Abstract
When brassinosteroid (BR) levels are low, the GSK3-like kinase BIN2 phosphorylates and inactivates the BZR1 transcription factor to inhibit growth in plants. BR promotes growth by inducing dephosphorylation of BZR1, but the phosphatase that dephosphorylates BZR1 has remained unknown. Here we identified protein phosphatase 2A (PP2A) as BZR1-interacting proteins using tandem affinity purification. Genetic analyses demonstrated a positive role of PP2A in BR signalling and BZR1 dephosphorylation. Members of the B'regulatory subunits of PP2A directly interact with BZR1's putative PEST domain containing the site of the bzr1-1D mutation. Interaction with and dephosphorylation by PP2A are enhanced by the bzr1-1D mutation, reduced by two intragenic bzr1-1D suppressor mutations, and abolished by deletion of the PEST domain. This study reveals a crucial function of PP2A in dephosphorylating and activating BZR1 and completes the set of core components of the BR-signalling cascade from cell surface receptor kinase to gene regulation in the nucleus.
INTRODUCTION
Brassinosteroids (BRs) are growth-enhancing hormones that regulate large numbers of genes and a wide range of developmental processes in plants1-3. BR deficient or insensitive mutants show multiple defects throughout the life cycle, including reduced seed germination, photomorphogenesis in the dark, dwarfism, and male sterility3. BRs are perceived by the cell surface receptor-like kinase BRI1, and the downstream signal transduction pathway leads to activation of the BZR family transcription factors, which control BR-responsive gene expression4-7. In the absence of BR, BZR1 and its homolog BZR2 (also named BES1) are phosphorylated and inactivated by the GSK3/SHAGGY-like protein kinase BIN28-10, similar to inactivation of β-catenin by GSK3 in the Wnt signalling pathway in animals11. Phosphorylation inactivates the BZR factors through abolishing DNA-binding activity12, 13, and increasing cytoplasmic retention by the phosphopeptide-binding 14-3-3 proteins12, 14. BR binding to the extracellular domain of BRI115 induces disassociation of the inhibitory BRI1-binding protein BKI1 from its intracellular kinase domain16, allowing association with the co-receptor kinase BAK117. Autophosphorylation and sequential transphosphorylation between BRI1 and BAK1 fully activate the BRI1 kinase17-19, initiating a cascade of signal transduction events, including BRI1 phosphorylation of the BR-signalling kinase 1 (BSK1) at its serine 230 residue20, BSK1 binding to the PP1-related protein phosphatase BSU1, and BSU1 dephosphorylation of BIN2 at phospho-tyrosine 200, which inactivates BIN2 kinase4. Upon BIN2 inactivation, BZR1 and BZR2 are dephosphorylated by an unknown phosphatase9, and they translocate into the nucleus to regulate a large number of BR-target genes, which represent a wide range of functions1, 7(see Supplementary Information, Fig. S1a online).
The BR signalling pathway is distinct from animal steroid signalling pathways, which use nuclear receptor transcription factors21, but shares similarities with the metazoan Wnt signalling pathway, in which the GSK3 kinase inactivates β-catenin and a PP1 phosphatase inactivates GSK311. In the Wnt pathway, PP2A dephosphorylates β-catenin, counteracting the effect of GSK311, 22, 23. The phosphatase that dephosphorylates BZR1/2 is the last major component of the BR signalling pathway yet to be identified1, 9 (see Supplementary Information, Fig. S1a online). In this study, we demonstrate that PP2A dephosphorylates BZR1 in the BR signalling pathway.
RESULTS
BZR1 interacts with PP2A
To identify the BZR1-dephosphorylating phosphatase, we performed tandem affinity purification of BZR1-interacting proteins followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS; see Methods). Protein extracts of transgenic Arabidopsis seedlings expressing BZR1-7Myc-6His or bzr1-1D-7Myc-6His were purified using Ni affinity column followed by immunoprecipitation using anti-cMyc antibody. The purified proteins were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Proteins specifically co-purified with BZR1 or bzr1-1D include several known BZR1-interacting proteins such as 14-3-3λ12, BIN2, AtSK12 and AtSK314, indicating that our purification was successful (Supplementary Information, Fig. S2a). In addition, several peptides that match different subunits of PP2A, including PP2A A subunits (RCN1/PP2AA1, PP2AA2 and PP2AA3) and PP2A B subunits (PP2AB’η, PP2AB’β, and PP2AB’θ), were also identified in the purification of BZR1 or bzr1-1D complexes (Supplementary Information, Fig. S2a-c).
As a heterotrimeric Ser/Thr phosphatase, PP2A holoenzyme is composed of a catalytic C subunit, a scaffolding A subunit, and a regulatory B subunit that determines substrate specificity and PP2A subcellular localization24. Recent structural studies of holoenzyme complexes suggest that the B subunits provide an exposed concave surface for substrate recruitment adjacent to the active site of the C subunit25. As in other multicellular eukaryotes, Arabidopsis PP2A B subunits can be further grouped into B, B’ and B” subfamilies26.
Because mass spectrometry analysis of the BZR1 complex identified partial sequences that match several isoforms of PP2A A and B subunits, we used yeast two hybrid assays to determine which PP2A subunits directly interact with BZR1. We found that specific members of the B’ subunit subfamily interacted directly with BZR1 in yeast two-hybrid assays (Fig. 1a), whereas no interaction was detected between BZR1 and PP2A A subunits or C subunits (Supplementary Information, Fig. S3a). BZR2, the close homolog of BZR1, also interacts with PP2A B’α and β Supplementary Information, Fig. S3b). Based on the interaction strength with BZR1, PP2AB’ family proteins can be further classified into three groups: strong BZR1 interactors (B’α, β, γ, and κ), weak interactors (B’η, θ and ζ) and non-interactors (B’δ and ε) (Fig. 1a). In vitro overlay assays showed that recombinant PP2AB’α binds to phosphorylated BZR1 more strongly than to unphosphorylated BZR1, whereas BIN2 binds to unphosphorylated BZR1 more strongly, consistent with stronger binding to substrates than products (Fig. 1b). These interactions were further confirmed in vivo by Bi-Molecular Fluorescence Complementation (BiFC) assays. Tobacco epidermal cells co-transformed with PP2AB’α, β, or η fused to the N-terminal half of YFP (B’α, β, η-nYFP) and BZR1 fused to the C-terminal half of CFP (BZR1-cCFP) showed strong fluorescence signals, whereas tobacco cells co-transformed with PP2AB’ε-nYFP and BZR1-cCFP showed no fluorescence signal (Fig. 1c). These results indicate that BZR1 directly interacts with several isoforms of PP2A B’. The PP2A scaffolding A subunits were most likely purified together with BZR1 as constituents of PP2A complexes recruited by B’ subunit binding. To further confirm that B’ subunit binding brings BZR1 to the PP2A holoenzyme in vivo, we used co-immunoprecipitation assays to detect BZR1 association with PP2A catalytic subunits (Fig. 1d). These results together indicate that direct binding to the B’ subunit in vivo promotes BZR1 association with PP2A holoenzymes containing A, B’ and C subunits.
