Abstract
Metallopeptidases (MPs) are among virulence factors secreted by pathogenic bacteria at the site of infection. One such pathogen is Tannerella forsythia, a member of the microbial consortium that causes peridontitis, arguably the most prevalent infective chronic inflammatory disease known to mankind. The only reported MP secreted by T. forsythia is karilysin, a 52-kDa multidomain protein comprising a central 18-kDa catalytic domain (CD), termed Kly18, flanked by domains unrelated to any known protein. We analyzed the 3D structure of Kly18 in the absence and presence of Mg2+ or Ca2+, which are required for function and stability, and found that it evidences most of the structural features characteristic of the CDs of mammalian matrix metalloproteinases (MMPs). Unexpectedly, a peptide was bound to the active-site cleft of Kly18 mimicking a left-behind cleavage product, which revealed that the specificity pocket accommodates bulky hydrophobic side chains of substrates as in mammalian MMPs. In addition, Kly18 displayed a unique Mg2+ or Ca2+ binding site and two flexible segments that could play a role in substrate binding. Phylogenetic and sequence similarity studies revealed that Kly18 is evolutionarily much closer to winged-insect and mammalian MMPs than to potential bacterial counterparts found by genomic sequencing projects. Therefore, we conclude that this first structurally-characterized non-mammalian MMP is a xenolog co-opted through horizontal gene transfer during the intimate coexistence between T. forsythia and humans or other animals, in a very rare case of gene shuffling from eukaryotes to prokaryotes. Subsequently, this protein would have evolved in a bacterial environment to give rise to full-length karilysin that is furnished with unique flanking domains that do not conform to the general multidomain architecture of animal MMPs.
INTRODUCTION
Metallopeptidases (MP) are mostly zinc-dependent protein and peptide hydrolases with major endogenous roles in the physiology and pathology of living organisms. In addition, MPs have exogenous functions in the venoms used by predators, such as in poisonous snakes, scorpions and spiders, or, conversely, in poison-mediated defense strategies against predators such as in scorpion fish (King, 2007; Fox and Serrano, 2009; Carrijo et al., 2005). Another source of exogenous MPs is bacteria, which secrete them as virulence factors during host infection. In this condition, MPs damage host tissues by cleaving cell-surface and tissue proteins, either directly or indirectly by activating host peptidases. MPs also inactivate key proteins in host defense, recruit nutrients, and activate other bacterial virulence factors. These strategies are required for bacterial invasion, survival, proliferation, and colonization of the host in a hostile environment (Miyoshi and Shinoda, 2000; Lantz, 1997; Potempa and Pike, 2009). One such human infection is periodontitis, which causes chronic inflammation of the gums and affects 10–15% of adults worldwide, leading to gingival tissue destruction and tooth loss (Fox, 1992; Cutler et al., 1995). Due to their inflammatory and infective character, severe forms of the disease further contribute to the development of systemic pathologies such as cardiovascular disease, stroke, rheumatoid arthritis, and diabetes (Jordan, 2004; Pihlstrom et al., 2005; Behle and Papapanou, 2006; Persson, 2006; Wegner et al., 2010). Current treatment and curettage of severe periodontal disease is only partially effective. Furthermore, it entails the intensive usage of antibiotics, which contributes to the spread of antibiotic resistance (Haffajee et al., 2003; Pihlstrom et al., 2005; Suvan, 2005). Consequently, there is a need for innovative and specific therapeutic approaches to this pathology, and detailed structural information of participating virulence factors may contribute to this development (Mittl and Grütter, 2006).
Periodontitis is caused by bacteria that grow on tooth surfaces and in the gingival crevice, and three species, Porphyromonas gingivalis, Treponema denticola, and Tannnerella forsythia, are the major periodontopathogens, collectively referred to as the red complex (Socransky et al., 1998). These pathogens secrete a vast armamentarium of proteolytic enzymes, which has been thoroughly characterized for the first two organisms (Potempa et al., 2000; Eley and Cox, 2003). In contrast, the only peptidases identified to date in T. forsythia are a cysteine proteinase, PrtH (Saito et al., 1997), and the MP karilysin (Karim et al., 2010). The latter is a 472-residue secretory protein comprising a 20-residue signal sequence, a 14-residue pro-peptide, an 18-kDa catalytic peptidase domain (CD), and a 30-kDa C-terminal domain of unknown function. The latter has been found only in other putative secretory proteins of T. forsythia (Karim et al., 2010). The recombinant full-length enzyme gives rise to karilysin CD—hereafter Kly18—through sequential autolysis. The first cut at Asn34-Tyr35 (numbering according to the complete gene sequence, see UniProt D0EM77) removes the pro-peptide and generates a fully active 48-kDa variant. Subsequent autolytic cleavage events happen downstream of the CD and do not affect activity (Karim et al., 2010).
Whereas the flanking sequences do not resemble any other structurally or functionally analyzed protein, Kly18 is similar to mammalian matrix metalloproteinases (MMPs) (see Fig. 2 in (Karim et al., 2010)). These constitute a separate family within the “metzincin” clan of MPs (Visse and Nagase, 2003; Murphy and Nagase, 2008; Tallant et al., 2010b; Bode et al., 1993; Gomis-Rüth, 2003; Gomis-Rüth, 2009; Stöcker et al., 1995) and participate in both broad-spectrum turnover of extracellular-matrix components and selective limited proteolysis to activate or inactivate other proteins and enzymes. When dysregulated, their degrading potential leads to uncontrolled proteolysis and tissue destruction, apoptosis, and inflammation, as observed in periodontitis among other conditions (Overall and López-Otín, 2002). MMPs have mainly been studied in mammals, and 23 paralogs are present in humans (see http://degradome.uniovi.es/met.html#M10; (Puente et al., 2003; Quesada et al., 2009)). They are secreted as soluble or membrane-bound mosaic proteins comprising several inserts and domains, which include a signal peptide for secretion, an ~80-residue zymogenic pro-domain including a conserved cysteine engaged in latency maintenance (Rosenblum et al., 2007), a ~165-residue zinc- and calcium-dependent CD, a linker region, and a ~200-residue hemopexin-like C-terminal domain for collagen binding, pro-MMP activation, and dimerization (Gomis-Rüth, 2004; Maskos, 2005). In addition to mammals, MMPs have been studied (at least at the mRNA level) in several classes of animals and plants (Table 1). As to lower eukaryotes and prokaryotes, sequences similar to MMP CDs have been found in the genomes of fungi, viruses, archaea, and bacteria, but to our knowledge they have not been studied (Gomis-Rüth, 2003). At the structural level, only human, mouse, and pig MMP CDs have been analyzed so far (see (Tallant et al., 2010b) and http://pfam.sanger.ac.uk/family/PF00413#tabview=tab8).
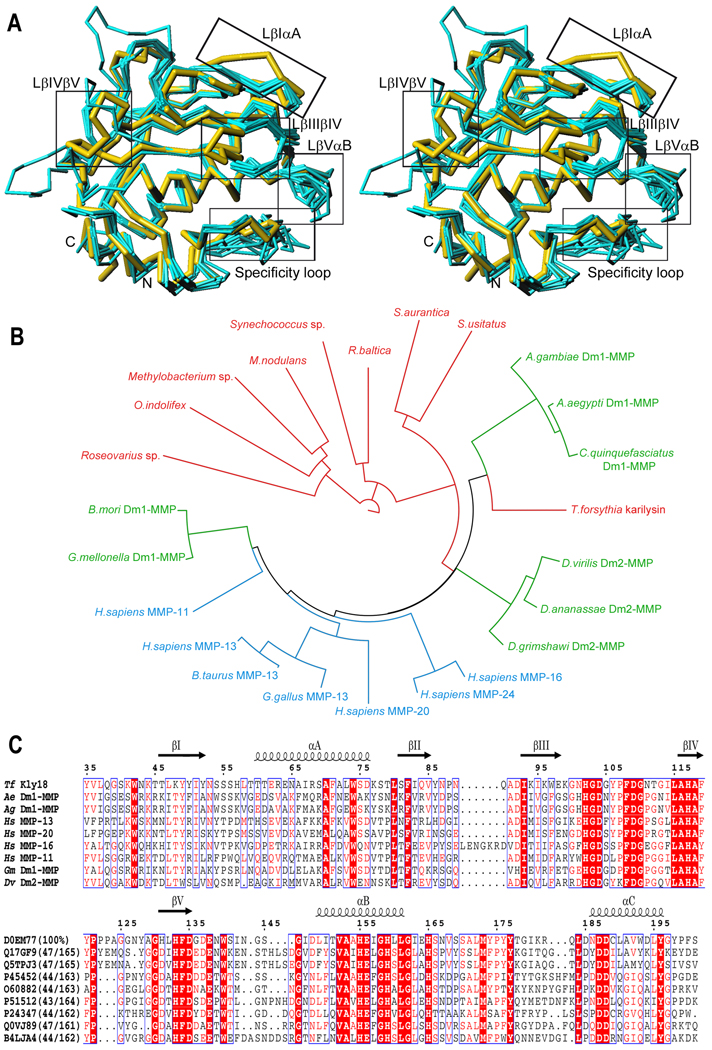
Figure 2. Structural, evolutionary, and sequence relationships of Kly18.
