Abstract
Induced pluripotent stem (iPS) cells can be generated from various embryonic or adult cell types upon expression of a set of few transcription factors, most commonly consisting of Oct4, Sox2, c-Myc and Klf4, following a strategy originally published by Takahashi and Yamanaka in 2006 (Takahashi and Yamanaka, 2006). Since iPS cells are molecularly and functionally similar to embryonic stem (ES) cells, they provide a source of patient-specific pluripotent cells for regenerative medicine and disease modeling, and therefore have generated enormous scientific and public interest. The generation of iPS cells also presents a powerful tool for dissecting mechanisms that stabilize the differentiated state and are required for the establishment of pluripotency. In this review, we discuss our current view of the molecular mechanisms underlying transcription factor-mediated reprogramming to induced pluripotency.
Introduction
Somatic cells can be reprogrammed to iPS cells by the delivery of a few pluripotency-related transcription factors. Since the original description of iPS cells in Shinya Yamanaka’s landmark 2006 report (Takahashi and Yamanaka, 2006), studies of transcription factor-induced reprogramming to the iPS cell state have branched into two explosive fields of research. First, no longer hindered by the technical and ethical limitations associated with somatic cell nuclear transfer (SCNT) and cell fusion, reprogramming via the Yamanaka approach provides a new avenue to investigate basic questions of cellular plasticity and pluripotency. Secondly, the iPS cell technology enables the derivation of patient-and disease specific pluripotent stem cell lines, which has opened the door to disease modeling, drug discovery, and cell replacement strategies.
Both of these branches of iPS cell research are affected by the inefficiency of the reprogramming process (Table 1). Despite the variety of recent publications reporting DNA-free or integration-free reprogramming via protein delivery of the reprogramming factors or the use of RNA viruses, the most efficient generation of iPS cells is based on genomic integration of DNA encoding the reprogramming factors, most commonly through lenti-or retroviral transduction (Table 1). The use of most iPS cells is therefore thought to be affected by genomic alterations that could lead to phenotypic artifacts arising from insertional mutagenesis or expression of the oncogenic reprogramming factors (Hochedlinger et al., 2005; Nakagawa et al., 2008; Okita et al., 2007; Wernig et al., 2008b). The hope is that a better understanding of the reprogramming process will lead to improved, more efficient reprogramming technologies that don’t require genomic integration, linking the two major avenues of reprogramming research. Similarly, a better general understanding of how a small set of transcription factors can reset the epigenetic landscape of cells, gained from the reprogramming process, could also further the development of rational differentiation strategies for pluripotent cells, which will be important for disease modeling and therapeutic applications of iPS and ES cells.
Table 1.
Summary of reprogramming methods and efficiencies
| Reprogramming scheme | Delivery/expression method | Advantage | Disadvantage | Donor cell type | Reprogramming efficiency (%) | Criteria used to score efficiency | References |
|---|---|---|---|---|---|---|---|
| Integrative | Retrovirus |
|
|
Various cell types | 0.05–0.1 | Drug resistance from pluripotency marker | Takahashi and Yamanaka 2006, Okita et al. 2007, Wernig et al. 2007 |
| Constitutive lentivirus |
|
|
N/D | N/D | Brambrink et al. 2008 | ||
| Inducible lentivirus |
|
|
0.5 | GFP reporter expression from pluripotency marker | Brambrink et al. 2008, Stadtfeld et al. 2008b, Sommer et al. 2009 | ||
| Secondary inducible lentiviral system |
|
|
0.02 –28* *myeloid progenitors |
Wernig et al. 2008a, Hanna et al. 2008, Eminli et al. 2009 | |||
| Inducible, reprogrammable mouse |
|
Mouse fibroblasts and blood | 0.01–40* *hematopoietic stem cells |
ES-like morphology, Nanog expression | Carey et al. 2009, Stadtfeld et al. 2010 | ||
| Integrative/excisable | LoxP-flanked reprogramming cassettes in lentivirus |
|
|
Mouse and human fibroblasts | 0.005–0.01 (0.5 for mouse) | GFP reporter expression from pluripotency marker, Tra-1-60 and NANOG staining | Soldner et al. 2009, Sommer et al. 2010 |
| Transposons (piggyBac) |
|
|
Mouse and human fibroblasts | 0.04–2.5 | ES-like morphology, GFP reporter expression from pluripotency marker | Woltjen et al. 2009, Kaji et al. 2009, Yusa et al. 2009 | |
| Non-integrative, DNA-based | Minicircle |
|
|
Human adult adipose stem cells | 0.005 | ES-like morphology | Jia et al. 2010 |
| Plasmid transfection | Mouse fibroblasts | 0.000015 | GFP reporter expression from pluripotency marker | Okita et al. 2008 | |||
| Episomal vector | Human foreskin fibroblasts | 0.000006 | ES-like morphology | Yu et al. 2009 | |||
| Adenovirus | Mouse hepatocytes | 0.0005 | GFP reporter expression from pluripotency marker | Stadtfeld et al. 2008c | |||
| Human embryonic fibroblasts | 0.0002 | ES-like morphology | Zhou and Freed, 2009 | ||||
| Non-integrative, DNA-free | ES protein extract |
|
|
Mouse cardiac and skin fibroblasts | 0.001 | ES-like morphology | Cho et al. 2010 |
| HEK293 expressed transducible protein | Human newborn fibroblasts | 0.001 | Alkaline phosphotase staining | Kim et al. 2009a | |||
| Bacterially expressed transducible protein | Mouse fibroblasts | 0.006 | GFP reporter expression from pluripotency marker | Zhou et al. 2009 | |||
| Sendai vectors (RNA) |
|
Human foreskin fibroblasts | 0.001–0.1 | Alkaline phosphotase staining | Fusaki et al. 2009 |
Reproramming efficiency is typically determined 2–4 weeks after reprogramming factor induction. By and large, efficiency is defined by the number of iPS clones (scored by different criteria) generated from a starting number of somatic cells. The method of efficiency calculation varies widely across labs.
Despite the numerous reports demonstrating tactics to boost the efficiency of reprogramming, the molecular requirements as well as barriers of the reprogramming process are only beginning to be defined. Many studies are looking for small molecules, miRNAs, siRNAs, or growth factors in efforts to substitute individual reprogramming factors to lower the need for genomic integration while allowing efficient reprogramming (Table 2). Others aim at uncovering pathways that are essential for the induction of pluripotency and contribute to overcoming reprogramming barriers. Perhaps the biggest question underlying the mechanism of reprogramming is how such a small set of transcription factors can destabilize the somatic program and eventually lead to the establishment of an ES cell- specific transcriptional network. Our review aims to summarize the most recent studies describing the molecular events taking place during the reprogramming process, and to discuss the mechanistic obstacles proposed to limit the rate and efficiency of faithful conversion to pluripotency.
Table 2.
Reprogramming factor replacements
| Original reprogramming factor(s) | Substitute factor | Proposed mechanism | References |
|---|---|---|---|
| cMyc | nMyc or LMyc | Homologues that may be redundant in function. LMyc has lower transformation activity compared with cMyc. | Nakagawa et al. 2003, 2010 |
| Wnt3a | c-Myc is a target of Wnt signaling. | Marson et al. 2008 | |
| miR-291-3p, -294, or-295 | miR290 cluster is a c-Myc target. | Judson et al. 2009 | |
| Klf4 | Klf1, Klf2, or Klf5 | Homologues that may be redundant in function. | Nakagawa et al. 2008 |
| Esrrb or Esrrg | Esrrb regulates Klf4 expression, has similar binding targets as Oct4, Sox2, and Klf4, and forms a protein complex with Qct4. | Feng et al. 2009a, van den Berg et al. 2008, Chen et al. 2008 | |
| Sox2 | Sox1, Sox3, Sox15, or Sox18 | Homologues that may be redundant in function. | Nakagawa et al. 2008 |
| Nanog | Nanog has similar binding targets as Sox2. | Ichida et al. 2009 | |
| Sox2 and c-Myc | Smad7 | Inhibits Tgfβ signaling, promoting MET. | Li et al, 2010, Maherali and Hochedlinger 2009, Ichida et al. 2009 |
| Oct4 | Nr5a1 or Nr5a2 | Binds enhancer and promoter of Oct4 and regulates its expression. | Heng et al. 2010 |
Listed are RNAs and proteins that are able to replace individual reprogramming factors. For a comprehensive review of small molecule replacers of reprogramming, see Feng et al. 2009.
