Abstract
Association analysis has led to identification of many genetic variants for complex diseases. While assessing the association between genes and a disease, other factors can play an important role. The consequence of not considering covariates (such as population stratification and environmental factors) is well-documented in genetic studies. We introduce a nonparametric test of association that adjusts for covariate effects. Specifically, the adjustment is realized through weights that are constructed from genomic propensity scores that summarize the contribution of all covariates. The benefit of our test is demonstrated through an important dataset on bipolar disorder (BD) collected by the Wellcome Trust Case Control Consortium (WTCCC). When compared to other tests, our test identified an un-reported region with three single nucleotide polymorphisms (SNPs) on chromosome 16 that show strong evidence of association (p-value < 5×10−7). This region is near the RPGRIP1L gene known to be associated with bipolar disorder. A haplotype block including these three SNPs was further discovered to be strongly associated with bipolar disorder. It is also interesting to note that our nonparametric test did not reveal strong signals at two SNPs that were detected by a covariate-adjusted parametric test. This suggests that different methods of covariate adjustment can complement each other. Thus, we recommend using both parametric and nonparametric testing. Additionally, we performed simulation studies to compare our proposed test with the unadjusted test and an adjusted parametric test. Our finding underscores the importance of accommodating and controlling for covariate effects in discovering genetic variants associated with complex disorders.
Keywords: bipolar disorder, covariate adjustment, haplotype analysis, propensity score
INTRODUCTION
Over the past several years, genome-wide association studies (GWAS) have been successful in localizing and identifying important genes that contribute to common human diseases [Klein et al., 2005; Arking et al., 2006; Duerr et al., 2006; Chen et al., 2007]. To test the association between a genetic variant and a disease, it has become increasingly clear that environmental factors may have important confounding effects [Amos et al., 1996; Lu and Cantor, 2007; Vansteelandt et al., 2009]. For example, population stratification is a special case of confounding caused by ethnicity or ancestry differences that a failure to account for this stratification can lead to a spurious association. There are many methods specifically designed to address population stratification such as genomic control, structured association, and the principal component method [Devlin and Roeder, 1999; Pritchard and Rosenberg, 1999; Price et al., 2006]. It remains important, however, to develop a general method to assess association in the presence of covariates.
A common method of accounting for covariates is to regress phenotypic values against the covariates, and then to perform an association analysis using the residuals against the genotypes [Wang et al., 2006; Lu and Cantor, 2007]. While this two-step approach is convenient, the theoretical properties of this approach are difficult to study due to the lack of an integrated model. Additionally, this framework may not be suitable when the covariates themselves are correlated with the genes. Another useful approach is to fit a parametric model of the relationship between phenotype and all risk factors. Here, the risk factors include both genotypes and covariates. For example, logistic regressions are typically used to test genetic contribution while adjusting for the covariates in a case-control study. The underlying assumptions of parametric models however, can be difficult to verify, especially so for discrete outcomes that usually involve the choice of link function. The shortcomings of these parametric methodologies motivate our nonparametric approach that tests the association between the phenotype and a genetic variant, while adjusting for the covariates.
Score tests are particularly useful in genetic studies and many of them are unified under the U-statistics [Laird et al., 2000; Zhang et al., 2010]. Most of these statistics evaluate the direct association between the phenotype and a genetic variant without adjusting for the covariates. Meanwhile, the concept of propensity score has been proven useful to remove the bias of the treatment effect caused by confounding variables, enabling causal inference [Rosenbaum and Rubin, 1983; Zhao et al., 2009]. Recall that the balance property of propensity scores [Rosenbaum and Rubin, 1983, Theorems 1–2] indicates that if a subclass of units is homogeneous in propensity score, then the treated and control units in that subclass will have the same distribution of covariates.
Thus, we propose a weighted nonparametric statistic of association by using propensity scores. Instead of exactly matching the samples using propensity scores, we choose to weigh each sample pair in the U-statistic in Zhang et al. (2010) according to their propensity scores. In this way, we are aiming to achieve the balance property without losing power of the test. In particular, we use propensity scores to summarize the contribution of all covariates, and then derive the weights from these scores. From the viewpoint of U-statistic, the weights increase the contribution of each sample pair if they share similar propensity scores, and they reduce that contribution otherwise. Taking into account the covariates, we use this weighted statistic to test the null hypothesis that there is no association between the phenotype and a genetic variant conditional on the covariates.
MATERIALS AND METHODS
Bipolar Disorder Dataset and Quality Control
We were given permission to use the bipolar disorder data set collected by WTCCC to demonstrate the utility of our test. The data contain 1998 cases and 3004 controls. The control samples came from two sources: half from the 1958 Birth Cohort (58C) and the remainder from a new UK Blood Service (UKBS) sample. The genotyping was performed using GeneChip 500K arrays at the Affymetrix Services Lab (California). After proper quality control/quality assurance (QC/QA), WTCCC (2007) identified a strong signal (p-value < 5×10−7) for bipolar disorder at SNP rs420259 on chromosome 16p12, and 13 other SNPs with moderate association (p-value between 10−5 and 5×10−7) genome-widely.
Following the same QC/QA procedure as in WTCCC (2007), we used a quality score threshold for inclusion of SNPs at 0.9, treating the genotype as missing when the most probable call fell below this threshold. Furthermore, we removed 130 samples from the BD cohort, 24 samples from the 58C cohort, and 42 samples from the UKBS cohort, due to low call rates, overall heterozygosity, and evidence of non-European ancestry. Additionally, SNPs were excluded from our analysis according to the exclusion list provided by WTCCC, including missing data rates and departures from Hardy-Weinberg equilibrium. We performed further quality controls based on minor allele frequency (MAF > 5%) and Hardy-Weinberg equilibrium in both cases and controls (p-value > 0.0001). As the WTCCC investigators, we set 5×10−7 as the genome-wide significance threshold for strong association, and p-values between 10−5 and 5×10−7 as moderate association.