(A) Superpositionin in stereo of the Cα chain traces of Kly18 (yellow) and the catalytic domains of MMP-1, -3, -7, -8, -9 (w/o fibronectin-type II insertions), -10, -11, -12, -13, and -16 (all in cyan). Kly18 segments showing significant deviations from the common chain trace are framed and labeled. (B) 24 selected catalytic domain sequences of described and potential MMPs were subjected to phylogenetic analysis in relation with Kly18. Chosen sequences included: sequences of hitherto uncharacterized bacterial proteins of Synechococcus sp. (UniProt B4WJ70), Rhodopirellula baltica (Q7UJQ3), Stigmatella aurantiaca DW4/3-1 (Q098Y1), Solibacter usitatus strain Ellin6076 (Q029Q8), Methylobacterium nodulans strain ORS2060/LMG 21967 (B8IRC9), Methylobacterium sp. strain 4–46 (B0UK88), Oceanibulbus indolifex HEL-45 (A9EGF2), Roseovarius sp. 217 (A3W4J0); insect sequences (designated as either Dm1-MMP-like or Dm2-MMP-like on the basis of sequence comparison with the two Drosophila melanogaster proteins, the two only biochemically characterized insect forms (Llano et al., 2002; Llano et al., 2000)) of Galleria mellonella (wax moth; Q0VJ89), Drosophila grimshawi (Hawaian fruitfly; B4JVX6), Drosophila virilis (fruitfly; B4LJA4), Drosophila ananassae (fruitfly; B3MCI2), Bombyx mori (silk moth; B2CI49), Aedes aegypti (yellow fever mosquito; Q17FG9), Anopheles gambiae (African malaria mosquito; Q5TPJ3), and Culex quinquefasciatus (Southern house mosquito; B0W3F6); and vertebrate MMPs, i.e. Homo sapiens MMP-16 (MT3-MMP) (P51512), H. sapiens MMP-24 (MT5-MMP) (Q9Y5R2), H. sapiens MMP-20 (enamelysin) (O60882), H. sapiens MMP-13 (collagenase-3) (P45452), H. sapiens MMP-11 (stromelysin-3) (P24347), Gallus gallus (chicken) MMP-13 (O93363), and Bos taurus (bovine) MMP-13 (O77656). Sequences and tree branches are colored red (bacteria), green (winged insects), and blue (vertebrates). (C) Sequence alignment of Kly18 with selected catalytic domains depicted in (B). The abbreviations are: Tf, T. forsythia; Ae, A. aegypti; Ag, A. gambiae; Hs, H. sapiens; Gm, G. mellonella; and Dv, D. virilis. The UniProt sequence access code, as well as the percentage of sequence identity with Kly18 and the common sequence (both in parenthesis), are shown for each sequence in the lower sequence block. The numbering and the regular secondary structure elements (strands βI-βV and helices αA-αC) correspond to the Kly18 structure.
Table 1.
MMPs reported from non-mammalian organisms.
| Kingdom Animalia | |
| Amphibia | |
| African clawed frog Xenopus laevis (Yang et al., 1997); bullfrog Rana catesbeiana (Oofusa et al., 1994; Gross and Lapière, 1962); Japanese newt Cynops pyrrhogaster (Suzuki et al., 2001); Eastern newt Notophthalmus viridescens (Vinarsky et al., 2005); Mexican axolotl Ambystoma mexicanum (Yang and Bryant, 1994); green-striped burrowing frog Cyclorana albogutta (Hudson et al., 2007); Pacific tree frog Pseudacris regilla (Veldhoen et al., 2006). | |
| Reptilia | |
| Spectacled caiman Caiman crocodilus apaporiensis (Shintani et al., 2007); tiger keelback snake Rhabdophis tigrinus tigrinus (Komori et al., 2006). | |
| Aves | |
| Chicken Gallus gallus (Yang and Kurkinen, 1998); common canary Serinus canaria (Kim et al., 2008). | |
| Actinopterygii (fish) | |
| Rainbow trout Oncorhynchus mykiss (Saito et al., 2000); zebrafish Danio rerio (Wyatt et al., 2009); Atlantic salmon Salmo salar (Skugor et al., 2008); Japanese pufferfish Takifugu rubripes (Tsukamoto et al., 2007); Amur catfish Silurus asotus (Cho et al., 2002); medaka fish Oryzias latipes (Matsui et al., 2000); channel catfish Ictalurus punctatus (Yeh and Klesius, 2008). | |
| Hydrozoa | |
| Hydra Hydra vulgaris (Leontovich et al., 2000) | |
| Echinoidea | |
| Sea urchins Paracentrotus lividus and Hemicentrotus pulcherrimus (Gache et al., 2004). | |
| Secernentea (nematodes) | |
| Microbivorous roundworm Caenorhabditis elegans (Wada et al., 1998); parasitic nematode Gnathostoma spinigerum (Uparanukraw et al., 2001); soybean cyst nematode Heterodera glycines (Kovaleva et al., 2004); potato cyst nematode Globodera rostochiensis (Kovaleva et al., 2004). | |
| Insecta | |
| Fruitfly Drosophila melanogaster (Llano et al., 2002; Llano et al., 2000); wax moth Galleria mellonella (Altincicek and Vilcinskas, 2008); red flour beetle Tribolium castaneum (Knorr et al., 2009); red flour beetle Tribolium castaneum (Knorr et al., 2009). | |
| Bivalvia (mollusks) | |
| Eastern oyster Crassostrea virginica (M. Gomez-Chiarri, personal communication); abalone (sea snail) Haliotis diversicolor (Wang et al., 2008). | |
| Kingdom Plantae | |
| Soybean Glycine max (Ragster and Chrispeels, 1979); thale cress Arabidopsis thaliana (Maidment et al., 1999); cucumber Cucumis sativus (Delorme et al., 2000); sugarcane Saccharum spp. (Ramos and Selistre-de-Araujo, 2001); barrel medic Medicago truncatula (Combier et al., 2007); loblolly pine Pinus taeda (Ratnaparkhe et al., 2009); tobacco plant Nicotiana tabacum cv. BY-2 (Schiermeyer et al., 2009). | |
In order to explore the mechanisms of action of karilysin and to provide a high-resolution scaffold for the design of specific inhibitors against this bacterial virulence factor, we studied structure and function of Kly18, in the absence and presence of alkaline-earth metal ions. In addition, phylogenetic studies and evolutionary considerations rendered a plausible explanation for the origin of this MP.
RESULTS AND DISCUSSION
Endopeptidolytic activity of Kly18
The distinct (autolytic) variants of T. forsythia karilysin have been previously characterized for their proteolytic potential (Karim et al., 2010; Koziel et al., 2010). These studies revealed that the enzyme requires calcium for activity and thermal stability and that it cleaves bonds preferentially N-terminally of bulky hydrophobic residues. It efficiently degraded elastin, fibrinogen, and fibronectin, thus pointing to possible roles in periodontitis progression. In addition, karilysin inactivated the antimicrobial peptide, LL-37, which is a component of the immune system that targets bacteria and other microorganisms, thus further supporting a role of this MP in T. forsythia virulence. Here, we further studied the endopeptidolytic potential of Kly18 in front of two fluorogenic peptide substrates of seven and ten residues, respectively. These were developed to be cleaved by distinct mammalian MMPs, with kcat/Km values ranging between 1E4 and 6E5 M−1 s−1 (Fields, 2001; Nagase et al., 1994; Knight et al., 1992). These substrates were efficiently cleaved by Kly18, with kcat/Km values of 4,895 and 14,709 M−1 s−1 for the two substrates, respectively, thus indicating that the bacterial enzyme and its mammalian counterparts evince comparable substrate specificity and efficiency.
Structure of Kly18
The structure of Kly18 was solved both in the absence and in the presence of magnesium, hereafter “unbound” and “magnesium-bound” variants, respectively. These variants crystallized in distinct space groups and contained two and one protein molecules, respectively, in the crystallographic asymmetric unit. Calcium had been reported to be important for karilysin activity and thermal stability (Karim et al., 2010), and so this cation was added to the protein prior to crystallization to produce a calcium-bound form. However, in the crystal structure, which was obtained to significantly higher resolution than the unbound form (Table 2), only one partially-occupied magnesium site was found as inferred from electron density maps and distances to ligands, probably owing to the excess of the latter over calcium in the crystallization conditions (see Experimental procedures).
Table 2.