Reprogramming Basics
iPS cells have been generated upon ectopic expression of Oct4, Sox2, cMyc and Klf4 from a number of species including human (Lowry et al., 2008; Park et al., 2008; Takahashi et al., 2007; Yu et al., 2007), mouse (Maherali et al., 2007b; Okita et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007), rat (Li et al., 2009b), pig (Wu et al., 2009), and rhesus monkey (Liu et al., 2008), and many different cell types such as fibroblasts, terminally differentiated lymphocytes and other blood cells, stomach and liver cells, neural progenitors, keratinocytes, melanocytes, and pancreatic β-cells (Aasen et al., 2008; Aoi et al., 2008; Hanna et al., 2008; Kim et al., 2008b; Stadtfeld et al., 2008a; Utikal et al., 2009a). While cMyc, Klf4, and Sox2 can be replaced in the reprogramming process by close homologs and small molecules, Oct4 appears more pivotal and so far could only be efficiently replaced by its upstream regulator, the orphan nuclear receptorNr5A2 (Table 2 and references therein). The diversity of cell types and species that have been reprogrammed and the general applicability of the four original reprogramming factors suggests a generic fashion in which the four factors act and indicates that there probably is no cell-type specific barrier that cannot be overcome by the action of the reprogramming factors leading to a evolutionary conserved pluripotency network. Nevertheless, the starting cell type can alter the dependence on the reprogramming factors, efficiency, and kinetics. For example, the high expression level of endogenous Sox2 and moderate levels of cMyc and Klf4 in neural precursor cells (NPCs) allow reprogramming with only Oct4, albeit very slowly (Kim et al., 2009b). Addition of ectopic Sox2 may even interfere with the reprogramming of NPCs (Eminli et al., 2008; Silva et al., 2008), suggesting that there are ideal levels of the reprogramming factors in relation to each other to induce pluripotency.
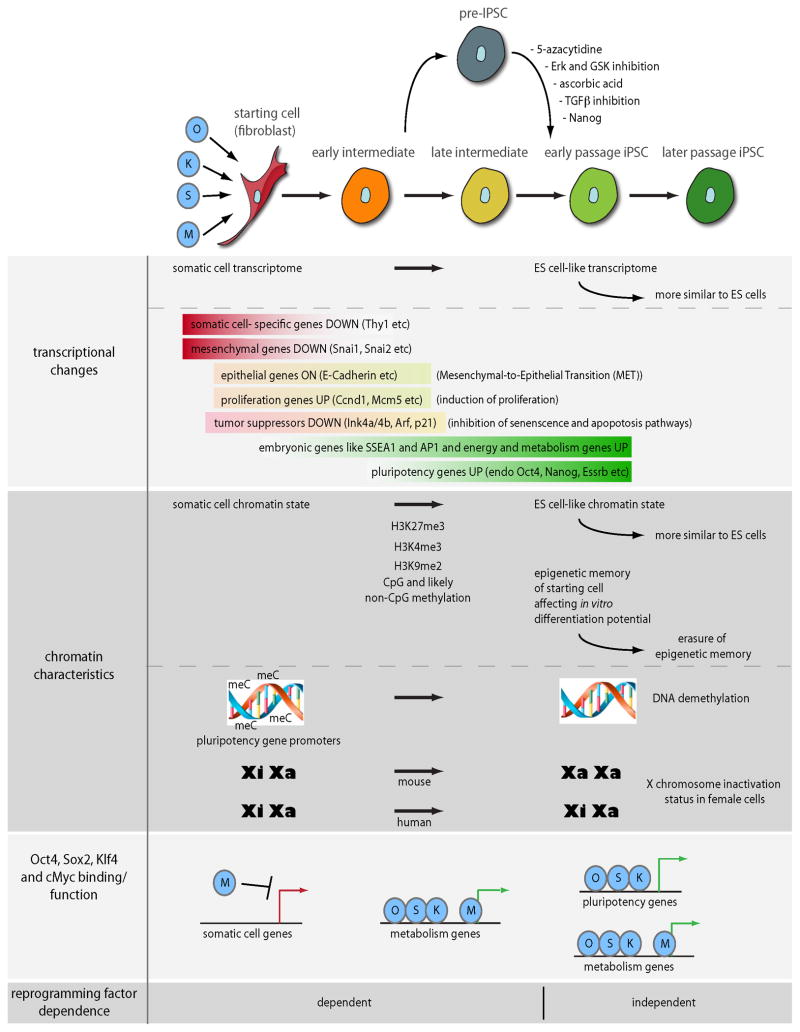
In a typical reprogramming experiment, ectopic expression of Oct4, Sox2, cMyc, and Klf4 in the starting cell type leads to downregulation of somatic genes and formation of ES cell-like colonies, culminating in the upregulation of an ES cell-like gene expression program and pluripotent capabilities after two to four weeks (Figure 1) (Brambrink et al., 2008; Chan et al., 2009; Li et al., 2010; Mikkelsen et al., 2008; Samavarchi-Tehrani et al., 2010; Sridharan et al., 2009; Stadtfeld et al., 2008b; Takahashi and Yamanaka, 2006). As indicated by the low reprogramming efficiency (Table 1), most cells that receive and express the reprogramming factors, and their daughter cells, do not progress to the faithfully reprogrammed state indicating major epigenetic barriers to reprogramming. It should be noted though, that the reported efficiencies depend hugely on the criteria with which iPS cell colonies are scored, whether all starting cells are considered or only those cells carrying all four reprogramming factors, and whether proliferation is taken into consideration.
Figure 1. Landmark events on the path to induced pluripotency.
It is thought that expression of the four reprogramming factors Oct4 (O), Klf4 (K), Sox2 (S), and cMyc (M) triggers a cascade of events within two to three weeks that leads to the iPS cell state. Stable and clonally expandable pre-iPS cells can also be generated as a by-product of this process and can enter the fully re-programmed state upon addition of indicated molecules.
To identify and/or quantify faithful reprogramming events, the best strategy generally is to observe the induced expression of endogenously encoded pluripotency markers such as Oct4 and Nanog (Maherali and Hochedlinger, 2008). In the mouse system, researchers often take advantage of pluripotency reporter cell lines generated via knockin approaches in ES cells. In human reprogramming experiments, staining for pluripotency surface markers have been applied successfully to identify faithful reprogramming events (Chan et al., 2009; Lowry et al., 2008). Alternatively, it has been proposed that iPS cell colony number can be assessed using morphological criteria or alkaline phosphatase staining when combined with inducible vectors coding for the reprogramming factors, as only cells that have entered the pluripotent state become independent of the ectopic factors while cells that have not progressed into pluripotency remain dependent on transgene expression and regress to their somatic state upon transgene silencing (Brambrink et al., 2008; Stadtfeld et al., 2008b; Wernig et al., 2008a).
