax/Bak activation, loss of mitochondrial membrane potential and cytochrome c release. The apoptosis induced by WRAP53 knockdown could moreover be blocked by Bcl-2 overexpression. Interestingly, human tumor cells are more sensitive to WRAP53 depletion as compared with normal human cells indicating that cancer cells in particular depends on WRAP53 expression for their survival. In agreement with this, we found that high levels of WRAP53 correlate with poor prognosis of head and neck cancer. Together these observations propose a role of WRAP53 in carcinogenesis and identify WRAP53 as a novel molecular target for a large fraction of malignancies.
Keywords: WRAP53, Wdr79, TCAB1, mitochondrial-dependent apoptosis, oncogene, head and neck cancer
The WRAP53 gene is located on chromosome 17p13 and partly overlaps the p53 tumor suppressor gene in opposite direction. The name WRAP53 (for WD40-encoding RNA antisense to p53) was recently approved by HUGO Gene Nomenclature Committee as the official name of this gene (also denoted TCAB1 or WDR79). We found that transcription of WRAP53 gives rise to p53 antisense transcripts that regulates p53 mRNA and is required for p53 action upon DNA damage.1 WRAP53 transcripts can also be translated into the WRAP53 protein. This protein belongs to the WD40 protein family and is highly conserved during evolution. The WD40 family is a large family of proteins involved in important processes such as apoptosis, cell cycle regulation, proteasomal degradation and RNA metabolism. We found that the WRAP53 protein is an essential component for Cajal body maintenance and that without WRAP53 Cajal bodies collapse.2 Cajal bodies are nuclear organelles involved in a variety of nuclear functions including ribonucleoprotein maturation, spliceosome formation, histone mRNA processing, RNA polymerase assembly, telomerase biogenesis and histone gene transcription.3, 4, 5 We also showed that the WRAP53 protein interacts with the survival of motor neuron (SMN) protein, which is a key regulator of splicing. WRAP53 recruits the SMN complex from the cytoplasm to Cajal bodies in the nucleus by mediating interactions between SMN, importinβ and coilin.2 Recent studies also show that the WRAP53 protein bind certain RNA species in the nucleus called small Cajal body-specific (sca) RNAs and recruits them to Cajal bodies.6, 7 ScaRNAs mediate posttranscriptional modifications of splicing RNAs, which occurs in Cajal bodies and is important for the function of the splicing machinery. A well-known member of the scaRNA family is the telomerase RNA, which is part of the telomerase holoenzyme. The telomerase enzyme extends telomeres and is activated in the large majority of cancer cells (90%) as a way to escape senescence and making cancer cells immortal. The WRAP53 protein was also found to be a new subunit of the telomerase enzyme, essential for the recruitment of telomerase to Cajal bodies and for telomere elongation in human cancer cells.7
The WRAP53 gene has moreover been implicated in primary human cancers. Single-nucleotide polymorphisms (SNPs) in WRAP53 were found to be overrepresented in women with breast cancer, in particular estrogen receptor-negative breast cancer.8 The same SNPs were also associated with aggressive ovarian cancer.9 The SNPs are located in the coding region of WRAP53 and results in the amino acid change R68G, suggesting that alterations of the WRAP53 protein could contribute to cancer. Here, we describe for the first time that the WRAP53 protein is upregulated in cancer and that WRAP53 overexpression promotes cellular transformation. WRAP53 knockdown specifically triggers apoptosis in cancer cells and increased WRAP53 levels are associated with poor prognosis in head and neck cancer. Our findings highlight the impact of WRAP53 in cancer and exposes WRAP53 as a new interesting therapeutic target.
Results
WRAP53 expression is elevated in cancer cell lines
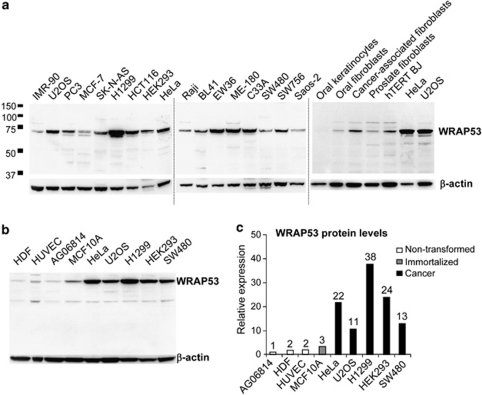
The WRAP53 protein has 548 amino acids and migrates as a 75 kDa species on SDS polyacrylamide gels. Western blot (WB) analysis of WRAP53 in a series of human cells showed ubiquitous expression of the protein (Figure 1a). Further examination of WRAP53 in non-transformed primary cells, in immortalized but noncancerous cells and in cancer cell lines revealed that WRAP53 protein expression is increased in immortalized cells and up to 20 times higher in cancer cells compared with primary cells (Figures 1b and c). These data suggest a role for WRAP53 in the pathogenesis of human cancer.
Figure 1.
