Abstract
The centromere is a unique chromosomal locus that ensures accurate segregation of chromosomes during cell division by directing the assembly of a multiprotein complex, the kinetochore1. The centromere is marked by a conserved variant of conventional histone H3 termed CenH3 or CENP-A2. A conserved motif of CenH3, the CATD, defined by loop 1 and helix 2 of the histone fold, is necessary and sufficient for specifying centromere functions of CenH33, 4. The structural basis of this specification is of outstanding interest. Yeast Scm3 and human HJURP are conserved nonhistone proteins that interact physically with the (CenH3-H4)2 heterotetramer and are required for the deposition of CenH3 at centromeres in vivo5, 6, 7, 8, 9, 10, 11, 12, 13. Here we have elucidated the structural basis for recognition of budding yeast CenH3 (Cse4) by Scm3. We solved the structure of the Cse4-binding domain (CBD) of Scm3 complexed with Cse4 and H4 in a single chain model. An α-helix and an irregular loop at the conserved N-terminus and a shorter α-helix at the C-terminus of Scm3-CBD wraps around the Cse4-H4 dimer. Four Cse4-specific residues in the N-terminal region of helix 2 are sufficient for specific recognition by conserved and functionally important residues in the N-terminal helix of Scm3 through formation of a hydrophobic cluster. Scm3-CBD induces major conformational changes and sterically occludes DNA binding sites in the structure of Cse4 and H4. These findings have implications for the assembly and architecture of the centromeric nucleosome.
Unlike other eukaryotic species that have complex regional centromeres with multiple centromeric nucleosomes14, budding yeast has a single centromeric nucleosome that is necessary and sufficient to mediate the accurate segregation of chromosomes during mitosis and meiosis15, 16, 17, 18. The simple centromeres of budding yeast provide an attractive system for investigating outstanding topics in centromere biology, including the pathway of CenH3 deposition and the architecture of the centromeric nucleosome19, 20.
Yeast Scm3 and human HJURP are binding partners of CenH3-H4 and functionally required for their deposition at centromeres in vivo5, 6, 7, 8, 9, 10, 11, 12, 13. A conserved domain of Scm3 dictates specific and stoichiometric binding of CenH3/H4 (Fig. 1a), forming a (Scm3-Cse4-H4)2 hexamer in 2 M NaCl5. This Cse4-binding domain (CBD) of Scm3 is mapped to residues 84–1875. To investigate the structural basis for the recognition of Cse4 by Scm3, we first analyzed the CBD of Scm3 by NMR and found that it is intrinsically disordered (Supplementary Fig. 1). To overcome instability inherent in complexes of individual Scm3, Cse4 and H4 fragments (Supplementary Figs. 2–4), we engineered a single chain molecule in which Scm3 is inserted between Cse4 and H4 to assemble a stably folded molecule (Supplementary Fig. 5). For convenience, we termed the single-chain molecule scSCH (Scm3, Cse4, H4).
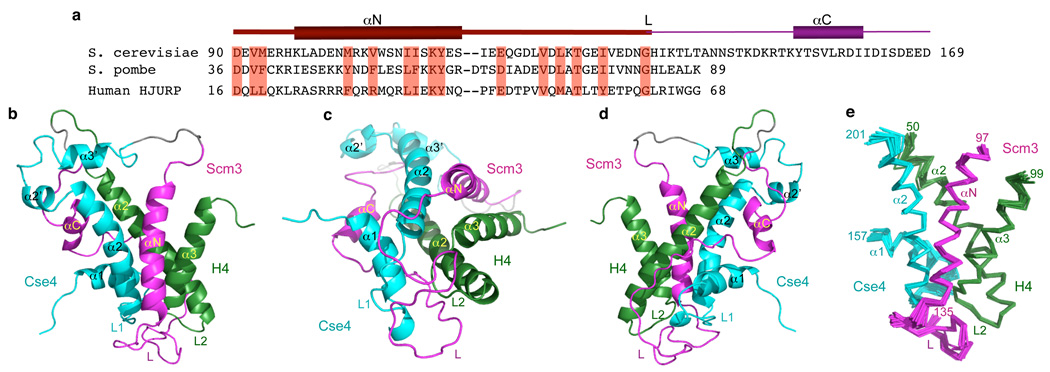
Figure 1. Overall structure and dynamics of scSCH.
a, The amino acid sequence and secondary structures of the Cse4-binding domain of Scm3 in scSCH. Also shown are the conserved regions in the Scm3 of S. pombe and human HJURP. The highly conserved residues are highlighted in red. The region in the folded core is shown in dark magenta (see e). b–d, Front, bottom, and back view of the scSCH structure shown in ribbon. Scm3, Cse4, and H4 are in magenta, cyan, and dark green, respectively. The full sequence of scSCH is M-His6-KK-Cse4150–227-LVPRGS-Scm393–169-GDK-H442–103.
