Abstract
Infection by human papillomaviruses (HPVs) leads to the formation of benign lesions, warts, and in some cases, cervical cancer. The formation of these lesions is dependent upon increased expression of pro-angiogenic factors. Angiogenesis is linked to tissue hypoxia through the activity of the oxygen sensitive hypoxia inducible factor 1α (HIF-1α). Our studies indicate that the HPV E7 protein enhances HIF-1 transcriptional activity while E6 functions to counteract the repressive effects of p53. Both high and low risk HPV E7 proteins were found to bind to HIF-1α through a domain located in the the N terminus. Importantly, the ability of E7 to enhance HIF-1 activity mapped to the C terminus and correlated with the displacement of the histone deacetylases HDAC1, HDAC4, and HDAC7 from HIF-1α by E7. Our findings describe a novel role of the E7 oncoprotein in activating the function of a key transcription factor mediating hypoxic responses by blocking the binding of HDACs.
Introduction
Human papillomaviruses (HPVs) are small, non-enveloped DNA viruses that persistently infect stratified squamous epithelia. A subset of viral types are the causative agents of a variety of malignancies, including over 99% of cervical cancers (1, 2). HPVs infect stratified epithelia and link their life cycles to the differentiation program of the host cell (3). Following entry into the basal layer, HPVs establish themselves as low copy number episomes. As basal keratinocytes divide, one daughter cell detaches from the basement membrane and begins the process of squamous differentiation. In HPV infection, viral proteins block normal cell cycle exit upon differentiation and activate expression of host DNA replication enzymes in suprabasal cells to replicate its genome (3). The oncogenes E6 and E7 from high-risk (cancer-associated) HPVs are responsible for maintaining differentiating cells active in the cell cycle (4). These two proteins promote the degradation of cellular tumor suppressors: pRb family members in the case of E7 and p53 in the case of E6. Binding and degradation of pRb family members by E7 results in the release of E2F transcription factors that drive the cell into S phase. The abrogation of pRb function by high risk E7 proteins induces a stress response leading to elevated levels of p53 which can induce apoptosis. The high risk E6 proteins degrade p53, thus preventing apoptosis and allowing continued proliferation. In addition to these well known activities, both proteins have a range of other targets (4), and the extent to which these additional interactions contribute to HPV associated carcinogenesis is not fully understood. Among the additional factors bound by E7 are histone deacetylases (HDACs), which catalyze the deacetylation of histones and other transcriptional regulatory proteins (5–7). The binding of HDACs by E7 results in the activation of cellular promoters and is necessary for the differentiation-dependent phase of the virus life cycle (5, 6).
One important characteristic of both benign and malignant lesions is the promotion of angiogenesis, or the formation of new blood vessels, which allows a growing lesion to access nutrients and oxygen for growth (8). Angiogenesis is triggered by hypoxia, or reduced tissue oxygen levels. The cellular response to hypoxia is primarily regulated through the activity of the transcription factor hypoxia inducible factor-1 (HIF-1) (9, 10). Under normal oxygen conditions (normoxia), the HIF-1α subunit has a very short half-life due to oxygen-dependent hydroxylation and consequent degradation through the von Hippel-Lindau (VHL)/proteasome pathway. Under hypoxic conditions, reduced oxygen levels result in the accumulation of HIF-1α, which activates expression of HIF-1 target genes, including a range of pro-angiogenic factors and enzymes that favor glycolytic over aerobic metabolism (10).
HIF-1 is regulated by a number of factors including p53, p300/CBP and HDACs. Although HDACs frequently inhibit transcription, HDAC activity is necessary for HIF-1 activity (11–16) and angiogenesis (17) as treatment with inhibitors reduces HIF-1 mediated transcription. Which HDAC(s) are involved in this activation and the mechanisms responsible are controversial. HDAC1 (11), HDAC4 (14, 18), HDAC5 (18), HDAC6 (14, 15), HDAC7 (19), and SIRT-6 (20) have each been reported to bind and/or regulate HIF-1 activity. This activation has been reported to occur through a variety of mechanisms including direct deacetylation of HIF-1α (11, 14), facilitation of nuclear localization (19), increasing p300 binding to HIF-1 (16, 18, 19), or through altering interactions with HSP70/90 (15). HIF-1 can also act as a repressor of some promoters such as the cyclin D promoter, through a mechanism dependent on HDAC7 (21).
Recently, several studies have reported that HPV gene products can induce the production of angiogenic factors by infected cells (22–25). Our previous work demonstrated that cells maintaining HPV genomes show enhanced levels of HIF-1α and increased expression of HIF-1 target genes under hypoxic conditions (26). In the present study, we demonstrate that E7 is responsible for enhanced HIF-1α activity. Our studies indicate that E7 enhances HIF-1 dependent transcription by inducing the dissociation of HDACs from HIF-1α. These findings shed light on the mechanisms by which HPV contributes to tumor angiogenesis and describe a novel role of E7 in the activation of host gene expression.
Materials and Methods
Cell Culture and creation of cell lines
Keratinocytes were isolated from neonatal foreskins. Keratinocytes and cell lines containing HPV genomes were cultured in E-medium NIH 3T3 J2 fibroblast feeders as described previously (27). Cells stably expressing HPV16 E7 from retroviral vectors were established by infection of keratinocytes followed by selection with G418 as described previously (28). Cell lines containing HPV episomes (HFK16 cells) were created by transfection of cloned recircularized viral DNA as described previously (29). U2OS cells were obtained from ATCC and cultivated in DMEM (Invitrogen) supplemented with 10% bovine growth serum (VWR). Hypoxia was mimicked by treatment for 6–16 hours with 100 μM deferoxamine mesylate (DFO, Sigma). Trichostatin A (300 nM TSA, Sigma) was dissolved in DMSO, and cells were treated for 16 hours.
