Abstract
Keratinocytes in skin epidermis, which have bright cytoplasmic contrast and dark nuclear contrast in reflectance confocal microscopy (RCM), were modeled with a simple error function reflectance profile: erf( ). Forty-two example keratinocytes were identified as a training set which characterized the nuclear size a = 8.6±2.8 μm and reflectance gradient b = 3.6±2.1 μm at the nuclear/cytoplasmic boundary. These mean a and b parameters were used to create a rotationally symmetric erf( ) mask that approximated the mean keratinocyte image. A computer vision algorithm used an erf( ) mask to scan RCM images, identifying the coordinates of keratinocytes. Applying the mask to the confocal data identified the positions of keratinocytes in the epidermis. This simple model may be used to noninvasively evaluate keratinocyte populations as a quantitative morphometric diagnostic in skin cancer detection and evaluation of dermatological cosmetics.
Keywords: reflectance confocal microscopy, skin, keratinocyte morphometry
Introduction
Confocal microscopy was invented in 1957 by Minsky to image in whole brain tissue,1, 2 a scattering environment that necessitated the invention of the optical section. Confocal microscopy was developed for dermatological imaging3 by Corcuff in 1993. The diagnostic implications in melanocytic lesions were investigated in 2007 by Pellacani.4 In particular, the absence of the normal honeycomb pattern of epidermal keratinocytes has sensitivity and specificity of 87.5% and 52.1%, respectively, for melanoma detection with the human eye.4 The pattern of keratinocytes is even more important in distinguishing between actinic keratoses and normal skin.5
Keratinocytes are born spherical in the deep basal layer and die flattened with a larger en face diameter in the superficial stratum corneum. Since chromatin filaments in nuclei are too small to scatter light efficiently, nuclei in keratinocytes appear dark.6 In the spinous and granular layers of the epidermis, the en face distribution of keratinocytes is gridlike, with a regular spatial frequency. Nuclei are present as dark centroids in the grid. Since organelles and intercellular tissue constituents are comparably sized to the illumination wavelength and are of high refractive index relative to the surrounding medium, they provide a bright contrast to the grid between nuclei in a characteristic honeycomb pattern. A method for automatically specifying the coordinates of keratinocytes is presented.
Materials and Methods
A single confocal stack [three-dimensional (3D) image] acquired at the site of a melanocytic nevus with the Vivascope 1500 (Lucid, Inc.) spanned 500 μm×500 μm in the en face plane and extended 175-μm axially to include the superficial stratum corneum, middle granular and spinous epidermis, the deeper basal layer, and the underlying dermis. The digital size of the image was 1000 pixels×1000 pixels in the en face plane and 175 optical sections deep.
Keratinocyte Identification Algorithm Development
An optical section was chosen in the midlevel epidermis [Fig. 1a] 45-μm deep in the 3D confocal image. This section contained mostly granular and spinous keratinocytes of various sizes and occasionally (due to spatially undulating tissue a-planarity) regions of bright7 pigmented basal cells and stratum corneum. This section was chosen to represent important tissue regions: stratum corneum and granular/spinous/basal epidermis. Fifty spinous or granular keratinocytes were manually selected to represent various en face diameters, and were averaged [Fig. 1b]. Surrounding each cell in the en face plane, a square window of 15 μm (30 pixels) was extracted. The 30 pixel window size was chosen to safely encapsulate the larger keratinocytes while not extending into adjacent keratinocytes for smaller sizes.
Figure 1.
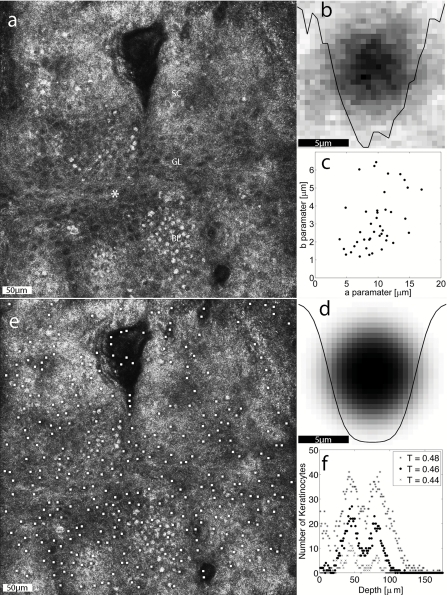
The automatic detection of keratinocytes. (a) An en face optical section through the undulating epidermal layers. Where the section is deepest, super-nuclear melanin caps appear bright in the basal layer, granular keratinocytes appear with dark nuclei in the granular layer, and the superficial stratum corneum appears with constant bright contrast. The site of the axial study is marked (*). (b) The mean image of 50 single keratinocytes chosen to represent the range of sizes. The data plotted through the image is the bisecting reflectance profile. (c) The a and b parameters for the fits to 42 of the 50 representative keratinocytes shown with a scatter plot. (d) The analytical mask that approximates the average keratinocyte using 〈a〉 and 〈b〉. (e) The result of mask application where the white marks identify the centers of identified keratinocytes. (f) The axial profile of keratinocyte populations as identified by threshold T = 0.44, 0.46, and 0.48 [-] (WMV, 9 MB). .