Figure 1. Identification of PP2A as a BZR1-interacting phosphatase.
(a) Yeast two-hybrid assays showing the interaction between BZR1 and PP2A B’ family members. (b) Gel blots of phosphorylated (pBZR1) and unphosphorylated (BZR1) MBP-BZR1 were probed with GST-PP2AB’α (B’α) or GST-BIN2 (BIN2) followed by HRP labelled anti-GST antibody, or stained with Ponceau S (Stain) to show equal loading. (c) BiFC assays showing that BZR1 interacts with PP2A B’α, B’β, B’η but not B’ε in Nicotiana Benthamiana leaf epidermal cells. Left column shows bright field and right column BiFC fluorescence. Scale bars represent 50 μm. (d) Co-immunoprecipitation of BZR1 with PP2A. Arabidopsis plants expressing BZR1::BZR1-CFP were treated with 100 nM brassinolide (BL) or mock solution for 15 min. BZR1-CFP was immunoprecipitated using anti-YFP antibody, and the immunoblots were probed with antibodies against YFP or the PP2A C subunits. Full scans of immunoblots are shown in Supplementary Information, Fig. S9.
Overexpression of PP2A increases BZR1 activity and suppresses BR-insensitive mutants
To test if PP2A can dephosphorylate BZR1 and activate BR responses in planta, we generated transgenic Arabidopsis plants overexpressing either PP2A B’α or PP2A B’β, B subunits that showed strong interaction with BZR1 in yeast. These plants showed reduced sensitivity to the BR biosynthetic inhibitor brassinazole (BRZ), with long hypocotyls on BRZ similar to the bzr1-1D mutant (Fig. 2a). Overexpression of PP2A B’α and PP2A B’β also partially suppressed the phenotypes of weak allele bri1-5 (Fig. 2b-2d, and Supplementary Information, Fig. S4a-c online). Quantitative RT-PCR showed the transcript level of BZR1-repressed target gene CPD was increased in the bri1-5 mutant, but reduced by overexpression of PP2A B’α and B’β (Fig. 2c, and Supplementary Information, Fig. S4c online). Consistent with the effects on growth and target gene expression, BZR1 accumulation was increased in plants overexpressing PP2AB’β or PP2AB’α (Fig. 2e). Interestingly, both unphosphorylated and phosphorylated BZR1 were increased, suggesting that binding to PP2A B’ protects BZR1 from degradation but may not be sufficient for dephosphorylation. It is possible that some of the overexpressed B’ proteins are not associated with the catalytic subunits.
Figure 2. Overexpression of PP2AB’α and PP2AB’β activates BR signalling and partially suppresses BR signalling mutants.
(a) Wild-type (Col), bzr1-1D, and seedlings overexpressing PP2AB’α-YFP or PP2AB’β-YFP fusion protein in the Col background were grown in the dark for 4 days on regular MS medium (MS) or MS medium containing 2 μM brassinazole (BRZ). Scale bars are 5 mm. (b) Overexpression of PP2AB’α-YFP and PP2AB’β-YFP partially suppresses the bri1-5 mutant. Wild type (WS), bri1-5, and bri1-5 transformed with 35S::PP2AB’α-YFP (B’α-OX) or PP2AB’β-YFP (B’β-OX) were grown in the dark for 4 days. Scale bar are 5 mm. Inset shows anti-YFP immunoblot (upper band) of the transgenic plants and Ponceau S staining as loading control (lower band). (c) Expression levels of the CPD gene in seedlings shown in panel b were analyzed by quantitative RT-PCR. Values shown indicate fold-increase over normalized wild-type (Ws) expression levels. The difference between bri1-5 and B’-OX or wild type plants is statistically significant (P<0.05, n=3). Error bars represent SE. (d) Four-week old light-grown plants of wild type (WS), bri1-5, and bri1-5 overexpressing PP2AB’β (B’β OX/bri1-5). Scale bars = 10 mm. (e) Immunoblot analysis of BZR1 in transgenic plants overexpressing PP2AB’α or PP2AB’β in the bri1-5 mutant or wild type background. Arrows mark the phosphorylated (+P) and unphosphorylated (-P) BZR1, and the asterisk marks a non-specific band that serves as loading reference. Full scan of immunoblot is shown in Supplementary Information, Fig. S9. (f) Overexpression of PP2AB’β-YFP partially suppresses the homozygous bri1-116 null mutant. Scale bars = 10 mm. (g) Overexpression of PP2AB’β-YFP partially suppresses the homozygous bin2-1 mutant phenotype. The bin2-1 mutant was crossed with the bzr1-1D mutant and with transgenic plants overexpressing PP2AB’β, BSK3, and BSU1. Scale bars = 10 mm.
Overexpression of PP2A B’β and PP2A B’α also partly suppressed the null mutant bri1-116 (Fig. 2f, and Supplementary Information, Fig. S4d online) and the homozygous gain-of-function bin2-1 mutant (Fig. 2g). The bin2-1 mutant is suppressed by bzr1-1D, but not by overexpression of upstream components BSK3 and BSU14 (Fig. 2g). In the dark-grown bin2-1 mutant, only phospho-BZR1 was detected, whereas a small amount of unphosphorylated BZR1 was detectable in wild-type plants. The transgenic bin2-1 plants overexpressing PP2AB’β accumulated unphosphorylated BZR1 at a level similar to that observed in the wild type (Supplementary Information, Fig. S4e online). These data are consistent with PP2A acting on BZR1 downstream of or parallel to BIN2 in the BR signal transduction pathway.