Plasmids and Cloning
pcDNA HIF-1α was a gift from Eric Huang, National Cancer Institute, Bethesda, MD. pGWI HA 18, 16, and 11 E7 were cloned by removing the 31E7 from pHA31E7 and ligating inserts created by PCR using primers shown in Supplementary Table 1. pcDNA TapC 16E7 was created by PCR using primers OZ5′ and OZ HA Bam3′ with an HA-flag double tagged HPV16 E7 retrovirus vector (a gift from Margaret McLaughlin-Drubin, Brigham and Women’s Hospital/Harvard Medical School, Cambridge, MA) as template. pcDNA TapN 16E7 was created by PCR using 16E7 Xho frame 5′ and 16E7 Not stop 3′ as primers and pUC HPV16 as template. The product was ligated into pOZN at the Xho1/Not1 sites to create pOZN 16E7. PCR was performed again using pOZN 16 E7 using OZN 5′ Nhe and 16E7 Not stop 3′ primers and ligating into the Nhe1/Not1 site of pcDNA 3.1- to create pcDNA TapN 16E7. Deletions were created by PCR using primers listed in Supplementary Table 1 and ligating into the Not1/Xho1 site of pcDNA TapN 16E7. Site directed mutagenesis was performed using the QuickChange II Site Directed Mutagenesis kit (Agilent) using the primers listed in Supplementary Table 1. pcDNA TapC 16E6 was cloned by removing E7 from pcDNA TapC 16E7 by digestion with XhoI and NotI and ligating a PCR product created using 16E6 pOZ5 and 16E6 pOZ3′ as primers and pUC HPV16 as template. All constructs were checked by sequencing before use. HPV31 E6 was expressed from pSG 31E6.
Transient transfection and western blot analysis
Transient transfections of keratinocytes and U2OS cells were performed using polyethyleneimine (PEI; Polysciences). Whole-cell extracts were prepared 24–36 hours after transfection and subjected to SDS-PAGE and Western blotting or immunoprecipitation as described previously (26, 28). Antibodies used in this study are shown in Supplementary Table 2.
Luciferase assays
A luciferase reporter consisting of a trimerized 24-mer containing 18 base pairs from the hypoxia response element (HRE) of the phosphoglycerate kinase promoter, HRE-TK-Luc, was a gift from Navdeep Chandel, Department of Medicine, Northwestern University, Chicago, IL. The pGL2 CA9 reporter was a gift from Stefan Kaluz, Emory University, Atlanta, GA. The pGL3 VEGF pro was a gift from Lee Ellis, MD Anderson Cancer Center, Houston, TX. 100,000 cells were plated per well in 6 well plates and transfected with PEI at 37 °C overnight. Fresh medium was added, and cells were subsequently treated with DFO and/or TSA overnight. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega), with Renilla luciferase as an internal control according to manufacturer’s instructions. Significance was determined using Student’s T test.
Quantitative real time RT-PCR
Total RNAs were harvested using RNA-STAT 60 (TelTest, Inc). cDNAs were synthesized using the SuperScript First Round Synthesis System (Invitrogen), and qPCR was performed using LightCycler 480 SYBR Green I Master mix (Roche) on a LightCycler 480 instrument. Primers used to detect CA9, TSP-1, PGK-11, and actin (as a reference) are shown in Supplementary Table 1.
Results
E7 activation of HIF-1-dependent transcription
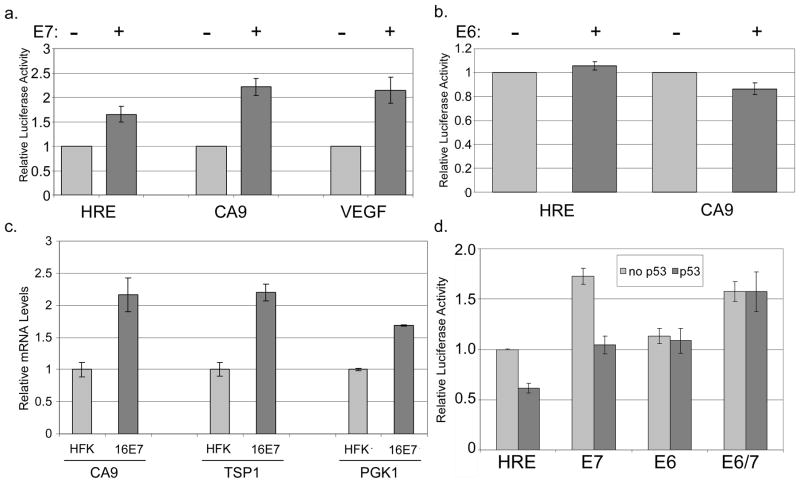
In previous studies, we demonstrated that cells harboring HPV genomes display an enhanced HIF-1 dependent transcriptional response to hypoxia (26). We investigated which viral protein is responsible for this effect and first examined the role of E7. For these studies, an expression vector for 16E7 was co-transfected into U2OS cells together with a luciferase reporter, HRE-TK-Luc, containing a trimerized hypoxia response element (HRE) from the phosphoglycerate kinase promoter, and cells were treated with the hypoxia mimic DFO. We found that activity of the HIF-1 reporter was enhanced upon co-transfection with 16E7 (p<0.01, Figure 1a). This effect was seen only upon DFO treatment and not in normoxia (not shown). Enhancement of activity was also seen using the HIF-1 dependent carbonic anhydrase 9 (CA9) and vascular endothelial growth factor (VEGF) promoters (p<0.01, Figure 1a). When similar transient expression experiments were performed in human foreskin keratinocytes (HFKs), a comparable enhancement in HIF-1 activity was seen (not shown).
Figure 1. E7 increases HIF-1 dependent promoter activity, but E6 does not.
U2OS cells were transfected overnight with the HRE-tk-Luc reporter, CA9 promoter reporter, or VEGF promoter reporter with or without an expression vector for (a) 16E7 or (b) 16E6. After 24 hours, samples were treated with 100 μM DFO for an additional 16 hours. Lysates were prepared, and the activities of firefly and Renilla luciferase were measured. Values were normalized to the Renilla luciferase activity in each sample, and the reporter alone with DFO was set to 1. Each point represents the mean of 4–15 experiments and error bars represent ±1 standard error of the mean. (c) Total RNAs from HFK cells or HFKs stably expressing HPV16 E7 were subjected to qRT-PCR using primers specific for the indicated HIF-1 target genes. Each point represent the means of 2–3 experiments and bars represent ±1 standard error of the mean. (d) U2OS cells were cotransfected overnight with HRE-tk-Luc with or without expression vectors for p53, 16E6, or 16E7. Luciferase activities were assayed and calculated as above.
It was next important to determine whether E6 would have an effect on HIF-1 dependent transcriptional activity. In contrast to E7, co-transfection of an expression vector for HPV 16 E6 in U2OS cells, which express wild type p53, had no effect on transcription from the HRE or CA9 reporters (Figure 1b). We conclude that E7 is the primary viral protein responsible for enhancing HIF-1 transcriptional activity.