Unadjusted Association Test
First, the SNP association is evaluated using the unadjusted association test in Zhang et al. (2010). To introduce the notation, suppose that we have data from n subjects, and for subject i, let Yi denote the trait and Gi a genotypic score. Here, Y can be the case/control status or a quantitative trait, and G can be the number of mutant alleles. The association test is based on the U-statistic generalized from Kendall’s tau to measure the association between Y and G, namely, . In the absence of covariates, the test statistic U follows an asymptotically normal distribution under the null hypothesis conditioning on all phenotypes [Rabinowitz and Laird, 2000]. Once the asymptotic mean and variance of U are evaluated, it can be used to test the null hypothesis H0 that there is no association between Y and G.
Covariate-Adjusted Nonparametric Association Test
Before we propose the covariate-adjusted nonparametric test of association, we provide a brief review of genomic propensity scores [Zhao et al., 2009]. For a SNP marker with two alleles A and a, the genomic propensity score is the probability of an individual having a particular test-locus genotype (AA, Aa, or aa) based on each specific individual’s covariates Z, pg(z) = P(G = g | Z = z), where g and z are observed values of G and Z. Specifically, for a dominant or a recessive model with g = 0 or 1, the propensity score becomes p(z) = p1(z) = P (G = 1|Z = z), where p0(z) = 1 − p1(z). For an additive model with g = 0, 1, or 2, the propensity score p(z) is a vector p(z) = (p1(z),p2(z)) = (P(G = 1|Z = z),P(G = 2|Z = z)), where p0(z) = 1 − p1(z) − p2(z).
As in matching subjects according to their propensity scores [Rosenbaum and Rubin, 1984], we propose a weighted U-statistic by imposing weights to each observation pair (i, j). In particular, the propensity scores are used to summarize the contribution of all covariates; weights are then derived from these scores. Sample pairs that share similar propensity scores are more heavily weighed than sample pairs that do not share similar propensity scores. That is, for the sample pair (i, j), we first evaluate p(zi) and p(zj), then a weight function is derived using a positive weight function, W : w(p(zi),p(zj)) = W(p(zi) − p(zj)). Thus, the proposed weighted test statistic is: .
To account for covariates, this weighted test statistic is used to test the null hypothesis, H0′, that there is no association between the phenotype and the genetic marker conditional on the covariates, that is, Y and G are independent conditional on the covariates, Z. The mean and variance of the weighted test statistic under this null hypothesis can be calculated conditioning on all phenotypes and covariates. Then the calculations become similar to those in Zhang et al. (2010) except that we need to evaluate P(Gi = g|Zi = zi) for each subject i, which are exactly the genomic propensity scores pg(zi). After calculating the mean and variance, a χ2 test statistic can be obtained from Uw for testing purpose.
When analyzing the bipolar disorder data set, the propensity scores were estimated from the data using proportional odds logistic model as log it(P(Gi ≤ g | Zi = zi)) = λg + β'zi, i = 1,…,n, using covariates gender and age at recruitment, with g = 0,1. The weight function was chosen as after standardizing the corresponding propensity scores such that they have a unit sample standard deviation.
RESULTS
Genome-wide Association Results
Our analyses confirmed previous results by WTCCC (2007) and further revealed other regions associated with bipolar disorder. Using the non-weighted test statistic U under the null hypothesis H0, Table 1 suggests that three SNPs show strong evidence of association. One of these three SNPs is rs420259 on chromosome 16 which was detected by WTCCC (2007); the other two SNPs are rs9378249 on chromosome 6 and rs12938916 on chromosome 17, which to the best of our knowledge, have not been reported previously using the same data.
Table 1.
SNPs Showing Strong Evidence of Association (P < 5×10−7)
| Chr | SNP | Position | P1a (addd) | P1 (dome) | P1 (recf) | P2b (add) | P2(dom) | P2 (rec) | P3c (add) | P3 (dom) | P3 (rec) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | rs9378249 | 31435680 | 1.21E-08 | 5.11E-08 | 5.06E-03 | 1.39E-08 | 3.41E-08 | 7.55E-03 | 1.71E-09 | 7.70E-09 | 8.87E-04 |
| 16 | rs4202591 | 23541527 | 2.19E-04 | 9.66E-02 | 8.51E-09 | 6.47E-04 | 1.20E-01 | 6.59E-08 | 4.26E-04 | 1.51E-01 | 3.33E-09 |
| 16 | rs2387823 | 51445620 | 2.90E-06 | 4.63E-05 | 3.98E-04 | 1.30E-07 | 5.32E-05 | 2.98E-04 | 1.77E-06 | 3.11E-05 | 3.11E-04 |
| 16 | rs1344485 | 51469833 | 1.78E-06 | 3.09E-05 | 3.77E-04 | 1.79E-07 | 1.34E-05 | 6.29E-04 | 1.41E-06 | 1.80E-05 | 5.12E-04 |
| 16 | rs11647459 | 51473252 | 2.93E-06 | 3.64E-05 | 6.33E-04 | 2.76E-07 | 4.16E-06 | 8.39E-04 | 1.89E-06 | 1.83E-05 | 7.59E-04 |
| 17 | rs12938916 | 53221286 | 8.15E-07 | 4.80E-07 | 2.81E-01 | 2.05E-06 | 1.11E-06 | 3.75E-01 | 1.47E-06 | 8.89E-07 | 1.82E-01 |
| 20 | rs48156032 | 3720527 | 7.51E-05 | 3.00E-06 | 2.05E-01 | 9.33E-04 | 1.42E-05 | 6.39E-02 | 1.82E-05 | 4.80E-07 | 1.69E-01 |
| 20 | rs37612181 | 3724175 | 4.44E-05 | 1.13E-06 | 2.14E-01 | 5.30E-04 | 3.27E-06 | 7.13E-02 | 1.16E-05 | 2.16E-07 | 1.73E-01 |
P-value from non-weighted test under H0.
P-value from weighted test under H0′.
P-value from a logistic regression model.
Additive genotype coding.
Dominant genotype coding.
Recessive genotype coding.
The SNPs identified by WTCCC with strong or moderate association.
The SNPs identified by WTCCC with p-value between 10−4 and 10−5.