PP2A activity is required for normal BZR1 dephosphorylation and plant growth
To determine if PP2A is required for BZR1 dephosphorylation and BR signalling in planta, we obtained T-DNA insertion mutants for several PP2A B’ and C subunits27. Of the pp2ab’ family mutants screened (α, β, γ, η, θ and ζ), only the pp2ab’β mutant showed slightly reduced responses to BR in hypocotyl elongation and root inhibition in the light (Supplementary Information, Fig. S5a-b online). When grown in the dark on regular medium or on BRZ, hypocotyls of the pp2ab’β mutant seedlings were slightly shorter than wild type (Supplementary Information, Fig. S5c online), consistent with reduced BR sensitivity. The T-DNA insertion mutant of PP2A catalytic subunit C5 (pp2ac5) also had shorter hypocotyls when grown on medium containing BRZ (Supplementary Information, Fig. S5d and S5e) and showed increased expression of the BZR1-repressed target genes DWF4 and CPD, consistent with reduced BR signalling (Supplementary Information, Fig. S5f).
The phenotypes of these B’ and C subunit single gene mutants are subtle, presumably because of redundant functions of other gene family members. Therefore we generated pp2ab’αβ double mutants lacking the two major BZR1-binding PP2A B’ isoforms. In the pp2ab’αβ double mutant populations, a variety of phenotypes were observed ranging from near wild type to severe dwarfism (Fig. 3a). The levels of PP2A B’β RNA are inversely correlated with the degree of dwarfism (Supplementary Information, Fig. S6), suggesting that loss of both PP2A B’α and B’β causes dwarfism. The variation of PP2A B’β RNA level in the double mutant is likely due to variable efficiencies of splicing of the T-DNA together with the intron. Immunoblot analysis showed that the pp2ab’αβ double mutant plants exhibiting the dwarf phenotype accumulate an increased level of phosphorylated BZR1 protein and slightly reduced level of unphosphorylated BZR1 (Fig. 3b). When grown in the dark, many pp2ab’αβ seedlings showed an obvious short-hypocotyl phenotype whereas wild type seedlings are uniformly tall (Fig. 3c). These results indicate that loss of two BZR1-interacting B’ subunits of PP2A (B’α and B’β) reduces BZR1 dephosphorylation and plant growth.
Figure 3. PP2A is essential for BR signalling and BZR1 dephosphorylation.
(a) A wild type plant and two pp2ab’αβ double mutant plants grown in the light for four weeks. Scale bars = 10 mm. (b) Immunoblotting analysis of phosphorylated (+P) and unphosphorylated (-P) BZR1 in plants shown in panel a, using the anti-BZR1 antibody. St, Ponceau S staining of blot shows loading. The ratio between phosphorylated and unphosphorylated BZR1 bands (+P/-P) and the level of unphosphorylated BZR1 relative to that observed in the wild type (-P%) were calculated after normalization against the intensity of Ponceau S staining and are shown beneath the gel images. (c) Six-day old dark-grown seedlings of wild type and the pp2ab’αβ double mutant with short hypocotyls. Scale bar = 5 mm. (d) Protein extracts were isolated from seven-day old BZR1::BZR1-CFP transgenic Arabidopsis seedlings grown in the absence (-OA) or presence of 250 nM okadaic acid (+OA) and treated with 100 nM BL for 30 min. After SDS-PAGE and transfer, the immunoblot was probed with anti-YFP antibody. +P/-P, ratio between phosphorylated and unphosphorylated BZR1. (e) Nine-day old light-grown seedlings of homozygous bin2-1 mutant were treated with mock solution or 1 μM okadaic acid for 1 hr, then with 50 μM bikinin for 15 min. BZR1 phosphorylation was analyzed by immunoblotting using the anti-BZR1 antibody. +P/-P, ratio between phosphorylated and unphosphorylated BZR1. Full scans of immunoblots are shown in Supplementary Information, Fig. S9.
BR-induced BZR1 dephosphorylation also is incomplete in the rcn1 pp2aa3 A subunit double mutant, which showed severe dwarfism, with some similarity to bri1-116 and bin2-1 mutants (Supplementary Information, Fig. S5g-h online). These results support a crucial role of PP2A in BZR1 dephosphorylation. However, the rcn1 pp2aa3 double mutant lacks two of the three scaffolding subunits, is severely defective in regulation of auxin transport28, 29, and is expected to grossly overproduce ethylene30, 31. The dwarf phenotypes of the rcn1 pp2aa3 double mutant reflect a drastic reduction in overall PP2A activity level, however, this double mutant combination does not directly affect the subunits involved in recruiting BZR1 for dephosphorylation. The radial expansion exhibited by rcn1 pp2aa3 hypocotyls is not characteristic of BR-insensitive dwarfs (Figure S5g), but is characteristic of ethylene overproducers. Similarly, the primary root meristem collapse exhibited by rcn1 pp2aa3 double mutants is not characteristic of brassinosteroid insensitivity, but is a hallmark of severe defects in root auxin transport29. In contrast, the B' subunit mutants described above have stronger effects on BZR1 phosphorylation patterns and more specific effects on BR signalling (compare Figs 3b and S5g-h).
Treatment with the PP2A inhibitor okadaic acid (OA) inhibited BR-induced BZR1 dephosphorylation (Fig. 3d) and nuclear localization32. Because OA can also inhibit the BSU1 phosphatase33 and thus lead to increased BIN2 activity, the increase in phosphorylated BZR1 after OA treatment could be due to increased phosphorylation by BIN2 or reduced dephosphorylation by PP2A. To avoid effects on BSU1-mediated regulation of BIN2, we used bin2-1 mutant plants, in which the bin2-1 kinase is constitutively active and insensitive to BSU1 regulation4. We also took advantage of the recently identified GSK3 kinase inhibitor bikinin34, which inhibits both BIN2 and bin2-1 kinases. Bikinin treatment caused accumulation of unphosphorylated BZR1 by inhibiting bin2-1 phosphorylation without affecting the dephosphorylating phosphatase. In the presence of OA, the bikinin-induced accumulation of dephosphorylated BZR1 is significantly decreased (Fig. 3e), indicating that OA affects BZR1 dephosphorylation downstream of BIN2, consistent with inhibition of PP2A-mediated BZR1 dephosphorylation.