The structure of scSCH was determined using multi-dimensional NMR and verified by structural analysis of its mutants (Fig. 1b–d and Supplementary Fig. 6). The structure of the folded core of scSCH, which includes residues 97–135 of Scm3, 157–202 of Cse4, and 50–99 of H4, is well-defined with RMSDs of 0.54 Å for backbone atoms and 1.06 Å for all heavy atoms (Fig. 1e and Supplementary Table 1). Importantly, linker residues inserted between Scm3, Cse4, and H4 do not alter the structure of the folded region. Proteolytic cleavage of the two linkers in the folded scSCH only affects chemical shifts of neighboring residues close to the cutting sites (Supplementary Fig. 7). Moreover, the folded structure of the above tertiary complex is unchanged by refolding after denaturation in 6M GdmCl to liberate the three components as individual polypeptides (Supplementary Fig. 8). Backbone amide 15N-{1H} hetero-nuclear Overhauser effects (NOE) reflect dynamic motions. The folded core shows small dynamic motions (NOE > 0.7) except for loop 1 of Cse4 and the small loop region following the N-terminal α-helix (αN) in Scm3 (Supplementary Fig. 9a, b). In contrast, other regions display larger dynamic motions (NOE < 0.7), corresponding to less-well defined structures (Supplementary Figs. 9–11).
In the structure of scSCH, Scm3 interacts broadly with both Cse4 and H4. The αN helix of Scm3 makes close contacts with both the α2 helix of Cse4 and the α3 helix of H4 through multiple hydrophobic interactions (Fig. 1b and Fig. 2a–d and Supplementary Figs. 9c and 13a). Following the αN helix, the loop region of Scm3 (residues 121 to 144) mainly interacts with loop 1 of Cse4, except that a bulge (Scm3 residues 125–130) in the middle of the loop lies on top of loop 2 of H4 (Fig. 1c and Supplementary Figs. 9d and 13b–d). Scm3 loop residues 140–144 also interact with the C-terminal portion of the α2 helix of H4 (Supplementary Figs. 9d and 13e). Interestingly, Scm3 residues 145–154 are completely disordered (Fig. 9b, e and Supplementary Fig. 10). Finally, the C-terminal α-helix (αC) of Scm3 (155–161) makes interactions with the N-terminal region of the α2 helix of H4 (Supplementary Fig. 13f).
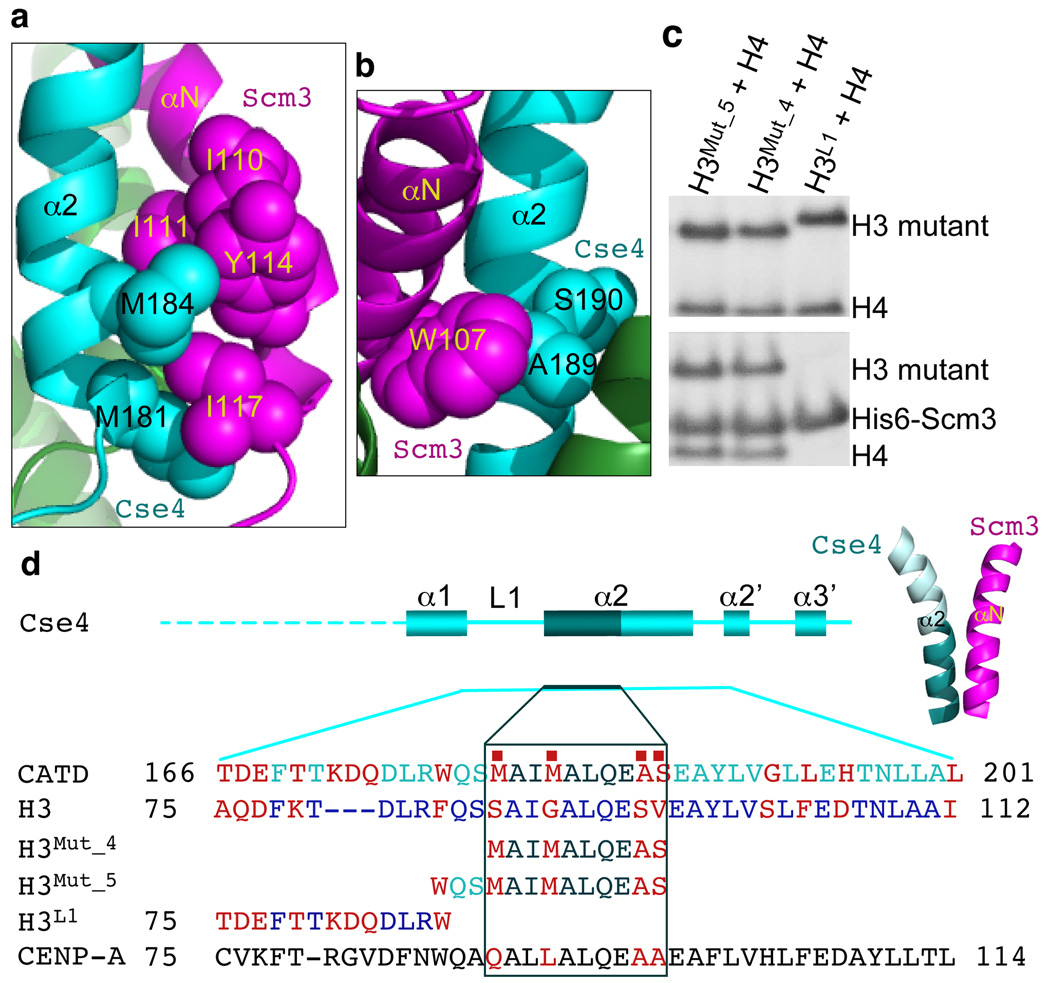
Figure 2. The N-terminal region (181–190) of the α2 helix of Cse4 is the Scm3 recognition motif.
