Abstract
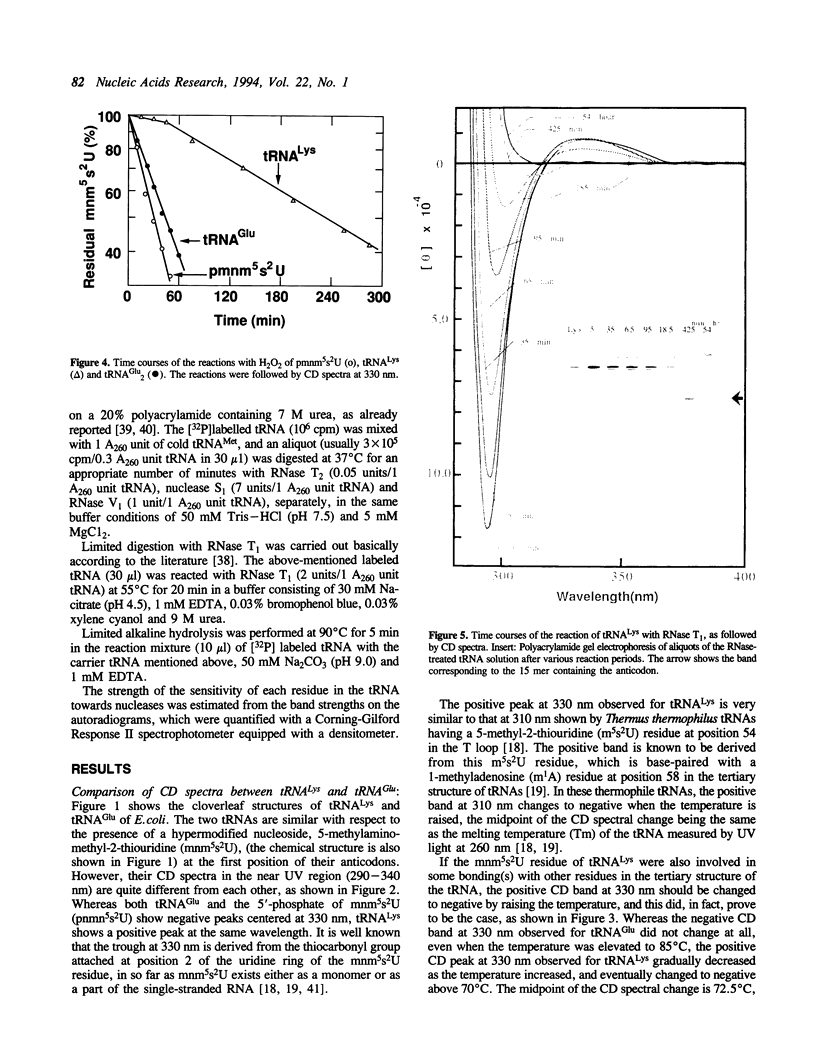
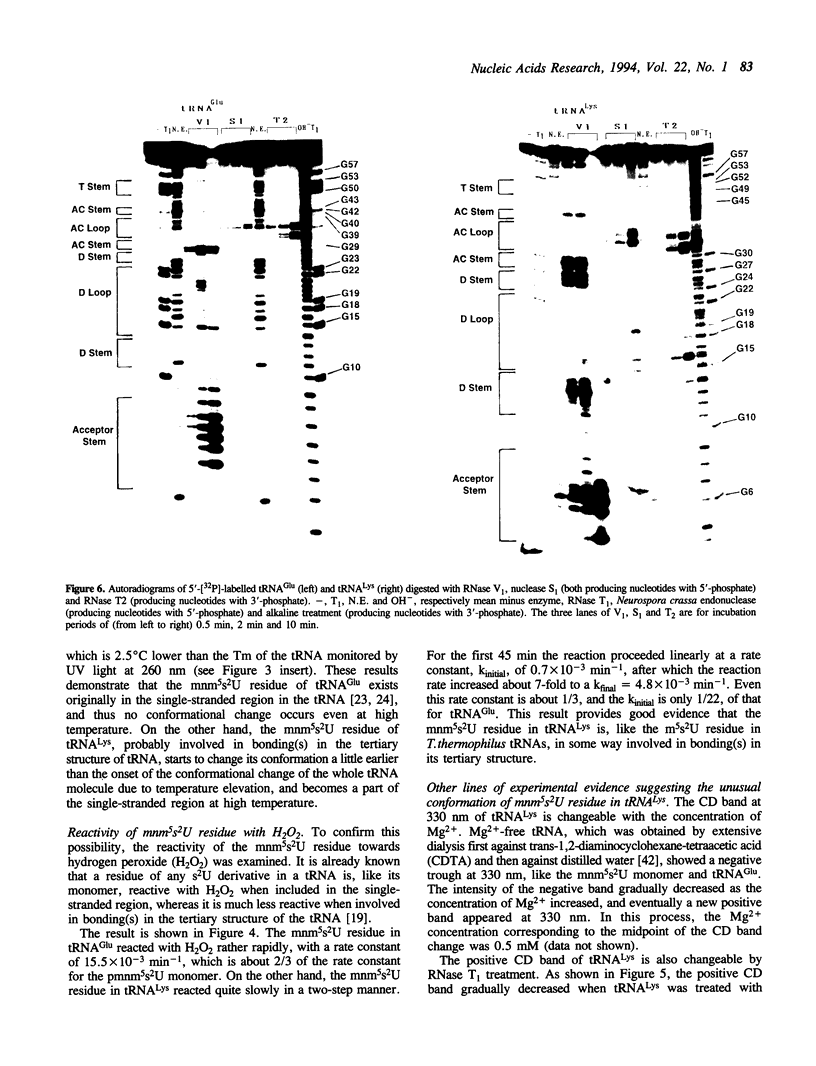
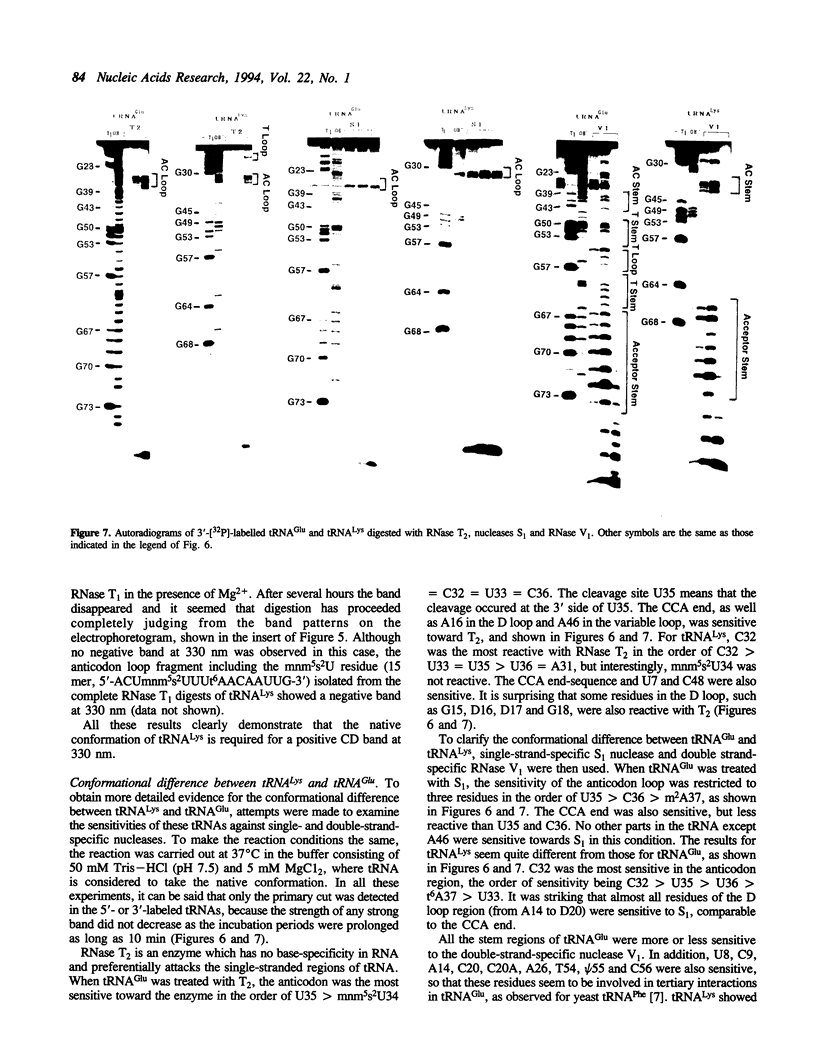
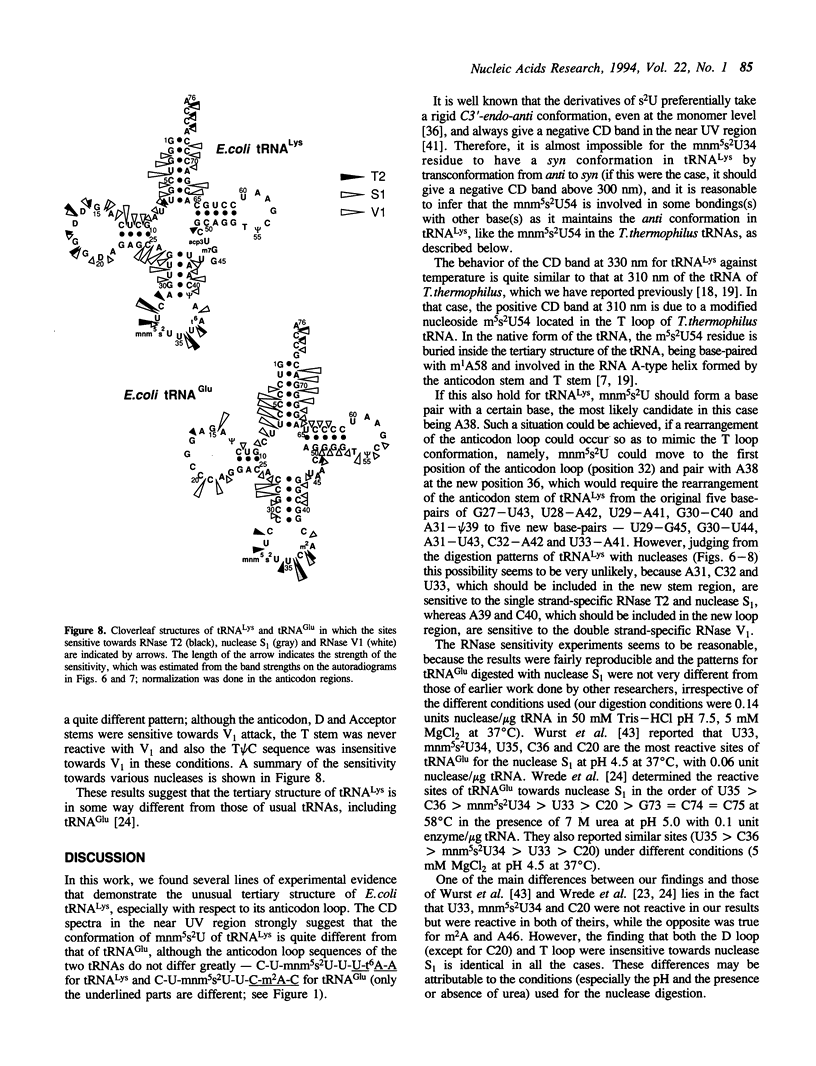
Although both tRNA(Lys) and tRNA(Glu) of E. coli possess similar anticodon loop sequences, with the same hypermodified nucleoside 5-methylaminomethyl-2-thiouridine (mnm5s2U) at the first position of their anticodons, the anticodon loop structures of these two tRNAs containing the modified nucleoside appear to be quite different as judged from the following observations. (1) The CD band derived from the mnm5s2U residue is negative for tRNA(Glu), but positive for tRNA(Lys). (2) The mnm5s2U monomer itself and the mnm5s2U-containing anticodon loop fragment of tRNA(Lys) show the same negative CD bands as that of tRNA(Glu). (3) The positive CD band of tRNA(Lys) changes to negative when the temperature is raised. (4) The reactivity of the mnm5s2U residue toward H2O2 is much lower for tRNA(Lys) than for tRNA(Glu). These features suggest that tRNA(Lys) has an unusual anticodon loop structure, in which the mnm5s2U residue takes a different conformation from that of tRNA(Glu); whereas the mnm5s2U base of tRNA(Glu) has no direct bonding with other bases and is accessible to a solvent, that of tRNA(Lys) exists as if in some way buried in its anticodon loop. The limited hydrolysis of both tRNAs by various RNases suggests that some differences exist in the higher order structures of tRNA(Lys) and tRNA(Glu). The influence of the unusual anticodon loop structure observed for tRNA(Lys) on its function in the translational process is also discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amitsur M., Morad I., Kaufmann G. In vitro reconstitution of anticodon nuclease from components encoded by phage T4 and Escherichia coli CTr5X. EMBO J. 1989 Aug;8(8):2411–2415. doi: 10.1002/j.1460-2075.1989.tb08371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinkowa A. L., Walker J. R. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 1990 Apr 11;18(7):1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A. D., Uhlenbeck O. C., Tinoco I., Jr Circular dichroism study of nine species of transfer ribonucleic acid. Biochemistry. 1972 Aug 15;11(17):3248–3256. doi: 10.1021/bi00767a019. [DOI] [PubMed] [Google Scholar]