WRAP53 is overexpressed in cancer cells. (a) WB analysis of WRAP53 in a panel of human cell lysates; including cancer cells (U2OS, PC3, MCF-7, SK-N-AS, H1299, HCT116, HEK293, HeLa, Raji, BL41, EW36, ME-180, C33A, SW480, SW756 and Saos-2), immortalized foreskin fibroblast (hTERT BJ) and primary cells (the rest). IMR-90 is a non-transformed lung fibroblast. β-Actin is used as loading control for all WB. (b) WB analysis of WRAP53 levels in non-transformed (HDF, HUVEC and AG06814), immortalized (MCF10A) and cancer (HeLa, U2OS, H1299, HEK293 and SW480) cells. (c) Densitometric quantifications of the relative WRAP53 levels in cancer versus non-transformed cells shown in (b). Levels of WRAP53 have been normalized against β-actin levels
WRAP53 overexpression transforms NIH 3T3 cells
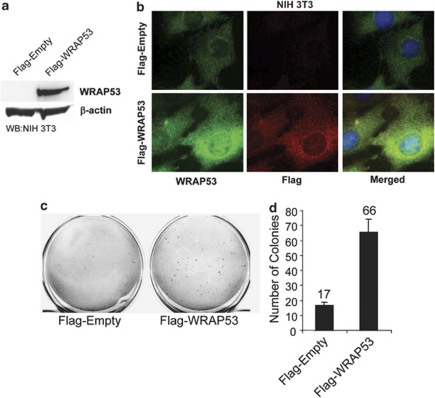
The finding that WRAP53 is overexpressed in human cancer cells encouraged us to investigate if WRAP53 has oncogenic properties and can induce cellular transformation. To determine this, we overexpressed Flag-tagged wild-type WRAP53 or Flag-empty vectors in immortal murine fibroblasts (NIH 3T3 cells). WB analysis showed high WRAP53 expression in Flag-WRAP53 transfected NIH 3T3 cells (Figure 2a) and immunofluorescence (IF) revealed enhanced WRAP53 expression both in the cytoplasm and in the nucleus (Figure 2b), in agreement with the normal localization of endogenous WRAP53.2 The amount of Flag-WRAP53-expressing cells was estimated to around 65% after 9 days of selection. Interestingly, WRAP53 overexpression induced anchorage-independent colony growth after 3 weeks, whereas the empty vectors failed to transform the cells. Whereas control cells gave rise to some small colonies, WRAP53 overexpression induced four times the number of colonies that were also larger in size (Figures 2c and d). These results indicate that WRAP53 is an oncogene, whose overexpression can induce cell transformation.
Figure 2.
WRAP53 overexpression leads to cell transformation. (a) WB analysis of WRAP53 protein levels in NIH 3T3 cells transfected with the indicated vectors. (b) IF of NIH 3T3 cells using WRAP53 and Flag-specific antibodies. Cells were transfected with the indicated plasmids and selected in the presence of 1 μg/ml G418 for 9 days before staining. Nuclei were stained with DAPI in all IF stainings. (c) Anchorage-independent transformation assay of NIH 3T3 cells expressing Flag-tagged WRAP53 or Flag-Empty. The cells were transfected with the indicated plasmids and selected in the presence of 1 μg/ml G418 for 9 days. Selected cells were suspended in soft agar and were photographed after 19–23 days incubation at 37°C. (d) The graph shows the mean of two independent experiments; error bars represent standard error
Depletion of WRAP53 triggers apoptosis in cancer cells
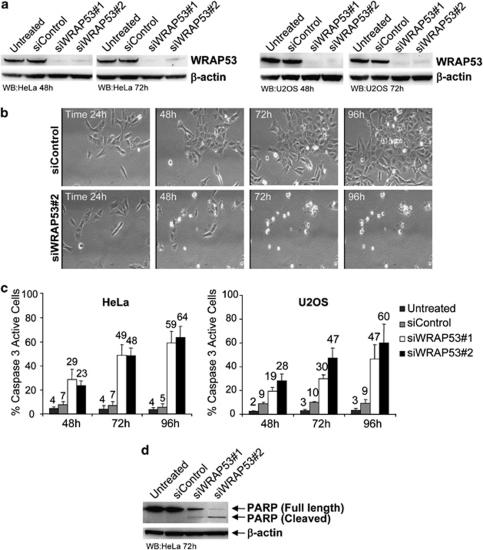
To investigate the impact of WRAP53 in cancer cells, we knocked down WRAP53 expression in HeLa and U2OS cells (Figure 3a). Interestingly, WRAP53-depleted cells started to die around 2–3 days post small interfering RNA (siRNA) treatment (Figure 3b). To better understand the mechanism and the dynamics of this cell death, we performed time-lapse video microscopy of siRNA-treated cells, recording cells in 4-min intervals for up to 96 h post transfection. We observed massive cell death in siWRAP53-treated cells starting at 48 h post transfection (Figure 3b and Supplementary Movies S1 and S2), along with several known morphological changes associated with apoptosis, for example, cell rounding, membrane blebbing and cell shrinkage. To confirm apoptosis, we assessed activation of cellular caspase 3 using FITC-conjugated active caspase 3 antibody, followed by FACS analysis. WRAP53 depletion triggered caspase 3 activation in around 50–60% of cells, 72–96 h post siRNA transfection (Figure 3c). Apoptosis was also verified by PARP (Poly ADP-ribose polymerase) cleavage. WB analysis of PARP showed a 116 kDa band corresponding to intact PARP in untreated and siControl-treated cells. In siWRAP53-treated cells, the 116 kDa band was weaker and the antibody detected an 85 kDa PARP species corresponding to cleaved PARP (Figure 3d). These data show that silencing of WRAP53 expression induces cell death by apoptosis.
Figure 3.