Upon faithful reprogramming, mouse and human iPS cells are similar to their respective ES cell counterparts in terms of gene expression and genome-wide distribution of epigenetic marks (Chin et al., 2009; Hawkins et al., 2010; Maherali et al., 2007b; Mikkelsen et al., 2008; Okita et al., 2007; Takahashi et al., 2007; Wernig et al., 2007). In agreement with their molecular similarity to ES cells, reprogrammed cells also satisfy a range of functional pluripotency assays (Jaenisch and Young, 2008). These include differentiation into the three germlayers in vitro and in teratomas, and, for mouse iPS cells, contribution to chimera with germline transmission and the most stringent pluripotency assay of all, tetraploid (4N) complementation, which allows the derivation of adult mice completely derived from iPS cells (Boland et al., 2009; Jaenisch and Young, 2008; Kang et al., 2009; Zhao et al., 2009). However, not all mouse iPS cell lines support tetraploid complementation and the inability to do so has recently been associated with inappropriate silencing of the imprinted Dlk1–Dio3 gene cluster on mouse chromosome 12qF1 (Stadtfeld et al., 2010), indicating that reprogramming also results in aberrant epigenetic programming. Interestingly, germline competence of mouse iPS cells appears improved upon expression of additional transcription factors like Tbx3 during the reprogramming process (Han et al., 2010), although it remains unclear why this would be the case. Of course, the analysis of the developmental potential of human iPS cells, as with human ES cells, is limited to teratoma formation and in vitro differentiation.
Differentiation Potential influences Reprogramming Efficiency and Kinetics
In the early days of reprogramming to induced pluripotency, it was thought that the low reprogramming efficiency seen within a couple of weeks after delivery of the reprogramming factors was due to the lack of expression of all four factors in the same cell, as the retroviruses coding for each reprogramming factor were individually packaged. However, the current use of polycistronic lentiviral cassettes argues against the idea of heterogeneous transgene expression as the main cause of the low reprogramming efficiency as it only slightly increases the number of faithfully reprogrammed colonies over the system that uses individual retroviruses (Chang et al., 2009; Sommer et al., 2009) (Table 1).
Similar results were obtained with secondary reprogramming systems (Carey et al., 2009; Hockemeyer et al., 2008; Maherali et al., 2008; Stadtfeld et al., 2009; Wernig et al., 2008a; Woltjen et al., 2009)(Table 1). This method entails the generation of primary iPS cell clones with inducible reprogramming cassettes and subsequent differentiation in vitro, or chimera formation upon blastocyst injection in vivo, in the absence of transgene expression to obtain genetically modified, homogeneous somatic cell populations. These cells can then be induced to re-express the reprogramming factors across the entire population to generate secondary iPS cell clones. Even the establishment of the “reprogrammable” mouse with a single defined integration site for an inducible, polycistronic reprogramming factor cassette enabled only slightly more efficient reprogramming of fibroblasts compared to viral or transposon methods (Carey et al., 2009; Stadtfeld et al., 2009). The inducible secondary reprogramming system or reprogrammable mouse have several benefits as they allow reprogramming of cell types that are typically difficult to transduce and enable the comparison of reprogramming efficiencies of different somatic cell types from the same mouse.
Notably, experiments with cells from “reprogrammable” mice support the conclusion that insertional mutagenesis is not a key driver of reprogramming as predicted from non-integrative reprogramming studies and mapping of viral insertion sites (Aoi et al., 2008; Varas et al., 2009; Winkler et al., 2010). Consequently, one of the key questions in the reprogramming field has been whether only a particular subset of cells within the starting population, for example only those cells with a more undifferentiated state such as adult stem cells or progenitors, possess the ability to reach the iPS cell status.
To test whether fully differentiated cell types can be reprogrammed, the generation of iPS cell lines from mature mouse B cells was successfully achieved. Resulting iPS cell lines contained specific immunoglobulin re-arrangements reflecting their origin from mature B cells and gave rise to mouse progeny with a monoclonal immune system (Hanna et al., 2008). However, up-regulation of the myeloid transcription factor CCAAT/enhancer-binding proteins-α (CeBPα), which can reprogram B cells into macrophage like cells (Xie et al., 2004), or downregulation of the transcription factor Pax5, an essential regulator of mature B cell development (Cobaleda et al., 2007), were necessary for iPS cell induction from mature B cells. Subsequently, the establishment of iPS lines from mature human and mouse B and T lymphocytes without the need to modulate these blood-specific transcription factors was reported (Eminli et al., 2009; Loh et al., 2010; Seki et al., 2010; Staerk et al., 2010). The reasons behind the different requirements for mature B cell reprogramming in these studies remain unclear but may be related to the particular secondary reprogramming system used, the expression levels of the reprogramming factors, and/or culture conditions.
In one of these studies, the authors then investigated how the developmental state affects reprogramming efficiency by isolating blood cells at various differentiation stages and found that progenitor and hematopoietic stem cells give rise to iPS colonies with a much higher efficiency and in less time compared to differentiated cells of the same lineage (10–28% versus 0.03%–0.5%)(Eminli et al., 2009). As the enhancement in reprogramming was independent of cell division rate, these data argue against a model positing strictly that only few starting cell types, particularly more undifferentiated states, are susceptible to reprogramming, but rather suggest that the degree of differentiation influences reprogramming efficiency and kinetics. A similar correlation of differentiation state and reprogramming potential has been made in SCNT experiments (Hochedlinger and Jaenisch, 2006).
Almost all cells in a population can give rise to daughter cells that form iPS cells
From the results described above, it still remains unclear why only a select number of cells, even when expressing similar levels of the reprogramming factors, become iPS cells within two to three weeks. An interesting question was therefore to elucidate whether every cell in the starting population has the potential to eventually, i.e. after longer exposure to the reprogramming factors, give rise to iPS cells. By clonally expanding individual pre-B cells or monocytes from a secondary reprogramming system in 96-well plates and screening the progeny derived from each cell for its ability to reprogram, it was shown that almost every starting cell ultimately gives rise to daughter cells that can reach the iPS cell state (Hanna et al., 2009b). In agreement with the previously observed low reprogramming efficiency, the first reprogramming events could only be observed after eight to ten days in 3–5% of wells. However, prolonged exposure of doxycycline for up to 18 weeks and constant passaging led to around 92% of wells to become Nanog positive at variable times.
The finding that the timing of faithful reprogramming varies widely among cells indicates that at least one event driving the reprogramming process, if not more, is stochastic in nature (Hanna et al., 2009b)(Figure 2A). The authors further found that an experimentally induced, increased proliferation rate of pre-B cells, through p53 or p21 inhibition or Lin28 overexpression, accelerates the reprogramming process (Hanna et al., 2009b). However, the authors also showed that Nanog overexpression in the same pre-B cells increased their reprogramming rate without altering cell cycle kinetics, indicating that the reprogramming rate per cell cycle can be enhanced. It is important to note though that, even by the end of these clonal reprogramming experiments at around 18 weeks, not all daughter cells within each clonal population was faithfully reprogrammed, but typically just a few, irrespective of an identical genetic background. This findings highlights that there are major epigenetic barriers that interfere with the reprogramming of most cells in the culture and can be overcome by events that are stochastic in nature (Figure 2B).
Figure 2. Considering stochastic events in reprogramming.