Crystallographic data.
| Dataset | Kly18 with Mg2+ | Kly18 without Mg2+ |
|---|---|---|
| Space group / cell constants (a, b, c, in Å; β in ° if ≠90) | I4 / 82.79, 85.79, 53.48 | C2 / 121.4, 53.1, 86.3, 134.6 |
| Wavelength (Å) | 0.9790 | 0.8726 |
| No. of measurements / unique reflections | 153,136 / 21,225 | 62,645 / 15,411 |
| Resolution range (Å) (outermost shell) a | 30.3 – 1.70 (1.79 – 1.70) | 45.3 – 2.40 (2.53 – 2.40) |
| Completeness (%) | 99.0 (95.9) | 99.3 (95.9) |
| Rmerge b | 0.034 (0.121) | 0.088 (0.511) |
| Rr.i.m. (= Rmeas) b / Rp.i.m. b | 0.036 (0.139) / 0.013 (0.065) | 0.101 (0.608) / 0.050 (0.322) |
| Average intensity (<[<I> / σ(<I>)]>) | 37.1 (10.8) | 14.8 (2.5) |
| B-Factor (Wilson) (Å2) / Average multiplicity | 18.9 / 7.2 (4.4) | 38.4 / 4.1 (3.4) |
| Resolution range used for refinement (Å) | ∞ – 1.70 | ∞ – 2.40 |
| No. of reflections used (test set) | 20,403 (820) | 14,703 (708) |
| Crystallographic Rfactor (free Rfactor) b | 0.158 (0.186) | 0.189 (0.259) |
| No. of protein atoms / solvent molecules / | 1,328 / 196 / | 2,644 / 115 / |
| ligands / ions | 1 tripeptide (AFT)/ 2 Zn2+, 1 Mg2+, 1 Cl− | 1 tris, 1 tetrapeptide (AFTS) / 2 Zn2+ |
| Rmsd from target values | ||
| bonds (Å) / angles (°) | 0.006 / 1.03 | 0.013 / 1.32 |
| bonded B-factors (main chain / side chain) (Å2) | 0.46 / 1.23 | 0.60 / 1.46 |
| Average B-factors for protein / peptide ligand atoms (Å2) | 16.6 / 19.6 | 33.3 / 50.2 |
| Main-chain conformational angle analysis c | ||
| Residues in favored regions / outliers / all residues | 160 / 0 / 166 | 315 / 0 / 332 |
Values in parentheses refer to the outermost resolution shell.
For definitions, see Table 1 in (Mallorquí-Fernández et al., 2008).
According to MOLPROBITY (Davis et al., 2007).
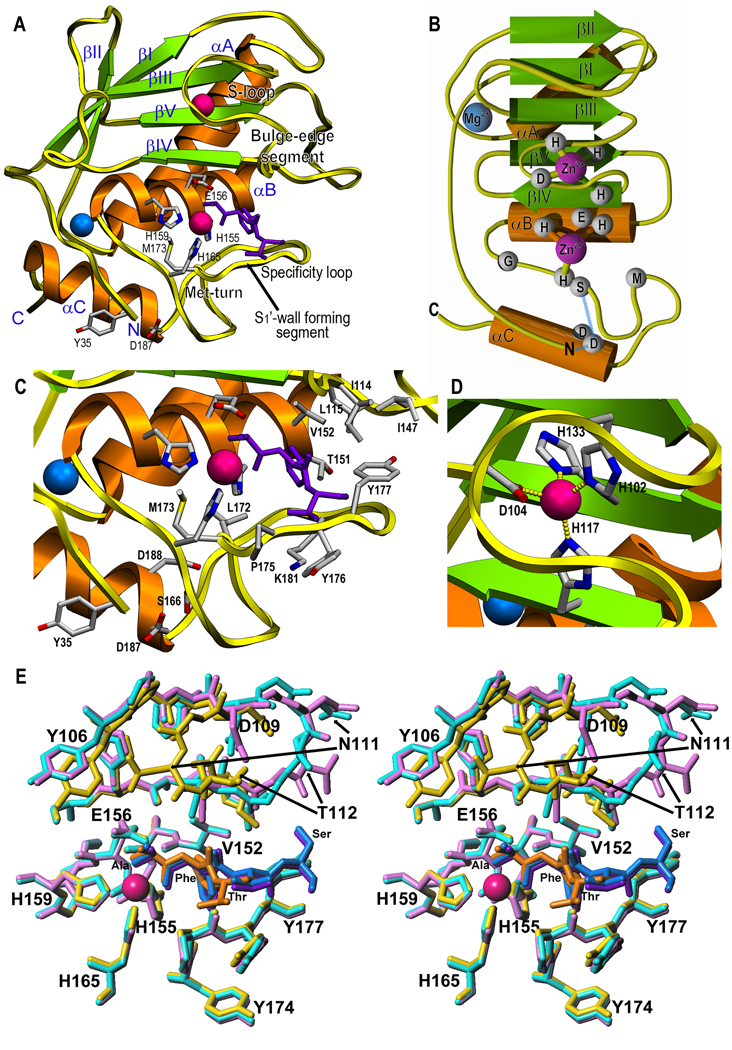
The Kly18 molecule is roughly spherical, with a diameter of ~40Å. Carved into its frontal surface, a shallow active-site cleft traverses the molecule left (non-primed side) to right (primed side) and divides it into a large N-terminal (NTS, Tyr35-Gly162) and a small C-terminal sub-domain (CTS; Ile163-Ser201; see Fig. 1a,b). Both the N- and the C-terminus are on the lower left of the molecule surface. The N-terminal α-amino group, in the front, is salt-bridged to the side chain of Asp187 within the “C-terminal helix” αC of CTS and thus anchored to the peptidase moiety. The NTS consists of a twisted five-stranded β-sheet (strands βI-βV), whose strands are all parallel to a substrate bound to the cleft except for outermost strand βIV, which forms the upper rim of the crevice and runs antiparallel. Two helices, the “backing helix” (αA) and the “active-site helix” (αB) nestle into the concave side of the sheet (Fig. 1a,c). On the convex side of the latter, three loops protrude from the molecular surface: the loop segment connecting βII and βIII (LβIIβIII), LβIVβV, and LβIIIβIV. The latter, termed “S-loop” due to its sinuous chain trace, encompasses a structural zinc-binding site, at which the cation is tetrahedrally co-ordinated by three histidines and an aspartate (His102, Asp104, His117, and His133; Fig. 1d). The final stretch of the S-loop is termed “bulge-edge segment” and it shapes the upper rim of the active-site cleft on its primed side (Fig. 1a). This loop opens out into the active-site helix, which includes the first half of a zinc-binding consensus sequence, HEXXHXXG/NXXH/D (amino-acid one-letter code; X for any residue), which is characteristic of MMPs in particular and metzincins in general (Tallant et al., 2010b; Gomis-Rüth, 2003; Gomis-Rüth, 2009). The first two histidines of the motif, His155 and His159, bind the catalytic zinc ion through their Nε2 atoms. Imbedded between them is the general-base/acid required for catalysis, Glu156. After this helix, the NTS ends at the glycine of the consensus sequence (Gly162), and the polypeptide enters the CTS. The latter residue allows for a sharp turn in the trajectory of the polypeptide to reach the third zinc ligand of the motif, His165. After this residue, Ser166 forms a hydrogen bond with Asp187, which also binds the N-terminus of the molecule (see above). Thereafter, four residues lead to a tight 1,4-turn of type I (Ala171-Tyr174), termed the “Met-turn” due to an invariable methionine found at the third turn position in all MMPs and metzincins (Tallant et al., 2010b; Gomis-Rüth, 2003; Gomis-Rüth, 2009; Tallant et al., 2010a). This turn is stabilized by a double main-chain interaction with the side chain of Asp188 within helix αC (Fig. 1c). Between the Met-turn and this helix, segment Pro175-Ty177, termed “S1’-wall forming segment,” shapes the lower rim of the cleft on its primed side. Thereafter the chain enters the “specificity loop” (Gly179-Gln183), which delimitates the back of the S1’ subsite or specificity pocket of Kly18 (Fig. 1a,c). At Asn186, the chain enters the C-terminal helix, αC, which leads into the C-terminus on the molecular surface.
Figure 1. Structure of Kly18.
(A) Richardson-type plot of Kly18 in the commonly accepted standard orientation for MPs, i.e. with the view into the active site cleft, which runs horizontally from left (non-primed side; it accommodates substrate residues upstream of the cleavage bond at subsites S1, S2, S3, etc. according to (Schechter and Berger, 1967)) to right (primed side; it binds downstream residues at S1’, S2’, S3’, etc.). The regular secondary structure elements are shown as green arrows (β-strands βI to βV) and orange ribbons (helices αA to αC) and labeled. Residues participating in catalytic-zinc binding, as well as the general base/acid glutamate, the Met-turn methionine, and the residues engaged in anchoring of the N-terminus to the protein moiety are displayed as sticks and labeled. The cleavage-product peptide bound to the primed-side of the active-site cleft is shown as deep purple sticks. The zinc and magnesium ions are shown as magenta and blue spheres, respectively. Chain segments characteristic of MMPs and discussed in the text are further pinpointed. Helix αA is inserted after strand βI and runs across the back surface of the molecule from top right to bottom left. Loop LβIαA is flexible at Ser54-His57 and was traced on the basis of weak electron density maps. Helix αB follows strand βV and runs horizontally across the center of the molecule, contributing to the back wall of the active-site cleft. The two aforementioned helices contact each other through the side chains of Ala70 (αA) and Ala154 (αB), which participate in a central hydrophobic core together with residues from the sheet. In addition, a significant part of the “bulge-edge segment” was likewise flexible in Kly18 and was traced on the basis of weak electron density maps at Asp109-Thr112. After the S-loop and strand βIV, LβIVβV includes three prolines in a row (Pro121-Pro123), which adopt a 1,4-turn structure and are followed by an alanine and two glycine residues. Thereafter, the polypeptide enters strand βV and a large loop, LβVαB, which contributes to cleft delimitation and substrate binding. (B) Topology scheme of Kly18 illustrating the regular secondary structure elements and cations with the same color code as in (A). Hydrogen bonds and salt bridges are displayed as blue dashed lines. (C) Close-up view of (A) showing the active site and residues framing the specificity pocket or S1’-subsite. (D) Close-up view of (A) centered on the structural zinc ion. (E) Superposition in stereo of the unbound (the two molecules in the asymmetric unit are shown as cyan and purple sticks, respectively) and magnesium-bound (yellow sticks) Kly18 structures with the corresponding cleavage-product peptides as blue/deep purple and orange sticks, respectively. Variation between the magnesium-bound and –unbound structures was found at LβIαA (slight variation) and within the bulge-edge segment of LβIIIβIV (large variation), which appears as essentially two conformers. The latter segment is folded inward in the magnesium-bound structure and outward in the unbound structure molecules, leading to a maximal displacement of ~13Å (measured at Asn111 Cα). As the bulge-edge segment contributes to substrate binding on the primed side of the cleft, an outswung conformation should enable binding of longer substrates. This resulted in that the peptide found in the cleft could be traced for one more residue, a tentative serine, in the unbound structure due to additional electron density.