PP2A interacts with BZR1 through its PEST domain, which is essential for BR-induced BZR1 dephosphorylation in vivo
Although the loss-of-function mutants of various PP2A subunits provided strong evidence for a role of PP2A in BZR1 dephosphorylation, these mutants either show a weak phenotype due to redundant function of a large numbers of isoforms35 or pleotropic phenotypes due to multiple functions of PP2A in a wide range of processes. We thus asked whether the interaction with and dephosphorylation by PP2A could be disrupted by mutations in BZR1. First we performed yeast two-hybrid assays using fragments of BZR1 containing known functional domains (Fig. 4a and 4b) to identify the region that is necessary and sufficient for binding to PP2AB’α (amino acids 215-251). This region contains the putative PEST domain proposed to be involved in protein degradation8, 10. Deletion of amino acids 232-251 in the full-length BZR1 (ΔPEST) abolished the interaction with PP2AB’α in yeast (Fig. 4b). We then generated transgenic Arabidopsis plants expressing either wild-type BZR1-YFP or a mutant BZR1ΔPEST-YFP fusion protein. In the absence of exogenous BR, the BZR1ΔPEST protein accumulated exclusively in the phosphorylated form, whereas a significant portion of the wild-type BZR1 was unphosphorylated or hypophosphorylated (Fig. 4c). BR treatment caused complete dephosphorylation of the wild type BZR1-YFP protein but very little change in the mutant BZR1-YFP proteins (Fig. 4c). Deletion of a large domain (aa 215-251) had similar effects on PP2A binding and in vivo dephosphorylation (Supplementary Information Fig. S7a-c). Thus dephosphorylation of BZR1 by PP2A is greatly compromised by the ΔPEST deletion. These results demonstrate that PP2AB’ binds to BZR1 through a specific PP2A-binding domain, and this binding is essential for BR-induced dephosphorylation of BZR1 in vivo. Interestingly, the transgenic plants that overexpress the BZR1ΔPEST protein showed reduced plant size with dark green leaves (Supplementary Information Fig. S7d), similar to BR-deficient mutants, suggesting that phosphorylated BZR1 may have an inhibitory effect on BR response. This would be consistent with the dwarf phenotype of the pp2ab’αβ double mutant, in which the levels of phospho-BZR1 increase dramatically but dephosphorylated BZR levels decrease only slightly (Fig. 3a). Phospho-BZR1 may inhibit cell elongation via a cytoplasmic process or via interference with nuclear localization of unphosphorylated BZR1 protein.
Figure 4. PP2A B’ subunit binds to the PEST domain of BZR1 to promote BZR1 dephosphorylation in vivo.
(a) Sequence of the region of BZR1 (aligned with the corresponding sequences of BZR2 and rice OsBZR1), containing the PP2A-binding domain and the bzr1-1D, bzs247 and bzs248 mutations (amino acid substitutions shown in boxes). (b) Yeast two-hybrid assay of PP2A binding by various fragments of BZR1 shown by box diagrams. Yeast growth on –AH medium indicates interaction between the BZR1 fragment and PP2AB’α. (c) Deletion of the PP2A-binding domain abolishes BR-induced BZR1 dephosphorylation in plants. Two-week old plants of a 35S::BZR1-YFP transgenic line expressing the wild type BZR1-YFP, or three independent transgenic lines expressing the mutant BZR1-YFP containing deletion of amino acids 232-251 (ΔPEST-A, -B, and -C lines), were treated with mock solution or 250 nM brassinolide (BL) for 1 hr. (d) The bzr1-1D mutation increases dephosphorylation of BZR1 in planta. Upper panel shows an immunoblot of BZR1/bzr1-1D protein extracted from wild type (BZR1) and bzr1-1D mutant plants. In each sample, the upper band is phosphorylated (pBZR1) and the lower band is unphosphorylated (BZR1) protein. Stain, Ponceau S staining shows equal loading. (e) PP2A binds more strongly to the bzr1-1D protein. MBP-BZR1 and MBP-bzr1-1D proteins were gel-blotted on a nitrocellulose membrane and probed sequentially with GST-PP2AB’α and anti-GST antibody. Lower panel shows Ponceau S staining of the gel blot. (f) From left to right are wild type, bzr1-1D, bzr1-1D bzs247 and bzr1-1D bzs248 seedlings grown in the dark on 2 μM BRZ for 5 days. Scale bar = 2.5 mm. (g) Anti-BZR1 immunoblot shows the phosphorylated BZR1 (pBZR1) and unphosphorylated BZR1 (BZR1) in the plants shown in panel f. (h) The bzs mutations reduce PP2A binding. Equal amounts of MBP, MBP-BZR1, MBP-bzr1-1D, and MBP-bzr1-1D containing bzs247 and bzs248 mutations were gel blotted to nitrocellulose membrane and probed with GST-PP2AB’α or GST-BIN2 and anti-GST antibody, and the blots were subsequently stained with Ponceau S (Stain). Full scans of immunoblots are shown in Supplementary Information, Fig. S9.
Binding to PP2A is enhanced by the bzr1-1D mutation and reduced by two intragenic bzr1-1D suppressor mutations
The PP2A-binding domain of BZR1 defined above includes the residue altered by the bzr1-1D mutation, Pro2348 (Fig. 4a). We thus tested whether the bzr1-1D mutation (Pro234Leu) affects the interaction with PP2A and dephosphorylation of BZR1. Although the bzr1-1D mutation subtly alters the migration of BZR1 protein9, a larger fraction of the mutant bzr1-1D protein is present in the unphosphorylated form compared to wild type BZR1 (Fig. 4d) . Furthermore, in vitro binding assays revealed that the bzr1-1D mutation increased binding to PP2AB’α (Fig. 4e). These results suggest that bzr1-1D increases BZR1 dephosphorylation by PP2A. To test this hypothesis, we identified two intragenic bzr1-1D suppressors (bzs247 and bzs248) that suppressed the BRZ-resistant phenotypes of the bzr1-1D mutant (Fig. 4f), due to mutations near the original bzr1-1D mutation (Thr236 to Ile and Glu242 to Lys) (Fig. 4a). Both bzs mutations also partly reversed the dephosphorylation change caused by bzr1-1D (Fig. 4g), and reduced binding of bzr1-1D to PP2AB’α (Fig. 4h). In contrast, the binding of BIN2 was not affected by bzr1-1D or the bzs247 and bzs248 mutations (Fig. 4h). These genetic and biochemical results demonstrate that the effect of the bzr1-1D mutation on BR signalling is due to increased binding between BZR1 and PP2A, which leads to accumulation of dephosphorylated BZR1.