- Chakraburtty K., Steinschneider A., Case R. V., Mehler A. H. Primary structure of tRNA-Lys of E. coli B. Nucleic Acids Res. 1975 Nov;2(11):2069–2075. doi: 10.1093/nar/2.11.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P., Sprinzl M., Watanabe K., Albani M., Kersten H. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res. 1979 Apr;6(4):1571–1581. doi: 10.1093/nar/6.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhuff T. J., Atkins J. F., Gesteland R. F. Characterization of ribosomal frameshift events by protein sequence analysis. J Biol Chem. 1986 Jun 5;261(16):7491–7500. [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower A. M., McHenry C. S. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci U S A. 1990 May;87(10):3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLLEY R. W., APGAR J., EVERETT G. A., MADISON J. T., MARQUISEE M., MERRILL S. H., PENSWICK J. R., ZAMIR A. STRUCTURE OF A RIBONUCLEIC ACID. Science. 1965 Mar 19;147(3664):1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- HOSKINSON R. M., KHORANA H. G. STUDIES ON POLYNUCLEOTIDES. XLI. PURIFICATION OF PHENYLALANINE-SPECIFIC TRANSFER RIBONUCLEIC ACID FROM YEAST BY COUNTERCURRENT DISTRIBUTION. J Biol Chem. 1965 May;240:2129–2134. [PubMed] [Google Scholar]
- Hare D. R., Ribeiro N. S., Wemmer D. E., Reid B. R. Complete assignment of the imino protons of Escherichia coli valine transfer RNA: two-dimensional NMR studies in water. Biochemistry. 1985 Jul 30;24(16):4300–4306. doi: 10.1021/bi00337a008. [DOI] [PubMed] [Google Scholar]
- Heerschap A., Mellema J. R., Janssen H. G., Walters J. A., Haasnoot C. A., Hilbers C. W. Imino-proton resonances of yeast tRNAPhe studied by two-dimensional nuclear Overhauser enhancement spectroscopy. Eur J Biochem. 1985 Jun 18;149(3):649–655. doi: 10.1111/j.1432-1033.1985.tb08973.x. [DOI] [PubMed] [Google Scholar]