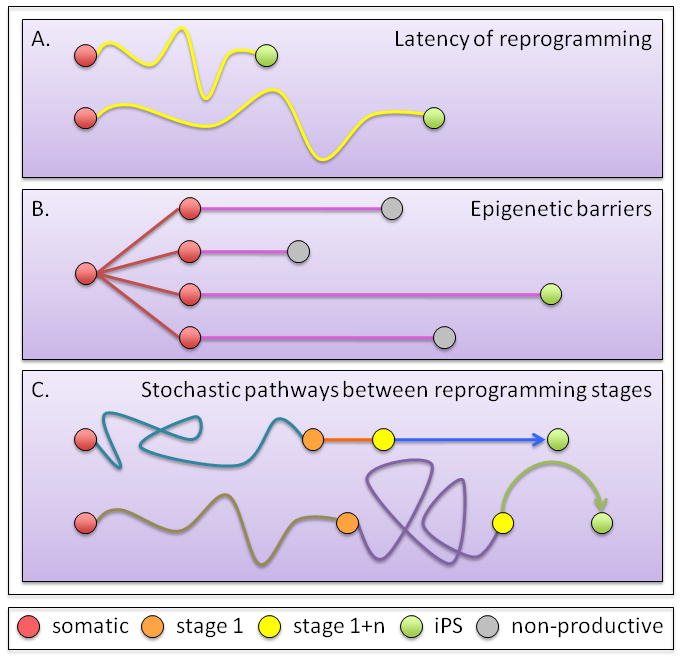
(A) Latency of faithful reprogramming. All cells have the potential to give rise to daughter cells that can faithfully reprogramming, but conversion to iPS cells occurs at divergent times - as early as two weeks (top) to as late as 18 weeks for pre-B cells (bottom).
(B) Epigenetic barriers. Within a clonal populations, only a few daughter cells reach the iPS cell status due to epigenetic barriers that need to be overcome in a stochastic manner.
(C)Pathways between reprogramming stages. It is thought that reprogramming occurs in defined steps (see Figure 1) but could use alternative pathways between these steps. The scenarios presented are not exhaustive, as other potential reprogramming profiles can exist.
Combining the fact that almost every pre-B cell in the culture has the potential to give rise to at least a few daughter cells that faithfully reprogramming, with the result that, at least for the blood lineage, differentiation state influences both reprogramming efficiency and kinetics, all somatic cells may be amendable to reprogramming, but more undifferentiated cells in the population have a higher probability to overcome reprogramming barriers. However, experiments with clonal reprogramming assays of cells at different differentiation stages, which determines proliferation rate, plating efficiency, and transgene levels, need to be performed to eventually test this hypothesis. Such experiments may be complicated by the fact that different cell types require different cytokines in vitro that could directly alter the reprogramming process and many other variables.
Molecular events during reprogramming
The detailed events occurring between the time of exogenous expression of the reprogramming factors and the establishment of the iPS cell state remain largely uncharacterized. This is primarily due to the low efficiency and slow kinetics of the process, and the fact that cells that will successfully complete the reprogramming process cannot be pre-selected. However populations that give rise to iPS cells with higher efficiencies can be enriched from intermediate stages of reprogramming (Stadtfeld et al., 2008b).
Several groups have chronologically traced events that occur during the first two to three weeks upon induction of the reprogramming factors in mouse embryonic fibroblasts (MEFs) (Figure 1). The first change in gene expression is the downregulation of somatic markers including key mesenchymal genes (Li et al., 2010; Mikkelsen et al., 2008; Samavarchi-Tehrani et al., 2010; Stadtfeld et al., 2008b). Concomitantly, epithelial genes like E-cadherin become upregulated as cells undergo a mesenchymal-to-epithelial transition (MET) and start proliferating (Li et al., 2010; Mikkelsen et al., 2008; Samavarchi-Tehrani et al., 2010). Undergoing MET is an essential early step of reprogramming as activation of Tgfβ signaling, inhibition of BMP signaling, or depletion of MET genes such as E-cadherin interfere with reprogramming to induced pluripotency (Li et al., 2010; Samavarchi-Tehrani et al., 2010). Using high resolution time-lapse imaging to backtrack faithful reprogramming events, an increase in proliferation rate and a concomitant decrease in cell size were detected in all successful reprogramming cases as early events, followed by stereotypic colony formation 4–8 days later leading to iPS cell clones (Smith et al., 2010).
It has also been reported that embryonic markers such as alkaline phosphatase (AP) and the stage specific embryonic antigen-1 (SSEA-1) cell surface marker are induced relatively early in the reprogramming process in a subset of cells (Brambrink et al., 2008; Stadtfeld et al., 2008b). Some cells from the SSEA-1 positive subpopulation then give rise to faithfully reprogrammed cells and activate the expression of endogenously encoded Oct4, Sox2, and Nanog, which are considered the most stringent markers of complete reprogramming (Maherali et al., 2007b; Stadtfeld et al., 2008b). Meanwhile, the SSEA-1 negative cells are depleted for iPS cells as judged at a parallel time point, suggesting a defined order of events in the reprogramming process. In the faithfully reprogramming cells, many other pluripotency-related genes, i.e. genes highly expressed in ES cells, often even specific for this cell type, and/or functionally important for the establishment and maintenance of the pluripotent state, are upregulated at around this point (Chan et al., 2009; Mikkelsen et al., 2008; Samavarchi-Tehrani et al., 2010).
At this point, cells can sustain the pluripotent state independently of ectopic reprogramming factor expression indicating a stable conversion of cell fate and the establishment of the pluripotency network (Brambrink et al., 2008; Stadtfeld et al., 2008b). Depletion of the exogenous factors at earlier times of reprogramming leads the cells to revert back to a differentiated cell phenotype (Brambrink et al., 2008; Stadtfeld et al., 2008b). When using retroviruses, silencing of the integrated retroviral transgenes occurs rather efficiently (Maherali et al., 2007b), possibly due to the actions of Trim28, Zfp806 and histone and DNA methyltransferases as the pluripotent state is established (Lei et al., 1996; Matsui et al., 2010; Rowe et al., 2010; Wolf and Goff, 2009). Retroviral silencing is not absolutely necessary to establish an autonomous, self-renewing ES cell state, since iPS cells can be generated using constitutively active lentiviral vectors to deliver the reprogramming factors (Brambrink et al., 2008; Sommer et al., 2010). However, differentiation of these cells can be severely impaired if the expression levels of the reprogramming factors remain high (Brambrink et al., 2008; Sommer et al., 2010). In female mouse cells, the somatically silenced X chromosome is reactivated during the reprogramming process (Maherali et al., 2007b), while in human iPS cells the inactive X chromosome is maintained from the somatic state (Tchieu et al., 2010), pointing at differences between the mouse and human reprogramming process (Figure 1).
Collectively, these data suggest that successful reprogramming leading to an iPS cell clone within two to three weeks upon initial induction of the reprogramming factors follows a defined sequence of events with a series of steps, each only taken successfully by few cells (Figure 2B and 2C). Why it typically takes at least 8–10 days to detect the first complete reprogramming event and whether the same path from one to the next reprogramming event is taken by each cell that ultimately will undergo faithful reprogramming remains unclear (Figure 2C). A finer resolution of intermediate cell states will address these question and reveal whether reprogramming simply reverses normal development and follows through a line of progenitor steps. Single cell studies will be key to address this problem (Chan et al., 2009; Smith et al., 2010). Studies of X chromosome reactivation during reprogramming may be particularly helpful as chromatin changes accompanying X chromosome silencing during development and the conversion of the euchromatic active to a heterochromatinized inactive state are well defined and because this process affects an entire chromosome and can therefore be easily visualized.
Even though the first reprogramming events occur in two to three week, the pre-B cell experiment described above demonstrated that most reprogramming events will occur much later, even as late as 18 weeks post induction of the reprogramming factors (Figure 2A). To explain this variable latency of induction of pluripotency, one can then propose a model in which one early, stochastically timed step determines the overall kinetics of each individual reprogramming event and all subsequent steps occur in a stereotypic fashion. Alternatively, each step may be stochastically timed, and transitions from one step to the next may not follow the same molecular path. Of course, these models need not to be mutually exclusive.