Kly18 was obtained as a serendipitous complex with a peptide, which mimics a slightly shifted C-terminal cleavage product bound to the primed side of the cleft (Fig. 1a,c,e). The high-resolution of the electron density maps enabled us to assign a tripeptide of sequence Ala-Phe-Thr in the magnesium-bound structure. The N-terminus of this peptide is bound to both the catalytic zinc ion and the general-base/acid glutamate, and the carbonyl oxygen of the first residue also binds the zinc. This arrangement is strongly reminiscent of hydroxamate-based MP inhibitors, which bind zinc through the oxygen atoms of an NH(OH)-CO warhead (Betz et al., 1997). The downstream phenylalanine occupies the S1’-pocket, which is delimited by Leu115, Ile116, Thr151, Val152, His155, Leu172, Tyr177, and Lys181 (Fig. 1c). These residues confer an overall hydrophobic character to this rather large cavity and explain the preference of Kly18 for bulky hydrophobic residues in position P1’ of substrates (Karim et al., 2010).
A magnesium site was found on the back of the molecule, close to the C-terminus. The ion is co-ordinated by Ser75 O of αA and Ser78 O of LαAβII. Three solvent molecules defined by proper electron density and a position occupied by a bulk solvent complete an octahedral ligand sphere characteristic for alkaline-earth metal cations. Superposition of the magnesium-bound structure over the two crystallographically-independent molecules of the unbound structure revealed that they are practically equivalent, with pairwise rmsd values of ~0.5Å (Fig. 1e). No significant differences were observed at the magnesium site apart from the lack of the cation. In contrast, variations were observed within two flexible segments, LβIαA and the bulge-edge segment (see Fig. 1e and its legend). However, as the magnesium site is far away from the active-site cleft and the latter segments (Fig. 1a,c), there is no evident structural explanation for the these differences or for the requirement of alkaline-earth ions for activity and stability (Karim et al., 2010).
Structural similarities
As expectable, structural similarity searches identified mammalian MMP CDs as the closest relatives of Kly18 (Table 3). Structurally-analyzed MMP CDs comprise only mammalian representatives of MMP-1 to MMP-3, MMP-7 to MMP-14, MMP-16, and MMP-20, which all span 160–173 residues (Tallant et al., 2010b; Maskos, 2005; Rao, 2005). Other MPs were identified as more distant relatives, among them serralysins, snapalysin, astacins, adamalysins/ADAMs, ulilysin, and leishmanolysin. This is consistent with the general similarity observed across these families, which justifies that they are grouped in the metzincin clan of MPs (Gomis-Rüth, 2003; Gomis-Rüth, 2009). Superimposition of the structures of Kly18 onto several MMPs revealed similar chain traces for the overall CDs, including all regular secondary structure elements, the catalytic residues, and the Met-turn (Fig. 2a). No disulfide bonds are present in any structure. In addition, Kly18 possesses most of the structural details characteristic of MMP CDs that distinguish them from other metzincins: (i) the residue downstream of the third zinc-binding histidine is always a serine, and it hydrogen bonds the aspartate residue in helix αC that binds the N-terminus in physiologically-relevant “superactive” MMP variants (Reinemer et al., 1994; Tallant et al., 2010b); (ii) after this conserved serine, the downstream chain trace, which leads to the Met-turn and is characteristic for MMPs, and the latter turn itself, are perfectly superimposable; (iii) the Met-turn is stabilized through hydrogen bonds of its main chain with a conserved aspartate residue immediately downstream of the aforementioned one within helix αC; (iv) the characteristic loop structures found in Kly18, the S-loop, the bulge-edge segment, the S1’-wall forming segment—which includes the sequence Pro175-Tyr-Tyr177—, and the specificity loop, are essential hallmarks of mammalian MMP CDs (Tallant et al., 2010b); (v) the S1’ pocket is hydrophobic and designed to accommodate bulky hydrophobic side chains; (vi) in addition, Kly18 accomplishes with the structural requirements of another characteristic feature of the active-site cleft of MMPs, i.e. the selectivity for proline in position P3 of a substrate, through an S3 pocket framed by His117 and Phe119 of strand βIV and Tyr106 of the S-loop; (vii) the first part of the S-loop includes an essential structural zinc site (Williams and Murray, 1994) formed by residues contained in conserved sequence stretches equivalent to Pro107-His-Asp-Gly-(Xxx)3-Phe-Asp-Gly110 of Kly18 (Fig. 2c); (viii) finally, the sequences found in Kly18 within the upper-rim strand βIV, Leu115-Ala-His-Ala-Phe-Xxx-Pro121, and within strand βV, His133-Phe-Asp135, are likewise conserved among MMPs.
Table 3.
Structural similarities of Kly18 with metzincins according to DALI (Holm and Sander, 1998).
| Metzincin family | Z-score | rmsd (Å) | Common sequence stretch (residues) |
Sequence identity (%) |
|---|---|---|---|---|
| MMPs | 20.5–25.6 | 1.3–1.8 | 147–156 | 42–46 |
| Serralysins | 14.9–16.3 | 2.3–2.8 | 132–158 | 20–24 |
| Snapalysin | 13.5 | 2.3 | 119 | 20 |
| Astacins | 11.5–11.8 | 2.4–2.7 | 123–131 | 14–20 |
| Adamalysins/ADAMs | 9.8–11.4 | 2.6–3.4 | 129–141 | 13–19 |
| Ulilysin | 9.1–9.2 | 2.9–3.1 | 135–138 | 12–13 |
| Leishmanolysin | 8.7 | 3.2 | 129 | 17 |
There are, however, minor deviations from an average MMP structure in Kly18, but they are found mostly in regions that are also variable among mammalian structures (Fig. 2a). Kly18 does not have any of the three characteristic calcium-binding sites featured by the bulge-edge segment plus the first part of LβVαB, by LβIVβV, and by LβIαA/LβVαB, respectively, in mammalian MMPs. This is particularly noteworthy because Kly18 has most of the conserved residues participating in the first site (Fig. 2c), and this could explain the flexibility of the bulge-edge segment in the bacterial enzyme (see above and legend to Fig. 1). Here it must be mentioned that mammalian MMPs are variable as to these calcium sites: they may comprise just the bulge-edge-segment site (MMP-11), two sites (MMP-7, MMP-8, MMP-16, and MMP-20) or three sites (MMP-3, MMP-9 and MMP-10). Some proteins have even been solved with either two or three calcium ions (MMP-1, MMP-2, MMP-12, and MMP-13). Further differences in length and conformation between Kly18 and mammalian MMP CDs are found within the specificity loop. This element, however, also deviates widely among the latter structures and it results in hydrophobic S1’ pockets of varying depth, i.e. with the capacity to allocate P1’ chains of disparate size. The same scenario is observed at LβVαB, which is highly variable among MMPs and contributes to substrate specificity. LβIαA also displays a slight variation, probably owing to the flexible nature of this segment in Kly18. Finally, LβIVβV is unique to Kly18 due to a three-residue insertion, which gives rise to a stretch of sequence Pro121-Pro2-Ala-Gly-Gly126 (Fig. 2c).
In the light of all these findings, we conclude that Kly18 is a true MMP CD, which like all its mammalian counterparts shows small hallmark deviations from the common average structure.
Phylogenetic and sequence similarity analyses of Kly18
Previous and current sequence similarity searches revealed that Kly18 is close to animal MMP sequences despite belonging to a bacterium from the phylum Bacteroidetes (Karim et al., 2010). Highest sequence identities (>40%) and shortest evolutionary distances are observed with respect to potential proteins from mosquitoes (order Diptera, family Culicidae) within the superorder endopterigota (Fig. 2b,c). The former include the transmitting insect vectors of the parasite that causes malaria (Anopheles gambiae); of the viruses that cause dengue fever, Chikungunya and yellow fever (Aedes aegypti); and of West Nile virus (Culex quinquefasciatus). All these mosquitoes have intimate contact with mammalian hosts, on whose blood they feed, mostly in developing countries, regions that also present the highest incidence of odontopathogenic bacterial infections (Axelsson et al., 2002). Next in evolutionary distance are vertebrate MMPs (e.g. MMP-11, -13, -16, and -20), sequences from other endopterigota including fruitflies (order Diptera, family Drosophilidae), and moths and butterflies (order Lepidoptera). Clearly farthest away are several putative bacterial proteins from cyanobacteria, planctomycetes, acidobacteria, and proteobacteria (Fig. 2b).