Dephosphorylation by PP2A abolishes BZR1 binding to the 14-3-3 proteins
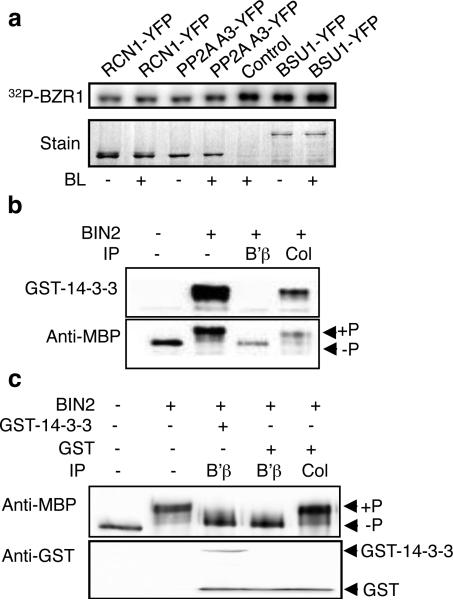
The preceding experiments identified the PP2A binding site of BZR1, but did not identify the target site[s] for PP2A-mediated BZR1 dephosphorylation. BZR1 contains twenty-five putative BIN2 phosphorylation sites12, 14. Phosphorylation of one of these sites, S173, promotes 14-3-3 binding and cytoplasmic retention12. To determine whether PP2A dephosphorylates all residues phosphorylated by BIN2 and affects 14-3-3 binding, we treated BIN2-phosphorylated BZR1 with PP2A immunopurified from transgenic plants. Immunoprecipitation of the scaffolding A subunit (PP2A A1-YFP or PP2A A3-YFP) yielded immunocomplexes that dephosphorylated BZR1. BSU1 immunoprecipitates showed no BZR1-dephosphorylating activity (Figure 5a). PP2A immunoprecipitated from BR-treated and untreated plants showed similar activity against phospho-BZR1 in vitro (Fig. 5a). In vitro phosphorylation by BIN2 dramatically alters SDS-PAGE migration of BZR1 and promotes strong binding to the 14-3-3λ protein. Both the mobility shift and 14-3-3 protein binding were abolished after incubation with immuno-purified PP2A (Fig. 5b), indicating that PP2A can dephosphorylate many or all BIN2 phosphorylation sites, including the 14-3-3 binding site at Ser173. Pre-incubation with 14-3-3 had little effect on PP2A dephosphorylation of BZR1 (Fig. 5c), which is consistent with the separate binding sites for 14-3-3 and PP2A. These data suggest that one critical effect of PP2A action on BZR1 is relief of the 14-3-3-mediated cytoplasmic retention of S173-phosphorylated BZR1. PP2A action very likely relives other inhibitory effects of BIN2 phosphorylation on BZR1 function, such as DNA binding. PP2AB’α and B’β are localized in the cytoplasm (Supplementary Information, Fig. S8 online), consistent with the hypothesis that PP2A acts on cytoplasmically retained phospho-BZR1. These results demonstrate that PP2A antagonizes BIN2 effects in the BR signalling pathway by dephosphorylating BIN2-phosphorylated BZR1.
Figure 5. PP2A dephosphorylates BZR1 and abolishes its binding by the 14-3-3 proteins.
(a) MBP-BZR1 protein was phosphorylated by BIN2 using 32P-γATP, and then incubated at 30 degree for 3 hr with PP2A (RCN1-YFP, PP2AA3-YFP) or BSU1-YFP, which were immunoprecipitated from BL-treated (+, 1 hr) or untreated (-) transgenic Arabidopsis plants. The control reaction used anti-YFP immunoprecipitate from non-transgenic plants. Lower panel shows the Coomassie Blue staining of the precipitated YFP fusion proteins. (b) Recombinant MBP-BZR1 (-BIN2) was phosphorylated by BIN2 in vitro (+BIN2), and then incubated with anti-YFP immunoprecipitate from wild type control plants (Col) or from the 35S::PP2AB’β-YFP transgenic plants (B’β) for 3.5 hr in the presence of 30 μM bikinin. The proteins were separated by SDS-PAGE, blotted, and incubated with GST-14-3-3 protein plus anti-GST antibody to detect binding by 14-3-3, or with anti-MBP antibody to detect phosphorylated (+P) and unphosphorylated (-P) MBP-BZR1. (c) Pre-incubation of BIN2-phosphorylated BZR1 with the 14-3-3 protein did not interfere with its dephosphorylation by PP2A. BIN2-phosphorylated BZR1 was pre-incubated with GST or GST-14-3-3 for 1 hr and then dephosphorylated by anti-YFP immunoprecipitation product from Col control (Col) or 35S::PP2A-B’β-YFP (B’β) for another 1 hr. Phosphorylated (+P) and unphosphorylated (-P) BZR1 are marked by arrows. Full scans of immunoblots are shown in Supplementary Information, Fig. S9.
DISCUSSION
This study not only reveals a key role of the PP2A phosphatase in steroid response in plants but also elucidates a detailed mechanism of PP2A regulation of BZR1 function. The in vivo interaction between PP2A and BZR1 was demonstrated by affinity purification, BiFC and co-immunoprecipitation. Direct interactions with specific regulatory B’ subunits were verified by both in vitro and in vivo assays, and the interaction was shown to be mediated by direct binding to the PEST domain of BZR1. The function of PP2A in dephosphorylating BZR1 and activating BR responses in plants was demonstrated by overexpression and loss-of-function mutants of PP2A, as well as by mutations of the PP2A-binding domain in BZR1. Interaction in yeast suggests that PP2A also regulates BZR2/BES1 through similar mechanisms. The combination of these biochemical and genetic data demonstrate that PP2A is a key component of the BR signalling pathway, activating BR responses by dephosphorylating BZR transcription factors.