Barriers of Reprogramming
Exploring the pre-iPS cell state
A key task during the last couple years has been to identify molecular barriers of reprogramming that explain why only few cells, even in a clonal population, reprogram. A few groups, including our own, have been able to isolate a relatively stable, intermediate population of cells coined partially induced or reprogrammed pluripotent stem (pre-iPS) cells. These cells arise from fibroblasts two to three weeks after induction of the reprogramming factors as SSEA1-positive colonies with an ES cell like morphology and are capable of clonal expansion (Mikkelsen et al., 2008; Silva et al., 2008; Sridharan et al., 2009). Comparison of the transcription profiles of pre-iPS and ES/iPS cells revealed that many endogenous pluripotency genes such as Oct4 and Nanog are not reactivated, while somatic markers are already efficiently silenced (Mikkelsen et al., 2008; Sridharan et al., 2009). In agreement with the notion that reactivation of the somatically silenced X chromosome occurs with similar timing as that of Oct4 and Nanog, the X is still inactive in pre-iPS cells (Silva et al., 2008; Sridharan et al., 2009). Furthermore, pre-iPS cells are likely stabilized by ectopic expression of the four reprogramming factors and express a subset of genes that are neither active in MEFs or ES/iPS cells. Interestingly, pre-iPS cells obtained from neural precursor and B cell reprogramming experiments appear stalled at a similar stage as those derived from fibroblasts suggesting that the reprogramming pathways from different somatic cells channel into comparable pre-iPS states(Mikkelsen et al., 2008).
While it is not absolutely clear that pre-iPS cells represent an intermediate that occurs transiently during the reprogramming process, they are not simply an aborted reprogramming artifact because pre-iPS cells can convert into iPS cells upon addition of ERK and GSK inhibitors (termed 2i), which also leads to the stabilization of Nanog protein levels(Silva et al., 2008; Sridharan et al., 2009). Similarly, the DNA demethylating agent 5-azacytidine (Mikkelsen et al., 2008), ascorbic acid addition (Esteban et al., 2010), or Tgfβ-inhibition (Ichida et al., 2009), each facilitate the conversion of pre-iPS cells into fully reprogrammed clones. Given that all these treatments also improve the efficiency and kinetics of the reprogramming process when starting from somatic cells and the fact that pre-iPS cells transcriptionally mirror a late intermediate of the reprogramming process, these findings support the use of pre-iPS as a useful platform for the identification of pathways that will allow the enhancement of final steps of reprogramming (Figure 1). Interestingly, various pre-iPS cell clones react differently to a range of small molecule stimuli in their ability to convert to iPS cells (Ichida et al., 2009), suggesting that there are so far unappreciated molecular differences among them that will be interesting to uncover in the future.
Reprogramming factor binding in pre-iPS and iPS cells
An analysis of Oct4, Sox2, Klf4, and cMyc target genes by chromatin immunoprecipitation in combination with microarrays (ChIP-chip) in mouse iPS and pre-iPS cells provided significant insight into the action of the reprogramming factors (Sridharan et al., 2009). In iPS cells, just like in ES cells, cMyc and the trio of pluripotency transcription factors Oct4, Klf4, and Sox2 form largely separable transcriptional networks in ES cells, based on the finding that they target largely non-overlapping sets of genes in these cells (Chen et al., 2008; Kim et al., 2008a; Sridharan et al., 2009). cMyc binds many genes involved in cellular metabolism, cell cycle regulation, and biosynthetic pathways, while the majority of Oct4, Sox2, and Klf4 targets encode developmental, transcriptional regulators. Interestingly, many ES/iPS cell targets of Oct4, Sox2, and Klf4 are not bound by these transcription factors in pre-iPS cells (Sridharan et al., 2009). A correlation of binding with expression data suggested that the lack of Oct4, Sox2, and Klf4 binding to pluripotency-related genes in pre-iPS cells is responsible for the lack of reactivation of these genes at this state. On the contrary, cMyc targets of ES/iPS cell are often already bound and strongly expressed in pre-iPS cells (Sridharan et al., 2009) (Figure 1). Collectively, these findings suggest that transcriptional network downstream of cMyc becomes induced early in the reprogramming process and that the activation of key pluripotency-associated genes occurs via binding of Oct4, Sox2 and Klf4 only at the end of the reprogramming process.
In agreement with this conclusion, expression of only cMyc in MEFs induces a gene expression profile most similar to that of ES cells when compared to the profiles of MEFs induced to express any of the other reprogramming factors individually (Sridharan et al., 2009). Additionally, upregulation of cMyc and Klf4 in fibroblasts before induction of Oct4 and Sox2 positively enhances reprogramming while pre-expression of Oct4 and Sox2 has no such effect (Markoulaki et al., 2009). Interestingly, cMyc is not essential as reprogramming factor but enhances the efficiency and kinetics of the process (Nakagawa et al., 2008; Wernig et al., 2008b). Thus, the main functions of ectopically expressed cMyc can, directly or indirectly, be taken over by Oct4, Sox2, and Klf4, and may occur via induction of endogenously encoded cMyc genes. Even though ectopic cMyc function may have a powerful role early in the reprogramming process, it is likely that it is also essential for later steps as well as it needs to maintain expression of its downstream target network in the pluripotent state.
Nanog expression lowers barriers during final steps of reprogramming
The inability of Oct4, Sox2, and Klf4 to bind to their iPS/ES cell target gene promoters until late in the reprogramming process was proposed to present a major obstacle to faithful reprogramming to pluripotency (Sridharan et al., 2009)and it is interesting to speculate on the causes. One explanation may be that additional ES cell-specific transcription factors are necessary to synergize with Oct4, Sox2, and Klf4 to allow binding and activation of pluripotency gene promoters. The key pluripotency regulator Nanog would be a great example for such a factor as it binds many of the Oct4, Sox2, and Klf4 targets in ES cells (Boyer et al., 2005; Loh et al., 2006), biochemically interacts with Oct4 and other transcription factors in ES cells (Wang et al., 2006), and is only upregulated during the pre-iPS to iPS cell transition at the end of the reprogramming process (Mikkelsen et al., 2008; Silva et al., 2009; Sridharan et al., 2009). Nanog’s absence in pre-iPS cells could therefore affect the binding pattern of Oct4, Sox2, and Klf4 in these cells. Inagreement with this idea, Nanog positively affects reprogramming by cell fusion and facilitates human and mouse reprogramming (Silva et al., 2006; Yu et al., 2007; Hanna et al., 2009b). Similarly, Tgfβ-inhibition, which enhances the pre-iPS to iPS cell transition, may also occur through upregulation of Nanog expression (Ichida et al., 2009). Importantly, Smith and colleagues demonstrated that Nanog is absolutely essential for the full induction of pluripotency and required only during the final stages of the reprogramming process (Silva et al., 2009). Similar to Nanog, other pluripotency transcription factors not yet strongly expressed in pre-iPS cells such as Sall4 also boost reprogramming efficiency (Tsubooka et al., 2009) and biochemically interact with the Oct4 transcriptional network (Liang et al., 2008; Wang et al., 2006; Yang et al., 2008). Future studies aimed at understanding why the four reprogramming factors target different genes in pre-iPS cells in comparison to ES/iPS cells should yield insights into the transcriptional regulation of the pluripotency program, the role of chromatin (see below), and perhaps lead to new methods of increasing the conversion efficiency into iPS cells.