WRAP53 depletion triggers apoptosis. (a) WB analysis of WRAP53 and β-actin levels in HeLa and U2OS cells treated with siControl, siWRAP53 no.;1 and siWRAP53 no.;2 for 48 h and 72 h. (b) Time-lapse analysis of U2OS cells treated with siControl (Supplementary Movie S1) and siWRAP53 no.;2 (Supplementary Movie S2) for 24–96 h. (c) The bars show the percentage of active caspase 3-positive HeLa and U2OS cells treated with siControl, siWRAP53 no.;1 and siWRAP53 no.;2 for the indicated time periods. Error bars represent standard error of three independent experiments. (d) WB analysis of PARP on untreated, siControl, siWRAP53 no.;1- and siWRAP53 no.;2-treated HeLa cells, 72 h post treatment
WRAP53 knockdown induces apoptosis through the mitochondrial pathway
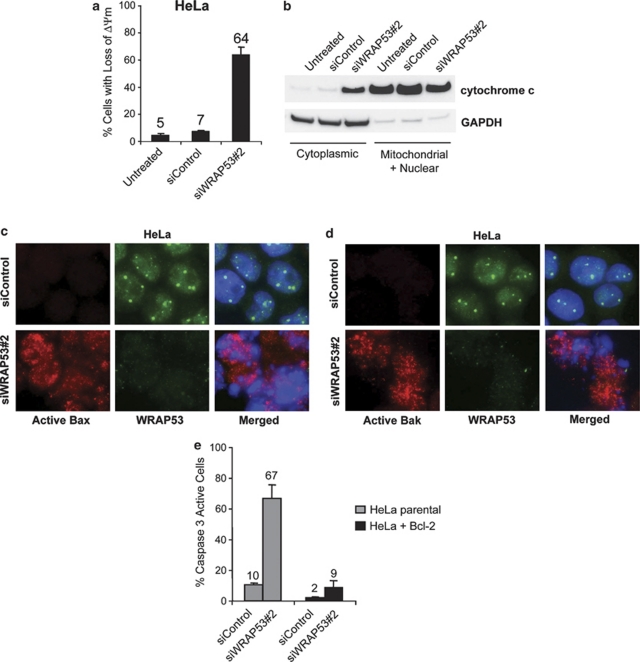
Mitochondria have an important role in apoptosis by releasing several proapoptotic proteins upon apoptotic signals, such as cytochrome c, that in turn activate caspases and other apoptotic effectors leading to cell death. To examine whether the mitochondrial pathway is activated by WRAP53 depletion, we looked at changes in mitochondrial membrane potential (Δψm) upon siWRAP53 knockdown. Treatment with siWRAP53 oligos for 72 h resulted in loss of Δψm in 64% of cells (Figure 4a). WRAP53 depletion also triggered cytochrome c release from the mitochondria. This was as demonstrated by increased levels of cytochrome c in the cytoplasm following siWRAP53 treatment (Figure 4b).
Figure 4.
WRAP53 knockdown induces mitochondrial-dependent apoptosis. (a) The bars show the percentage of TMRE-negative HeLa cells treated with siControl or siWRAP53 no.;2 for 72 h. Error bars represent standard error of three independent experiments. (b) WB analysis of cytochome c and GAPDH levels in cytoplasmic and mitochondrial/nuclear fractions of HeLa cells treated with the indicated siRNA oligos for 72 h. GAPDH was used as a cytoplasmic marker. (c and d) IF staining of WRAP53 and active Bax (c) or WRAP53 and active Bak (d) in HeLa cells treated with siControl and siWRAP53 no.;2 oligos for 72 h. Cytospin preparations have been used for Bax and Bak IF experiments resulting in a skewed subcellular distribution of WRAP53. Normally, the majority of WRAP53 protein is concentrated in the cytoplasm of HeLa cells similar to the staining pattern in NIH 3T3 cells (Figure 2b) (e) The bars show the percentage of active caspase 3-positive parental HeLa or Bcl-2 overexpressing (HeLa+Bcl-2) HeLa cells, treated with siControl and siWRAP53 no.;2 for 72 h. Error bars represent standard error of three independent experiments
We next investigated whether the proapoptotic proteins Bax and Bak were activated and involved in the apoptosis triggered by WRAP53 depletion. A significant activation of both Bax and Bak was observed after siWRAP53 treatment for 48 and 72 h (Figures 4c and d), suggesting a role of these proteins in the loss of Δψm and cytochrome c release following WRAP53 knockdown. To validate the importance of the mitochondria in siWRAP53-induced apoptosis, we investigated whether Bcl-2 overexpression could protect cells from apoptosis triggered by WRAP53 silencing. Bcl-2 is an anti-apoptotic member of the Bcl-2 protein family that controls the permeability and integrity of the mitochondrial membrane. HeLa cells that stably overexpress Bcl-2 (HeLa+Bcl-2) were transfected with siWRAP53 oligos for 72 h and apoptosis was assessed by caspase 3 activation. Notably, caspase 3 activation was reduced by 87% in the Bcl-2-overexpressing cells (Figure 4e). Thus, apoptosis induced by WRAP53 depletion is almost entirely blocked by Bcl-2 overexpression. Altogether, these results provide strong evidence for a critical role of the mitochondrial pathway in the apoptosis triggered by WRAP53 silencing.
Tumor cells are more sensitive to WRAP53 depletion than normal cells
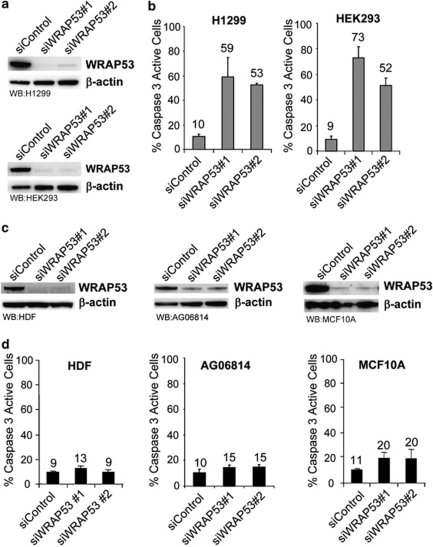
Next, we extended our phenotype analysis of WRAP53 depletion to other tumor cells. A panel of human tumor cell lines, including H1299 (lung cancer), HEK293 (embryonic kidney cancer) and HCT116 (colon cancer) were treated with siWRAP53 oligos and analyzed for cell death. Treatment with siWRAP53 oligos clearly reduced WRAP53 levels and induced cell death in all tumor cells to a similar degree, as previously shown for HeLa and U2OS (Figures 3 and 5a, b and data not shown). We also silenced WRAP53 in normal human diploid fibroblasts (HDF and AG06814) and in non-transformed but spontaneously immortalized mammary epithelial cells (MCF10A). WB analysis showed a substantial reduction in WRAP53 expression after siWRAP53 treatment (Figure 5c). However, in contrast to the tumor cells, the normal cells did not show any significant increase, and in case of MCF10A only a minor increase, in apoptosis upon WRAP53 depletion (Figure 5d). This indicates that cancer cells, but not normal cells, are dependent on WRAP53 expression for their survival.
Figure 5.