Since visually, the reflectance profile (R) of a line sampled to bisect the average of the 50 keratinocyte images [Fig. 1b] appears to transition from dark nucleus to bright cytoplasm in the form of an error function, the following analytical expression was adopted:
| (1) |
a and b are fitting parameters that shape the contour of the error function. a defines the diameter of the putative cell. b defines the slope at the half max of the nuclear-to-cytoplasm brightness transition region.
By substituting r = √(x2+y2) for x, the analytical expression [Eq. 1] can be adapted to represent a two-dimensional (en face) cell image, which can be compared to an equally sized window of the image as a mask. Before the least squares difference was computed between the mask and the image window, each image window was normalized so that the minimum brightness was zero and the sum of brightness counts (pixel intensity) across all the pixels in the image window was unity. The iterative least-squares fitting used fminsearch( ) in MATLAB, which is a multidimensional unconstrained nonlinear minimization (Nelder–Mead), to find the a and b parameters that fit each of the 50 keratinocytes. Of the 50 fitted a,b pairs, the 84% (42) that reached convergent fits were selected as a cluster for analysis.
The clustered a,b pairs [Fig. 1e] yielded a mean a,b pair that was used to create an analytical mask [Fig. 1c] that was reapplied to the data to identify keratinocytes. A threshold was chosen during this data analysis of T = 0.46 (with standard cross-validation techniques) such that if the difference between the power-normalized mask and image window was less than the threshold T, the region was designated as a keratinocyte. The mask was applied to the en face optical section [Fig. 1a] as well as to an en face subregion of a 100 μm×100 μm2 centered at the asterisk (*) in Fig. 1a over the full depth range of 175 μm. The restriction to the 100 μm×100 μm2 minimized the axial spread of cell populations due to tissue axial undulation of the Rete ridges.
Results
The fit parameters a and b for the 42 convergent fits were normally distributed [Fig. 1e]. The eight outliers, where the fitting routine did not converge, yielded unreasonable a,b results and had no apparent order. The resulting cell size parameter was a = 8.6±2.8 μm and the transition slope parameter was b = 3.6±2.1 μm. Figure 1c shows the analytical model mask constructed from the mean of the a,b parameters shown in Fig. 1e. Figure 1d shows the mask-scanning result where the analytical model was applied as a mask to the entire optical section. White dots indicate identified keratinocytes.
In the axial scan centered at the marked asterisk (*) in Fig. 1a, the number of automatically identified keratinocytes was scored for each optical section throughout the 3D confocal image stack, i.e., as a function of depth (see the video supplement to Fig. 1). This axial analysis in Fig. 1f showed that the number of keratinocytes identified by the mask at each depth is large for the epidermis and zero in the deeper dermis. Results were scored for various threshold values (T = 0.44, T = 0.46, and T = 0.48) to illustrate the range from loose criteria (T = 0.48) where false-positives are scored in the dermis, to strict criteria (T = 0.44) where false negatives are scored in the granular and basal layers. For validation, expert confocal reviewer Giovanni Pellacani analyzed the data (see video supplement to Fig. 1) blinded to the algorithm outputs and reported the surface-depth of the stratum corneum, stratum granulosum, basal layer, and dermis as 24, 39, 59, and 70 μm, respectively.
Discussion
The number of keratinocytes reported here [10 per en face 0.01 mm2 for T = 0.44, Fig. 1f] agrees with visual counts of 1000 per en face mm2 first reported.8 The algorithm presented here identifies dark nuclei, hence the subtype of keratinocyte detected lacks heavy pigment. Basal keratinocytes with dense pigment that present solid bright contrast will not be detected, unless an optical section slightly deeper than the nuclear melanin cap shows a dark nucleus.
Keratinocyte presence and distribution of keratinocytes may be important metrics for a number of applications. Potential applications include: (1) the absence of a healthy keratinocyte pattern indicates melanoma,4 (2) a score for dyskeratosis may objectively identify actinic keratosis versus normal skin and versus squamous cell carcinoma, and (3) grading of mild dyskeratotic keratinocytes may be useful for the identification of early signs of aging skin and risk of developing skin cancers. Future directions for this technique include applying the analysis to characterize the average en face diameter of nuclei as a function of depth. Making the reasonable assumption that the nucleus diameter is slightly less than half that of the entire cell the mean nuclear diameter (a = 8.6 μm) reported here compares well to the published diameter8 for the entire cell: 15 to 35 μm. The maturation cycle, where spherical spinous keratinocytes near the basal layer develop into wider and flatter granular keratinocytes indicates healthy skin. In squamous cell carcinoma, the atypical pattern of anaplasia constitutes reversal of differentiation in maturation. Therefore, the axial size distribution of keratinocytes is a valuable diagnostic.