a, Ile110, Ile111, Tyr114, and Ile117 (balls in magenta) in the αN helix of Scm3 form a hydrophobic cluster with Cse4-specific residues Met181 and Met184 (balls in cyan) in the α2 helix of Cse4. b, Trp107 (balls in magenta) in the αN helix of Scm3 has close interactions with the Cse4-specific residue Ala189 (balls in cyan) in the α2 helix of Cse4. Ser190 is also a Cse4-specific residue (balls in cyan). c, SDS-page gels showing the pull-down results with mutants of H3. Upper panel shows the input of H3 mutants and H4. Lower panel shows the molecules eluted from His6-Scm3 (Scm365–169)-bound beads with 250 mM imidazole. d, Illustration of the secondary structures in Cse4 and the Scm3 recognition motif (dark cyan), CATD, and the mutants used in the pull-down experiments. The red squares indicate the four residues that are sufficient for specific recognition of Cse4 by Scm3. The sequences swapped from Cse4 to H3 in the mutations are shown. The sequences that are not changed in the swap are omitted.
Next, we analyzed the effects of mutations on the formation of Scm3-Cse4-H4 complexes with isothermal titration calorimetry. The results reveal that the Scm3 recognition motif resides in the N-terminal region (181–190) of the α2 helix of Cse4. Double mutations Met181Ser/Met184Gly and Ala189Ser/Ser190Val in Cse4 that change the Cse4-specific residues to the corresponding residues in H3 reduced the binding affinity by a factor of 5.5 and 9, respectively (Fig. 2a, b and Supplementary Table 2 and Supplementary Fig. 14). A double mutation Ile110Asp/Ile117Asn in the αN helix of Scm3 decreased the binding affinity by a factor of 85 (Fig. 2a and Supplementary Fig. 14 and Supplementary Table 2). These residues are important for cell growth: mutation of the three residues (Met184, Ala189, and Ser190) in Cse4 to corresponding residues in H3 leads to growth defect (small colony)21 and mutation Ile110Asp/Ile117Asn in Scm3 abrogates cell viability7, consistent with the effects of these mutations on the binding affinity between Scm3 and Cse4/H4 (Supplementary Table 2). Met181 should be important for cell function as well since it interacts with Ile117 of Scm3 (Fig. 2a). It is possible that simultaneous mutation of the four residues in Cse4 to the corresponding residues in H3 would abrogate cell viability.