Cancer cells are more sensitive to WRAP53 depletion compared with normal cells. (a) WB analysis of WRAP53 and β-actin levels in H1299 and HEK293 cells treated with siControl, siWRAP53 no.;1 and siWRAP53 no.;2 for 72 h or 48 h, respectively. (b) The bars show the percentage of active caspase 3-positive H1299 and HEK293 cells treated with siControl, siWRAP53 no.;1 and siWRAP53 no.;2 for 96 h. Error bars represent standard error of three independent experiments. (c) WB analysis of WRAP53 and β-actin levels in HDF, AG06814 and MCF10A cells treated with siControl, siWRAP53 no.;1 and siWRAP53 no.;2 for 72–96 h. (d) The bars show the percentage of active caspase 3-positive HDF, AG06814 and MCF10A cells treated with siControl, siWRAP53 no.;1 and siWRAP53 no.;2 for 96 h. Error bars represent standard error of three independent experiments
WRAP53 overexpression correlates with poor prognosis in head and neck cancer
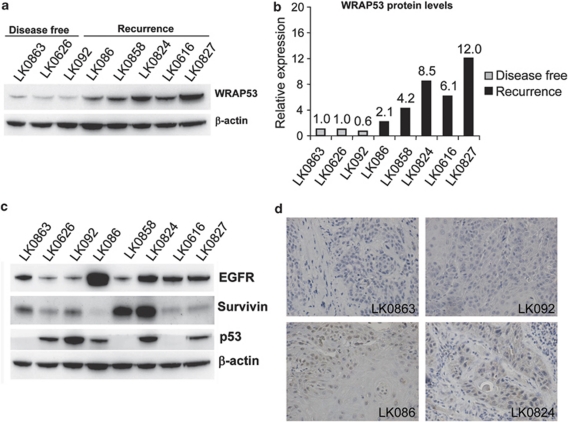
Our finding that WRAP53 overexpression promotes survival of several well-known cancer cell lines encouraged us to investigate the impact of WRAP53 in primary tumors. Eight recently established head and neck squamous-cell carcinoma (HNSCC) cell lines derived from primary tumors (Table 1) were analyzed for correlation between WRAP53 expression and patient prognosis. Strikingly, high expression of WRAP53 was observed in HNSCC cells from patients with poor outcome, whereas low expression of WRAP53 was observed in cells from patients with a more beneficial outcome (Figures 6a and b). Moreover, WRAP53 was the strongest prognostic factor when compared with the other investigated proteins: EGFR, Survivin and p53 in the same cells (Figure 6c). Expression of WRAP53 was also assessed in the patient tumor samples confirming that WRAP53 levels are higher in recurrent patients compared with patients with good outcome (Figure 6d).
Table 1. Characteristics of the HNSCC tumor cell lines.
| Cell line | Site | TNMa | SF2 | Prognosis |
|---|---|---|---|---|
| LK0863 | Larynx | T1N0M0 | 0.51 | Disease free. |
| LK0626 | Gingiva | T2N0M0 | 0.48 | Disease free after 1 year. Dead in lung cancer after 2 years. |
| LK092 | Tongue | T1N0M0 | 0.57 | Disease free. |
| LK086 | Gingiva | T4aN0M0 | 0.59 | DOD, 6 months after diagnosis. |
| LK0858 | Tongue | T3N0M0 | 0.73 | DOD, 16 months after diagnosis |
| LK0824 | Tongue | T2N1M0 | 0.75 | DOD, 7 months after diagnosis. |
| LK0616 | Base of tongue | T4N2M0 | 0.79 | DOD, 3 months after diagnosis. |
| LK0827 | Tongue | T2N0M0 | 0.89 | DOD, 12 months after diagnosis. |
Abbreviation: DOD, dead of disease.
TNM classification according to the International Union against Cancer (UICC, 2002), SF2 (Surviving Fraction at 2 Gy).
Figure 6.
WRAP53 overexpression correlates with poor prognosis in head and neck cancer. (a) WB analysis of WRAP53 and β-actin protein levels in HNSCC cells. (b) Densitometric quantifications of WRAP53 protein levels in the HNSCC cells shown in (a). Levels of WRAP53 have been normalized against β-actin levels. (c) WB analysis of EGFR, Survivin, p53 and β-actin protein levels in the same set of HNSCC cells as in (a). (d) Immunohistochemical staining of WRAP53 in primary HNSCC tumors
Radiotherapy in combination with surgery is the primary mode of treatment for HNSCC. To strengthen the prognostic value of WRAP53 in head and neck cancer, we next analyzed the correlation between WRAP53 levels and the intrinsic radiosensitivity of the tumors. The intrinsic radiosensitivity was determined by measuring the surviving fraction of cells after treatment with a therapeutic dose of two Gray (SF2) using a clonogenic assay and crystal violet staining. The correlation between WRAP53, EGFR, Survivin and p53 expression and the intrinsic radiosensitivity of the HNSCC cells was analyzed and the only protein with a significant correlation to radiosensitivity was WRAP53 (r=0.929, P=0.001; Pearson's correlation test). Enhanced WRAP53 expression was observed in HNSCC cells with low intrinsic radiosensitivity and low expression was observed in cells with high intrinsic radiosensitivity (Table 1). Together these results show a clear correlation between WRAP53 expression and patient prognosis. This supports a role of WRAP53 in the survival of cancer cells in vivo and indicates that WRAP53 may be a novel marker for the prognosis of head and neck cancer.