To determine whether E7 alone had an effect on endogenous HIF-1 transcription in keratinocytes, transcripts from HFKs or HFKs stably expressing HPV16 E7 from an integrated retrovirus vector were examined for the levels of three HIF-1 target genes: CA9, TSP1, and PGK1. As shown in Figure 1c, each of these transcripts was expressed at elevated levels in 16E7 cells providing further support that E7 expression is sufficient to increase levels of HIF-1 targets in keratinocytes.
The effect of p53 on transcriptional activity
In order to determine the mechanism of transcriptional enhancement by E7, we asked whether p53 might be involved. p53 has been reported to inhibit HIF-1 transcriptional activity (10, 30–35) and E7 can alter the levels as well as several functions of p53 (36–40). We therefore investigated whether E7 could enhance HIF-1 activity by counteracting p53-mediated inhibition of HIF-1α. We first observed that co-transfection of a vector expressing p53 with the reporter reduced activity by approximately one-half. When an expression vector for E7 was co-transfected along with p53 expression plasmids, HIF-1 activity was increased but still reduced from that seen in the absence of p53 (Figure 1d). We conclude that E7 and p53 act independently to alter HIF-1 activity, and the effect of each is not altered by the other. This indicates that E7’s enhancement of HIF-1 activity is likely not due to an effect on p53-mediated inhibition.
Although E6 cannot directly activate HIF-1 mediated transcription, we suspected that E6 might augment HIF-1 activity in the presence of additional p53 through its ability to induce degradation of p53. To investigate this possibility, an expression vector for E6 was transfected along with the p53 vector and we observed that E6 was able to block p53-mediated inhibition. While E6 does not directly synergize or interfere with the ability of E7 to enhance HIF-1 activity, it can overcome the repressive effects of high levels of p53 (Figure 1d).
E7 binds HIF-1α
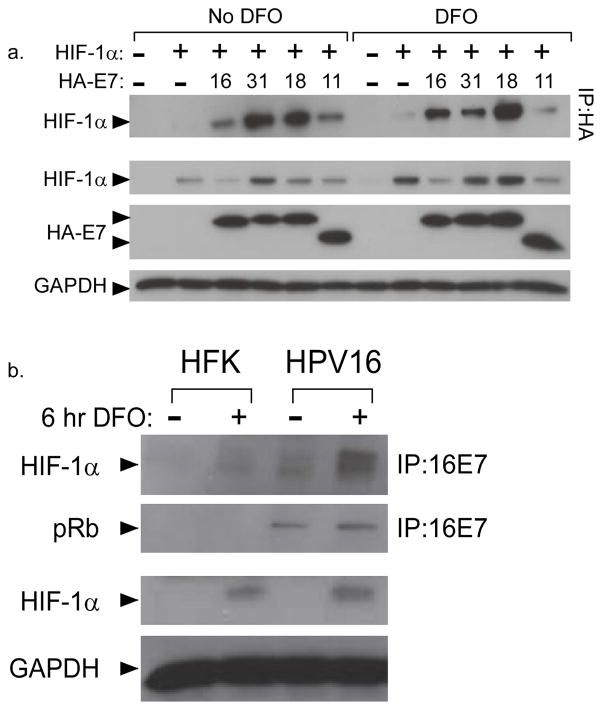
In order to determine the mechanism by which E7 enhances HIF-1 mediated transcription, we next investigated whether E7 could form a complex with HIF-1α. For this analysis, U2OS cells were transiently transfected with expression plasmids for HIF-1α and HA-tagged E7 from HPV16, 31, and 18. Cell extracts were then immunoprecipitated with antibodies to the HA tag and screened for the presence of HIF-1α by western blot analysis of the immunoprecipitates. E7 from all three HPV types was able to co-immunoprecipitate HIF-1α when HIF-1α was overexpressed in normoxia or upon treatment of cells with DFO (Figure 2a). In these experiments, the HA tag was located at the N terminus of E7, but similar effects were seen when the HA tag was placed on the C terminus (not shown). When the reverse immunoprecipitation was performed using anti-HIF-1α antibodies, complex formation with E7 was also detected (not shown). In additional studies, we observed that bacterially expressed and purified GST-31E7 could bind to HIF-1α translated in wheat germ extracts, suggesting that the interaction may be direct (not shown). Previous studies indicated that stabilization of HIF-1α in hypoxia was also enhanced in keratinocytes containing low risk HPV11 genomes (26). To test whether low risk HPV11 E7 could associate with HIF-1α, we performed similar co-transfection experiments using an expression vector for HA-11E7 and found that 11E7 was able to form a complex with HIF-1α at a level comparable to the high risk E7 proteins (Figure 2a). These results indicate that both high and low risk HPV E7 proteins can form complexes with HIF-1α in transient assays. Unlike 11E7, high risk E7s migrated slower than their predicted molecular weights, as previously observed by others (41).
Figure 2. E7 co-immunoprecipitates with HIF-1α.
(a) Expression vectors encoding HA-tagged E7s from HPV types 31, 16, 18, and 11 were cotransfected overnight into U2OS cells with an expression vector for HIF-1α. Cells were treated with 100 μM DFO for 6 hours or left untreated. E7-containing complexes were immunoprecipitated from total cell lysates with anti-HA antibodies. HIF-1α was detected by SDS/PAGE and western blotting of the immunoprecipitates. (b) Human foreskin keratinocytes (HFK) or HFK stably maintaining HPV16 genomes (HFK16 cells) were treated or untreated with 100 μM DFO for 6 hours and immunoprecipitation was performed from total cell lysates using anti-16E7. HIF-1α and pRb were detected in the immunoprecipitates or in total lysates by SDS/PAGE and western blotting.
To determine whether complex formation can be detected when E7 and HIF-1α are expressed at physiological levels, we created cell lines that stably maintain HPV16 episomes by transfecting human foreskin keratinocytes (HFK) with recircularized HPV16 genomes (HFK16). Following drug selection, these cell lines were expanded in culture and found to maintain episomal HPV16 at approximately 100 copies per cell (not shown). HFK16 cells were then treated with DFO, lysates were immunoprecipitated with antibodies directed against 16E7, and the immunoprecipitates were analyzed by Western blot analysis for the presence of HIF-1α. Consistent with results from transient transfection studies, 16E7 expressed from episomally replicating HPV16 genomes was able to co-immunoprecipitate endogenous HIF-1α (Figure 2b). Levels of HIF-1α were increased in HPV-containing cells treated with DFO relative to those in HFK cells, in agreement with previous studies (26). As a control, we confirmed that 16E7 was able to form complexes with pRb in the same cell lines. We conclude that the interaction between 16E7 and HIF-1α occurs under physiological conditions in cell culture.