PEST domains in many proteins are known to mediate protein degradation. While the PEST domains in BZR1 and BZR2 were previously proposed to mediate their degradation9, 10, our data show that this region functions as a PP2A-binding domain. The stabilizing effect of the bzr1-1D mutation in the PEST domain could be due to increased dephosphorylation by PP2A, as previous studies suggested that phosphorylation promotes BZR1 degradation by the proteasome9. However, there have been conflicting observations of whether BR treatment increases or decreases the levels of BZR1 and BZR2/BES1 9, 10, 13. Alternatively, binding to PP2A might directly affect BZR1 stability and phosphorylation status, as suggested by the accumulation of higher levels of both phosphorylated and unphosphorylated BZR1 in plants overexpressing PP2A B’α or B’β, and increased level of phosphorylated BZR1 in the pp2ab’αβ double mutant. Deletion of the PEST domain reduced dephosphorylation of BZR1 in vivo, but had little effect on total BZR1 accumulation. Whether the PEST domain also plays a direct role in BZR1 turnover remains to be analyzed in future studies.
Together with the previously characterized components3, 4, PP2A completes a core signalling module that relays the extracellular BR cue to the nucleus, regulating BZR1 activity and gene expression in plants (Supplementary Information, Fig. S1b online). When BR levels are low, BZR1 is phosphorylated by BIN2 in the nucleus9, 13, 32, and consequently exported from the nucleus and retained in the cytoplasm by the 14-3-3 proteins12, 36. Upon BR binding to BRI1, sequential activation of BSK1 and BSU1 leads to tyrosine dephosphorylation and inactivation of BIN24, 20, while BZR1 is rapidly dephosphorylated by PP2A in the cytoplasm and moves into the nucleus to activate and repress different target genes7. Consistent with their functions in driving BZR1 in and out of the nucleus, PP2AB’α and B’β are localized predominantly in the cytoplasm (Supplemental Information, Fig. S8), and BIN2 has been shown to act more effectively in the nucleus than in the cytoplasm13, 32. In contrast, BSU1 is present in both cytoplasm and nucleus4, 32, presumably to be activated by BSK1 at the plasma membrane and to inactivate BIN2 in the nucleus. It remains unclear whether BR only inactivates BIN2 or also activates PP2A to promote accumulation of unphosphorylated BZR1. As an abundant cellular phosphatase, PP2A is involved in many processes, such as regulation of gravitropic response28, auxin transport29, and phototropic responses37. Whether PP2A is constitutively active or regulated by any signal remains to be demonstrated in future studies.
This study provides further evidence for intriguing similarities between the BR pathway in plants and the Wnt pathway in metazoans11, because recent studies demonstrated a role of PP2A in dephosphorylating and activating β-catenin23. In both BR and Wnt pathways, GSK3 phosphorylates and inactivates transcription factors (BZR1 and β-catenin) and PP2A counteracts these effects by dephosphorylating the GSK3 substrates23, whereas PP1 and related phosphatases (BSU1) play a positive role upstream of GSK34, 11, 38(Supplementary Information, Fig. S1b). Another similar interplay between PP2A and 14-3-3 was reported recently for the regulation of FOXO3a nuclear localization and transcription activation in humans39. Further studies of the detailed signalling mechanisms of each system will be required to understand whether there was an evolutionary convergence or conservation of signalling modules of these pathways in the two kingdoms.
METHODS
Methods and any associated references are available in the online version of the paper at attp://www.nature.com/naturecellbiology/.
Supplementary Material
Acknowledgement
This work was funded by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through Grant DE-FG02-08ER15973, and by NIH (R01GM066258), National Science Foundation of China (30870213, 90917008), NSF (IOS-0724688 and IOS-0846282), and the Herman Frasch Foundation. R.W and M.Y were supported by the China Scholarship Council. The UCSF Mass Spectrometry Facility (A.L. Burlingame, Director) is supported by the Biomedical Research Technology Program of the National Centre for Research Resources, NIH NCRR RR01614, RR012961 and RR019934.
Methods
Plant materials, growth, and treatments
The bri1-5 mutant is in WS ecotype background, and all other Arabidopsis thaliana plants are in Columbia ecotype background. For hypocotyl and root growth assays, seedlings were grown on vertical agar plate containing half strength Murashige and Skoog (1/2MS) medium supplemented with 1% sucrose. For short duration hormone treatments, the seedlings were submerged in hormone or mock solution for the times of treatment shown.
Tandem affinity purification of BZR1-interacting proteins
Arabidopsis thaliana Columbia transformed with 35S::BZR1-7Myc-6His, 35S::bzr1-1D-7Myc-6His or pBZR1::BZR1-CFP, pBZR1::bzr1-1D-CFP were grown on solid Murashige and Skoog medium supplemented with 3% sucrose for two weeks, then harvested and ground to fine powder in liquid nitrogen. The tissues were extracted using a buffer containing 50 mM PBS, 150 mM NaCl, 1% Triton X-100, 15% glycerol, 5 mM imidazole and protease inhibitor cocktails (Sigma) at 4 °C The homogenate was centrifuged at 14,000 g for 10 minutes and the supernatant was incubated with Ni-NTA Agarose beads (Qiagen) for 30 minutes at 4 °C. The Beads were washed on a column with 20 bed volumes of wash buffer 1 (50 mM PBS, 150 mM NaCl, 0.1% Triton X-100, 10% glycerol, 5 mM imidazole) and then eluted with elution buffer (50 mM PBS, 150 mM NaCl, 0.1% Triton X-100, 10% glycerol, 80 mM imidazole). Proteins eluted from the Ni-NTA beads were incubated with anti-Myc antibody agarose (SIGMA) for 30 minutes at 4 °C After Washing with 200 bed volume of washing buffer 2 (50 mM PBS, 150 mM NaCl, 0.1% Triton X-100 and 10% glycerol), the proteins were eluted twice by boiling in 1% SDS for 5 minutes, and then separated in a 5-20% gradient SDS-PAGE gel. Each gel lane was cut into 20 pieces, then in-gel digested with trypsin and analyzed by LC-MS-MS as described previously40. Proteins identified in the BZR1-Myc-His samples but not in the BZR1-CFP samples are considered real BZR1 interacting proteins.