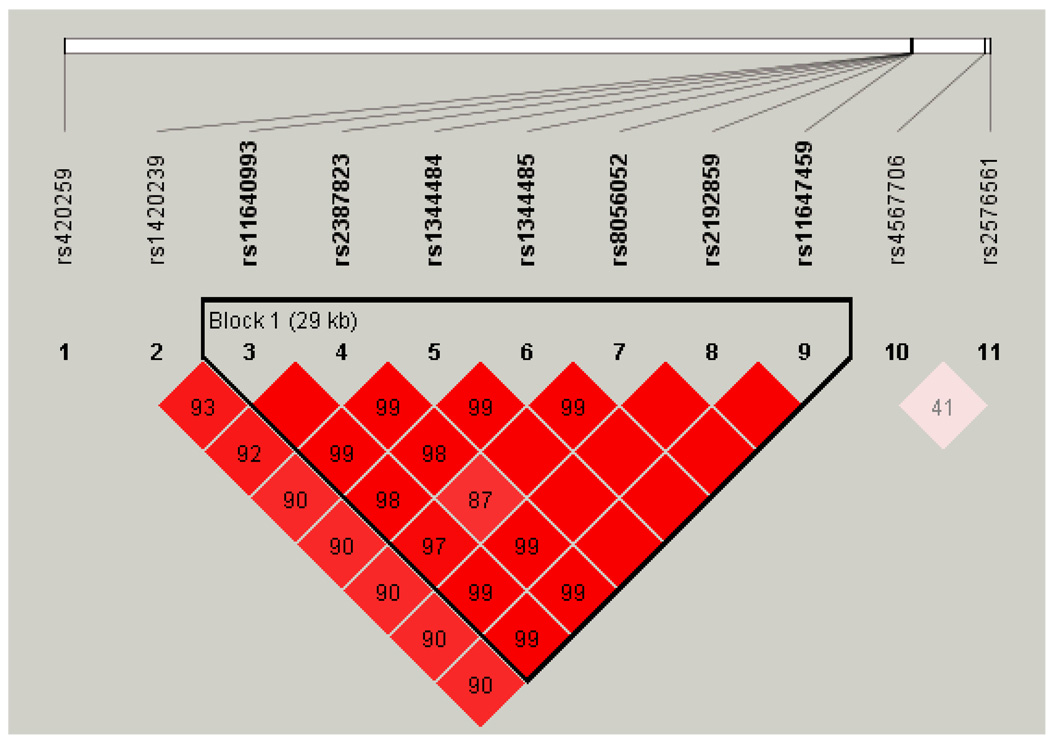
Next, we considered gender and age at recruitment as covariates. Compared with the previous test, the adjusted nonparametric test confirmed rs9378249 and rs420259 as signals with strong association, but did not pick up rs12938916. Moreover, the adjusted nonparametric test identified three additional SNPs (rs2387823, rs1344485, and rs11647459, all on chromosome 16) with strong evidence of association. These three SNPs were neither detected by the unadjusted test nor reported by WTCCC (2007). It is noteworthy that the three additional SNPs are in strong linkage disequilibrium, but not with the reported rs420259 (Figure 1).
Figure 1.
Linkage disequilibrium heat map for the identified SNPs on chromosome 16
To make our comparisons more comprehensive, we further analyzed the bipolar disorder data set using a logistic regression model, adjusting for gender and age at recruitment. The results corroborated the results of the nonparametric method. The results of the logistic regression confirmed the genome-wide significance of exactly the same two SNPs and excluded the other SNP identified by the unadjusted test (see Table 1).
Two additional strong signal SNPs were detected using the logistic regression as opposed to the nonparametric adjusted test: rs4815603 and rs3761218, both on chromosome 20. Note that WTCCC (2007) identified rs3761218 as showing moderate evidence of association and rs4815603 had a p-value between 10−4 and 10−5. Additionally, both SNPs showed only moderate associations using unadjusted and nonparametric adjusted tests. Nonetheless, the logistic regression model missed the three SNPs (rs2387823, rs1344485, rs11647459) on chromosome 16 that were identified by our nonparametric approach as discussed above. These findings show that different methods of adjusting for covariates can lead to different insights. Although parametric adjustment is commonly used in the literature, nonparametric adjustment can be more robust to the model specification. Thus, these two approaches serve as complements to each other in the identification of important SNPs.
We further examined the three new SNPs rs2387823, rs1344485, and rs11647459 (all on chromosome 16) that were identified by the covariate-adjusted nonparametric test, and also the two new SNPs rs4815603 and rs3761218 (both on chromosome 20) that were identified by the parametric test. Recall that we fitted the proportional odds logistic model to estimate the genomic propensity scores. Thus we can evaluate the correlations between the genotype and the covariates through this model. On one hand, the three SNPs on chromosome 16 did not appear to be associated with the covariates, implying that almost no confounding effect exists. The adjustment was mainly for the effects of covariates on trait, producing a more significant p-values than the unadjusted test (this phenomenon is supported by the simulations reported here). One the other hand, the two SNPs on chromosome were associated with the covariates, resulting confounding effects and increasing the p-values of the weighted test (see Table 1). However the logistic regression achieved the genome-wide significance, possibly because of the efficiency of the parametric method when the underlying model assumption is valid (see more discussions for the simulation results).
In addition to Table 1, Table 2 lists all SNPs showing moderate evidence of association identified by any of the above three methods (with p-values between 10−5 and 5×10−7). Although there exist some non-overlapping results, most findings are consistent among these three methods.
Table 2.