Beyond cancer, the cosmetic industry lacks a noninvasive method to quantify the effects of treatment on keratinocyte populations. The overall melanin concentration is expected to change across skin types and in the cosmetic bleaching process for post-inflammatory pigmentation and melasma. However, the melanin distribution in cells is not expected to change, so the masking technique is expected to be easily adapted to a wide range of dermatological applications provided that assumptions and ad-hoc thresholds remain minimized.
In this study, the size of the keratinocyte mask window (15 μm) and the threshold for power difference between the mask and the image window for cell identification were the only chosen parameters. Future work will integrate the algorithm reported here with a previous algorithm9 that identifies pigmented cells to advance the breadth of digital morphometric cellular analysis.
In addition to the quantitative nature of the presented technique, vision is extended beyond the dynamic range of the human eye. An example is the void section in the top central portion of Fig. 1d, which appears to be completely outside the tissue. The subtle contrast of the superficial stratum corneum (which is “index-matched”10 by the crodomol oil used to couple the skin to the imaging window)6 has dropped below the dynamic range of detection for the human eye. However the algorithm reveals an important diagnostic finding: parakeratosis, the presence of nucleated cells in the stratum corneum. Both the quantification of patterns already perceived by the eye and the ability of the machine vision algorithm to rapidly change the dynamic range of the analysis may enhance morphometric analysis. In conclusion, automatic identification of epidermal keratinocytes is a promising technique that extends our diagnostic vision through reflectance confocal microscopy.
Acknowledgments
The author, whose salary is supported by Grant No. NIH 5-T32-CA106195, wishes to thank Steven Jacques, Eric Simpson, Kevin White, Alvin Solomon, Molly Kulesz-Martin, Milind Rajadhyaksha, Ashfaq Marghoob, Alon Scope, Allan Halpern, Itay Klaz, and Giovanni Pellacani for intellectual discussions that also supported this work.
References
- Minsky M., “Microscopy Apparatus,” U.S. Patent No. 3013467 (1957).
- Minsky M., “Memoir on inventing the confocal scanning microscope,” Scanning 10, 128–138 (1988). [Google Scholar]
- Corcuff P. and Leveque J. L., “In vivo vision of the human skin with the tandem scanning microscope,” Dermatology 186, 50–54 (1993). 10.1159/000247302 [DOI] [PubMed] [Google Scholar]
- Pellacani G., Guitera P., Longo C., Avramidis M., Seidenari S., and Menzies S., “The impact of in vivo reflectance confocal microscopy for the diagnostic accuracy of melanoma and equivacal melanocytic lesions,” J. Invest. Dermatol. 127, 2759–2765 (2007). [DOI] [PubMed] [Google Scholar]
- Ulrich M., Stockfleth E., Roewert-Huber J., and Astner S., “Noninvasive diagnostic tools for nonmelanoma skin cancer,” Br. J. Dermatol. 157, 56–58 (2007). 10.1111/j.1365-2133.2007.08275.x [DOI] [PubMed] [Google Scholar]
- Gonzalez S., Halpern A. C., and Gill M., Reflectance Confocal Microscopy of Cutaneous Tumors: An Atlas with Clinical, Dermoscopic and Histological Correlations, Informa U.K. Ltd., London: (2008). [Google Scholar]
- Rajadhyaksha M., Gonzalez S., Zavislan J. M., Anderson R. R., and Webb R. H., “In vivo confocal scanning laser microscopy of human skin II: Melanin provides strong contrast,” J. Invest. Dermatol. 104, 946–952 (1995). 10.1111/1523-1747.ep12606215 [DOI] [PubMed] [Google Scholar]
- Huzaira M., Rius F., Rajadhyaksha M., Anderson R. R., and González S., “Topographic variations in normal skin, as viewed by in vivo reflectance confocal microscopy,” J. Invest. Dermatol. 116(6), 846–852 (2001). 10.1046/j.0022-202x.2001.01337.x [DOI] [PubMed] [Google Scholar]
- Gareau D., Hennessey R., Wan E., Pellacani G., and Jacques S. L., “Automated detection of malignant features in confocal microscopy on superficial spreading melanoma versus nevi,” J. Biomed. Opt. 15(6), 061713 (2010). 10.1117/1.3524301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas G., Chan E. K., Barton J. K., Rylander H. G., and Welch A. J., “Use of an agent to reduce scattering in skin,” Lasers Surg. Med. 24(2), 133–141 (1999). [DOI] [PubMed] [Google Scholar]