Chromatin state as a reprogramming barrier
Reprogramming to pluripotency is associated with a major resetting of the chromatin landscape, which is deemed necessary in order to activate the ES cell-specific transcriptional program while silencing tissue-specific genes. Accordingly, genome-wide analyses of several histone methylation marks (H3K9me2, H3K4me3, H3K27me3) and DNA methylation in fibroblasts, pre-iPS and iPS cells indicated that substantial changes in these modifications occur during reprogramming largely being reset to an ES cell-like pattern (Chin et al., 2009; Doi et al., 2009; Hawkins et al., 2010; Maherali et al., 2007b; Mikkelsen et al., 2008; Sridharan et al., 2009). Likewise, asymmetric cytosine methylation that is only prevalent in ES cells is reestablished upon faithful reprogramming based on candidate gene analysis (Lister et al., 2009).
A wide range of publications suggests that re-establishing ES cell-like chromatin marks is critical for the reprogramming process. The analysis of promoter DNA methylation profiles of the Nanog and Oct4 loci revealed that these genes are hypermethylated in MEFs and pre-iPS cells in direct contrast to the unmethylated status observed in iPS and ES cells (Maherali et al., 2007a; Mikkelsen et al., 2008). Notably, this demethylation appears to occur very late in the reprogramming process and serves as a good indicator of faithful reprogramming events. In agreement with the notion that DNA methylation may interfere with efficient reprogramming, the addition of the DNA de-methylating drug 5-azacytidine or depletion of maintenance methyl transferase Dnmt1 increases the efficiency with which iPS cells are generated from pre-iPS cell lines (Mikkelsen et al., 2008). When added at different times during the reprogramming process, only 5-azacytidine treatment during the final phase enhanced the process suggesting that it largely is advantageous at the late intermediate stage (Mikkelsen et al., 2008). However, one should be careful when interpreting such results given that inhibition of DNA methylation induces apoptosis in differentiated cells (Jackson-Grusby et al., 2001), but not in the pluripotent state (Li et al., 1992), and hence perhaps acts by changing population dynamics at earlier stages of reprogramming.
Interestingly, the DNA demethylation machinery centered on the AID protein has recently been implicated as an essential player in ES cell–somatic cell fusion experiments (Bhutani et al., 2010), but direct targets and the question of how the enzyme is recruited still remain unclear, and its role in iPS cell production has not yet been tested. Nevertheless, this finding suggests that active DNA demethylation is important to allow reprogramming. Alternatively, passive mechanisms may be in place to lower the DNA methylation content in pluripotency promoters, potentially acting in S-phase when the DNA methylation mark gets copied during DNA replication.
Similar to DNA methylation, it has been suggested that histone modifications impact the reprogramming process. For example, a subset of pluripotency genes lacks the typically activating histone H3 lysine 4 methylation in MEFs and pre-iPS cells but not in ES/iPS cells (Sridharan et al., 2009). Since histone H3 K4 methylation and DNA methylation are typically mutually exclusive (Meissner et al., 2008; Zhang et al., 2009), the lack of this modification may correlate strongly with the presence of DNA methylation at pluripotency promoters in pre-iPS cells and MEFs. This repressive chromatin environment could preclude Oct4, Sox2, and Klf4 binding at pluripotency loci in pre-iPS cells.
Further evidence for the role of chromatin in reprogramming comes from the result that histone deacetylase inhibitors including TSA, valproic acid (VPA), suberoylanilide hydroxamic acid(SAHA), and butyrate (Huangfu et al., 2008a; Liang et al., 2010), as well as the small molecule BIX (Shi et al., 2008), supposed to inhibit the histone H3 K9 methyltransferase G9A, enhance the production of iPS cells. It should be noted though, that all these small molecules that target either histone or DNA modifiers likely also act on the global chromatin status. Therefore, they could not only directly affect the chromatin state at key promoters or enhancers, but be critical for the reprogramming process in a more indirect manner. However, addition of VPA enables replacement of cMyc in mouse and cMyc and Klf4 in human reprogramming experiments (Huangfu et al., 2008a; Huangfu et al., 2008b), suggesting a functional overlap between these reprogramming factors and histone acetylation. The fact that cMyc interacts with histone acetyl transferases further supports this idea (Frank et al., 2003; McMahon et al., 2000).
Of course, chromatin-based regulation is not limited to the direct modification of DNA and histones. Chd1, a chromatin remodeling enzyme belonging to the chromodomain family with a SNF2-like helicase domain, is essential for the production of iPS cells (Gaspar-Maia et al., 2009). Similarly, overexpression of chromatin remodelers with an ES cell-specific expression component, like the SWI/SNF-type BAF complex, enhances reprogramming efficiency and kinetics (Singhal et al., 2010), but again insights into the mechanisms involved are lacking at this point. Future studies of the localization of chromatin marks, of the enzymes that establish or remove these marks or remodel chromatin, and of reprogramming factor binding during the reprogramming are necessary to reveal the detailed path by which chromatin governs reprogramming. One interesting question will be whether the chromatin state at pluripotency genes changes first to allow reprogramming factor binding or whether the reprogramming factors bind first to then change the chromatin state by recruiting the respective chromatin modifying machinery.
Nuclear architecture as a roadblock to reprogramming
In the mouse system, distinct pluripotent stem cell populations have been generated from embryos at different developmental stages (Nichols and Smith, 2009). Epiblast stem cells (EpiSCs), obtained from the d5.5 egg cylinder express many pluripotency-associated genes alsopresent in blastocyst-derived ES cells (as for example Oct4 and Sox2) and, like ES cells, can differentiate into the three germ layers in vitro and in teratomas (Brons et al., 2007; Tesar et al., 2007). Notable differences between these two pluripotent stages, however, include differential signaling pathway dependence with EpiSCs relying on Activin A and bFGF and ES cells depending on Lif, the inability of EpiSCs to contribute to chimeras upon blastocyst injection, and silencing of one X chromosome in female EpiSCs but not in ES cells. EpiSCs have therefore represent a developmentally more advanced, “primed” pluripotent state, while ES cells display “naïve” pluripotency (Nichols and Smith, 2009). Upon switching from bFGF/activin to Lif- containing media, ectopic expression of Nanog, Klf4, or cMyc, addition of small molecules that can replace these reprogramming factors, or enhancement of Lif signaling, mouse EpiSCs reprogram to the pluripotent capabilities of ES cells (Bao et al., 2009; Guo et al., 2009; Hanna et al., 2009a; Silva et al., 2009; Yang et al., 2010). Notably, for the field of induced pluripotency, the conversion of EpiSCs to the ES-like state is also typically characterized by alow efficiency, with about 1% reprogramming efficiency. While the optimal set of reprogramming factors or culture conditions for efficient EpiSC to ES cell conversion may not yet be identified, this finding points to major barriers that even limit reprogramming from primed to naïve pluripotency and suggests that some aspects of the cellular identity of EpiSCs is more similar to somatic, differentiated cells than iPS/ES cells.
In agreement with this conclusion, the mapping of replication timing has revealed that EpiSCs are more similar to committed cells types than to ES/iPS cells (Hiratani et al., 2009). Furthermore, within the nucleus of EpiSCs, there is an accumulation of compacted chromatin near the periphery that is typically found in somatic cells but not in ES cells, and the Oct4 gene is localized more peripherally in EpiSCs than in ES cells (Hiratani et al., 2009). Together, these data suggest that a global event that can reorder replication timing profiles and subnuclear architecture is required to permit induction of naïve pluripotency. Interestingly, like EpiSCs, pre-iPS cells are more somatic cell- like in their genome organization and replication timing signature (Hiratani et al., 2009). Potentially, nuclear reorganization is difficult to achieve and represents a major barrier to the reactivation of pluripotency genes towards the end of the reprogramming process. Thus, EpiSCs, just like pre-iPS cells, are a useful tool to study the induction of naïve pluripotency, particularly since fewer factors are required for this conversion to iPS cells. As a side note, naïve human ES cells have so far not been derived from human blastocysts and the typical human ES cell is similar to primed mouse EpiSCs state. Recently, Jaenisch and colleagues demonstrated that human ES/iPS cells can also acquire a naïve pluripotent phenotype upon ectopic expression of KLF4 with OCT4 or KLF2 and culture in 2i media with LIF, but again, the efficiency of this conversion is low(Hanna et al., 2010).