In contrast, deleting the three extra residues (Lys172, Asp173, and Gln174) and mutating Thr170 in loop 1 (to Lys as in H3), all residues specific to Cse4 (Supplementary Fig. 12), had little effect on the binding affinity (a factor of 1.1) (Supplementary Table 2). Mutation of four residues (Val165, Thr166, Asp167, Glu168) at the C-terminal region of the α1 helix of Cse4 to corresponding residues in H3 (Ile, Ala, Gln, and Asp) also showed little effect on the binding affinity (a factor of 1.4) (Supplementary Table 2). In addition, we found that Scm3 is capable of pulling down the H3CATD-H4 chimera, in which the CATD of Cse4 is swapped to the corresponding region of H322 (Supplementary Fig. 15). Furthermore, Scm3 can pull down an H3 mutant with only four residues replaced by the corresponding residues in the α2 helix of Cse4 (Met181, Met184, Ala188, and Ser189) as well (Fig. 2c, d). Importantly, Scm3 residues that interact with the four Cse4-specific residues are well conserved in human HJURP (Fig. 1a). Indeed, Cse4 can also pull down the N-terminal region (residues 2–81) of HJURP (homologous to the Cse4-binding motif of Scm39, 22) and human H4 (Supplementary Fig. 16). This result is consistent with the ability of Cse4 to replace human CENP-A at centromeres and maintain centromere function in human cells23. In addition, the CENP-A residues that correspond to the four Cse4-specific residues are reasonably conserved (Fig. 2d).
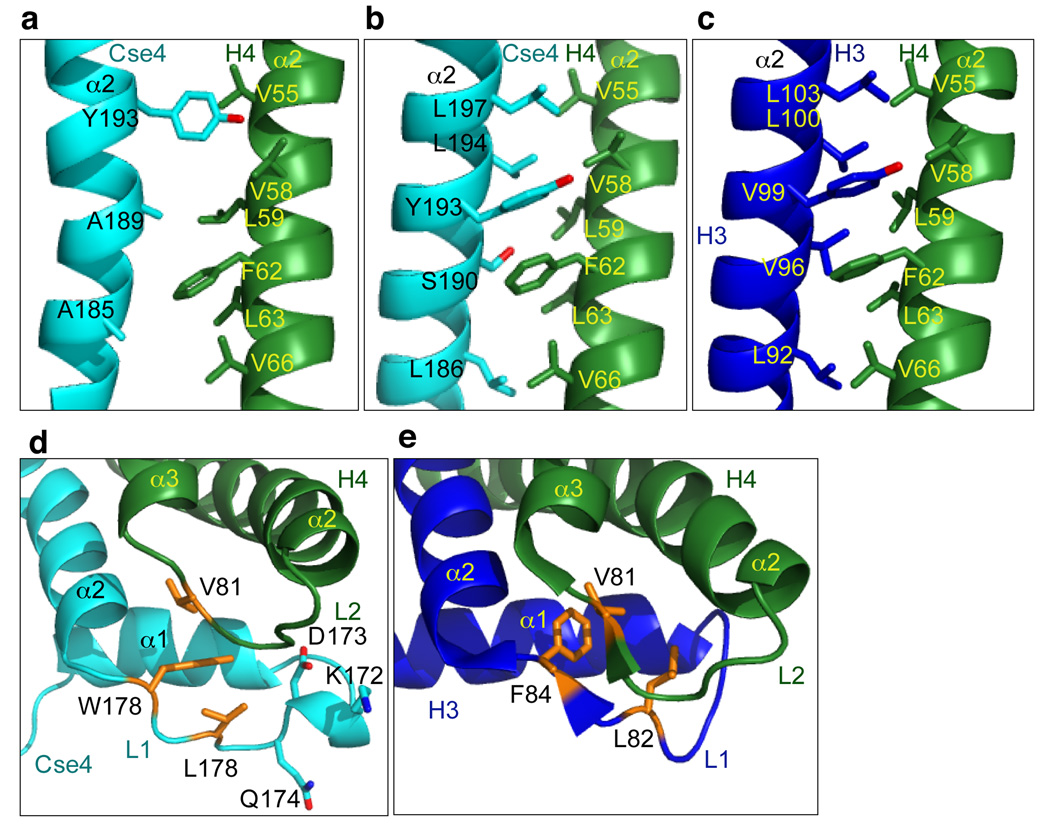
The structure of scSCH reveals the induction of major local conformational changes in the structure of Cse4 and H4 relative to the (CENP-A-H4)2 tetramer. First, the packing between the central α2 helices of Cse4 and H4 in the structure of scSCH is loose in comparison to tight hydrophobic interactions in the homology-modeled dimer based on the H3-H4 structure in the nucleosome (Fig. 3a, b), or in the CENP-A-H4 dimer in the (CENP-A-H4)2 tetramer24. Hydrophobic residues Leu59, Phe62, and Val66 in the α2 helix of H4 lose interacting partners Tyr193, Ser192, and Leu186 in the α2 helix of Cse4 (Fig. 3a, b), (Phe101, Ala98, and Leu94 in human CENP-A24). Second, due to the insertion of the Scm3 loop, loop 1 in Cse4 loses close contact with loop 2 of H4 (Supplementary Fig. 13b) when compared with the corresponding loops in the canonical histone octamer25 (Fig. 3c, d) or in the human (CENP-A-H4)2 tetramer24 (Supplementary Fig. 17). Third, the α2 helix of Cse4 kinks in the middle in scSCH (Fig. 4a), as forced by the side chain of residue Met103 of the αN helix of Scm3 (Fig. 4a). In contrast, the α2 helix of CENP-A or H3 is relatively straight in the CENP-A-H424 (Fig. 4b) or H3-H4 tetramer25 (Supplementary Fig. 18). Fourth, the C-terminal region (94–99) of H4 in scSCH adopts a striking helical conformation and extends the α3 helix of H4. The helical conformation is induced by the side chain of Leu98 in the αN helix of Scm3 through hydrophobic interactions with the side chains of Leu98 and Tyr99 of H4 (Fig. 4a, Supplementary Fig. 14 and Supplementary Table 2). This region is presumably disordered in the (CENP-A-H4)2 tetramer24 (Fig. 4b). Interestingly, the same region forms a mini β-strand that pairs with a β-strand of H2A in the canonical histone octomer or with a β-strand of histone chaperone Asf1 in the Asf1-H3-H4 complex26, 27(Supplementary Fig. 18).