Discussion
The dependence of a cancer cell on one overactive gene or pathway for its survival and/or growth is called ‘oncogene addiction' and provides cancer-specific weaknesses that can be targeted in anticancer therapy.10 Here, we report such properties for the WRAP53 protein, that is, WRAP53 protein is a potential oncogene essential for cancer cell survival, exposing WRAP53 as an interesting target for therapeutic intervention. In this study, we show that the WRAP53 protein is overexpressed in a broad range of human cancer cell lines in comparison with non-transformed cells. Functional analysis revealed that WRAP53 overexpression confers anchorage-independent growth in noncancerous NIH 3T3 mouse embryonic fibroblasts. This shows oncogenic properties of WRAP53 and indicates that enhanced WRAP53 has a role in tumor development by contributing to malignant transformation. Conversely, we found that knockdown of WRAP53 impairs cancer cell growth by inducing massive apoptosis. Characterization of the apoptosis showed activation of the intrinsic mitochondrial pathway, as concluded by the following findings upon WRAP53 depletion: (1) morphological changes associated with apoptosis including cell rounding, membrane blebbing and cell shrinkage, (2) caspase 3 activation and PARP cleavage, (3) cytochrome c release and loss of mitochondrial membrane potential, (4) Bax and Bak activation and (5) apoptotic rescue by Bcl-2 overexpression. We also found that silencing of WRAP53 specifically kills cancer cells, as depletion of WRAP53 in different normal human cells had no effect on cell survival. This indicates that cancer cells, but not normal cells, depend on WRAP53 expression for their survival in agreement with the definition of oncogenic addition.
We previously identified WRAP53 as a natural p53 antisense gene that has a critical role in the regulation of p53 at the RNA level.1 Although this raises the possibility that the oncogenic properties of WRAP53 are mediated by its p53-regulating activities, several observations suggest that this is not the case. First, WRAP53 regulates p53 through the WRAP53-encoded antisense transcript and not through the WRAP53 protein.1 The WRAP53 gene has different start exons (α, β and γ) and only α-containing transcripts have the capacity to interact with p53 mRNAs and regulate p53 levels/action. Neither β- or γ-transcripts nor the WRAP53 protein had any effects on p53 when overexpressed or knocked down. Second, the WRAP53 protein is mainly produced from WRAP53β-transcripts, which has a separate promoter from α-transcripts. Finally, the apoptosis triggered by WRAP53 knockdown is p53-independent as cells lacking or having inactivated p53 (H1299, HCT116 p53−/− and HeLa) die to the same extent as wild-type p53-harboring cells (U2OS, HEK293 and HCT116 p53+/+) (Figures 3c and 5b and data not shown). Together this indicates that the oncogenic properties of WRAP53 are not mediated through p53.
We recently showed that the WRAP53 protein is crucial for Cajal body formation and that loss of WRAP53 disrupt Cajal bodies.2 Even though many processes in this organelle are important for survival, Cajal bodies per se are not essential for viability of cells, and should thus not trigger apoptosis upon disruption.11, 12, 13, 14, 15 WRAP53 also targets the SMN complex to Cajal bodies.2 Reduced expression of SMN has been shown to induce the mitochondrial apoptotic pathway involving cytochrome c release and caspase activation.16, 17, 18, 19 However, as cells survive without Cajal bodies, the function of WRAP53 in directing the SMN complex to these nuclear structures is not likely to be the underlying reason for the observed apoptosis upon WRAP53 depletion. Moreover, the SMN complex has not previously been implicated in cancer and SMN levels were not found elevated in the cancer cells of this study (data not shown). This suggests that other functions of WRAP53 are essential for cancer cell survival.
The WRAP53 protein was also recently shown to be a subunit of the telomerase holoenzyme and important for telomere maintenance in human cancer cells.7 WRAP53's involvement in the telomerase complexes is another link to tumorigenesis, which may provide mechanistic explanation to some of our observations, however, not to all of them. It is possible that factors involved in enhanced telomerase expression (found in 90% of tumors) also contribute to WRAP53 overexpression in cancer. Indeed MCF10A cells, which have increased telomerase activity compared with primary cells, also have higher levels of WRAP53 protein compared with primary cells. Telomerase activity is mainly regulated at the level of hTERT gene transcription, and number of activators and inhibitors influence the hTERT promoter, including c-Myc, Sp1, estrogen receptor (ERalpha) and p53.20, 21 WRAP53 mRNA is increased in cancer cells22, 23 indicating that upregulation of WRAP53 protein is caused by enhanced transcription, however, the responsible factors influencing the WRAP53 promoter remain to be identified. Notably, MCF10A cells were also more sensitive to WRAP53 depletion in comparison with the primary cells, suggesting that as the cells become more cancerous the more important WRAP53 levels are for their survival.
WRAP53's role in the telomerase enzyme may also, at least partly, explain the oncogenic properties of WRAP53. Evidence suggests that telomerase can promote tumorigenesis in mice and human independently of telomere elongation by inducing cell proliferation and facilitating cell growth under conditions of physiologic stress.24, 25, 26, 27 Thus, it is possible that overexpression of WRAP53 facilitates the cell transformation activities of telomerase or vice versa. The ability of WRAP53 to transform NIH 3T3 cells, which already have enhanced telomerase activity, however, indicates that WRAP53's contribution in the transformation process goes beyond WRAP53's function in the telomerase complex. Moreover, the dynamics of the cell death triggered by WRAP53 inhibition is not in-line with previous reports of telomerase inhibition. Knockdown of telomerase (hTERT) by RNAi induces apoptosis only after several weeks of cell culture,28 pointing against WRAP53's role in telomere elongation as the underlying cause of WRAP53-induced apoptosis. Furthermore, U2OS cells maintain telomere length though the alternative lengthening of telomeres (ALT) pathway and still undergo apoptosis upon WRAP53 silencing. This suggests that the cancer-specific dependence of WRAP53 is additional to WRAP53's role in the telomerase enzyme. Thus, none of the currently known functions of the WRAP53 protein can satisfyingly explain the precise mechanisms underlying the apoptotic protective functions of WRAP53 or its oncogenic properties. Further investigations are required to elucidate the tumorigenic properties of WRAP53.