Binding of E7 to HIF-1α is not necessary for transcriptional activation
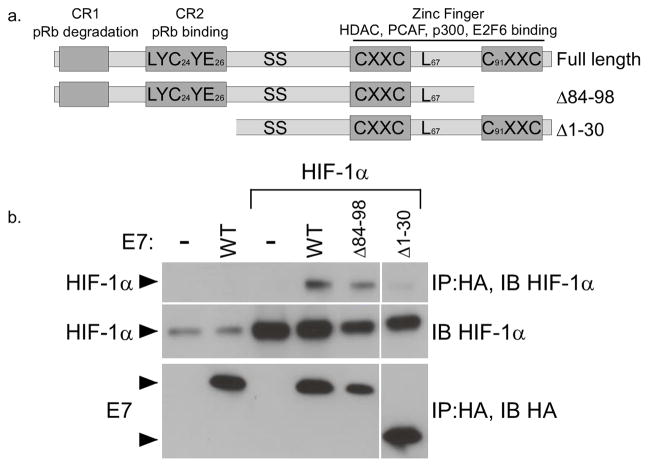
Several important regulatory domains of E7 have been identified through extensive functional and biochemical analyses by many groups (42; Figure 3a). The CR1 and CR2 domains in the N terminus are involved in pRb binding and degradation while the C terminus contains motifs that bind zinc, as well as histone deacetylases (HDACs), pCAF, E2F6, and other factors. In order to determine which region of E7 is responsible for binding HIF-1α, we generated deletions of E7 and tested their ability to bind HIF-1α in transient transfection/co-immunoprecipitation analyses. The plasmid pΔ1–30 contains a deletion of the first 30 amino acids of E7 which includes the pRb binding and degradation domains. The plasmid pΔ84–98 contains a deletion of the last 14 amino acids from the C terminus of E7, thereby disrupting the zinc binding C terminal domain. Following transient transfection of U2OS cells, the mutant E7Δ1–30 protein was expressed at high levels and was substantially reduced in its ability to associate with HIF-1α (Figure 3b). This indicates that the binding site for HIF-1α in E7 maps to the N terminal 30 amino acids. Although E7Δ84–98 was expressed at levels somewhat below that of full length E7, it formed complexes with HIF-1α at a level comparable to full length. We conclude that the C terminus of E7 is dispensable for binding HIF-1α and that binding activity is localized to the N-terminus.
Figure 3. Mapping the HIF-1α binding domain of E7.
(a) A schematic of the E7 protein, showing important functional domains and amino acids that were mutated in this study. (b) U2OS cells were cotransfected with the indicated 16E7 deletion construct and an expression vector for HIF-1α. Following 24 hours incubation, cells were treated with 100 μM DFO for 16 hours and total lysates prepared. E7-containing complexes were immunoprecipitated with anti-HA antibodies. HIF-1α was detected by SDS/PAGE and western blotting of the immunoprecipitates.
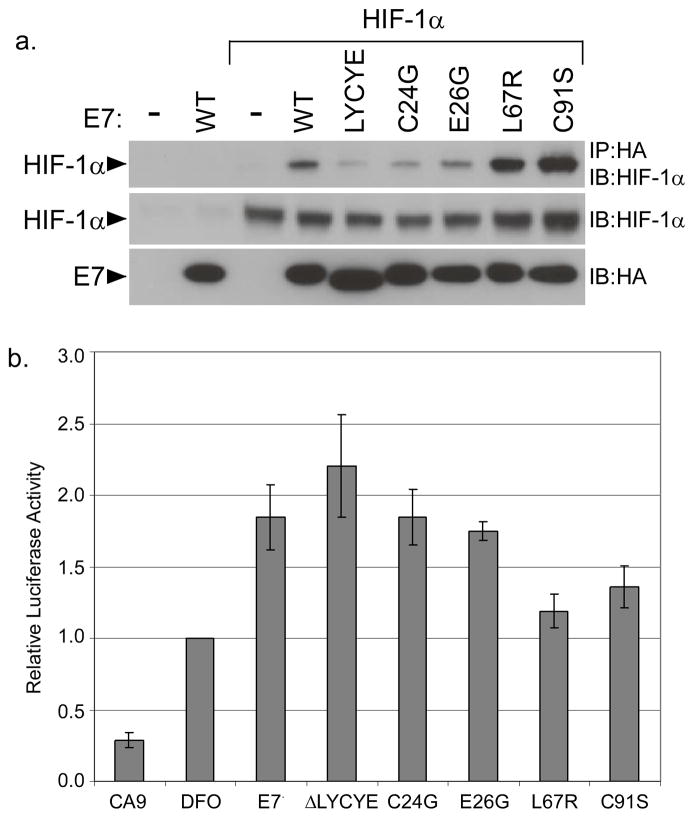
We next tested several previously characterized point mutants and small deletions of E7 for their ability to form complexes with HIF-1α. The LYCYE mutant consists of an in-frame deletion of the whole pRb binding domain, while C24G and E26G are point mutations within this motif (43, 44). The L67R and C91S substitution mutations disrupt the C terminal domain, and have been shown to abrogate, among other things, E7’s ability to interact with HDACs (5). We found that all three mutations in the pRb-binding domain reduced the ability of E7 to associate with HIF-1α, suggesting a role for this domain in the interaction between E7 and HIF-1α. Interestingly, both mutations in the C terminus not only retained the ability to bind HIF-1α, they showed a clear and reproducible increase in binding as compared to wild type (Figure 4a).. Furthermore, mutants in the CR1 domain bound to HIF-1α as well as wild type (not shown). We conclude that the N-terminus of E7 and the pRb binding domain in particular is responsible for binding to HIF-1.
Figure 4. Mapping the HIF-1 transcriptional enhancement domain of E7.
(a) U2OS cells were cotransfected with the indicated 16E7 deletion construct and an expression vector for HIF-1α. Following 24 hours incubation, cells were treated with 100 μM DFO for 16 hours and total lysates prepared. E7-containing complexes were immunoprecipitated with anti-HA antibodies. HIF-1α was detected by SDS/PAGE and western blotting of the immunoprecipitates. (b) U2OS cells were cotransfected overnight with the indicated E7 mutant construct and a reporter for the CA9 promoter. After 24 hours, samples were treated with 100 μM DFO for an additional 16 hours. Lysates were prepared, and the activities of firefly and Renilla luciferase were measured. Values were normalized to the Renilla luciferase activity in each sample, and the reporter alone with DFO was set to 1. Each point represents the mean of 6 experiments and error bars represent ± 1 standard error of the mean.