Yeast two-hybrid assays, co-immunoprecipitation, and BiFC experiments were performed as described previously4, 12. Anti-BZR1 antibody was produced by immunizing rabbits with an MBP-tagged full-length BZR1 protein. The antibody was affinity purified using a GST-tagged BZR1 fragment (aa 91-336) without the conserved DNA binding domain. Because BZR1 and BZR2 share 88% amino acid sequence identity, this antibody does detect recombinant BZR2 protein; it also detects increased signal in the dominant bzr1-1D and bes1-D mutants. Therefore the signal detected by this antibody includes both BZR1 and BZR2. Anti-PP2A C antibody was described previously30.
Protein expression and purification
GST tagged PP2AB’α, PP2AB’β, BIN2 and MBP tagged BZR1, bzr1-1D, bzs247 and bzs248 were expressed in E. coli and purified by standard procedure using glutathione agarose beads (GE healthcare) or amylose agarose beads (New England Biolabs). BIO-RAD Profinity eXact fusion-tag system was used to generate tag-free BIN2 for in vitro MBP tagged BZR1 phosphorylation. In brief, a polymerase chain reaction (PCR) amplified BIN2 cDNA was cloned into BamHI/EcoRI site of pPAL7 (BIO-RAD) expression vector, and the plasmid was introduced into BL21 codon plus E. coli cells. Tag-free BIN2 protein was induced and purified according to manufacture's protocol using a Profinia protein purification system (BIO-RAD).
In vitro interaction assay using gel blot overlay assays
To prepare phosphorylated BZR1, 2 μg MBP-tagged BZR1 was incubated with 2μg tag-free BIN2 in the presence or absence of 200 μM ATP for 16 hrs at 30°C. Phosphorylated and unphosphorylated BZR1 protein were then separated on a SDS-PAGE gel and blotted to nitrocellulose membrane. The membrane was incubated in blocking solution (5% non-fat milk and 0.1% Tween-20 in PBS) overnight at 4 °C. The membrane was then incubated in blocking solution plus 2 μg/ml GST tagged PP2AB’α, PP2AB’β, 14-3-3λ, or BIN2 for 2 hrs at room temperature (RT), washed briefly for 30 min, followed by 1 hr incubation at room temperature with HRP-labelled anti-GST antibody in blocking solution. Overlay signal was detected using the SuperSignal West Dura chemiluminescence reagent (Pierce).
Overexpression of PP2A B’α and B’β
Full-length cDNAs of PP2AB’α and PP2AB’β without stop codon were amplified by PCR using gene specific primers (B’α -For, caccATGTTTAAGAAGATCATGAAAG; B’α -Rev, AGAAGTGATCATAGGATCTTCTCTTTC; B’β -For, caccATGTTTAAGAAAATCATGAAAG; B’β -Rev, GGAAGTGATCATATGATCTTCTTCTC). The cDNAs were cloned into pENTR/SD/D-TOPO vectors (Invitrogen, Carlsbad, CA), and subcloned into gateway compatible binary vectors pEarleyGate 104 or BiFC vectors12 by LR clonase (Invitrogen). To create the ΔPEST construct, site-directed mutagenesis was performed according to the manufacturer's protocol (Stratagene) to delete the sequence coding for amino acids 232 to 251 from the BZR1 entry clone. The primers used for mutagenesis were: Forward: 5’-CACATCGCCACCAGTTTCATTGGATAAGCTTTCAG-3’; reverse: 5’-CTGAAAGCTTATCCAATGAAACTGGTGGCGATGTG-3’). The mutagenized cDNA was cloned into the gateway compatible binary vectors pEarleyGate 101 by LR clonase (Invitrogen).
The constructs were introduced into Agrobacterium strain GV3101 by electroporation and transformed into Arabidopsis by the floral dipping method. To test genetic interactions between BRI1, BIN2 and PP2AB’α or PP2AB’β, single 35S:: PP2AB’α or 35::PP2AB’β -YFP transgenic plant was crossed into the bri1-116 and bin2-1 mutants. The phenotypes of F3 double homozygous plants were analyzed. Real time reverse transcription polymerase chain reaction (qRT-PCR) analysis of the CPD and DWF4 gene expression was performed as described previoiusly10.
Isolation of PP2A mutants
T-DNA knockout mutants SALK_052612 and SALK_077700 (pp2ab’α), SALK_103167 (pp2ab’β), SALK_039172 (pp2ab’γ), SALK_057440 (pp2ab’η), SALK_039168 (pp2ab’η), SAIL-300-B01 (pp2ab’θ), SALK_107944c (pp2ab’ ζ) and SALK_139822 (pp2ac5) were obtained from the Arabidopsis Biological Resource Centre (ABRC, www.arabidopsis.org)27. Mutants were first genotyped with gene specific primers and T-DNA specific primer to confirm the position of T-DNA insertion site. Then PCR was performed using gene specific primers to confirm loss of the wild type gene in homozygous mutants. The primers used for genotyping are: PP2ABα-F: 5’TCGTGTCTTTGGTTCTGATTTG-3’ and PP2ABα-R: 5’-AAGGGCCTGTGAACCATAAAC-3’ for SALK _077700; Beta GF: 5’-CTAGTGCTTCATCAAGTTTAGTG-3’ and Beta R: 5’-GGAAGTGATCATATGATCTTCTTCTC-3’ for Salk_103167; PP2ABb3-PF: 5’-caccGCAAATTACTTATATCCTTGGACTCGTG-3’ and PP2ABb3-PR: 5’-AATTACAGAAACCCTAAATTATCTCC-3’ for Salk_039172; PP2ABb-F: 5’-caccATGTGGAAACAGATTCTAAGTAAGCTTC-3’ and PP2ABb1-GR: 5’-ACTCGTTCAGAAACATGACTTCTTTGG-3’ for SALK_057440; PP2ABb1-GF: 5’-CTTGCAGCTTGTGTATGAGCTTTTCTT-3’ and PP2ABb1-GR: 5’-ACTCGTTCAGAAACATGACTTCTTTGG-3’ for SALK_039168; PP2ABb2-GF: 5’-AACGAAGATGGTAGCGGTTAATCTGTT-3’ and PP2ABb2-GR: 5’-TCTAATAACGGTATCAGCGAGCTTGAA-3’ for SALK_107944c; At1g13460-GF: 5’-TGGGATTTATCTCCATTTCAGG-3’ and At1g13460-GR: 5’-GAAACCTGGCAATGCTTCAT-3’ for SAIL-300-B01; PP2A5F:5’-CACCATGCCGCCGGCGACCGGAGATATCGA-3’ and PP2A5-GR: 5’-CTTTCAGTGCTACCAAAAGTGAGACTG-3’ for SALK_139822.