We also found that overexpression of WRAP53 is a marker of poor prognosis in HNSCC. WRAP53 expression was analyzed in eight recently established cell lines from patients with HNSCC, which were selected according to prognosis and intrinsic radiosensitivity. The corresponding patient tumor samples were also analyzed for WRAP53 expression. In this material, high expression of WRAP53 was associated with aggressive disease, shorter disease-free survival and resistance to radiotherapy. In contrast, low expression was observed in cells sensitive to radiotherapy from patients with a more beneficial outcome. Our findings indicate that WRAP53 is upregulated during head and neck carcinogenesis and contributes to disease progression. This discovery not only identifies WRAP53 as a prognostic marker candidate for head and neck cancer but also support a role of WRAP53 in the development and progression of human cancer cells in vivo. However, these findings should be verified in a larger tumor collection, and the correlation between WRAP53 expression, tumor size and treatment outcome should be investigated. In-line with this, previous studies found common genetic variations (SNPs) in the WRAP53 gene associated with aggressive breast and ovarian cancer.8, 9
In conclusion, we have observed elevated levels of WRAP53 in a variety of tumor cell lines compared with non-transformed cells and furthermore detected tumorigenic properties of WRAP53. Furthermore, we have found that WRAP53 knockdown leads to massive apoptosis through the intrinsic mitochondrial pathway. Human tumor cells are more sensitive to WRAP53 inhibition as compared with normal cells. Strikingly, we found a clear correlation between WRAP53 expression and prognosis in eight recently established head and neck carcinoma cell lines derived from primary tumors. Together, these data highlight a role of WRAP53 in cancer in vitro and in vivo and identify WRAP53 as a viable target for cancer therapy.
Materials and Methods
Cells and culture conditions
U2OS, HeLa, H1299, HEK293, HCT116 and HDF cells were maintained in Dulbecco's modified medium supplemented with 10% fetal bovine serum (Invitrogen Corporation, Paisley, UK), 2 mM -glutamine (Invitrogen Corporation) and 2.5 μg/ml Plasmocin (InvivoGen, Toulouse, France) at 37°C in 5% CO2 humidified incubators. AG06814 cells were maintained in MEM supplemented with 10% FBS. MCF10A cells were maintained in a 1 : 1 mixture of MEBM (Clonetics/Lonza, Cologne, Germany) and Nutrient mixture F12 Ham (Sigma-Aldrich, Stockholm, Sweden) medium supplemented with 5% horse serum (Sigma-Aldrich), MEGM SingleQuotes (Clonetics/Lonza) and 2.5 μg/ml Plasmocin (InvivoGen) at 37°C in 5% CO2 humidified incubators.
Fresh tumor samples from patients with HNSCC obtained during surgery were immersed immediately in Ca2+- and Mg2+-free phosphate-buffered saline (PBS; approved by the Linköping University ethical committee). The tissues were washed, minced into 1–2 mm pieces and placed in culture flasks. Thereafter, growth medium (Keratinocyte-SFM; GIBCO, Paisly, UK, Invitrogen Corporation) supplemented with amphotericin B (Fungizone, 2.5 μg/ml), antibiotics (penicillin 100 U/ml, streptomycin 100 μg/ml; all from GIBCO) and 20% fetal bovine serum (FBS, Integro B.V., Leuvenheim, the Netherlands) was added. Fibroblasts contaminating the cultures were continuously removed by differential trypsinization (0.25% trypsin+0.02% EDTA (ethylenediaminetetraacetic acid). After 3–6 weeks, the tumor cells had reached 50% confluence.
Following establishment, the cultures was grown in Keratinocyte-SFM supplemented with antibiotics (penicillin 100 U/ml, streptomycin 100 μg/ml) and 10% FBS, and were subcultured at confluence using 0.25% trypsin+0.02% EDTA with a weekly split ratio of about 1 : 2. Exchange of medium were made every third day. Cultures from passages 4 to 10 were used in all experiments. Typical epithelial morphology under phase contrast microscope was verified by IF staining of cytokeratin using a mouse-anti-cytokeratin antibody (Novacastra Laboratories Ltd., Newcastle, UK, dilution 1 : 50) as earlier described.29 The staining demonstrated variable but consistent expression of cytokeratin among all eight tumor cell lines used in this paper.
Western blotting
Cell extracts for WB analysis: cells were harvested, washed and lysed in ice cold WB lysis buffer (100 mM Tris-HCL, pH 8, 150 mM NaCl, 1% NP-40, 1% PMSF and 1% protease inhibitor cocktail) for 30 min on ice. Lysates were centrifuged at 14 000 r.p.m. for 15 min at 4°C and protein concentrations were determined using Bradford assay (Bio-Rad Laboratories AB, Sundbyberg, Sweden). WB was performed according to standard procedures.
Transformation assay
For soft agar colony formation assays, NIH 3T3 cells were transfected by Lipofectamine 2000 with 1–3 μg of Flag-empty or Flag-WRAP53 plasmid DNA. The transfected cells were selected for in G418 for 9 days and then suspended in DMEM (10% FBS) containing 0.4% agar, and overlaid onto a 0.5% agar base at a density of 1 × 104 cells per 60-mm dish. After 19–23 days, dishes were stained for with 0.01% crystal violet and the number of colonies was counted.
Immunofluorescence microscopy
For active Bax and Bak IF experiments, cells were harvested, washed and spun onto glass slides by Cytospin (Hettich Zentrifugation) for 5 min at 2500 r.p.m. and then fixed with 4% Formalin at room temperature (RT). For IF analysis of WRAP53 overexpressing NIH 3T3 cells, cells were grown on sterilized cover slips and fixed with 100% MeOH for 20 min at −20°C. Both formalin- and MeOH-fixed cells were then permeabilized with 0.1% Triton X-100 for 5 min at RT, followed by 30 min of blocking in blocking buffer (2% BSA, 5% glycerol, 0.2% Tween20, 0.1% NaN3). Cover slips were subsequently incubated for 1 h in primary antibody and 40 min in secondary antibody diluted in blocking buffer. The cover slips were mounted with Vectorshield mounting medium with DAPI (Vector Laboratories Ltd. United Kingdom, Peterborough, UK). Images were acquired with a Zeiss Axioplan 2 microscope, equipped with an AxioCam HRm Camera (Carl Zeiss AB, Stockholm, Sverige) using 43 or 60 oil immersion lenses, and processed using Axiovision Release 4.7.