We next investigated if binding of E7 to HIF-1 was important for its ability to enhance transcriptional activity. Surprisingly, we found that mutants in the pRb-binding domain were as effective as wild type at activating HIF-1 transcription (p>0.3 vs wild type). In contrast mutations that disrupted the C terminal domain, L67R and C91S, were severely attenuated for transcriptional activation, and this was similar to the reporter levels seen in the absence of E7 (Figure 4b). Therefore, even though the C terminus is dispensable for binding of E7 to HIF-1α, it is necessary for enhancement HIF-1 transcriptional activity by E7. These data therefore indicate that an activity of E7 other than binding to HIF-1α is necessary for enhancement of transcriptional activity.
Enhancement of HIF-1 activity by E7 requires HDAC activity
Since E7 binding to HIF-1α is not necessary for its activation, we considered other possible mechanisms focusing on activities mediated through the C-terminus of E7. In transient reporter assays, the ability of E7 to enhance HIF-1 transcription was reduced by the C91S and L67R mutations. These two mutations have been reported to block the binding of E7 with a number of cellular factors, including HDACs (5, 45) and HDACs have been implicated in enhancing HIF-1 activity (11–16). We investigated whether E7’s enhancement of HIF-1 activity requires HDAC activity by treating transfected U2OS cells with trichostatin A (TSA), a broad-spectrum HDAC inhibitor. Exposure of these cells to TSA abrogated activation of HIF-1 dependent transcriptional activity, confirming that in our system HDACs are important for HIF-1 function (p<0.01, Figure 5a). Significantly, E7 had no enhancing effect on HIF-1 activity in the absence of HDAC activity in cells treated with TSA.
Figure 5. HDACs are needed for HIF-1 activity and E7’s enhancement.
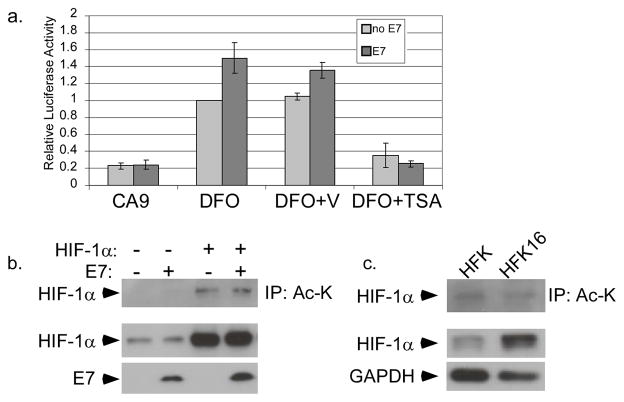
(a) U2OS cells were cotransfected overnight with a reporter for the CA9 promoter with or without a 16E7 expression vector. After 24 hours, samples were treated with 100 μM DFO with or without 300 nM TSA or DMSO vehicle for an additional 16 hours. Lysates were prepared, and the activities of firefly and Renilla luciferase were measured. Values were normalized to the Renilla luciferase activity in each sample, and the reporter alone with DFO was set to 1. Each point represents the mean of 5 experiments and error bars represent ± 1 standard error of the mean. (b) U2OS cells were cotransfected overnight with 16E7 and/or HIF-1α expression vectors. Following 24 hours incubation, cells were treated with 100 μM DFO for 16 hours and total lysates prepared. Immunoprecipitation was performed using anti-acetylated lysine (Ac-K) antibody. E7 and HIF-1α were detected by SDS/PAGE and western blotting. (c) HFK or HFK16 cells were treated with 100 μM DFO for 16 hours. Total lysates were immunoprecipitated with Ac-K antibody. HIF-1α was detected by SDS/PAGE and western blotting.
We next tested whether HIF-1α is itself acetylated and whether acetylation is affected by E7. Using lysates from U2OS cells transiently transfected with a HIF-1α expression vector, we performed immunoprecipitations with an antibody against acetylated lysine and then screened for HIF-1α by Western analysis. As shown in Figure 5b, HIF-1α was clearly detected by this analysis indicating that either HIF-1α itself or a protein associated with HIF-1α is acetylated. Importantly the level of acetylation was unchanged by the presence of E7. When a similar co-immunoprecipitation experiment was performed using lysates from HFKs or HFK16 cells that stably maintain HPV episomes, we found no differences in the amount of HIF-1α immunoprecipitated with anti acetyl-lysine antibody (Figure 5c). These results indicate that E7 does not alter the level of HIF-1α acetylation and it is unlikely to be the mechanism by which E7 modulates HIF-1 activity. Additionally, cell fractionation analyses indicated that E7 had no effect on the subcellular localization of either HIF-1α or HDAC1 (not shown).
E7 displaces HDACs from HIF-1α
Another mechanism to explain how association of E7 with HDACs results in HIF-1 activation could involve the modulation of HIF-1α binding to HDACs by E7. We therefore investigated the state of HDAC1 complexes with HIF-1α in the presence of E7. For these studies, we first transfected HIF-1α expression vectors into U2OS cells, immunoprecipitated HDAC1 containing complexes, and screened for the association of HIF-1α by Western blot analysis. Consistent with previous reports we confirmed that HIF-1α and HDAC1 form complexes in cells (Figure 6a) (11). When E7 was cotransfected along with HIF-1α, a dramatic reduction in the amount of HIF-1α bound to HDAC1 was observed. Importantly, both of the C terminal mutants of E7, which were defective in activating HIF-1, were unable to displace HDAC1 from HIF-1α (Figure 6a). Furthermore, the LYCYE mutant, which was able to enhance HIF-1 activity despite reduced binding to HIF-1α, was able to mediate displacement of HDAC1 from HIF-1α (Figure 6c). We also examined the interaction of HIF-1α with two additional HDACs, HDAC4 and HDAC7, that have been implicated in other studies to be important in HIF-1 regulation and confirmed that both formed complexes with HIF-1α(14, 18), (19). When E7 was included in the transfections, both HDAC4 and HDAC7 were displaced from HIF-1α in a manner dependent on the C terminal domain of E7, similar to HDAC1. We conclude that the ability of E7 to displace HDACs from HIF-1α correlates with transcriptional enhancement by E7. Binding of HIF-1α to p300/CBP is also reported to be an important regulator of HIF-1 activity (9, 46, 47), but we did not observe any consistent changes in the association between HIF-1α and p300 or with pCAF due to the presence of E7 (Figure 6a). It was next important to confirm that similar effects would be seen in cells that stably express E7 at physiological levels. For this study, we immunoprecipitated HDAC1 from HFKs or HFK16 cells and then analyzed the resultant complexes for the levels of bound HIF-1α by Western analysis. Similar to our observations in transient transfections, we found that HIF-1α/HDAC1 association is markedly reduced in cells maintaining HPV16, despite increased total levels of HDAC1 (Figure 6c). This indicates that the E7 acts to reduce the binding of HDACs to HIF-1 and this correlates with enhanced ability of HIF-1 to activate target promoters. In initial chromatin immunoprecipitation studies, the presence of E7 did not significantly alter the levels of HIF-1α bound to the CA9 promoter (Supplementary Figure 1). This further supports the idea that displacement of HDACs rather than increasing the binding of HIF-1α is the target of E7’s activating effect.