SNPs Showing Moderate Evidence of Association (5×10−7 < P < 10−5)
| Chr | SNP | Position | P1 (add) | P1 (dom) | P1 (rec) | P2 (add) | P2 (dom) | P2 (rec) | P3 (add) | P3 (dom) | P3 (rec) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs1461356 | 60749036 | 2.76E-05 | 4.63E-06 | 3.91E-02 | 3.05E-05 | 4.85E-06 | 6.37E-02 | 3.92E-05 | 6.74E-06 | 4.20E-02 |
| 1 | rs29894761 | 60771280 | 2.27E-05 | 2.20E-06 | 4.61E-02 | 2.66E-05 | 2.25E-06 | 7.27E-02 | 4.03E-05 | 3.23E-06 | 6.06E-02 |
| 1 | rs107792792 | 213314367 | 3.99E-05 | 6.08E-05 | 7.89E-02 | 8.93E-06 | 2.29E-05 | 9.67E-02 | 2.65E-05 | 3.62E-05 | 9.11E-02 |
| 2 | rs40271321 | 11988090 | 1.31E-05 | 9.20E-03 | 3.09E-06 | 1.74E-05 | 7.03E-03 | 1.42E-05 | 1.35E-05 | 6.56E-03 | 5.61E-06 |
| 2 | rs75706821 | 104441785 | 3.12E-06 | 3.37E-05 | 9.06E-04 | 8.75E-06 | 1.60E-04 | 1.95E-04 | 8.01E-06 | 1.44E-04 | 4.14E-04 |
| 2 | rs11123306 | 115948011 | 2.79E-06 | 7.60E-05 | 2.24E-04 | 3.76E-05 | 9.80E-05 | 8.86E-04 | 2.45E-06 | 5.67E-05 | 2.75E-04 |
| 2 | rs13751441 | 115957416 | 2.44E-06 | 5.83E-05 | 2.68E-04 | 4.71E-05 | 2.63E-04 | 1.10E-03 | 2.02E-06 | 4.24E-05 | 3.11E-04 |
| 2 | rs2421104 | 115958495 | 9.55E-06 | 3.32E-04 | 1.72E-04 | 4.76E-05 | 1.03E-03 | 1.57E-04 | 5.37E-06 | 2.04E-04 | 1.27E-04 |
| 2 | rs6741692 | 116020161 | 4.65E-06 | 4.01E-05 | 1.36E-03 | 7.48E-05 | 1.39E-04 | 4.56E-03 | 5.85E-06 | 5.00E-05 | 1.47E-03 |
| 2 | rs2953146 | 241235242 | 6.73E-06 | 4.42E-04 | 1.21E-05 | 3.42E-05 | 1.01E-03 | 4.73E-05 | 1.57E-05 | 8.64E-04 | 1.17E-05 |
| 2 | rs2953174 | 241242257 | 6.43E-06 | 1.34E-04 | 1.15E-04 | 5.30E-06 | 1.27E-04 | 1.40E-04 | 7.82E-06 | 1.26E-04 | 1.75E-04 |
| 3 | rs42762271 | 32305690 | 4.59E-06 | 2.64E-05 | 2.04E-03 | 1.12E-05 | 3.41E-05 | 5.62E-03 | 5.15E-06 | 4.65E-05 | 1.24E-03 |
| 3 | rs4627791 | 32322828 | 2.38E-06 | 7.73E-06 | 3.50E-03 | 6.34E-06 | 1.59E-05 | 1.91E-03 | 4.60E-06 | 2.18E-05 | 2.84E-03 |
| 3 | rs130745752 | 61558323 | 3.49E-05 | 1.16E-05 | 7.90E-01 | 8.74E-05 | 2.00E-05 | 9.58E-01 | 3.17E-05 | 8.77E-06 | 9.50E-01 |
| 3 | rs589445 | 184351697 | 4.45E-06 | 8.34E-07 | 8.27E-01 | 5.56E-05 | 5.14E-06 | 8.17E-01 | 4.99E-06 | 1.22E-06 | 6.75E-01 |
| 3 | rs6833951 | 184352520 | 2.30E-06 | 8.57E-07 | 5.44E-01 | 2.29E-05 | 3.88E-06 | 4.72E-01 | 2.74E-06 | 1.21E-06 | 4.56E-01 |
| 3 | rs514636 | 184353138 | 2.50E-06 | 7.45E-07 | 7.22E-01 | 2.64E-05 | 4.70E-06 | 9.49E-01 | 4.45E-06 | 1.31E-06 | 6.74E-01 |
| 3 | rs6414498 | 184353916 | 4.73E-06 | 8.95E-07 | 8.76E-01 | 5.36E-05 | 4.85E-06 | 7.25E-01 | 5.38E-06 | 1.17E-06 | 7.94E-01 |
| 3 | rs6414500 | 184354001 | 5.14E-06 | 8.10E-07 | 9.75E-01 | 6.59E-05 | 4.22E-06 | 9.00E-01 | 5.86E-06 | 1.06E-06 | 9.46E-01 |
| 4 | rs6844851 | 181063149 | 3.76E-05 | 9.41E-07 | 2.71E-01 | 1.98E-03 | 6.25E-05 | 2.96E-01 | 4.15E-05 | 1.24E-06 | 2.67E-01 |
| 5 | --- | 169013376 | 1.42E-04 | 1.79E-04 | 2.44E-01 | 8.11E-06 | 2.22E-05 | 9.97E-02 | 1.89E-04 | 2.36E-04 | 2.56E-01 |
| 6 | rs64583071 | 42839093 | 3.43E-01 | 2.11E-01 | 2.82E-05 | 1.67E-01 | 2.60E-01 | 7.87E-06 | 2.84E-01 | 2.44E-01 | 1.90E-05 |
| 7 | rs22864922 | 22758250 | 5.06E-01 | 6.98E-01 | 8.40E-06 | 3.56E-01 | 8.80E-01 | 8.58E-06 | 4.73E-01 | 7.36E-01 | 9.35E-06 |
| 8 | rs26096531 | 34356534 | 6.87E-06 | 2.10E-05 | 1.41E-02 | 3.48E-05 | 4.11E-05 | 3.83E-02 | 1.57E-05 | 4.62E-05 | 1.54E-02 |
| 8 | rs4739466 | 37060700 | 1.99E-01 | 6.56E-01 | 3.60E-06 | 6.17E-01 | 9.15E-01 | 1.76E-05 | 2.00E-01 | 6.19E-01 | 1.14E-05 |
| 9 | rs914715 | 79540452 | 2.28E-06 | 1.26E-06 | 9.56E-01 | 1.71E-06 | 2.14E-06 | 7.66E-01 | 1.02E-06 | 7.05E-07 | 7.29E-01 |