Figure 3. Altered interactions in the CATD region in scSCH.
a, The region of the α2 helices of Cse4 and H4 in scSCH, showing that there is little side chain interaction between the two helices. b, The corresponding region of the α2 helices of Cse4 and H4 in the Cse4-H4 dimer structure obtained by homology modeling based on the structures of H3 and H4 in the nucleosome, showing that there are many hydrophobic interactions. c, The region of the α2 helices of H3 and H4 in the nucleosome. d, Region of loop 1 of Cse4 and the loop 2 of H4. The side chains of the hydrophobic residues are shown in stick and orange. The extra three residues in loop 1 of Cse4 are shown in stick. e, The corresponding loop 1 of H3 and loop 2 of H4 in the nucleosome structure. The hydrophobic residues are shown in stick and orange.
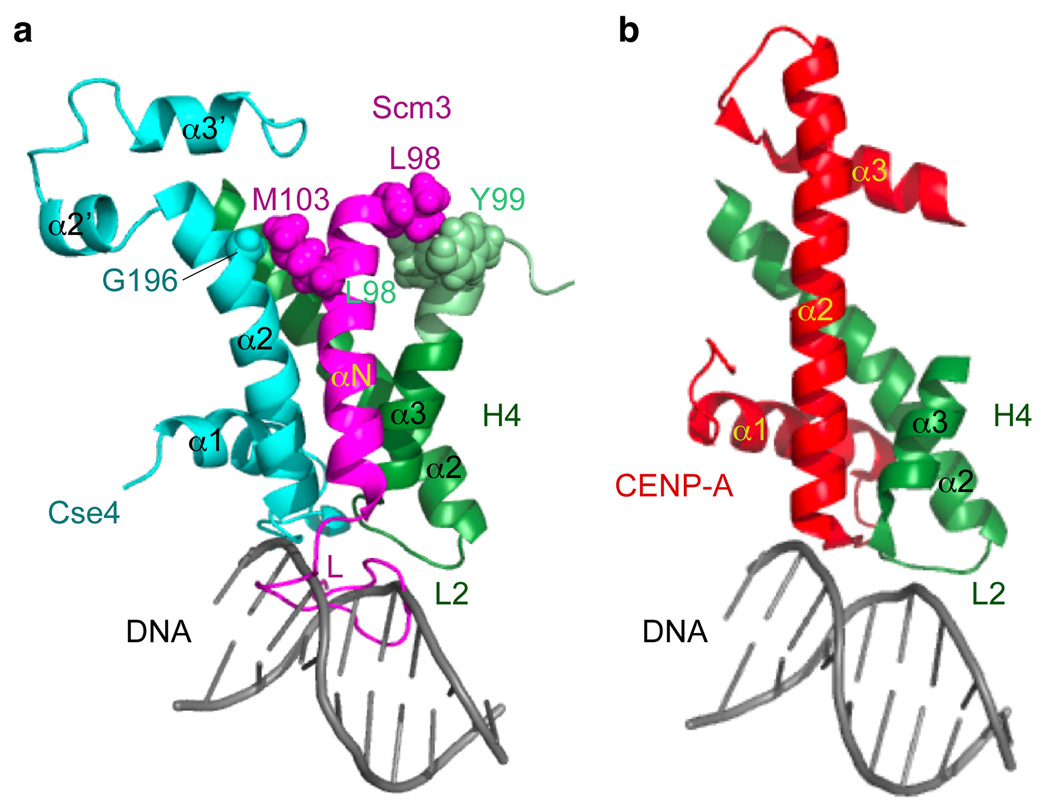
Figure 4. Scm3 induces large conformational changes in Cse4 and H4 and prevents loop 2 of H4 from binding to DNA.