Evolutionary implications
Horizontal or lateral gene transfer (HGT) entails shuffling of genes—xenologs—between different species and it contributes to the diversification of life by complementing the Darwinian tree-based pathway that is based on reproductive isolation between species (Koonin et al., 2001; Gogarten, 1994). Such xenologs are defined as homologs of genes that have evolved within a donor species before being transferred to a receptor, i.e. they have not been inherited from the last universal common ancestor and are thus incongruent with the species tree. HGT is only successful if the co-opted gene becomes incorporated into the genome of the acceptor and if selective advantage is conferred. Mechanisms of HGT include transformation, transduction, and conjugation of nucleic acid chains through several pathways that may include viral and bacteriophage vectors. The direction of HGT probably followed the “out of Africa” principle, which pinpoints the most crowded taxon as the most probable source (Cavalli-Sforza, 1998; Koonin et al., 2001). HGT is widespread and well documented within and between bacteria and archaea, as exemplified by the sharing of antibiotic resistance plasmids, often across species barriers (Macrina and Archer, 1993). In contrast, the transfer from bacteria to eukaryotic cells is less common, and examples include the interaction between Agrobacterium tumefaciens and plant cells, which induces Crown-Gall disease in the latter (Sprague, 1991; Gomis-Rüth et al., 2004). Finally, HGT from eukaryotes to bacteria is of particular importance given the possible roles of transferred genes in bacterial pathogenicity (Koonin et al., 2001; Ochman et al., 2000). This transfer is very rare and restricted to few cases proposed on the basis of phylogenetic and comparative genome analyses—“lateral genomics” (Koonin et al., 2001; Doolittle, 1999). Direct proof of such gene shuffling is unavailable for there is no footprint of such evolutionary events. For this reason, additional evidence as provided by structural relatedness may help to uncover new potential cases of xenology.
Karilysin is a unique protein of T. forsythia and its natural habitat is the gingival crevice of humans, where it contributes to periodontal disease. Its CD, Kly18, shows closest structural similarity and comparable S1’-subsite specificity to mammalian MMPs and shorter evolutionary distance to MMP sequences of winged insect and mammals than to other bacterial yet uncharacterized sequences. The MMP family is currently described only in animals and plants, with 23 paralogs found in humans. The existence of sequences in mosquitoes related to the only two insect forms present in Drosophila melagonaster, Dm1-MMP and Dm2-MMP (Llano et al., 2002; Llano et al., 2000), and to sequences found in other fruitflies, moths and butterflies, strongly suggests that they likely correspond to expressed functional proteins and that MMPs within winged insects result from Darwinian evolution, as is the case for the presence of MMPs within vertebrates. Coincidently, winged-insect MMPs follow the general domain architecture of mammalian MMPs, which is found in all MMPs studied, except for a protein from the nematode Gnathostoma spinigerum, and plant MMPs. Notwithstanding, the latter possess at least a pro-domain with a characteristic cysteine possibly participating in latency as in animal forms. In contrast, karilysin has only a short pro-peptide between the signal peptide and the beginning of the mature catalytic domain and a C-terminal domain, and neither bears any relationship to any other characterized protein. The pro-peptide lacks the characteristic cysteine residue (Springman et al., 1990; Van Wart and Birkedal-Hansen, 1990; Gonzales et al., 2004; Rosenblum et al., 2007), but contains an aspartate that could contribute to latency maintenance as in proastacin and profragilysin-3 (Goulas et al., 2010; Guevara et al., 2010). In addition, no gene potentially encoding a Kly18-relative was found either in the other two red complex constituents, P. gingivalis and T. denticola, or in any bacterium of the phylum Bacteroidetes. Taken together, these facts strongly suggest that Kly18 is an MMP xenolog that has been co-opted from an external source. Likely donor candidates are mammals or mosquitoes feeding on human blood, due to the intimate coexistence between T. forsythia and the human blood-irrigated gingival crevice. This hypothesis is further backed by the existence of a sequence within the genome of invertebrate iridescent virus 3 (UniProt Q196W5), which infects mosquitoes of the genus Aedes and Culex, that shows 40% sequence identity to Kly18 within 146 common residues. This would suggest participation of a viral vector in the HGT. Subsequently, Kly18 would have evolved in a bacterial environment, where it was furnished with unique flanking domains to give rise to full-length karilysin. This proposed example of HGT entailing the shuffling of a metazoan MMP to a human pathogenic bacterium, which involves distinct domains of life, is both a short-circuit of Darwinian evolution and a testimony to the compatibility of proteins and cellular mechanisms despite evolutionary divergence over billions of years.
EXPERIMENTAL PROCEDURES
Protein production and purification and endopeptidolytic assays
Karilysin catalytic domain (Kly18) was obtained by autolysis of full-length karilysin, which was produced by recombinant overexpression as a fusion construct with glutathione-S-transferase in Escherichia coli BL21 cells, as described elsewhere (Karim et al., 2010). Activity of Kly18 in front of the fluorogenic peptide substrates, Mca-Arg-Pro-Lys-Pro-Val-Glu-Nva-Trp-Arg-Lys(Dnp)-NH2 (Mca, 7-methoxycoumarin-4-acetate; Nva, norvaline; Dnp, dinitrophenyl) and Mca-Pro-Leu-Gly-Leu-Dap(Dnp)-Ala-Arg-NH2 (Dap, L-diaminopropionyl; both from Bachem, with λEX=328nm and λEM=393nm) was assayed at 37°C in 100mM Tris·HCl, 5mM calcium chloride, pH8.0 by using an enzyme concentration of 25nM for the former substrate and 50nM for the latter substrate. Fluorescence was recorded for 12h in a fluorimetric plate reader (Tecan Infinite M200). Substrate concentrations varied between 1 and 8µM to avoid quenching effects due to high substrate concentrations (Fields, 2001; Nagase et al., 1994; Knight et al., 1992). The kcat/Km parameter was estimated using the minimum concentration that permitted to reliably determine the associated velocity according to equation, [E]min = vmin / (kcat/Km × [S]), which is a valid approximation when [S]≪Km (Knight et al., 1992).
Crystallization and structure analysis
Crystallization assays were performed by the sitting-drop vapor diffusion method. Reservoir solutions were prepared by a Tecan robot and crystallization drops were dispensed on 96x2-well MRC plates (Innovadyne) by a Cartesian (Genomic Solutions) nanodrop robot at the High-Throughput Crystallography Platform at Barcelona Science Park. Crystals suitable for structure analysis were obtained for unbound Kly18 in a Bruker steady-temperature crystal farm at 20°C from drops containing 100nl of protein solution (at 9mg ml−1 in 5mM Tris·HCl, pH8.0, 0.02% sodium azide) and 100nl of 45% 2-methyl-2,4-pentanediol, 0.2M ammonium acetate, 0.1M Tris·HCl, pH8.5 as reservoir solution. Crystals of magnesium-bound Kly18 were obtained at 4°C by mixing protein solution preincubated with calcium (at 7.3mg ml−1 in 5mM Tris·HCl, pH8.0, 5mM calcium chloride, 0.02% sodium azide) and 14% sodium polyacrylate 5100, 20mM magnesium chloride, 0.1M Hepes, pH7.5 in a volume ratio of 1:3. The latter conditions were scaled up to the microliter range with 24-well Cryschem crystallization dishes (Hampton Research). Crystals were cryo-protected with 50% 2-methyl-2,4-pentanediol, 15% glycerol, 0.2M ammonium acetate, 0.1M Tris·HCl, pH8.5 (unbound Kly18) and 30% sodium polyacrylate 5100, 20% glycerol, 0.02M magnesium chloride, 0.1M Hepes, pH7.5 (magnesium-bound Kly18). Complete diffraction datasets were collected for unbound and magnesium-bound Kly18 from liquid-N2 flash-cryo-cooled crystals at 100K (provided by an Oxford Cryosystems 700 series cryostream) at beam lines ID23-2 (equipped with a MarMosaic 225 CCD detector) and ID29 (with an ADSC 315R CCD detector), respectively, of the European Synchrotron Radiation Facility (ESRF, Grenoble, France) within the Block Allocation Group "BAG Barcelona.” Crystals were either monoclinic (unbound Kly18) or tetragonal (magnesium-bound Kly18), with two or one molecules per asymmetric unit, respectively. Diffraction data were integrated, scaled, merged, and reduced with programs XDS (Kabsch, 2001) and SCALA (Evans, 2006) within the CCP4 suite of programs (CCP4, 1994) (see Table 2).