Figure 6. Activation of HIF-1 transcription by E7 correlates with loss of HDAC1 association with HIF-1α.
(a and c) U2OS cells were cotransfected overnight with the indicated mutant or wild-type 16E7 and HIF-1α expression vectors. Following 24 hours incubation, cells were treated with 100 μM DFO for 16 hours and total lysates prepared. Immunoprecipitation was performed using antibodies specific for HDAC1, HDAC4, HDAC7, p300, and/or pCAF. HIF-1α was detected by SDS/PAGE and western blotting of the immunoprecipitates. (b) Western blots of input protein levels for the experiments shown in (a). (d) HFKs or two independently derived lines of HFK16 cells were treated with DFO for 16 hours, total lysates prepared, and immunoprecipitation was performed using anti-HDAC1 antibodies. HIF-1α was detected by SDS/PAGE and western blotting of the immunoprecipitates. The blots shown in panel (d) are from the same experiment.
Discussion
The development of benign warts as well as cancerous lesions following infection by human papillomaviruses requires the induction of angiogenesis. An important factor responsible for activation of angiogenesis is HIF-1, and our studies indicate that the HPV E7 proteins enhance HIF-1 activity. During long term HPV infections, these changes likely have significant effects on the development of vasculature in both benign and malignant lesions. Our studies further indicate that that both high and low risk E7 proteins form complexes with HIF-1α but that this is not the primary activity responsible for the enhancement of HIF-1 mediated transcription. Rather, E7’s ability to enhance HIF-1 mediated transcription correlated with the ability to dissociate HDACs from HIF-1α. Disruption of HIF-1α/HDAC binding was observed in transient assays as well as in keratinocytes containing the complete HPV16 genome. In addition, initial studies demonstrate a similar disruption of HIF-1α/HDAC binding in keratinocytes expressing E7 alone, indicating that E7 alone is sufficient to mediate this effect (J.B. and L.L., unpublished). The activation of transcription factor function by blocking HDAC binding is a novel mechanism of action for E7 and we believe that this activity is important for the promotion of angiogenesis and metabolic changes during HPV infections.
The ability of E7 to enhance HIF-1 transcriptional activity correlated with inhibition of the binding of HDACs to HIF-1α̃. HDACs have been shown to utilize multiple mechanisms to modulate HIF-1 activity. It is likely that different combinations of HDACs mediate these activities. Previous studies have suggested that binding of HDACs leads to increased activity of HIF-1 but this largely is based on effects of overexpression of HDACs in transient assays. Our mutational studies indicate the ability of E7 to bind to HDACs correlates with the ability of E7 to block binding of HDAC1, 4 and 7 to HIF-1α, and results in its increased activity. It is not clear which of these HDACS are responsible for E7’s effects and it is possible that multiple HDACS are involved.
E7 displaced several HDACs from HIF-1α including HDAC1, HDAC4, and HDAC7 and this correlated with activation of the CA9 promoter. We hypothesize that by displacing HDACs from HIF-1α, E7 reduces the inhibitory effects of these factors on transcriptional complexes bound to cellular promoters (Figure 7). This model of E7-mediated promoter activation is consistent with previous observations from our laboratory in which the binding of HDACs by E7 activated transcription of the E2F2 promoter through the displacement of HDAC1 and 3 from promoter sequences (6). Our current observations on HIF-1 activation provide a novel mechanism for E7 action in blocking HDAC binding directly to a transcriptional activator.
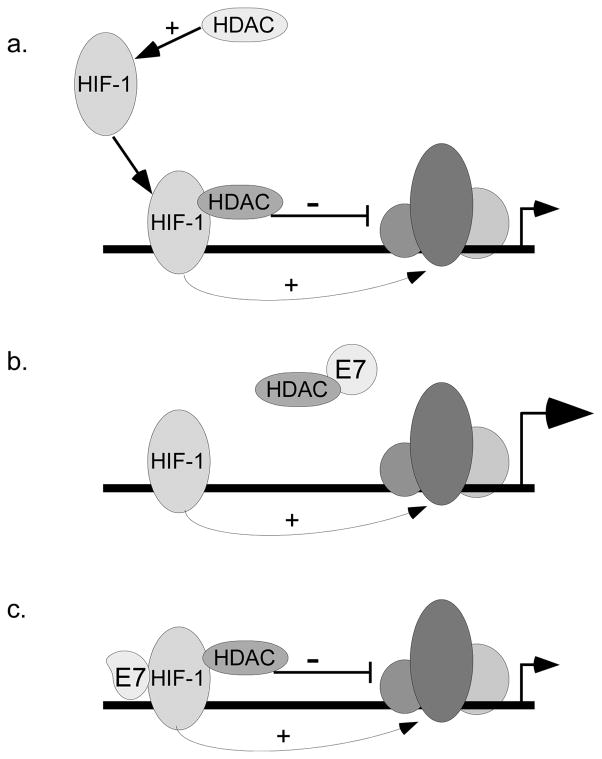
Figure 7. Model of E7’s activation of HIF-1 dependent transcription.
(a) HDACs provide a necessary positive signal activating HIF-1, but can have a negative impact on the surrounding transcriptional complex. (b) E7 binds HDACs and reduces the inhibitory effect of HDACs on transcription HIF-1 mediated transcription, while allowing the positive signal to persist. (c) In the presence of E7 mutants that cannot bind HDACs, enhancement of transcription is abolished, and interaction of E7 with HIF-1α is increased.