Cell fractionations
Mitochondrial fractionations were performed as follows; cells were harvested, washed and lysed in Mitochondrial buffer (5 mM Tris-HCl, pH 7.4, 125 mM KCl, 5 mM MgCl2, 1 mM EGTA, 0.05% Digitonin, 1% protease inhibitor cocktail) for 5 min at RT, followed by centrifugation at 13 000 r.p.m. for 5 min. Supernatant (cytosol) was transferred to a new eppendorf tube and the remaining pellet (nucleus+mitochondria) was lysed in WB lysis buffer. Equal volumes of each sample were loaded on an SDS-PAGE gel.
Antibodies
Three different WRAP53 antibodies were used: rabbit α-WRAP53-C2 (cat no.; PA-2020-100, Innovagen AB, Lund, Sweden, used for WB), rabbit α-WRAP53 (Wdr79, cat no.; A301-442A-1, Bethyl Laboratories, Inc., Montgomery, AL, USA, used for WB and IF) and rabbit α-WRAP53-483,1 used for WB and immunohistochemical staining. The following antibodies were used in IF and WB: mouse α-Bax (cat no.; 556467, BD Biosciences, Stockholm, Sweden), mouse α-active Bak (cat no.; AM03, Calbiochem, USA), mouse α-cytochrome c (cat no.; 556433, BD Biosciences), mouse α-PARP (cat no.; 556362, BD Biosciences), rabbit α-GAPDH (cat no.; sc-25778, Santa Cruz, Heidelberg, Germany), mouse α-β-actin (Sigma-Aldrich), mouse α-EGFR (cat no.; 610016, BD Biosciences), rabbit α-Survivin (cat no.; S18191, Sigma-Aldrich) and mouse α-p53 (cat no.; 554293, BD Biosciences). Secondary antibodies: sheep α-mouse HRP (cat no.; NA931V, GE Healthcare), donkey α-rabbit HRP (cat no.; NA934V, GE Healthcare, Stockholm, Sweden), goat α-rabbit Alexa Fluor 488 (cat no.; A11008, Invitrogen Corporation), donkey α-mouse Alexa Fluor 594 (cat no.; A21203, Invitrogen Corporation).
siRNA oligonucleotides, plasmids and transfections
siRNA oligonucleotides targeting WRAP53: siWRAP53 no.;1 (cat no.; SI00388941, Qiagen, Sollentuna, Sweden) and siWRAP53 no.;2 (cat no.; SI00388948) and a control siRNA (cat no.; 1027280, Qiagen) was used. A measure of 10–20 nM of siRNA was transfected into cells by either Oligofectamine (Invitrogen Corporation) or by HiPerfect (Qiagen) transfection reagents in accordance with the supplier's recommendations. Flag-WRAP53 was cloned as described in (Mahmoudi et al.1). Plasmid transfections were performed using Lipofectamine 2000 Reagent (Invitrogen Corporation) following the manufacturer's instructions.
Video time-lapse microscopy
For time-lapse microscopy, U2OS cells were cultured sparsely on 3 cm plates. The cells were transfected with 20 nM of either Control or WRAP53 siRNA oligos and incubated in 37°C incubator. At 24 h post transfection, the plates were placed in a 37°C heated microscope chamber. The cells were observed under an inverted Zeiss Axiovert 100 microscope equipped with charged-coupled device (CCD) camera using a × 10 magnification lens. Images were acquired every 4 min for the denoted time by a framegrabber card (PX-510-25E).
FACS analysis
For active caspase 3 stainings, cells were transfected with siRNA as described above and harvested at the indicated time points with trypsin, washed twice in PBS and fixed with 0.25% formaldehyde for 10 min at RT. Next, the samples were washed and incubated with PBS supplemented with FITC-conjugated rabbit active caspase 3 (cat no.; 51-68654X, BD Biosciences) and 0.1% digitonin for 30 min at 4°C. The cells were washed and then analyzed for active caspase 3 by flow cytometry on a FACS Calibur (BD Biosciences) using the Cell Quest software.
Changes in Δψm were detected by incubation of living cells with tetramethylrhodamine ethyl ester (TMRE) (Invitrogen Corporation) and analyzed by a FACS calibur flow cytometer (BD Biosciences) using the Cell Quest software.
Assessment of intrinsic radiosensitivity
Tumor cells were seeded into 12-well plates at densities of 300–800 cells per cm2 depending on the plating efficiency of each cell line. After 24 h, cells were irradiated (2 Gy) with 4 MeV photons generated by a linear accelerator (Clinac 4/100, Varian, Palo Alto, CA, USA), delivering a dose-rate of 2.0 Gy/min. The effect of radiotherapy was determined after another 9 days by crystal violet staining following fixation in 4% paraformaldehyde (20 min). Surviving (adherent) cells were stained with 0.04% crystal violet in 1% ethanol for 20 min at RT, and the plates were washed extensively under running tap water and air dried. The cells were solubilized in 1% SDS, and optical density values were recorded in a Victor plate reader (EG & G Wallac, Upplands Väsby, Sweden) at 550 nm, and corrected for background absorbance.
Immunohistochemical staining
Sections from tumor biopsies were mounted on positively charged slides, deparaffinized in xylene and rehydrated through decreasing concentrations of ethanol. Thereafter, the sections were pretreated with 10 mM citrate buffer (DakoCytomation epitope retrieval solution) in a hot water bath (up to 100°C) for 40 min, blocked with Envision peroxidase block (BCPX968) for 5 min, and incubated for 30 min at RT with rabbit α-WRAP53-4831 at a dilution of 1 : 1000. Staining was achieved with DakoCytomation EnVision System-HRP (DAB) kit. Sections were counterstained for 1 min with Tacha's hematoxylin.