a, Cse4-H4 in scSCH. The extended α3 helix in H4 is shown in light green. The loop of Scm3 pushes loop 1 of Cse4 away from loop 2 of H4 and prevents loop 2 of H4 from binding to DNA. DNA is modeled to bind the loop 2 region of H4 based on the canonical nucleosome structure29. b, CENP-A-H4 in the (CENP-A-H4)2 tetramer. DNA is modeled to bind to loop 2 region of H4 based on the canonical nucleosome struture29.
Furthermore, the C-terminal region of Cse4 has considerable disorder in scSCH (Fig. 4a, b and Fig. 1i). The same region is also disordered in the Cse4-H4 dimer and is not required for Cse4-H4 binding to Scm3 (Supplementary Fig. 5 and Supplementary Fig. 19). This “tetramerization domain” is well folded in the (CENP-A-H4)2 tetramer. Structure modeling shows that imposing this folded domain on the corresponding region of Cse4 in scSCH allows association as a (Scm3-Cse4-H4)2 hexamer without major structural incompatibility (Supplementary Fig. 20), consistent with the existence of (Scm3-Cse4-H4)2 hexamers in 2M NaCl5. However, in this context, histone topography in the scSCH structure outside the tetramerization domain displays dramatic global conformational changes when compared with the (CENP-A-H4)2 tetramer, making the modeled (Scm3-Cse4-H4)2 hexamer incompatible with DNA binding (Supplementary Fig. 20). Moreover, the Scm3 loop in the scSCH structure blocks loop 2 of H4, which makes contacts with DNA in the canonical nucleosome (Fig. 4a, b).
Thus, the structure of scSCH suggests that retention of Scm3 in association with centromere DNA is unlikely to occur via binding of Scm3-CBD to Cse4/H4, as binding of DNA and Scm3-CBD to Cse4/H4 are mutually incompatible. Instead, Scm3-CBD behaves as a specific histone chaperone, and the retention of Scm3 with Cse4/H4 on centromeric DNA requires its distinct DNA-binding domain (H. Xiao and C. Wu, manuscript in preparation).
The structure of scSCH reveals that a sub-region within the CATD, including four Cse4-specific residues in the N-terminal region of the α2 helix of Cse4, is necessary and sufficient for specific recognition by Scm3. Thus, the remainder of the CATD of Cse4 should be important for association with other proteins for Cse4 functions. The CBD of Scm3 uses both induced histone conformation changes26, 27 and direct steric occlusion28 to prevent Cse4-H4 in the Scm3-Cse4-H4 complex from DNA binding (Supplementary Fig. 21). Conversely, Cse4-H4, with a conformation similar to that of CENP-A-H4 in the (CENP-A-H4)2 tetramer, is unfavorable for Scm3-CBD binding but favors DNA binding, suggesting a competition mechanism for Scm3 and HJURP as CenH3-specific chaperones22.
Methods Summary
See Supplementary Information for detailed methods.
All the proteins used in the present study were over-expressed in Escherichia coli and purified using Ni-NTA column (Qiagen), ion exchange, gel filtration, and reverse-phase HPLC (Waters). Uniformly isotope-labeled proteins were produced using M9 medium with 15NH4Cl, 13C-D-Glucose, and D2O as the sole source of the isotopes. The molecular weight and stoichiometry of the complex were determined by velocity and equilibrium sedimentation experiments on a Beckman Coulter Proteome XL-I analytical ultracentrifuge at 20.0 °C. The multi-dimensional NMR spectra were collected on Bruker 500, 600, 800, and 900 MHz and Varian 600 and 800 MHz instruments. The structure was calculated using the distance constraints measured by NMR and the program Xplor-NIH. Mutations were made using a quick-change kit. The binding constants were measured on the MicroCal VP-ITC instrument.
Methods
Protein sample preparation
All proteins were expressed in E. coli (BL21-codonPlus(DE3)-RIL) with pET vectors (Stratagene). N-terminal His6-tagged fragments of Cse4 and Scm3 and their mutants were first purified via Ni-NTA (Qiagen) whereas H4 and non-His-tagged fragments of Cse4 and Scm3 were first purified via SP sepharose (GE Healthcare). They were next subjected to reverse phase HPLC purification using acetonitrile and water as solvents. Purified proteins were lyophilized. Isotope-labeled proteins for NMR studies were produced by growing E. coli cells in M9 media with 15NH4Cl, U-13C6-Glucose, and D2O as the sole source for nitrogen, carbon, and deuterium, respectively. For the measurement of side chain NOEs, specific methyl labeling (-13CH3) for Ile, Leu, and Val residues was also made following the protocol of Kay and coworkers30.