- Holmes W. M., Hurd R. E., Reid B. R., Rimerman R. A., Hatfield G. W. Separation of transfer ribonucleic acid by sepharose chromatography using reverse salt gradients. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1068–1071. doi: 10.1073/pnas.72.3.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houssier C., Degée P., Nicoghosian K., Grosjean H. Effect of uridine dethiolation in the anticodon triplet of tRNA(Glu) on its association with tRNA(Phe). J Biomol Struct Dyn. 1988 Jun;5(6):1259–1266. doi: 10.1080/07391102.1988.10506468. [DOI] [PubMed] [Google Scholar]
- Hyde E. I., Reid B. R. Assignment of the low-field 1H NMR spectrum of Escherichia coli tRNAPhe using nuclear Overhauser effects. Biochemistry. 1985 Jul 30;24(16):4307–4314. doi: 10.1021/bi00337a009. [DOI] [PubMed] [Google Scholar]
- Kawai G., Yokoyama S., Hasegawa T., Murao K., Ishikura H., Nishimura S., Miyazawa T. Two-dimensional NMR analyses of dynamic structures of tRNA and the regulation of codon recognition by post-transcriptional modifications. Nucleic Acids Symp Ser. 1988;(19):105–106. [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Ziro, Harada Fumio, Nishimura Susumu. Primary sequence of glutamic acid tRNA II from Escherichia coli. FEBS Lett. 1972 Feb 1;20(2):239–241. doi: 10.1016/0014-5793(72)80804-4. [DOI] [PubMed] [Google Scholar]
- Rich A., Kim S. H. The three-dimensional structure of transfer RNA. Sci Am. 1978 Jan;238(1):52–62. doi: 10.1038/scientificamerican0178-52. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Roy S., Redfield A. G. Assignment of imino proton spectra of yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1983 Mar 15;22(6):1386–1390. doi: 10.1021/bi00275a010. [DOI] [PubMed] [Google Scholar]
- Schevitz R. W., Podjarny A. D., Krishnamachari N., Hughes J. J., Sigler P. B., Sussman J. L. Crystal structure of a eukaryotic initiator tRNA. Nature. 1979 Mar 8;278(5700):188–190. doi: 10.1038/278188a0. [DOI] [PubMed] [Google Scholar]
- Sekine Y., Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Meissner F., Moll J., Vorderwülbecke T. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1987;15 (Suppl):r53–188. doi: 10.1093/nar/15.suppl.r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z., Brown P. O. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev. 1992 Mar;6(3):511–519. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z. Translational frameshifting in the Escherichia coli dnaX gene in vitro. Nucleic Acids Res. 1991 May 11;19(9):2457–2462. doi: 10.1093/nar/19.9.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary C. P., Vournakis J. N. RNA structure analysis using T2 ribonuclease: detection of pH and metal ion induced conformational changes in yeast tRNAPhe. Nucleic Acids Res. 1984 Sep 11;12(17):6763–6778. doi: 10.1093/nar/12.17.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Imahori K. The conformation difference between tRNA Met f and formylmethionyl-tRNA Met f from E. coli. Biochem Biophys Res Commun. 1971 Oct 15;45(2):488–494. doi: 10.1016/0006-291x(71)90845-x. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Miura K. Specific interaction between tRNA and its cognate amino acid as detected by circular dichroism and fluorescence spectroscopy. Biochem Biophys Res Commun. 1985 Jun 28;129(3):679–685. doi: 10.1016/0006-291x(85)91945-x. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Oshima T., Iijima K., Yamaizumi Z., Nishimura S. Purification and thermal stability of several amino acid-specific tRNAs from an extreme thermophile, Thermus thermophilus HB8. J Biochem. 1980 Jan;87(1):1–13. doi: 10.1093/oxfordjournals.jbchem.a132713. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Oshima T., Nishimura S. CD spectra of 5-methyl-2-thiouridine in tRNA-Met-f from an extreme thermophile. Nucleic Acids Res. 1976 Jul;3(7):1703–1713. doi: 10.1093/nar/3.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Oshima T., Saneyoshi M., Nishimura S. Replacement of ribothymidine by 5-methyl-2-thiouridine in sequence GT psi C in tRNA of an extreme thermophile. FEBS Lett. 1974 Jul 1;43(1):59–63. doi: 10.1016/0014-5793(74)81105-1. [DOI] [PubMed] [Google Scholar]
- Watanabe K. Reactions of 2-thioribothymidine ad 4-thiouridine with hydrogen peroxide in transfer ribonucleic acids from Thermus thermophilus and Escherichia coli as studied by circular dichroism. Biochemistry. 1980 Nov 25;19(24):5542–5549. doi: 10.1021/bi00565a013. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Yokoyama S., Hansske F., Kasai H., Miyazawa T. CD and NMR studies on the conformational thermostability of 2-thioribothymidine found in the T psi C loop of thermophile tRNA. Biochem Biophys Res Commun. 1979 Nov 28;91(2):671–677. doi: 10.1016/0006-291x(79)91574-2. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Shuh M., Atkins J. F., Gesteland R. F. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989 Nov;1(2):159–169. [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Loop stereochemistry and dynamics in transfer RNA. J Biomol Struct Dyn. 1983 Oct;1(2):337–355. doi: 10.1080/07391102.1983.10507446. [DOI] [PubMed] [Google Scholar]
- Willick G. E., Kay C. M. Magnesium-induced conformational change in transfer ribonucleic acid as measured by circular dichroism. Biochemistry. 1971 Jun 8;10(12):2216–2222. doi: 10.1021/bi00788a005. [DOI] [PubMed] [Google Scholar]
- Woo N. H., Roe B. A., Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980 Jul 24;286(5771):346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- Wrede P., Woo N. H., Rich A. Initiator tRNAs have a unique anticodon loop conformation. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3289–3293. doi: 10.1073/pnas.76.7.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrede P., Wurst R., Vournakis J., Rich A. Conformational changes of yeast tRNAPhe and E. coli tRNA2Glu as indicated by different nuclease digestion patterns. J Biol Chem. 1979 Oct 10;254(19):9608–9616. [PubMed] [Google Scholar]
- Wurst R. M., Vournakis J. N., Maxam A. M. Structure mapping of 5'-32P-labeled RNA with S1 nuclease. Biochemistry. 1978 Oct 17;17(21):4493–4499. doi: 10.1021/bi00614a021. [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Watanabe T., Murao K., Ishikura H., Yamaizumi Z., Nishimura S., Miyazawa T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4905–4909. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S., Yamaizumi Z., Nishimura S., Miyazawa T. 1H NMR studies on the conformational characteristics of 2-thiopyrimidine nucleotides found in transfer RNAs. Nucleic Acids Res. 1979 Jun 11;6(7):2611–2626. doi: 10.1093/nar/6.7.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]