Our studies showed that GST-E7 produced in bacteria could bind to HIF-1α generated in wheat germ extracts. This would suggest that the interaction between E7 and HIF-1α is direct, although we cannot exclude the possibility that the presence of plant pRb-like factors or other proteins in the wheat germ extract may mediate binding. The formation of complexes between E7 and HIF-1α was also shown in both transient assays and in stable cell lines, but given that binding of E7 to HIF-1α did not correlate with transcriptional activity, we do not yet understand the significance of this interaction. It is possible that this interaction serves some function in the differentiation-dependent phase of the viral life cycle, or in other activities of E7 not easily measured in our assays. Further studies will be necessary to understand the role of these interactions. Two mutants of E7 that lack the ability to bind HDACs, L67R and C91S, bound HIF-1α at higher levels then wild type E7. The HDAC binding activity maps to the C terminus of E7 while the ability to associate with HIF-1α localizes to the N terminus. We believe the two activities may be in competition with each other, so that elimination of HDAC binding activity may promote HIF-1α binding (Figure 7c).
In natural infections, E6 and E7 are expressed together, and their functions may be expected to complement one another. Our data demonstrate that E7 enhances HIF-1 dependent reporter activity and E6 abrogated p53 inhibitory activity. Together these two oncogenes would enhance HIF-1 dependent gene expression in growing lesions despite negative effects of p53. Our finding that E6 does not directly activate HIF-1 transcription is in contrast to reports indicating that E6 can activate HIF-1 mediated transcription (22, 23, 35). We do not know the basis for this difference but it may be due to the specific promoter studied or to levels of p53 in different cell lines.
In summary, our studies indicate that E7 can activate HIF-1 dependent transcription by blocking the interaction of HDACs with HIF-1α in a manner dependent on the HDAC binding domain of E7. E6 primarily blocks the inhibitory effects of p53 on HIF-1 activity. These findings shed light on the mechanisms by which HPV contributes to tumor angiogenesis and describes a novel function of the E7 proteins in the viral pathogenesis.
Supplementary Material
Acknowledgments
We thank Eric Huang, Navdeep Chandel, Stefan Kaluz, Lee Ellis, Bayar Thimmapaya, and Margaret McLaughlin-Drubin for materials; Keya Raychaudhuri and Kyle Jamison for technical assistance; and Kathy Rundell and members of the Laimins lab for helpful discussion. This work was supported by grants to LAL from the National Cancer Institute (R37CA74202 and R01CA59655).
References
- 1.zur Hausen H. Papillomaviruses in human cancers. Proc Assoc Am Physicians. 1999;111:581–7. doi: 10.1046/j.1525-1381.1999.99723.x. [DOI] [PubMed] [Google Scholar]
- 2.zur Hausen H. Papillomavirus infections--a major cause of human cancers. Biochimica et Biophysica Acta. 1996;1288:F55–78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 3.Longworth MS, Laimins LA. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev. 2004;68:362–72. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munger K, Baldwin A, Edwards KM, et al. Mechanisms of human papillomavirus-induced oncogenesis. Journal of Virology. 2004;78:11451–60. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longworth MS, Laimins LA. The binding of histone deacetylases and the integrity of zinc finger-like motifs of the E7 protein are essential for the life cycle of human papillomavirus type 31. Journal of Virology. 2004;78:3533–41. doi: 10.1128/JVI.78.7.3533-3541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longworth MS, Wilson R, Laimins LA. HPV31 E7 facilitates replication by activating E2F2 transcription through its interaction with HDACs. The EMBO Journal. 2005;24:1821–30. doi: 10.1038/sj.emboj.7600651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–8. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–64. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 9.Brat DJ, Kaur B, Van Meir EG. Genetic modulation of hypoxia induced gene expression and angiogenesis: relevance to brain tumors. Front Biosci. 2003;8:d100–16. doi: 10.2741/942. [DOI] [PubMed] [Google Scholar]
- 10.Bardos JI, Ashcroft M. Negative and positive regulation of HIF-1: a complex network. Biochimica et Biophysica Acta. 2005;1755:107–20. doi: 10.1016/j.bbcan.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Yoo YG, Kong G, Lee MO. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. The EMBO Journal. 2006;25:1231–41. doi: 10.1038/sj.emboj.7601025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toh Y, Nicolson GL. The role of the MTA family and their encoded proteins in human cancers: molecular functions and clinical implications. Clinical & Experimental Metastasis. 2009;26:215–27. doi: 10.1007/s10585-008-9233-8. [DOI] [PubMed] [Google Scholar]
- 13.Moon HE, Cheon H, Chun KH, et al. Metastasis-associated protein 1 enhances angiogenesis by stabilization of HIF-1alpha. Oncology Reports. 2006;16:929–35. [PubMed] [Google Scholar]
- 14.Qian DZ, Kachhap SK, Collis SJ, et al. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Research. 2006;66:8814–21. doi: 10.1158/0008-5472.CAN-05-4598. [DOI] [PubMed] [Google Scholar]
- 15.Kong X, Lin Z, Liang D, Fath D, Sang N, Caro J. Histone deacetylase inhibitors induce VHL and ubiquitin-independent proteasomal degradation of hypoxia-inducible factor 1alpha. Molecular and Cellular Biology. 2006;26:2019–28. doi: 10.1128/MCB.26.6.2019-2028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fath DM, Kong X, Liang D, et al. Histone deacetylase inhibitors repress the transactivation potential of hypoxia-inducible factors independently of direct acetylation of HIF-alpha. The Journal of Biological Chemistry. 2006;281:13612–9. doi: 10.1074/jbc.M600456200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MS, Kwon HJ, Lee YM, et al. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nature Medicine. 2001;7:437–43. doi: 10.1038/86507. [DOI] [PubMed] [Google Scholar]
- 18.Seo HW, Kim EJ, Na H, Lee MO. Transcriptional activation of hypoxia-inducible factor-1alpha by HDAC4 and HDAC5 involves differential recruitment of p300 and FIH-1. FEBS Letters. 2009;583:55–60. doi: 10.1016/j.febslet.2008.11.044. [DOI] [PubMed] [Google Scholar]
- 19.Kato H, Tamamizu-Kato S, Shibasaki F. Histone deacetylase 7 associates with hypoxia-inducible factor 1alpha and increases transcriptional activity. The Journal of Biological Chemistry. 2004;279:41966–74. doi: 10.1074/jbc.M406320200. [DOI] [PubMed] [Google Scholar]