Acknowledgments
We thank Bert Vogelstein, Johns Hopkins Oncology Center, for the HCT116 cells, Arne Östman, Karolinska Institutet, for the primary culture of prostate fibroblasts and Bertrand Joseph, Karolinska Institutet, for the Hela Bcl-2 cells. This work was supported by grants from the Swedish Cancer Society (Cancerfonden), the Swedish Childhood Cancer Society (Barncancerfonden), Radiumhemmets Fund's, Åke Wiberg's Fund, Mary Beve's Fund, Åke Olsson's Fund, Socialstyrelsen's Funds and the Karolinska Institutet. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Glossary
- WDR79
WD repeat domain 79
- TCAB1
telomerase Cajal body protein 1
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- Mahmoudi S, Henriksson S, Corcoran M, Mendez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Söderberg O, Strömblad S, et al. WRAP53 is essential for Cajal Body formation and for targeting the survival of motor neuron complex to Cajal Bodies. PLoS Biology. 2010;8:e1000521. doi: 10.1371/journal.pbio.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, et al. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell. 2007;27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, et al. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M, Kristensen V, Langerod A, Qi Y, Yeager M, Burdett L, et al. Common genetic variation in TP53 and its flanking genes, WDR79 and ATP1B2, and susceptibility to breast cancer. Int J Cancer. 2007;121:2532–2538. doi: 10.1002/ijc.22985. [DOI] [PubMed] [Google Scholar]
- Schildkraut JM, Goode EL, Clyde MA, Iversen ES, Moorman PG, Berchuck A, et al. Single nucleotide polymorphisms in the TP53 region and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2009;69:2349–2357. doi: 10.1158/0008-5472.CAN-08-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein IB, Joe A.Oncogene addiction Cancer Res 2008683077–3080.discussion 3080. [DOI] [PubMed] [Google Scholar]
- Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, et al. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw P. A distant coilin homologue is required for the formation of cajal bodies in Arabidopsis. Mol Biol Cell. 2006;17:2942–2951. doi: 10.1091/mbc.E05-12-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MP, Tian L, Matera AG. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS One. 2009;4:e6171. doi: 10.1371/journal.pone.0006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wu LZ, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, et al. Coilin is essential for Cajal body organization in Drosophila melanogaster Cajal body. Mol Biol Cell. 2009;20:1661–1670. doi: 10.1091/mbc.E08-05-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittom AA, Xu H, Hebert MD. Coilin levels and modifications influence artificial reporter splicing. Cell Mol Life Sci. 2008;65:1256–1271. doi: 10.1007/s00018-008-7587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilangovan R, Marshall WL, Hua Y, Zhou J. Inhibition of apoptosis by Z-VAD-fmk in SMN-depleted S2 cells. J Biol Chem. 2003;278:30993–30999. doi: 10.1074/jbc.M303763200. [DOI] [PubMed] [Google Scholar]
- Parker GC, Li X, Anguelov RA, Toth G, Cristescu A, Acsadi G. Survival motor neuron protein regulates apoptosis in an in vitro model of spinal muscular atrophy. Neurotox Res. 2008;13:39–48. doi: 10.1007/BF03033366. [DOI] [PubMed] [Google Scholar]
- Trulzsch B, Garnett C, Davies K, Wood M. Knockdown of SMN by RNA interference induces apoptosis in differentiated P19 neural stem cells. Brain Res. 2007;1183:1–9. doi: 10.1016/j.brainres.2007.09.025. [DOI] [PubMed] [Google Scholar]
- Vyas S, Bechade C, Riveau B, Downward J, Triller A. Involvement of survival motor neuron (SMN) protein in cell death. Hum Mol Genet. 2002;11:2751–2764. doi: 10.1093/hmg/11.22.2751. [DOI] [PubMed] [Google Scholar]
- Greenberg RA, O'Hagan RC, Deng H, Xiao Q, Hann SR, Adams RR, et al. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene. 1999;18:1219–1226. doi: 10.1038/sj.onc.1202669. [DOI] [PubMed] [Google Scholar]
- Kyo S, Takakura M, Fujiwara T, Inoue M. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99:1528–1538. doi: 10.1111/j.1349-7006.2008.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Fernandez D, Laurell C, Saqui-Salces M, Pantoja JP, Candanedo-Gonzalez F, Reza-Albarran A, et al. Differential RNA expression profile by cDNA microarray in sporadic primary hyperparathyroidism (pHPT): primary parathyroid hyperplasia versus adenoma. World J Surg. 2006;30:705–713. doi: 10.1007/s00268-005-0708-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Eberhard DA, Frantz GD, Dowd P, Wu TD, Zhou Y, et al. GEPIS – quantitative gene expression profiling in normal and cancer tissues. Bioinformatics. 2004;20:2390–2398. doi: 10.1093/bioinformatics/bth256. [DOI] [PubMed] [Google Scholar]
- Chang S, DePinho RA. Telomerase extracurricular activities. Proc Natl Acad Sci USA. 2002;99:12520–12522. doi: 10.1073/pnas.212514699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Shay JW. Actions of human telomerase beyond telomeres. Cell Res. 2008;18:725–732. doi: 10.1038/cr.2008.74. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Samper E, Ramirez A, Flores JM, Martin-Caballero J, Jorcano JL, et al. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 2001;20:2619–2630. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayuela ML, Flores JM, Blasco MA. The telomerase RNA component Terc is required for the tumour-promoting effects of Tert overexpression. EMBO Rep. 2005;6:268–274. doi: 10.1038/sj.embor.7400359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas MA, Koley H, Batchu RB, Bertheau RC, Protopopov A, Munshi NC, et al. Telomerase inhibition by siRNA causes senescence and apoptosis in Barrett's adenocarcinoma cells: mechanism and therapeutic potential. Mol Cancer. 2005;4:24. doi: 10.1186/1476-4598-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg K, Ceder R, Farnebo L, Norberg-Spaak L, Grafstrom RC. Multiple genotypic aberrances associate to terminal differentiation-deficiency of an oral squamous cell carcinoma in serum-free culture. Differentiation. 2008;76:868–880. doi: 10.1111/j.1432-0436.2008.00267.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.