To prepare the Cse4, H4 and Scm3 complexes, lyophilized proteins were first dissolved in H2O. Their concentrations were determined by measuring the absorbance at 280 nm. Equal amounts of each species were mixed together and dialyzed against 10 mM Tris-HCl and 2 M NaCl at pH 7.4 and 4°C. After centrifugation, the soluble fractions were subjected to gel filtration on Superdex 75 10/300 GL column (GE Healthcare, USA). The eluted complexes were concentrated with an Amicon with Ultra Ultracel-10 membrane (Millipore, USA) and exchanged to a final buffer of 50 mM MES at pH 5.6. The Cse4-H4 complexes were made in the same way. Scm380–211 samples for NMR study were prepared by dissolving them in 8M urea and dialysis against corresponding buffer. scSCH and all other single chain molecules derived from scSCH are purified with Ni-NTA (Qiagen) under native conditions (20 mM Tris-HCl and 0.5 M NaCl at pH 8.0), followed by gel filtration with Superdex 200 10/60 column at 4 °C (GE healthcare, USA). The fractions with target protein were concentrated and exchange to its final buffer.
Analytical ultracentrifugation
Sedimentation velocity experiments were conducted in duplicate at 20.0°C on a Beckman Coulter Proteome XL-I analytical ultracentrifuge. 400 µL of the sample of 35 µM in 50 mM Mes (pH, 5.6) was loaded in a double sector centerpiece cell and analyzed at a rotor speed of 50 krpm. 100 scans were acquired as single absorbance measurements (λ = 280 nm) at 7.1 minute intervals using a radial spacing of 0.003 cm. Data were analyzed in SEDFIT 11.71 in terms of a continuous c(s) distribution to obtain a sedimentation coefficient, s, and molecular mass M (ref 31). Solution densities ρ were measured at 20.00 °C on a Mettler Toledo DE51 density meter and solution viscosities η were measured using a Cannon-Ubbelohde viscometer and Cannon-CT 500 constant temperature bath set at 20.00 °C. The partial specific volume v of the complex was calculated in SEDNTERP 1.09 (ref 32). c(s) analyses were carried out using an s-value range of 0.5 to 6.0 with a linear resolution of 100 and a confidence level (F-ratio) of 0.68. The analyses, implemented using time independent noise corrections, returned root mean square deviation (rmsd) values for the best fits of 0.0040 absorbance units. Sedimentation equilibrium experiments were conducted at 20.0°C on a Beckman Optima XL-A. 135 µL volumes of the complex were studied at loading concentrations of 20, 39 and 78 µM, along with the sample recovered from the sedimentation velocity experiments. Experiments were carried out using six channel centerpiece cells at rotor speeds ranging from 18 to 34 krpm. In all cases data were acquired as an average of 4 absorbance measurements at wavelengths of 280 and 250 nm using a radial spacing of 0.001 cm. Sedimentation equilibrium at each speed was achieved within 40 hours. Data were analyzed globally in terms of a single ideal species using SEDPHAT 6.21 (refs 32, 33).
NMR experiments
NMR experiments were performed on Bruker 500, 600, 800, and 900 MHz and Varian 600 and 800 MHz spectrometers at 35°C. The following experiments were recorded. 2D: [1H, 1H]-NOESY, [1H, 15N]-TROSY, [1H, 13C]-HMQC, 15N-{1H} NOE; TROSY version 3D: HNCACB, HNCOCACB, HNCA, HNCOCA, HNCO, HNCACO; 3D HBHACONH, HCCH-TOCSY, CCH-TOCSY, CCC(CO)NH, [1H,15N]-NOESY-HSQC, [1H,15N] NOESY-HSQC (13C methyl-labeled sample), [1H, 13C]-NOESY-HSQC, [1H,13C]-NOESY-HSQC (13C methyl-labeled sample). The spectra were processed using NMRPipe34 and analyzed with NMRView35.
Structure calculation
Structure calculation was done using the program Xplor-NIH7. The NOE-derived restraints were subdivided into four classes, strong, medium, weak, and very weak by comparison with NOEs of protons separated by known distances as described in Zhou et al.37. Backbone dihedral angle restraints (ϕ and ψ angles) were obtained from analysis of 1Hα, HN, 13Cα, 13Cβ, 13CO, and 15N chemical shifts by using the program TALOS38. Two constraints per hydrogen bond (dNH-O ≤ 2.2 Å and dN-O ≤ 3.2 Å) were added in the final structure calculation after initial NOE-derived structures were obtained. The program PROCHECK_NMR39 was used to evaluate the quality of the calculated structures.