The structure of unbound Kly18 was solved by Patterson search with program PHASER (McCoy et al., 2007) and the coordinates of human MMP-8 (Protein Data Bank access code (PDB) 1MNC) as searching model. These calculations rendered two unique solutions at, respectively, 328.1, 106.1, 166.2, −0.27, 1.00, −0.55; and 237.5, 143.7, 46.9, −0.28, 0.99, 0.01 (Eulerian angles α, β, and γ, and x, y, z in fractional cell coordinates). The global value of log-likelihood gain was 384. The correspondingly rotated and translated searching models were used to compute initial phases, which were subjected to a run with program ARP/wARP (Perrakis et al., 2001). This process significantly improved the electron density map and enabled straightforward tracing of the entire polypeptide chain on a Silicon Graphics Octane2 workstation with TURBO-Frodo program (Carranza et al., 1999). Subsequent crystallographic refinement with REFMAC5 (Murshudov et al., 1997), which included TLS refinement, alternated with manual model building until completion of the model. The latter comprised residues Tyr35-Phe200 (numbering according to UniProt database entry D0EM77) and two zinc ions for each of the two chains, A and B. In addition, 115 solvent molecules, a tetrapeptide of tentative sequence Ala-Phe-Thr-Ser, and a Tris molecule from the crystallization conditions were assigned. The structure of magnesium-bound Kly18 was solved with program PHASER by using the protein chain coordinates of Kly18 as searching model. A single solution was obtained at 43.0, 89.6, 89.8, 0.01, 0.01, 0.01 with a global log-likelihood gain of 2,135. Phase improvement, model building, and refinement proceeded as stated above. The final model spanned residues Tyr35-Ser201, two zinc ions, a tripeptide of sequence Ala-Phe-Thr, tentatively assigned chloride and magnesium ions, and 196 solvent molecules (see Table 2).
Phylogenetic analysis
Selected MMP sequences obtained from sequence similarity searches were pairwise aligned with Kly18 to delimit the catalytic domains of the former with program MULTALIN (Corpet, 1988). These fragments were thereafter aligned altogether with Kly18 with MULTALIN, which computes parameters for a phylogenetic tree in rfd format. The latter was manually converted to dnd format and plotted as a circular tree with PHYLIP DRAWGRAM at http://mobyle.pasteur.fr/cgi-bin/portal.py?form=drawgram.
Miscellaneous
Figures were prepared with SETOR (Evans, 1993), GRASP (Nicholls et al., 1993), and TURBO-Frodo. Structure similarities were determined with DALI (Holm et al., 2006), and sequence alignments were performed with MULTALIN. Experimental model validation was performed with MOLPROBITY (Davis et al., 2007) and the WHATCHECK routine of WHATIF (Vriend, 1990). The final coordinates of unbound and magnesium-bound Kly18 are available from the PDB at www.pdb.org (access codes 2XS3 and 2XS4).
ACKNOWLEDGMENTS
We are grateful to the Automated Crystallography Platform (PAC) at PCB for assistance during crystallization experiments and to Robin Rycroft for helpful suggestions to the manuscript. This study was in part supported by grants from European, Spanish, Polish, and Catalan public agencies (FP7-HEALTH-F3-2009-223101 “AntiPathoGN”, FP7-HEALTH-2010-261460 “Gums&Joints”, BIO2008-04080-E, BIO2009-10334, CSD2006-00015, PSE-010000-2009-8, 2009SGR1036, and MNiSzW 1642/B/P01/2008/35) and by grant DE 09761 from the National Institutes of Health, USA. The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a beneficent of the structural funds from the European Union (grant No: POIG.02.01.00-12-064/08 – “Molecular biotechnology for health”). We acknowledge the help provided by EMBL and ESRF synchrotron local contacts. Funding was provided by ESRF for data collection.
ABBREVIATIONS
The following abbreviations are used:
- CD
catalytic domain
- HGT
horizontal gene transfer
- Kly18
Tannerella forsythia karilysin catalytic domain
- CTS
C-terminal sub-domain
- Dap
L-diaminopropionyl
- Dnp
dinitrophenyl
- Mca
7-methoxycoumarin-4-acetate
- MMP
matrix metalloproteinases
- MP
metallopeptidase
- NTS
N-terminal sub-domain
- Nva
norvaline
- PDB
Protein Data Bank at www.pdb.org
Footnotes
The authors state they have no competing financial interest.
REFERENCES
- Altincicek B, Vilcinskas A. Identification of a lepidopteran matrix metalloproteinase with dual roles in metamorphosis and innate immunity. Dev Comp Immunol. 2008;32:400–409. doi: 10.1016/j.dci.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Axelsson P, Albandar JM, Rams TE. Prevention and control of periodontal diseases in developing and industrialized nations. Periodontol 2000. 2002;29:235–246. doi: 10.1034/j.1600-0757.2002.290112.x. [DOI] [PubMed] [Google Scholar]
- Behle JH, Papapanou PN. Periodontal infections and atherosclerotic vascular disease: an update. Int Dent J. 2006;56:256–262. doi: 10.1111/j.1875-595x.2006.tb00110.x. [DOI] [PubMed] [Google Scholar]
- Betz M, Huxley P, Davies SJ, Mushtaq Y, Pieper M, Tschesche H, et al. 1.8-Å crystal structure of the catalytic domain of human neutrophil collagenase (matrix metalloproteinase-8) complexed with a peptidomimetic hydroxamate primed-side inhibitor with a distinct selectivity profile. Eur J Biochem. 1997;247:356–363. doi: 10.1111/j.1432-1033.1997.00356.x. [DOI] [PubMed] [Google Scholar]
- Bode W, Gomis-Rüth FX, Stöcker W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ´metzincins´. FEBS Lett. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-i. [DOI] [PubMed] [Google Scholar]
- Carranza C, Inisan A-G, Mouthuy-Knoops E, Cambillau C, Roussel A. AFMB Activity Report 1996–1999. Marseille: CNRS-UPR; 1999. Turbo-Frodo; pp. 89–90. CNRS-UPR 9039. [Google Scholar]
- Carrijo LC, Andrich F, de Lima ME, Cordeiro MN, Richardson M, Figueiredo SG. Biological properties of the venom from the scorpionfish (Scorpaena plumieri) and purification of a gelatinolytic protease. Toxicon. 2005;45:843–850. doi: 10.1016/j.toxicon.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL. The DNA revolution in population genetics. Trends Genet. 1998;14:60–65. doi: 10.1016/s0168-9525(97)01327-9. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite : programs for protein crystallography. Acta Crystallogr sect D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Combier JP, Vernie T, de Billy F, El Yahyaoui F, Mathis R, Gamas P. The MtMMPL1 early nodulin is a novel member of the matrix metalloendoproteinase family with a role in Medicago truncatula infection by Sinorhizobium meliloti. Plant Physiol. 2007;144:703–716. doi: 10.1104/pp.106.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucl Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler CW, Kalmar JR, Genco CA. Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol. 1995;3:45–51. doi: 10.1016/s0966-842x(00)88874-5. [DOI] [PubMed] [Google Scholar]
- Cho JH, Park IY, Kim MS, Kim SC. Matrix metalloproteinase 2 is involved in the regulation of the antimicrobial peptide parasin I production in catfish skin mucosa. FEBS Lett. 2002;531:459–463. doi: 10.1016/s0014-5793(02)03584-6. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucl Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme VGR, McCabe PF, Kim D-J, Leaver CJ. A matrix metalloproteinase gene is expressed at the boundary of senescence and programmed cell death in cucumber. Plant Physiol. 2000;123:917–927. doi: 10.1104/pp.123.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. Lateral genomics. Trends Cell Biol. 1999;9:M5–M8. [PubMed] [Google Scholar]
- Eley BM, Cox SW. Proteolytic and hydrolytic enzymes from putative periodontal pathogens: characterization, molecular genetics, effects on host defenses and tissues and detection in gingival crevice fluid. Periodontol 2000. 2003;31:105–124. doi: 10.1034/j.1600-0757.2003.03107.x. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr sect D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Evans SV. SETOR : hardware lighted three-dimensional solid model representations of macromolecules. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- Fields GB. Using fluorogenic peptide substrates to assay matrix metalloproteinases. In: Clark IM, editor. Matrix metalloproteinase protocols. Totowa, NJ: Humana Press Inc.; 2001. pp. 495–518. [Google Scholar]
- Fox CH. New considerations in the prevalence of periodontal disease. Curr Opin Dent. 1992;2:5–11. [PubMed] [Google Scholar]
- Fox JW, Serrano SM. Timeline of key events in snake venom metalloproteinase research. J Proteomics. 2009;72:200–209. doi: 10.1016/j.jprot.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Gache C, Lepage T, Croce J. 151. Envelysin. In: Barrett AJ, Rawlings ND, Woessner JF Jr, editors. Handbook of proteolytic enzymes. London: Elsevier Academic Press; 2004. pp. 575–578. [Google Scholar]