| 9 | rs109822561 | 114340388 | 8.82E-06 | 5.85E-04 | 1.21E-04 | 5.29E-06 | 3.51E-04 | 2.01E-04 | 1.05E-05 | 4.11E-04 | 2.39E-04 |
| 10 | rs17600642 | 72133000 | 5.19E-06 | 8.07E-06 | 3.34E-02 | 2.38E-05 | 3.78E-05 | 5.38E-02 | 5.01E-05 | 7.44E-05 | 5.43E-02 |
| 10 | rs78961312 | 94539655 | 1.56E-04 | 1.35E-05 | 8.25E-01 | 2.65E-04 | 2.40E-05 | 7.17E-01 | 1.14E-04 | 9.12E-06 | 8.29E-01 |
| 11 | rs1568889 | 27966039 | 1.45E-03 | 1.01E-01 | 2.32E-06 | 2.94E-03 | 1.52E-01 | 2.77E-05 | 3.05E-03 | 1.57E-01 | 2.28E-06 |
| 12 | rs7136898 | 23945839 | 1.32E-05 | 1.30E-05 | 8.33E-03 | 2.24E-05 | 7.44E-05 | 2.19E-02 | 6.84E-06 | 1.11E-05 | 4.34E-03 |
| 12 | rs11168839 | 47743902 | 2.38E-04 | 2.63E-05 | 1.80E-01 | 8.72E-05 | 8.75E-06 | 1.24E-01 | 7.06E-05 | 1.10E-05 | 9.48E-02 |
| 12 | rs7296288 | 47766235 | 4.92E-04 | 2.76E-05 | 1.72E-01 | 2.05E-04 | 9.60E-06 | 8.15E-02 | 1.91E-04 | 1.49E-05 | 9.92E-02 |
| 13 | rs28069222 | 45416060 | 7.90E-03 | 3.42E-01 | 1.82E-05 | 3.42E-02 | 4.00E-01 | 2.40E-05 | 5.54E-03 | 3.63E-01 | 4.61E-06 |
| 13 | rs9530460 | 75157071 | 1.86E-03 | 4.12E-01 | 1.40E-05 | 4.58E-04 | 2.87E-01 | 2.16E-06 | 7.88E-04 | 2.94E-01 | 7.02E-06 |
| 14 | rs101349441 | 57188949 | 3.22E-06 | 1.15E-06 | 4.79E-01 | 3.96E-06 | 7.45E-07 | 6.78E-01 | 2.46E-06 | 7.93E-07 | 5.07E-01 |
| 14 | rs12890287 | 57191398 | 2.06E-05 | 1.10E-05 | 3.85E-01 | 7.09E-06 | 2.63E-06 | 4.63E-01 | 1.52E-05 | 6.13E-06 | 4.91E-01 |
| 14 | rs11622600 | 57194142 | 1.70E-05 | 1.02E-05 | 3.66E-01 | 5.31E-06 | 2.06E-06 | 3.93E-01 | 1.18E-05 | 5.51E-06 | 4.61E-01 |
| 14 | rs6574988 | 87629745 | 5.66E-06 | 4.22E-06 | 3.70E-01 | 3.11E-05 | 1.57E-05 | 5.10E-01 | 1.59E-05 | 1.05E-05 | 4.77E-01 |
| 14 | rs8021692 | 103498329 | 2.98E-06 | 3.62E-06 | 1.67E-02 | 1.60E-05 | 1.97E-05 | 9.39E-03 | 3.64E-06 | 9.48E-06 | 7.58E-03 |
| 14 | rs116224751 | 103578829 | 2.10E-06 | 2.20E-06 | 1.95E-02 | 7.78E-06 | 9.07E-06 | 1.42E-02 | 2.41E-06 | 5.26E-06 | 9.26E-03 |
| 15 | rs8031347 | 31377909 | 7.82E-05 | 7.69E-05 | 6.46E-02 | 7.99E-06 | 1.93E-05 | 3.09E-02 | 2.53E-05 | 3.16E-05 | 3.66E-02 |
| 16 | rs14202392 | 51434112 | 4.48E-05 | 1.14E-04 | 1.38E-02 | 7.56E-06 | 5.12E-05 | 6.03E-03 | 5.49E-05 | 1.44E-04 | 1.43E-02 |
| 16 | rs11640993 | 51444106 | 3.42E-06 | 4.77E-05 | 5.57E-04 | 1.35E-06 | 1.25E-05 | 1.70E-04 | 2.05E-06 | 2.79E-05 | 5.01E-04 |
| 16 | rs13444841 | 51469800 | 1.65E-06 | 2.62E-05 | 3.97E-04 | 9.60E-07 | 7.49E-06 | 6.74E-04 | 1.07E-06 | 1.28E-05 | 4.98E-04 |
| 16 | rs8056052 | 51470239 | 4.23E-06 | 9.10E-05 | 3.57E-04 | 2.73E-06 | 1.10E-04 | 4.39E-04 | 4.47E-06 | 8.47E-05 | 4.27E-04 |
| 16 | rs2192859 | 51472250 | 1.88E-06 | 2.82E-05 | 4.41E-04 | 2.44E-06 | 3.51E-06 | 9.14E-04 | 1.26E-06 | 1.45E-05 | 5.46E-04 |
| 16 | rs45677062 | 53859772 | 1.55E-05 | 7.46E-05 | 4.88E-03 | 4.01E-06 | 2.63E-05 | 4.89E-03 | 6.22E-06 | 3.73E-05 | 2.46E-03 |
| 16 | rs2576561 | 54028475 | 2.99E-05 | 2.91E-05 | 1.26E-01 | 1.22E-05 | 4.50E-06 | 2.51E-01 | 7.81E-05 | 6.31E-05 | 1.90E-01 |
| 17 | rs203097 | 47368276 | 2.01E-03 | 7.33E-02 | 4.50E-05 | 3.72E-04 | 2.48E-02 | 9.59E-06 | 1.55E-03 | 6.64E-02 | 2.23E-05 |
| 18 | rs18931462 | 8977427 | 8.15E-05 | 1.71E-04 | 4.50E-02 | 5.83E-06 | 1.58E-05 | 7.92E-03 | 5.05E-05 | 1.00E-04 | 4.79E-02 |
| 19 | rs7247513 | 12552185 | 2.07E-06 | 8.94E-05 | 1.08E-04 | 5.54E-06 | 2.02E-04 | 1.10E-04 | 3.96E-06 | 1.90E-04 | 9.19E-05 |
| 19 | rs74081692 | 48485993 | 6.10E-05 | 2.07E-04 | 7.96E-03 | 4.32E-05 | 1.87E-04 | 3.31E-03 | 9.84E-06 | 3.66E-05 | 4.08E-03 |
| 19 | rs72484932 | 63402920 | 3.08E-05 | 4.30E-05 | 4.24E-02 | 1.40E-05 | 2.46E-05 | 1.08E-02 | 6.04E-06 | 2.31E-05 | 6.78E-03 |
The SNPs identified by WTCCC (2007) with strong or moderate association.