Apoptosis and senescence as barriers of fibroblast reprogramming
In contrast to many somatic cells that often have a limited proliferative potential and undergo stress- induced senescence, ES cells self-renew indefinitely in culture and are therefore considered immortal. Intriguingly, senescence appears to be incompatible with reprogramming to pluripotency and could represent another barrier to the process. In support of this idea, serial passaging of fibroblasts is associated with a dramatic decrease in reprogramming efficiency (Utikal et al., 2009b). Several reports have now demonstrated that the downregulation of tumor suppressor components such as p53, p21 (Cdkn1a), p16 (Ink4a) and p19 (Arf) in fibroblasts enhances the efficiency and kinetics by which iPS cells are generated, either via inducing immortalization or interfering with apoptosis(Banito et al., 2009; Hong et al., 2009; Kawamura et al., 2009; Li et al., 2009a; Marion et al., 2009; Utikal et al., 2009b). In agreement with these findings, iPS cells, like ES cells, normally express low levels of p53 and Arf. The fact that another study that used pre-B cells instead of fibroblasts argued that p53 depletion acts by enhancing cell cycle kinetics (Hanna et al., 2009b) may indicate that p53 acts differently in these cell types, or could be the result of the particular reprogramming scheme employed or differing reprogramming factor levels.
The use of knockout serum replacement (KSR) enhances the generation of mouse iPS cells in comparison to FBS-containing culturing media(Blelloch et al., 2007). Vitamin C (ascorbic acid), known for its antioxidant function, is contained in KSR and may be the key mediator of this effect since treatment of reprogramming cultures with vitamin C enhances both efficiency and kinetics of the process (Esteban et al., 2010). Surprisingly, this effect is largely independent of a modulation of reactive oxygen species, but may act through lowering p53 levels (Esteban et al., 2010). Given that reprogramming in the absence of p53, the guardian of genome integrity, enriches for damaged cells that are not desirable for clinical use (Marion et al., 2009), vitamin C treatment may be a safer choice to boost the reprogramming process, particularly when applying human cells for disease studies and cell replacement strategies. Similarly, hypoxic conditions can be applied during reprogramming to lower p53 levels in a subtle manner to enhance the reprogramming process (Utikal et al., 2009b; Yoshida et al., 2009).
Together, the studies of pre-iPS cells and the reprogramming process have yielded considerable insight into the mechanism and barriers of the reprogramming process, albeit the detailed steps often remain unclear.
Epigenetic memory and continuing changes of the iPS cell state
Recent evidence suggests that there are differences between ES and iPS cells that are indicative of an epigenetic memory of the starting cell in iPS cells and affect the differentiation potential of iPS cells (Kim et al., 2010; Polo et al., 2010). Specifically, when first derived, iPS cells still express genes and display a chromatin state that resemble aspects of the starting cell and appear to differentiate with higher efficiency into the originating lineage in comparison to other lineages.
These molecular and functional disparities between ES and iPS cells seem to disappear upon continued expansion of iPS cells (Chin et al., 2009; Chin et al., 2010; Polo et al., 2010), but whether this process selects for a more properly reprogrammed cell or whether reprogramming continues in culture is unclear. Similarly, the use of drugs that affect epigenetic function like TSA and 5-azacytidine erase the epigenetic memory (Kim et al., 2010). In comparison to SCNT-derived ES cells, iPS cells may carry a stronger epigenetic memory of their cell type of origin than SCNT-derived ES cells (Kim et al., 2010; Polo et al., 2010) suggesting that different mechanisms are responsible for those two reprogramming methods.
Conclusions
At present, the reprogramming field is paced by the synergistic relationship between studies of mechanisms and the development of improved reprogramming methods that are more conducive for clinical applications and disease modeling. While the exploration of intermediate states proves to be an invaluable tool for improving our understanding of the process, much work is needed in order to elucidate the exact mechanism of reprogramming and get a handle on whether each cell follows the same path to pluripotency, what controls the latency of reprogramming, and how different cellular origins affect the process. Recently, small molecule screens revealed modulators of the reprogramming process (Ichida et al., 2009; Lyssiotis et al., 2009) suggesting that reprogramming without protein factors may be feasible in the near future. Another question that remains to be answered is whether reprogramming selects for cells that genetically inactivate the p53/Arf pathway or similar pathways, that when inactivated permanently are potentially detrimental to the iPS cell state or their differentiated progeny. In any case, reprogramming to pluripotency via the Yamanaka approach has shown that a small set of transcription factors can dramatically modulate cell fate. We are beginning to see how new transcription factor combinations are being defined that can convert one somatic cell type to another without going through the pluripotent state (Ieda et al., 2010; Vierbuchen et al., 2010; Zhou et al., 2008) and it will be interesting to understand the similarities and differences of these processes.
Acknowledgments
We are grateful to Bill Lowry and Bernadett Papp for critical reading of the manuscript. RH is supported by a Training Grant of the National Institutes of Health 5T32AI060567-07, CC by a California Institute for Regenerative Medicine (CIRM) Training Award (TG2-01169), KP by the NIH Director’s Young Innovator Award (DP2OD001686) and a CIRM Young Investigator Award (RN1-00564).