- 20.Zhong L, D’Urso A, Toiber D, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via HIF1alpha. Cell. 2010;140:280–93. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen W, Ding J, Sun W, et al. Suppression of cyclin D1 by hypoxia-inducible factor-1 via direct mechanism inhibits the proliferation and 5-fluorouracil-induced apoptosis of A549 cells. Cancer Research. 2010;70:2010–9. doi: 10.1158/0008-5472.CAN-08-4910. [DOI] [PubMed] [Google Scholar]
- 22.Tang X, Zhang Q, Nishitani J, Brown J, Shi S, Le AD. Overexpression of human papillomavirus type 16 oncoproteins enhances hypoxia-inducible factor 1 alpha protein accumulation and vascular endothelial growth factor expression in human cervical carcinoma cells. Clin Cancer Res. 2007;13:2568–76. doi: 10.1158/1078-0432.CCR-06-2704. [DOI] [PubMed] [Google Scholar]
- 23.Clere N, Bermont L, Fauconnet S, et al. The human papillomavirus type 18 E6 oncoprotein induces Vascular Endothelial Growth Factor 121 (VEGF121) transcription from the promoter through a p53-independent mechanism. Exp Cell Res. 2007;313:3239–50. doi: 10.1016/j.yexcr.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 24.Chen W, Li F, Mead L, et al. Human papillomavirus causes an angiogenic switch in keratinocytes which is sufficient to alter endothelial cell behavior. Virology. 2007;367:168–74. doi: 10.1016/j.virol.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toussaint-Smith E, Donner DB, Roman A. Expression of human papillomavirus type 16 E6 and E7 oncoproteins in primary foreskin keratinocytes is sufficient to alter the expression of angiogenic factors. Oncogene. 2004;23:2988–95. doi: 10.1038/sj.onc.1207442. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura M, Bodily JM, Beglin M, Kyo S, Inoue M, Laimins LA. Hypoxia-specific stabilization of HIF-1alpha by human papillomaviruses. Virology. 2009;387:442–8. doi: 10.1016/j.virol.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyers C, Laimins LA. In vitro systems for the study and propagation of human papillomaviruses. Current Topics in Microbiology and Immunology. 1994;186:199–215. doi: 10.1007/978-3-642-78487-3_11. [DOI] [PubMed] [Google Scholar]
- 28.Hebner CM, Wilson R, Rader J, Bidder M, Laimins LA. Human papillomaviruses target the double-stranded RNA protein kinase pathway. The Journal of General Virology. 2006;87:3183–93. doi: 10.1099/vir.0.82098-0. [DOI] [PubMed] [Google Scholar]
- 29.Wilson R, Laimins LA. Differentiation of HPV-containing cells using organotypic “raft” culture or methylcellulose. Methods in Molecular Medicine. 2005;119:157–69. doi: 10.1385/1-59259-982-6:157. [DOI] [PubMed] [Google Scholar]
- 30.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Research. 2004;64:2627–33. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J, Schmid T, Schnitzer S, Brune B. Tumor hypoxia and cancer progression. Cancer Lett. 2006;237:10–21. doi: 10.1016/j.canlet.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Yu D, Hu M, et al. Wild-type p53 suppresses angiogenesis in human leiomyosarcoma and synovial sarcoma by transcriptional suppression of vascular endothelial growth factor expression. Cancer Research. 2000;60:3655–61. [PubMed] [Google Scholar]
- 33.Qin G, Kishore R, Dolan CM, et al. Cell cycle regulator E2F1 modulates angiogenesis via p53-dependent transcriptional control of VEGF. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11015–20. doi: 10.1073/pnas.0509533103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaluzova M, Kaluz S, Lerman MI, Stanbridge EJ. DNA damage is a prerequisite for p53-mediated proteasomal degradation of HIF-1alpha in hypoxic cells and downregulation of the hypoxia marker carbonic anhydrase IX. Molecular and Cellular Biology. 2004;24:5757–66. doi: 10.1128/MCB.24.13.5757-5766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravi R, Mookerjee B, Bhujwalla ZM, et al. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes & Development. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Disbrow GL, Yuan H, Tomaic V, Schlegel R. Myc and human papillomavirus type 16 E7 genes cooperate to immortalize human keratinocytes. Journal of Virology. 2007;81:12689–95. doi: 10.1128/JVI.00669-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seavey SE, Holubar M, Saucedo LJ, Perry ME. The E7 oncoprotein of human papillomavirus type 16 stabilizes p53 through a mechanism independent of p19(ARF) Journal of Virology. 1999;73:7590–8. doi: 10.1128/jvi.73.9.7590-7598.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones DL, Thompson DA, Suh-Burgmann E, Grace M, Munger K. Expression of the HPV E7 oncoprotein mimics but does not evoke a p53- dependent cellular DNA damage response pathway. Virology. 1999;258:406–14. doi: 10.1006/viro.1999.9733. [DOI] [PubMed] [Google Scholar]
- 39.Massimi P, Banks L. Repression of p53 transcriptional activity by the HPV E7 proteins. Virology. 1997;227:255–9. doi: 10.1006/viro.1996.8315. [DOI] [PubMed] [Google Scholar]
- 40.Eichten A, Westfall M, Pietenpol JA, Munger K. Stabilization and functional impairment of the tumor suppressor p53 by the human papillomavirus type 16 E7 oncoprotein. Virology. 2002;295:74–85. doi: 10.1006/viro.2002.1375. [DOI] [PubMed] [Google Scholar]
- 41.Heck DV, Yee CL, Howley PM, Munger K. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4442–6. doi: 10.1073/pnas.89.10.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munger K, Basile JR, Duensing S, et al. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene. 2001;20:7888–98. doi: 10.1038/sj.onc.1204860. [DOI] [PubMed] [Google Scholar]
- 43.Phelps WC, Munger K, Yee CL, Barnes JA, Howley PM. Structure-function analysis of the human papillomavirus type 16 E7 oncoprotein. Journal of Virology. 1992;66:2418–27. doi: 10.1128/jvi.66.4.2418-2427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edmonds C, Vousden KH. A point mutational analysis of human papillomavirus type 16 E7 protein. Journal of Virology. 1989;63:2650–6. doi: 10.1128/jvi.63.6.2650-2656.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brehm A, Nielsen SJ, Miska EA, et al. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. The EMBO Journal. 1999;18:2449–58. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death and Differentiation. 2008;15:678–85. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koh MY, Spivak-Kroizman TR, Powis G. HIF-1 regulation: not so easy come, easy go. Trends in Biochemical Sciences. 2008;33:526–34. doi: 10.1016/j.tibs.2008.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.