The SNPs identified by WTCCC (2007) with p-value between 10−4 and 10−5.
Haplotype Analysis Results
Among all SNPs showing strong or moderate evidence of association, we found that some SNPs are in close proximity within a chromosome. This motivates a haplotype analysis for these SNPs. We focus on the SNPs on chromosome 16 that are tabulated in Tables 1 and 2. The linkage disequilibrium (LD) heat map in Figure 1 displays that seven out of the eleven SNPs were in a 29kb region. They were identified as a haplotype block [Barrett et al., 2005]. Using the seven SNPs within the block, the haplotype frequency was estimated [Zhao, 2004]. The top haplotypes are “GGTTCGG”, “ACCCGAA”, “GCTTCGG”, and “GGTTGGG”, with probabilities 0.535, 0.381, 0.049, and 0.027, respectively. Next, a pair of haplotypes was estimated for each subject using EM algorithm, and the haplotype association analysis was performed. We chose to dichotomize haplotype H as 1 or 0 according to whether it is equal to “ACCCGAA”. Then, the previous association tests can be applied simply by replacing genotype G with haplotype H. Using the non-weighted test under H0, the p-value was 3.45×10−6. The weighted test determined the p-value at 2.64×10−7 as a genome-wide strong association between this block and bipolar disorder, highlighting the power of the weighted test and further confirming the importance of this region. Moreover, using the logistic regression model adjusting for the covariates determined the p-value of this block at 2.16×10−6, which did not achieve the genome-wide significance. We note that this region is close to the RPGRIP1L gene, which encodes the protein that localize to the basal body-centrosome complex or to primary cilia and centrosomes in ciliated cells. The RPGRIP1L gene was previously identified to be associated with schizophrenia as well as bipolar disorder [O’Donovan et al., 2008; Riley et al., 2009].
SIMULATIONS
Simulation Settings
To further evaluate our method, we performed a proof-of-principle simulation study to assess the influences of the weighted test to the confounding effects. First, a continuous covariate Zco was simulated from N(0,1) distribution, and a categorical covariate Zca was simulated as P(Zca = 1) = 1 − P(Zca = 0) = 0.7. To then introduce the correlation between the covariates and the test-locus genotype G, we generated G according to Binomial distribution Binomial(2, p) with probability p satisfying it(p) = μ +νco Zco + νcaZca where νco and νca control the correlation between the genotype and the covariates, and u ~ N(0,1) is random noise. Lastly, conditional on the genotype G and the covariates Zco and Zca, a binary trait Y was generated according to a logistic model including genetic effect, environmental effects, and gene-environment interactions, logit(P(Y = 1)) = α + βgg + βcozco + βcazca + βg,cogzco + βg,cagzca + ε, where ε ~ N(0,1). The interaction terms were added purposely to evaluate the robustness of different testing methods.
In the simulation, we set μ = α = −0.5. The choices of the coefficients (νco,νca) and (βg,βco,βca,βg,co,βg,ca) were provided as in Table 3 as different settings and models. The weight function was chosen as after standardizing the corresponding propensity scores such that they have a unit sample standard deviation. We compared our weighted test to the non-weighted test to assess the influence of adjustment for covariates. In addition, as in the analysis of bipolar disorder data, we provided results from logistic regression model including marginal effects from both gene and covariates.
Table 3.
Simulation Settings and Models
| Coefficients | |
|---|---|
| Setting S1 | νco = νca = 0 |
| Setting S2 | νco = νca = 0.5 |
| Setting S3 | νco = νca = −0.5 |
| Model N1 | βg = βco = βca = βg,co = βg,ca = 0 |
| Model N2 | βg = 0, βco = βca = 1, βg,co = βg,ca = 0 |
| Model A1 | βg = 0.5, βco = βca = 1, βg,co = βg,ca = 0 |
| Model A2 | βg = 0.5, βco = βca = 2, βg,co = βg,ca = 0 |
| Model A3 | βg = 0, βco = βca = 0, βg,co = βg,ca = 1 |
| Model A4 | βg = 0, βco = βca = 1, βg,co = βg,ca = 1 |
| Model A5 | βg = 0.5, βco = βca = 0, βg,co = βg,ca = 1 |
| Model A6 | βg = 0.5, βco = βca = 1, βg,co = βg,ca = 1 |
Simulation Results
Tables 4, 5 and 6 report the empirical type I error and power of the three tests: the non-weighted test using H0, the weighted test using H0′, and the test from the logistic regression. Tables 4, 5 and 6 correspond to Settings S1, S2 and S3 (see Table 3), respectively.
Table 4.