Isothermal titration calorimetric experiments
The ITC experiments were performed on a MicroCal VP-ITC by injecting Scm383–169 solution (250 µM) to a solution of single chain Cse4-H4 or their mutants (His6-KK-Cse4151–207-LVPRGS-H445–103) solution (20 µM) in a chamber of 1.4 ml at 25 °C in 50 mM MES (pH 5.4) and 0.1 M NaCl. Twenty-nine injections (each of 10 µl) were made and the heat released was analyzed. The data was analyzed as described by Houtman et al.40.
Pull-down experiments
Pull-down experiments were carried out in 50 mM sodium phosphate, 25 mM imidazole, 2 M NaCl, pH 8.0 at room temperature. Ni-NTA (Qiagen) beads were mixed with His6-Scm366–169 with a final concentration of 6 µM. Approximately 10 fold excess of (Cse4-H4)2 or (H3-H4)2 or their mutants was mixed with beads and incubated. The incubation was at 25 °C for 30 minutes. The beads were washed with the same buffer three times. The complex formed on the beads was eluted with 250 mM imidazole and analyzed by SDS-PAGE. Beads without His6-Scm3 were also incubated with corresponding (Cse4-H4)2 under identical conditions to assess background binding and the integrity of the tetramer. No non-specific binding was identified in 2 M NaCl. For molecules derived from thrombin digested single chain proteins, the complex was incubated with Ni-NTA (Qiagen) beads at 25 °C for 30 minutes and then washed 3 times. The final complex formed on the beads was eluted with either 8 M Urea or 250 mM imidazole. The eluted molecules were analyzed by SDS-PAGE.
Supplementary Material
Acknowledgements
We thank Drs. J. Ying, K. Varney, J. F. Ellena, and J. Gruschus for help collecting NMR spectra, A. Bax for discussion, C. Klee and M. Lichten for comments on the manuscript, and D. Cleveland for plasmids of human CENP-A and H4 histones. This work is supported by the intramural research programs of NCI, NIDDK, and NHLBI.
Footnotes
Author contributions Z.Z. and H.F. contributed equally to this work. Z.Z. performed protein engineering, biochemical, and ITC studies. B-R. Z. contributed to protein sample preparation. B-R.Z. and L.J. contributed to the analysis of ITC data. H.F., K.H., A.Z., and N.T. collected the NMR spectra. H.F. and Z.Z. analyzed the NMR data and H.F. solved the structure. R.G. performed the sedimentation experiments. H.X. provided initial plasmids and guidance in cloning. C.W. proposed the project and participated in manuscript writing. Y.B. contributed to the overall strategy, project management and writing of the manuscript. All authors read and commented on the manuscript.
Author Information The atomic coordinates have been deposited in the Protein Data Bank (PDB id: 2L5A). Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Cleveland DW, Mao Y, Sullivan KF. Centromeres and Kinetochores: From epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 2.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 3.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 4.Black BE, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 5.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 6.Camahort R, et al. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Stoler S, et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camahort R, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez-Pulido L, Pidoux AL, Pointing CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pidoux AL, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foltz DR, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–482. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 14.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 15.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 16.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 17.Cottarel G, Shero JH, Hieter P, Hegemann JH. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol. Cell Biol. 1989;9:3342–3349. doi: 10.1128/mcb.9.8.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 19.Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keith KC, et al. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shuaib M, Ouararhni K, Dimiyrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A–H4)2 reveals physical features that mark centromeres. Nature. 2010 doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood CM, et al. High-resolution structure of the native histone octamer. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 2005;61:541–545. doi: 10.1107/S1744309105013813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Natsume R, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Z, et al. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat. Struct. Mol. Biol. 2008;15:868–869. doi: 10.1038/nsmb.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 31.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity untracentrifugation and Lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole JL, Lary JW, Moody TP, Laue TM. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008;84:143–179. doi: 10.1016/S0091-679X(07)84006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 34.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Johnson BA, Blevins RA. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 36.Schwieters CD, Kuszewski J, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Z, et al. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nat. Struct. Mol. Biol. 2008;15:868–869. doi: 10.1038/nsmb.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, et al. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 40.Houtman JC, et al. Binding specificity of multiprotein signaling complexes is determined by both cooperative interactions and affinity preferences. Biochemistry. 2004;43:4170–4178. doi: 10.1021/bi0357311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.