- Gogarten JP. Which is the most conserved group of proteins? Homology-orthology, paralogy, xenology, and the fusion of independent lineages. J Mol Evol. 1994;39:541–543. doi: 10.1007/BF00173425. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX. Structural aspects of the metzincin clan of metalloendopeptidases. Mol Biotech. 2003;24:157–202. doi: 10.1385/MB:24:2:157. [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX. Hemopexin domains. In: Messerschmidt A, Bode W, Cygler M, editors. Handbook of metalloproteins. Chichester (UK): John Wiley & Sons, Ltd.; 2004. pp. 631–646. [Google Scholar]
- Gomis-Rüth FX. Catalytic domain architecture of metzincin metalloproteases. J Biol Chem. 2009:15353–15357. doi: 10.1074/jbc.R800069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Solà M, de la Cruz F, Coll M. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des. 2004;10:1551–1565. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- Gonzales PE, Solomon A, Miller AB, Leesnitzer MA, Sagi I, Milla ME. Inhibition of the tumor necrosis factor-a-converting enzyme by its pro domain. J Biol Chem. 2004;279:31638–31645. doi: 10.1074/jbc.M401311200. [DOI] [PubMed] [Google Scholar]
- Goulas T, Arolas JL, Gomis-Rüth FX. Structure and latency regulation of the Bacteroides fragilis enterotoxin, a potential adamalysin/ADAM xenolog. Submitted. 2010 [Google Scholar]
- Gross J, Lapière CM. Collagenolytic activity in amphibian tissues: a tissue culture assay. Proc Natl Acad Sci USA. 1962;48:1014–1022. doi: 10.1073/pnas.48.6.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara T, Yiallouros I, Kappelhoff R, Bissdorf S, Stöcker W, Gomis-Rüth FX. Proenzyme structure and activation of astacin metallopeptidase. J Biol Chem. 2010;285:13958–13965. doi: 10.1074/jbc.M109.097436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffajee AD, Socransky SS, Gunsolley JC. Systemic anti-infective periodontal therapy. A systematic review. Ann Periodontol. 2003;8:115–181. doi: 10.1902/annals.2003.8.1.115. [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Curr Protoc Bioinformatics. 2006;Chapter 5 doi: 10.1002/0471250953.bi0505s14. Unit 5 5. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucl Acid Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson NJ, Harper GS, Allingham PG, Franklin CE, Barris W, Lehnert SA. Skeletal muscle extracellular matrix remodelling after aestivation in the green striped burrowing frog, Cyclorana alboguttata. Comp Biochem Physiol A Mol Integr Physiol. 2007;146:440–445. doi: 10.1016/j.cbpa.2006.12.023. [DOI] [PubMed] [Google Scholar]
- Jordan RC. Diagnosis of periodontal manifestations of systemic diseases. Periodontol 2000. 2004;34:217–229. doi: 10.1046/j.0906-6713.2002.003433.x. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Chapter 25.2.9: XDS. In: Rossmann MG, Arnold E, editors. International Tables for Crystallography Volume F: Crystallography of Biological Macromolecules. Dordrecht (The Netherlands): Kluwer Academic Publishers (for The International Union of Crystallography); 2001. pp. 730–734. [Google Scholar]
- Karim AY, Kulczycka M, Kantyka T, Dubin G, Jabaiah A, Daugherty PS, et al. A novel matrix metalloprotease-like enzyme (karilysin) of the periodontal pathogen Tannerella forsythia ATCC 43037. Biol Chem. 2010;391:105–117. doi: 10.1515/BC.2010.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Lilliehook C, Roides B, Chen Z, Chang M, Mobashery S, Goldman SA. Testosterone-induced matrix metalloproteinase activation is a checkpoint for neuronal addition to the adult songbird brain. J Neurosci. 2008;28:208–216. doi: 10.1523/JNEUROSCI.3674-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King LE., Jr Common ground?: Tetracyclines, matrix metalloproteinases, pustular dermatoses, and loxoscelism. J Invest Dermatol. 2007;127:1284–1286. doi: 10.1038/sj.jid.5700724. [DOI] [PubMed] [Google Scholar]
- Knight CG, Willenbrock F, Murphy G. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
- Knorr E, Schmidtberg H, Vilcinskas A, Altincicek B. MMPs regulate both development and immunity in the tribolium model insect. PLoS One. 2009;4:e4751. doi: 10.1371/journal.pone.0004751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori K, Konishi M, Maruta Y, Toriba M, Sakai A, Matsuda A, et al. Characterization of a novel metalloproteinase in Duvernoy's gland of Rhabdophis tigrinus tigrinus. J Toxicol Sci. 2006;31:157–168. doi: 10.2131/jts.31.157. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaleva ES, Masler EP, Skantar AM, Chitwood DJ. Novel matrix metalloproteinase from the cyst nematodes Heterodera glycines and Globodera rostochiensis. Mol Biochem Parasitol. 2004;136:109–112. doi: 10.1016/j.molbiopara.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Koziel J, Karim AY, Przybyszewska K, Ksiazek M, Rapala-Kozik M, Nguyen KA, Potempa J. Proteolytic inactivation of LL-37 by karilysin, a novel virulence mechanism of Tannerella forsythia. J Innate Immun. 2010;2:288–293. doi: 10.1159/000281881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz MS. Are bacterial proteases important virulence factors? J Periodontal Res. 1997;32:126–132. doi: 10.1111/j.1600-0765.1997.tb01393.x. [DOI] [PubMed] [Google Scholar]
- Leontovich AA, Zhang J, Shimokawa K, Nagase H, Sarras MP., Jr A novel hydra matrix metalloproteinase (HMMP) functions in extracellular matrix degradation, morphogenesis and the maintenance of differentiated cells in the foot process. Development. 2000;127:907–920. doi: 10.1242/dev.127.4.907. [DOI] [PubMed] [Google Scholar]
- Llano E, Adam G, Pendas AM, Quesada V, Sánchez LM, Santamaria I, et al. Structural and enzymatic characterization of Drosophila Dm2-MMP, a membrane-bound matrix metalloproteinase with tissue-specific expression. J Biol Chem. 2002;277:23321–23329. doi: 10.1074/jbc.M200121200. [DOI] [PubMed] [Google Scholar]
- Llano E, Pendas AM, Aza-Blanc P, Kornberg TB, López-Otín C. Dm1-MMP, a matrix metalloproteinase from Drosophila with a potential role in extracellular matrix remodeling during neural development. J Biol Chem. 2000;275:35978–35985. doi: 10.1074/jbc.M006045200. [DOI] [PubMed] [Google Scholar]
- Macrina FL, Archer GL. Conjugation and broad host range plasmids in streptococci and staphylococci. In: Clewell DB, editor. Bacterial conjugation. New York: Plenum Press; 1993. pp. 313–329. [Google Scholar]
- Maidment JM, Moore D, Murphy GP, Murphy G, Clark IM. Matrix metalloproteinase homologues from Arabidopsis thaliana. Expression and activity. J Biol Chem. 1999;274:34706–34710. doi: 10.1074/jbc.274.49.34706. [DOI] [PubMed] [Google Scholar]
- Mallorquí-Fernández N, Manandhar SP, Mallorquí-Fernández G, Usón I, Wawrzonek K, Kantyka T, et al. A new autocatalytic activation mechanism for cysteine proteases revealed by Prevotella intermedia interpain A. J Biol Chem. 2008;283:2871–2882. doi: 10.1074/jbc.M708481200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskos K. Crystal structures of MMPs in complex with physiological and pharmacological inhibitors. Biochimie. 2005;87:249–263. doi: 10.1016/j.biochi.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Matsui H, Ogiwara K, Ohkura R, Yamashita M, Takahashi T. Expression of gelatinases A and B in the ovary of the medaka fish Oryzias latipes. Eur J Biochem. 2000;267:4658–4668. doi: 10.1046/j.1432-1327.2000.01513.x. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittl PR, Grütter MG. Opportunities for structure-based design of protease-directed drugs. Curr Opin Struct Biol. 2006;16:769–775. doi: 10.1016/j.sbi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Miyoshi S, Shinoda S. Microbial metalloproteases and pathogenesis. Microbes Infect. 2000;2:91–98. doi: 10.1016/s1286-4579(00)00280-x. [DOI] [PubMed] [Google Scholar]
- Murphy G, Nagase H. Progress in matrix metalloproteinase research. Mol Aspects Med. 2008;29:290–308. doi: 10.1016/j.mam.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr sect D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nagase H, Fields CG, Fields GB. Design and characterization of a fluorogenic substrate selectively hydrolyzed by stromelysin 1 (matrix metalloproteinase-3) J Biol Chem. 1994;269:20952–20957. [PubMed] [Google Scholar]
- Nicholls A, Bharadwaj R, Honig B. GRASP : graphical representation and analysis of surface properties. Biophys J. 1993;64(2, part 2):A166-A166. [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Oofusa K, Yomori S, Yoshizato K. Regionally and hormonally regulated expression of genes of collagen and collagenase in the anuran larval skin. Int J Dev Biol. 1994;38:345–350. [PubMed] [Google Scholar]
- Overall CM, López-Otín C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nature Rev Cancer. 2002;2:657–672. doi: 10.1038/nrc884. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr sect D. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- Persson GR. What has ageing to do with periodontal health and disease? Int Dent J. 2006;56:240–249. doi: 10.1111/j.1875-595x.2006.tb00108.x. [DOI] [PubMed] [Google Scholar]
- Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol 2000. 2000;24:153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. [DOI] [PubMed] [Google Scholar]
- Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J Innate Immun. 2009;1:70–87. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente XS, Sánchez LM, Overall CM, López-Otín C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- Quesada V, Ordoñez GR, Sánchez LM, Puente XS, López-Otn C. The Degradome database: mammalian proteases and diseases of proteolysis. Nucl Acids Res. 2009;37(Database issue):D239–D243. doi: 10.1093/nar/gkn570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragster LV, Chrispeels MJ. Azocoll-digesting proteinases in soybean leaves: characteristics and changes during leaf maturation and senescence. Plant Physiol. 1979;64:857–862. doi: 10.1104/pp.64.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos OHP, Selistre-de-Araujo HS. Identification of metalloprotease gene families in sugarcane. Genet Mol Biol. 2001;24:285–290. [Google Scholar]
- Rao BG. Recent developments in the design of specific matrix metalloproteinase inhibitors aided by structural and computational studies. Curr Pharm Des. 2005;11:295–322. doi: 10.2174/1381612053382115. [DOI] [PubMed] [Google Scholar]
- Ratnaparkhe SM, Egertsdotter EM, Flinn BS. Identification and characterization of a matrix metalloproteinase (Pta1-MMP) expressed during Loblolly pine (Pinus taeda) seed development, germination completion, and early seedling establishment. Planta. 2009;230:339–354. doi: 10.1007/s00425-009-0949-8. [DOI] [PubMed] [Google Scholar]
- Reinemer P, Grams F, Huber R, Kleine T, Schnierer S, Piper M, et al. Structural implications for the role of the N terminus in the 'superactivation' of collagenases. A crystallographic study. FEBS Lett. 1994;338:227–233. doi: 10.1016/0014-5793(94)80370-6. [DOI] [PubMed] [Google Scholar]
- Rosenblum G, Meroueh S, Toth M, Fisher JF, Fridman R, Mobashery S, Sagi I. Molecular structures and dynamics of the stepwise activation mechanism of a matrix metalloproteinase zymogen: challenging the cysteine switch dogma. J Am Chem Soc. 2007;129:13566–13574. doi: 10.1021/ja073941l. [DOI] [PubMed] [Google Scholar]
- Saito M, Sato K, Kunisaki N, Kimura S. Characterization of a rainbow trout matrix metalloproteinase capable of degrading type I collagen. Eur J Biochem. 2000;267:6943–6950. doi: 10.1046/j.1432-1033.2000.01807.x. [DOI] [PubMed] [Google Scholar]
- Saito T, Ishihara K, Kato T, Okuda K. Cloning, expression, and sequencing of a protease gene from Bacteroides forsythus ATCC 43037 in Escherichia coli. Infect Immun. 1997;65:4888–4891. doi: 10.1128/iai.65.11.4888-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Schiermeyer A, Hartenstein H, Mandal MK, Otte B, Wahner V, Schillberg S. A membrane-bound matrix-metalloproteinase from Nicotiana tabacum cv. BY-2 is induced by bacterial pathogens. BMC Plant Biol. 2009;9:83. doi: 10.1186/1471-2229-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani S, Kobata M, Kamakura N, Toyosawa S, Ooshima T. Identification and characterization of matrix metalloproteinase-20 (MMP20; enamelysin) genes in reptile and amphibian. Gene. 2007;392:89–97. doi: 10.1016/j.gene.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Skugor S, Glover KA, Nilsen F, Krasnov A. Local and systemic gene expression responses of Atlantic salmon (Salmo salar L.) to infection with the salmon louse (Lepeophtheirus salmonis) BMC Genomics. 2008;9:498. doi: 10.1186/1471-2164-9-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Sprague GF., Jr Genetic exchange between kingdoms. Curr Opin Genet Dev. 1991;1:530–533. doi: 10.1016/s0959-437x(05)80203-5. [DOI] [PubMed] [Google Scholar]
- Springman EB, Angleton EL, Birkedal-Hansen H, Van Wart HE. Multiple modes of activation of latent human fibroblast collagenase : evidence for the role of a Cys73 active-site zinc complex in latency and a "cysteine switch" mechanism for activation. Proc Natl Acad Sci USA. 1990;87:364–368. doi: 10.1073/pnas.87.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöcker W, Grams F, Baumann U, Reinemer P, Gomis-Rüth FX, McKay DB, Bode W. The metzincins - Topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Prot Sci. 1995;4:823–840. doi: 10.1002/pro.5560040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvan JE. Effectiveness of mechanical nonsurgical pocket therapy. Periodontol 2000. 2005;37:48–71. doi: 10.1111/j.1600-0757.2004.03794.x. [DOI] [PubMed] [Google Scholar]
- Suzuki AS, Tadano Y, Yamamoto T, Abe SI, Tajima T. Expression of a novel matrix metalloproteinase gene during Cynops early embryogenesis. Biochem Biophys Res Commun. 2001;288:380–384. doi: 10.1006/bbrc.2001.5784. [DOI] [PubMed] [Google Scholar]
- Tallant C, García-Castellanos R, Baumann U, Gomis-Rüth FX. On the relevance of the Met-turn methionine in metzincins. J Biol Chem. 2010a;285 doi: 10.1074/jbc.M109.083378. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallant C, Marrero A, Gomis-Rüth FX. Matrix metalloproteinases: fold and function of their catalytic domains. Biochim Biophys Acta - Mol Cell Res. 2010b;1803:20–28. doi: 10.1016/j.bbamcr.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H, Yokoyama Y, Suzuki T, Mizuta S, Yoshinaka R. Molecular cloning and expression of gelatinases (MMP-2 and MMP-9) in the pufferfish Takifugu rubripes. Comp Biochem Physiol B Biochem Mol Biol. 2007;148:295–302. doi: 10.1016/j.cbpb.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Uparanukraw P, Morakote N, Harnnoi T, Dantrakool A. Molecular cloning of a gene encoding matrix metalloproteinase-like protein from Gnathostoma spinigerum. Parasitol Res. 2001;87:751–757. doi: 10.1007/s004360100440. [DOI] [PubMed] [Google Scholar]
- Van Wart HE, Birkedal-Hansen H. The cysteine switch : a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc Natl Acad Sci USA. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen N, Boggs A, Walzak K, Helbing CC. Exposure to tetrabromobisphenol-A alters TH-associated gene expression and tadpole metamorphosis in the Pacific tree frog Pseudacris regilla. Aquat Toxicol. 2006;78:292–302. doi: 10.1016/j.aquatox.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Vinarsky V, Atkinson DL, Stevenson TJ, Keating MT, Odelberg SJ. Normal newt limb regeneration requires matrix metalloproteinase function. Dev Biol. 2005;279:86–98. doi: 10.1016/j.ydbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- Vriend G. WHAT IF : a molecular modelling and drug design program. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- Wada K, Sato H, Kinoh H, Kajita M, Yamamoto H, Seiki M. Cloning of three Caenorhabditis elegans genes potentially encoding novel matrix metalloproteinases. Gene. 1998;211:57–62. doi: 10.1016/s0378-1119(98)00076-6. [DOI] [PubMed] [Google Scholar]
- Wang KJ, Ren HL, Xu DD, Cai L, Yang M. Identification of the up-regulated expression genes in hemocytes of variously colored abalone (Haliotis diversicolor Reeve, 1846) challenged with bacteria. Dev Comp Immunol. 2008;32:1326–1347. doi: 10.1016/j.dci.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Wegner N, Wait R, Sroka A, Eick S, Nguyen KA, Lundberg K, et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 2010;62 doi: 10.1002/art.27552. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DH, Murray EJ. Specific amino acid substitutions in human collagenase cause decreased autoproteolysis and reveal a requirement for a second zinc atom for catalytic activity. FEBS Lett. 1994;354:267–270. doi: 10.1016/0014-5793(94)01136-2. [DOI] [PubMed] [Google Scholar]
- Wyatt RA, Keow JY, Harris ND, Hache CA, Li DH, Crawford BD. The zebrafish embryo: a powerful model system for investigating matrix remodeling. Zebrafish. 2009;6:347–354. doi: 10.1089/zeb.2009.0609. [DOI] [PubMed] [Google Scholar]
- Yang EV, Bryant SV. Developmental regulation of a matrix metalloproteinase during regeneration of axolotl appendages. Dev Biol. 1994;166:696–703. doi: 10.1006/dbio.1994.1348. [DOI] [PubMed] [Google Scholar]
- Yang M, Kurkinen M. Cloning and characterization of a novel matrix metalloproteinase (MMP), CMMP, from chicken embryo fibroblasts. CMMP, Xenopus XMMP, and human MMP19 have a conserved unique cysteine in the catalytic domain. J Biol Chem. 1998;273:17893–17900. doi: 10.1074/jbc.273.28.17893. [DOI] [PubMed] [Google Scholar]
- Yang M, Murray MT, Kurkinen M. A novel matrix metalloproteinase gene (XMMP) encoding vitronectin-like motifs is transiently expressed in Xenopus laevis early embryo development. J Biol Chem. 1997;272:13527–13533. doi: 10.1074/jbc.272.21.13527. [DOI] [PubMed] [Google Scholar]
- Yeh HY, Klesius PH. Complete structure, genomic organization, and expression of channel catfish (Ictalurus punctatus, Rafinesque 1818) matrix metalloproteinase-9 gene. Biosci Biotechnol Biochem. 2008;72:702–714. doi: 10.1271/bbb.70579. [DOI] [PubMed] [Google Scholar]