The pp2ab’αβ double mutant was generated by crossing the pp2ab’α with pp2ab’β single mutants, and the double homozygous mutant was identified by PCR genotyping in the F2 generation. Transcript levels of PP2AB’α (At5g03470), B’β (At3g09880) and ubiquitin (At3g02540) were determined using TaqMan assays (At02337056, At02291206 and At02163241) (Applied biosystems, CA).
BZR1 dephosphorylation assay
YFP-tagged PP2AA1 (RCN1), PP2AA3, BSU1, or PP2AB’β were immunoprecipitated from transgenic Arabidopsis seedlings using anti-YFP antibodies according to Gampala et al 200712. BIN2-phosphorylated MBP-BZR1 was separated from BIN2 using amylose resin and incubated with immunoprecipitated PP2A or BSU1 at 30 °C for indicated time. For figures 5b and 5c, BIN2 was not removed after kinase reaction, but 30 μM bikinin was added to inhibit BIN2 during dephosphorylation reaction. Phosphatase activity was stopped by adding an equal volume of 2x SDS sample buffer and boiling for 5 min. Dephosphorylation of BZR1 was detected either by autoradiography or immunoblotting using an anti-MBP antibody.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Kim TW, Wang ZY. Brassinosteroid Signal Transduction from Receptor Kinases to Transcription Factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 2.Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 3.Clouse SD. Brassinosteroids. Plant counterparts to animal steroid hormones? Vitam Horm. 2002;65:195–223. doi: 10.1016/s0083-6729(02)65065-4. [DOI] [PubMed] [Google Scholar]
- 4.Tang W, Deng Z, Wang ZY. Proteomics shed light on the brassinosteroid signaling mechanisms. Curr Opin Plant Biol. 2010;23:27–33. doi: 10.1016/j.pbi.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim T-W, et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Bio. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gendron JM, Wang ZY. Multiple mechanisms modulate brassinosteroid signaling. Curr Opin Plant Biol. 2007;10:436–441. doi: 10.1016/j.pbi.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y, et al. Integration of Brassinosteroid Signal Transduction with the Transcription Network for Plant Growth Regulation in Arabidopsis. Dev Cell. 2010 doi: 10.1016/j.devcel.2010.10.010. In Press, NIHMSID # 248553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 9.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 11.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gampala SS, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 14.Bai MY, et al. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci U S A. 2007;104:13839–13844. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinoshita T, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, et al. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell. 2005;8:855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thummel CS, Chory J. Steroid signaling in plants and insects--common themes, different pathways. Genes Dev. 2002;16:3113–3129. doi: 10.1101/gad.1042102. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, et al. PR55 alpha, a regulatory subunit of PP2A, specifically regulates PP2A-mediated beta-catenin dephosphorylation. J Biol Chem. 2009;284:22649–22656. doi: 10.1074/jbc.M109.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su Y, et al. APC is essential for targeting phosphorylated beta-catenin to the SCFbeta-TrCP ubiquitin ligase. Mol Cell. 2008;32:652–661. doi: 10.1016/j.molcel.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Janssens V, Longin S, Goris J. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail). Trends Biochem Sci. 2008;33:113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Mumby M. The 3D structure of protein phosphatase 2A: new insights into a ubiquitous regulator of cell signaling. ACS Chem Biol. 2007;2:99–103. doi: 10.1021/cb700021z. [DOI] [PubMed] [Google Scholar]
- 26.Farkas I, Dombradi V, Miskei M, Szabados L, Koncz C. Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 2007;12:169–176. doi: 10.1016/j.tplants.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 28.Rashotte AM, DeLong A, Muday GK. Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell. 2001;13:1683–1697. doi: 10.1105/TPC.010158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michniewicz M, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 30.Blakeslee JJ, et al. Specificity of RCN1-mediated protein phosphatase 2A regulation in meristem organization and stress response in roots. Plant Physiol. 2008;146:539–553. doi: 10.1104/pp.107.112995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muday GK, et al. RCN1-regulated phosphatase activity and EIN2 modulate hypocotyl gravitropism by a mechanism that does not require ethylene signaling. Plant Physiol. 2006;141:1617–1629. doi: 10.1104/pp.106.083212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryu H, Kim K, Cho H, Hwang I. Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol Cells. 2010;29:291–296. doi: 10.1007/s10059-010-0034-y. [DOI] [PubMed] [Google Scholar]
- 33.Mora-Garcia S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Rybel B, et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem Biol. 2009;16:594–604. doi: 10.1016/j.chembiol.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terol J, Bargues M, Carrasco P, Perez-Alonso M, Paricio N. Molecular characterization and evolution of the protein phosphatase 2A B' regulatory subunit family in plants. Plant Physiol. 2002;129:808–822. doi: 10.1104/pp.020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryu H, et al. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;19:2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng TS, Briggs WR. The Arabidopsis rcn1-1 mutation impairs dephosphorylation of Phot2, resulting in enhanced blue light responses. Plant Cell. 2010;22:392–402. doi: 10.1105/tpc.109.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo W, et al. Protein phosphatase 1 regulates assembly and function of the beta-catenin degradation complex. EMBO J. 2007;26:1511–1521. doi: 10.1038/sj.emboj.7601607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh A, et al. Protein phosphatase 2A reactivates FOXO3a through a dynamic interplay with 14-3-3 and AKT. Mol Biol Cell. 2010;21:1140–1152. doi: 10.1091/mbc.E09-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang W, et al. Proteomic studies of brassinosteroid signal transduction using prefractionation and Two-dimensional DIGE. Mol Cell Proteomics. 2008;7:728–738. doi: 10.1074/mcp.M700358-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.