Type I Error and Power (Setting S1)
| α | n | T a |
|
Tlc | T | Tl | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type I Error |
Model N1 | Model N2 | ||||||||
| 0.05 | 200 | 0.075 | 0.070 | 0.074 | 0.055 | 0.042 | 0.047 | |||
| 400 | 0.052 | 0.049 | 0.055 | 0.046 | 0.037 | 0.053 | ||||
| 0.01 | 200 | 0.016 | 0.019 | 0.018 | 0.013 | 0.004 | 0.009 | |||
| 400 | 0.006 | 0.010 | 0.007 | 0.002 | 0.007 | 0.010 | ||||
| Power | Model A1 | Model A2 | ||||||||
| 0.05 | 200 | 0.422 | 0.420 | 0.512 | 0.255 | 0.268 | 0.395 | |||
| 400 | 0.748 | 0.743 | 0.796 | 0.485 | 0.540 | 0.680 | ||||
| 0.01 | 200 | 0.212 | 0.196 | 0.254 | 0.103 | 0.104 | 0.175 | |||
| 400 | 0.501 | 0.494 | 0.589 | 0.254 | 0.269 | 0.420 | ||||
| Power | Model A3 | Model A4 | ||||||||
| 0.05 | 200 | 0.701 | 0.769 | 0.746 | 0.294 | 0.398 | 0.397 | |||
| 400 | 0.939 | 0.969 | 0.955 | 0.516 | 0.684 | 0.673 | ||||
| 0.01 | 200 | 0.448 | 0.555 | 0.493 | 0.116 | 0.149 | 0.180 | |||
| 400 | 0.822 | 0.878 | 0.853 | 0.296 | 0.433 | 0.428 | ||||
| Power | Model A5 | Model A6 | ||||||||
| 0.05 | 200 | 0.980 | 0.988 | 0.988 | 0.725 | 0.826 | 0.847 | |||
| 400 | 1.000 | 1.000 | 1.000 | 0.951 | 0.986 | 0.992 | ||||
| 0.01 | 200 | 0.899 | 0.936 | 0.928 | 0.456 | 0.591 | 0.650 | |||
| 400 | 0.999 | 0.999 | 1.000 | 0.832 | 0.931 | 0.951 | ||||
Non-weighted test under H0.
Weighted test under H0′.
Logistic regression.
Table 5.
Type I Error and Power (Setting S2)
| α | n | T a |
|
Tlc | T | Tl | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type I Error |
Model N1 | Model N2 | ||||||||
| 0.05 | 200 | 0.058 | 0.058 | 0.061 | 0.348 | 0.054 | 0.053 | |||
| 400 | 0.044 | 0.049 | 0.052 | 0.585 | 0.047 | 0.053 | ||||
| 0.01 | 200 | 0.012 | 0.013 | 0.008 | 0.150 | 0.008 | 0.008 | |||
| 400 | 0.010 | 0.005 | 0.005 | 0.332 | 0.014 | 0.014 | ||||
| Power | Model A1 | Model A2 | ||||||||
| 0.05 | 200 | 0.926 | 0.459 | 0.507 | 0.957 | 0.319 | 0.363 | |||
| 400 | 0.998 | 0.762 | 0.794 | 0.998 | 0.609 | 0.675 | ||||
| 0.01 | 200 | 0.798 | 0.227 | 0.261 | 0.854 | 0.133 | 0.173 | |||
| 400 | 0.988 | 0.527 | 0.583 | 0.993 | 0.366 | 0.418 | ||||
| Power | Model A3 | Model A4 | ||||||||
| 0.05 | 200 | 0.973 | 0.754 | 0.732 | 0.944 | 0.441 | 0.387 | |||
| 400 | 1.000 | 0.967 | 0.948 | 0.998 | 0.743 | 0.669 | ||||
| 0.01 | 200 | 0.882 | 0.545 | 0.497 | 0.824 | 0.220 | 0.193 | |||
| 400 | 0.995 | 0.866 | 0.826 | 0.993 | 0.518 | 0.426 | ||||
| Power | Model A5 | Model A6 | ||||||||
| 0.05 | 200 | 1.000 | 0.980 | 0.976 | 0.995 | 0.854 | 0.833 | |||
| 400 | 1.000 | 1.000 | 1.000 | 1.000 | 0.994 | 0.983 | ||||
| 0.01 | 200 | 0.993 | 0.927 | 0.919 | 0.975 | 0.660 | 0.637 | |||
| 400 | 1.000 | 0.998 | 0.998 | 1.000 | 0.963 | 0.938 | ||||
Non-weighted test under H0.
Weighted test under H0′.
Logistic regression.
Table 6.
Type I Error and Power (Setting S3)
| α | n | T a |
|
Tlc | T | Tl | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type I Error |
Model N1 | Model N2 | ||||||||
| 0.05 | 200 | 0.059 | 0.053 | 0.058 | 0.326 | 0.052 | 0.050 | |||
| 400 | 0.049 | 0.045 | 0.046 | 0.544 | 0.054 | 0.045 | ||||
| 0.01 | 200 | 0.012 | 0.014 | 0.015 | 0.138 | 0.005 | 0.006 | |||
| 400 | 0.006 | 0.009 | 0.007 | 0.328 | 0.010 | 0.007 | ||||
| Power | Model A1 | Model A2 | ||||||||
| 0.05 | 200 | 0.066 | 0.437 | 0.490 | 0.158 | 0.320 | 0.384 | |||
| 400 | 0.072 | 0.720 | 0.773 | 0.230 | 0.614 | 0.674 | ||||
| 0.01 | 200 | 0.013 | 0.205 | 0.261 | 0.044 | 0.136 | 0.167 | |||
| 400 | 0.010 | 0.470 | 0.542 | 0.084 | 0.330 | 0.416 | ||||
| Power | Model A3 | Model A4 | ||||||||
| 0.05 | 200 | 0.225 | 0.603 | 0.557 | 0.111 | 0.328 | 0.289 | |||
| 400 | 0.403 | 0.865 | 0.830 | 0.153 | 0.622 | 0.535 | ||||
| 0.01 | 200 | 0.081 | 0.367 | 0.323 | 0.024 | 0.134 | 0.112 | |||
| 400 | 0.186 | 0.705 | 0.646 | 0.049 | 0.343 | 0.284 | ||||
| Power | Model A5 | Model A6 | ||||||||
| 0.05 | 200 | 0.759 | 0.967 | 0.944 | 0.072 | 0.796 | 0.778 | |||
| 400 | 0.974 | 1.000 | 1.000 | 0.103 | 0.987 | 0.979 | ||||
| 0.01 | 200 | 0.547 | 0.868 | 0.840 | 0.023 | 0.598 | 0.549 | |||
| 400 | 0.898 | 0.998 | 0.996 | 0.026 | 0.943 | 0.909 | ||||
Non-weighted test under H0.