References
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilic J, Pekarik V, Tiscornia G, Edel M, Boue S, Izpisua Belmonte JC. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26(11):1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science (New York, NY) 2008 doi: 10.1126/science.1154884. (published online Feb 14) [DOI] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, Vallier L, Gil J. Senescence impairs successful reprogramming to pluripotent stem cells. Genes & development. 2009;23(18):2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461(7268):1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463(7284):1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R, Venere M, Yen J, Ramalho-Santos M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell. 2007;1(3):245–247. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461(7260):91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CG, Wernig M, Suh H, Jaenisch R. Sequential Expression of Pluripotency Markers during Direct Reprogramming of Mouse Somatic Cells. Cell Stem Cell. 2008;2(2):151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Beard C, Hanna J, Jaenisch R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nature methods. 2009;7(1):56–59. doi: 10.1038/nmeth.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, Daley GQ, Schlaeger TM. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27(11):1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Chang CW, Lai YS, Pawlik KM, Liu K, Sun CW, Li C, Schoeb TR, Townes TM. Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem cells (Dayton, Ohio) 2009;27(5):1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, Khvorostov I, Ott V, Grunstein M, Lavon N, Benvenisty N, Croce CM, Clark AT, Baxter T, Pyle AD, Teitell MA, Pelegrini M, Plath K, Lowry WE. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5(1):111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Pellegrini M, Plath K, Lowry WE. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010;7(2):263–269. doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Lee CS, Kwon YW, Paek JS, Lee SH, Hur J, Lee EJ, Roh TY, Chu IS, Leem SH, Kim Y, Kang HJ, Park YB, Kim HS. Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood. 2010;116(3):386–395. doi: 10.1182/blood-2010-02-269589. [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449(7161):473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, Miller J, Schlaeger T, Daley GQ, Feinberg AP. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature genetics. 2009;41(12):1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eminli S, Foudi A, Stadtfeld M, Maherali N, Ahfeldt T, Mostoslavsky G, Hock H, Hochedlinger K. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nature genetics. 2009;41(9):968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem cells (Dayton, Ohio) 2008;26(10):2467–2474. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, Chen K, Li Y, Liu X, Xu J, Zhang S, Li F, He W, Labuda K, Song Y, Peterbauer A, Wolbank S, Redl H, Zhong M, Cai D, Zeng L, Pei D. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6(1):71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, Vega VB, Cacheux-Rataboul V, Lim B, Lufkin T, Ng HH. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nature cell biology. 2009a;11(2):197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009b;4(4):301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4(6):575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(8):348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, Ramalho-Santos M. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460(7257):863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development (Cambridge, England) 2009;136(7):1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL, Lufkin T, Ng HH, Tam WL, Lim B. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463(7284):1096–1100. doi: 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(20):9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Mitalipova M, Cheng AW, Cassady JP, Staerk J, Carey BW, Lengner CJ, Foreman R, Love J, Gao Q, Kim J, Jaenisch R. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009a;4(6):513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, Lengner CJ, Dausman JA, Jaenisch R. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133(2):250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009b;462(7273):595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6(5):479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, Lim B, Ng HH. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6(2):167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, Rathjen PD, Gilbert DM. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2009;20(2):155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441(7097):1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121(3):465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3(3):346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460(7259):1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008a;26(7):795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008b;26(11):1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5(5):491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature genetics. 2001;27(1):31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132(4):567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, Longaker MT, Wu JC. A nonviral minicircle vector for deriving human iPS cells. Nature methods. 2010;7(3):197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27(5):459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458(7239):771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5(2):135–138. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460(7259):1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009a;4(6):472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008a;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hubner K, Bernemann C, Ortmeier C, Zenke M, Fleischmann BK, Zaehres H, Scholer HR. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009b;136(3):411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, Scholer HR. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008b;454(7204):646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010 doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development (Cambridge, England) 1996;122(10):3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009a;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7(1):51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009b;4(1):16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Liang G, Taranova O, Xia K, Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem. 2010;285(33):25516–25521. doi: 10.1074/jbc.M110.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nature cell biology. 2008;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K, Qu X, Xiang T, Lu D, Chi X, Gao G, Ji W, Ding M, Deng H. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3(6):587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Loh YH, Hartung O, Li H, Guo C, Sahalie JM, Manos PD, Urbach A, Heffner GC, Grskovic M, Vigneault F, Lensch MW, Park IH, Agarwal S, Church GM, Collins JJ, Irion S, Daley GQ. Reprogramming of T cells from human peripheral blood. Cell Stem Cell. 2010;7(1):15–19. doi: 10.1016/j.stem.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature genetics. 2006;38(4):431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(8):2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, Bollong M, Kunick C, Brinker A, Cho CY, Schultz PG, Jaenisch R. Reprogramming of murine fibroblaststo induced pluripotent stem cells with chemical complementation of Klf4. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(22):8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3(3):340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;3(6):595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol. 2009;19(20):1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007a;1(1):55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold S, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly Reprogrammed Fibroblasts Show Global Epigenetic Remodeling and Widespread Tissue Contribution. Cell Stem Cell. 2007b;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460(7259):1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoulaki S, Hanna J, Beard C, Carey BW, Cheng AW, Lengner CJ, Dausman JA, Fu D, Gao Q, Wu S, Cassady JP, Jaenisch R. Transgenic mice with defined combinations of drug-inducible reprogramming factors. Nat Biotechnol. 2009;27(2):169–171. doi: 10.1038/nbt.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3(2):132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464(7290):927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Molecular and cellular biology. 2000;20(2):556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454(7200):49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Takizawa N, Narita M, Ichisaka T, Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(32):14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4(6):487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science (New York, NY) 2008;322(5903):949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451(7175):141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28(8):848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463(7278):237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7(1):64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Seki T, Yuasa S, Oda M, Egashira T, Yae K, Kusumoto D, Nakata H, Tohyama S, Hashimoto H, Kodaira M, Okada Y, Seimiya H, Fusaki N, Hasegawa M, Fukuda K. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7(1):11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2(6):525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS biology. 2008;6(10):e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441(7096):997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138(4):722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Scholer HR. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141(6):943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28(5):521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, Isacson O, Jaenisch R. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136(5):964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Sommer AG, Longmire TA, Christodoulou C, Thomas DD, Gostissa M, Alt FW, Murphy GJ, Kotton DN, Mostoslavsky G. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem cells (Dayton, Ohio) 2010;28(1):64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem cells (Dayton, Ohio) 2009;27(3):543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136(2):364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465(7295):175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of pancreatic beta cells into induced pluripotent stem cells. Curr Biol. 2008a;18(12):890–894. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nature methods. 2009;7(1):53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining Molecular Cornerstones during Fibroblast to iPS Cell Reprogramming in Mouse. Cell Stem Cell. 2008b;2(3):230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science (New York, NY) 2008c;322(5903):945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staerk J, Dawlaty MM, Gao Q, Maetzel D, Hanna J, Sommer CA, Mostoslavsky G, Jaenisch R. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010;7(1):20–24. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, Zack JA, Clark AT, Pyle AD, Lowry WE, Plath K. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7(3):329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448(7150):196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Tsubooka N, Ichisaka T, Okita K, Takahashi K, Nakagawa M, Yamanaka S. Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells. 2009;14(6):683–694. doi: 10.1111/j.1365-2443.2009.01301.x. [DOI] [PubMed] [Google Scholar]
- Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009a;122(Pt 19):3502–3510. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009b;460(7259):1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg DL, Zhang W, Yates A, Engelen E, Takacs K, Bezstarosti K, Demmers J, Chambers I, Poot RA. Estrogen-related receptor beta interacts with Oct4 to positively regulate Nanog gene expression. Molecular and cellular biology. 2008;28(19):5986–5995. doi: 10.1128/MCB.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varas F, Stadtfeld M, de Andres-Aguayo L, Maherali N, di Tullio A, Pantano L, Notredame C, Hochedlinger K, Graf T. Fibroblast-derived induced pluripotent stem cells show no common retroviral vector insertions. Stem cells (Dayton, Ohio) 2009;27(2):300–306. doi: 10.1634/stemcells.2008-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444(7117):364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, Staerk J, Markoulaki S, Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008a;26(8):916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008b;2(1):10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448(7151):318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Winkler T, Cantilena A, Metais JY, Xu X, Nguyen AD, Borate B, Antosiewicz-Bourget JE, Wolfsberg TG, Thomson JA, Dunbar CE. No evidence for clonal selection due to lentiviral integration sites in human induced pluripotent stem cells. Stem cells (Dayton, Ohio) 2010;28(4):687–694. doi: 10.1002/stem.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458(7242):1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458(7239):766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Chen J, Ren J, Bao L, Liao J, Cui C, Rao L, Li H, Gu Y, Dai H, Zhu H, Teng X, Cheng L, Xiao L. Generation of pig induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol. 2009;1(1):46–54. doi: 10.1093/jmcb/mjp003. [DOI] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yang J, Chai L, Fowles TC, Alipio Z, Xu D, Fink LM, Ward DC, Ma Y. Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(50):19756–19761. doi: 10.1073/pnas.0809321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JC, Smith A. Stat3 Activation Is Limiting for Reprogramming to Ground State Pluripotency. Cell Stem Cell. 2010;7(3):319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5(3):237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science (New York, NY) 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science (New York, NY) 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nature methods. 2009;6(5):363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, Ragozin S, Reinhardt R, Groth M, Walter J, Jeltsch A. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet. 2009;5(3):e1000438. doi: 10.1371/journal.pgen.1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XY, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Guo CL, Ma QW, Wang L, Zeng F, Zhou Q. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461(7260):86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Scholer HR, Duan L, Ding S. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4(5):381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455(7213):627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem cells (Dayton, Ohio) 2009;27(11):2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]