Weighted test under H0′.
Logistic regression.
From the perspective of type I error, for model N1, since βg = βco = βca = βg,co = βg,ca = 0, there was neither genetic effect nor covariate effect. We found that all three tests behaved fairly well in terms of the accuracy for type I errors as noted in all three settings (Tables 4, 5 and 6). For model N2, since βg = 0, βco = βca = 1, βg,co = βg,ca = 0, there was no genetic effect but there were covariate effects. If there are correlations between the genotype and covariates, we expect that the non-weighted test using H0 cannot control the type I error at the significance level because of the confounding effects caused by the covariates. In Table 4, since there were no correlations between the genotype and the covariates, all three tests performed reasonably well. However, in Tables 5 and 6, the type I errors of the non-weighted test were always much higher than the significance level. By contrast, the weighted test using H0′ and the logistic regression performed well in controlling the type I errors.
From the perspective of power, we considered a collection of models representing different situations. Models A1 and A2 only have marginal effects from both genotype and covariates, while Models A3–A6 all include gene-environment interactions. Since the non-weighted test using H0 cannot control the type I error in Tables 5 and 6, the power is not truly comparable. Nonetheless, we tabulated the power results for all three tests. The non-weighted test using H0 sometimes had a higher power (as seen in Table 5), and sometimes had a lower power (as seen in Tables 4 and 6). In Setting 2, due to the confounding effects enhancing the overall genetic effects, a false positive occurred. We have the opposite situation in Setting 3 as the false negative occurred. This suggests that to adjust for confounders, the null hypothesis must be conditional, as is H0′. It is noteworthy that, even if there were no confounders as in Setting 1, we still observe the advantage of adjusting for covariates when there exist environmental effects to the trait.
The comparison between the weighted test using H0′ and the logistic regression falls into the classic framework of comparing nonparametric and parametric tests. In Models A1 and A2, due to the correctness of underlying model assumptions, the logistic regression performed slightly better than the weighted test. By contrast, when the underlying model assumptions were violated as in Models A3–A6, the nonparametric test performed better than the parametric test due to its robustness to model specifications. In summary, the weighted test is robust to the model assumptions compared with the logistic regression, and it performed consistently well in our simulation studies.
DISCUSSION
In conclusion, we applied a propensity score-based nonparametric test to analyze an important data set on bipolar disorder made available to us by the WTCCC. By analyzing the bipolar disorder data, we not only confirmed the results reported by the WTCCC (2007), but also identified other regions at the genome-wide significance level, including the haplotype block identified on chromosome 16. It is important to note that the identified haplotype block is near the RPGRIP1L gene that was reported to be associated with bipolar disorder. Simulation studies provide further supporting evidence for the weighted test. Compared with the non-weighted test, the weighted test can not only correctly control the type I error at the significance level to avoid false positives, but also increase the power when false negatives occur. Compared with other parametric test such as logistic regression, the weighted test has the merit of robustness to model specifications. Our approach provides a promising way of adjusting for general covariates in genome-wide association studies, and has demonstrated its usefulness in an existing GWAS data set. In this analysis, we examined age and gender as two potentially important covariates. There are likely other covariates that are worthy consider investigation.
ACKNOWLEDGMENTS
This work is supported in part by grant R01DA016750 from the National Institute on Drug Abuse. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475.
REFERENCES
- Amos CI, Zhu DK, BoerWinkle E. Assessing genetic linkage and association with robust components of variance approaches. Ann Hum Genet. 1996;60:143–160. doi: 10.1111/j.1469-1809.1996.tb01184.x. [DOI] [PubMed] [Google Scholar]
- Arking DE, Pfeufer A, Post W, Kao WHL, Newton-Cheh C, Ikeda M, West K, Kashuk C, Akyol M, Perz S, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu C-T, Zhang M, Zhang H. A forest-based approach to identifying gene and gene-gene interactions. Proc Natl Acad Sci USA. 2007;104:19199–19203. doi: 10.1073/pnas.0709868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19 Suppl 1:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Lu ATH, Cantor RM. Weighted variance FBAT: a powerful method for including covariates in FBAT analyses. Genet Epidemiol. 2007;31:327–337. doi: 10.1002/gepi.20213. [DOI] [PubMed] [Google Scholar]
- O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz D, Laird NM. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50:211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, McMichael GO, Fanous AH, Vladimirov V, O'Neill FA, Walsh D, Kendler KS. Replication of association between schizophrenia and ZNF804A in the Irish Case–Control Study of Schizophrenia sample. Mol Psychiatry. 2009;15:29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum PR, Rubin DB. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. [Google Scholar]
- Rosenbaum PR, Rubin DB. Reducing bias in observational studies using subclassification on the propensity score. J Am Stat Assoc. 1984;387:516–524. [Google Scholar]
- Vansteelandt S, Goetgeluk S, Lutz S, Waldman I, Lyon H, Schadt EE, Weiss ST, Lange C. On the adjustment for covariates in genetic association analysis: a novel, simple principle to infer direct causal effects. Genet Epidemiol. 2009;33:394–405. doi: 10.1002/gepi.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ye Y, Zhang H. Family-based association tests for ordinal traits adjusting for covariates. Genet Epidemiol. 2006;30:728–736. doi: 10.1002/gepi.20184. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu C-T, Wang X. An association test for multiple traits based on the generalized Kendall’s tau. J Am Stat Assoc. 2010;105:473–481. doi: 10.1198/jasa.2009.ap08387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang X, Ye Y. Detection of genes for ordinal traits in nuclear families and a unified approach for association studies. Genetics. 2006;172:693–699. doi: 10.1534/genetics.105.049122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Rebbeck TR, Mitra N. A propensity score approach to correction for bias due to population stratification using genetic and non-genetic factors. Genet Epidemiol. 2009;33:679–690. doi: 10.1002/gepi.20419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JH. 2LD, GENECOUNTING and HAP: computer programs for linkage disequilibrium analysis. Bioinformatics. 2004;20:1325–1326. doi: 10.1093/bioinformatics/bth071. [DOI] [PubMed] [Google Scholar]