Abstract
Hypoxia-inducible factors (HIFs) are stress-responsive transcriptional regulators of cellular and physiological processes involved in oxygen metabolism. Although much is understood about the molecular machinery that confers HIF responsiveness to oxygen, far less is known about HIF isoform-specific mechanisms of regulation, despite the fact that HIF-1 and HIF-2 exhibit distinct biological roles. We recently determined that the stress-responsive genetic regulator sirtuin 1 (Sirt1) selectively augments HIF-2 signaling during hypoxia. However, the mechanism by which Sirt1 maintains activity during hypoxia is unknown. In this report, we demonstrate that Sirt1 gene expression increases in a HIF-dependent manner during hypoxia in Hep3B and in HT1080 cells. Impairment of HIF signaling affects Sirt1 deacetylase activity as decreased HIF-1 signaling results in the appearance of acetylated HIF-2α, which is detected without pharmacological inhibition of Sirt1. We also find that Sirt1 augments HIF-2 mediated, but not HIF-1 mediated, transcriptional activation of the isolated Sirt1 promoter. These data in summary reveal a bidirectional link of HIF and Sirt1 signaling during hypoxia.
Keywords: Gene Expression, Hypoxia, Post-translational Modification, SIRT, Transcription Regulation, HIF-1, HIF-2, Acetylation, Deacetylation, Hypoxia-inducible Factor
Introduction
The ability to sense and respond to changes in oxygen content, conserved in almost all eukaryotic organisms, is conferred at the cellular level and is dictated by changes in gene expression, including by de novo transcriptional events (1). Members of the hypoxia-inducible factor (HIF)2 family of transcription factors are key regulators of genes whose expression is altered during hypoxia. HIFs, obligate heterodimeric protein complexes, are composed of an oxygen-labile α-subunit and a shared, oxygen-stable β-subunit also referred to as ARNT (2). Whereas invertebrates contain a single HIF-α member, mammals contain three HIF-α genes: HIF-1α, HIF-2α (also called endothelial PAS domain protein 1 (EPAS1)) (3–5), and HIF-3α.
HIF-α proteins have similar domain structures with conserved sequence identity in some regions, particularly for HIF-1α and HIF-2α. The N termini of HIF-α and HIF-β proteins contain the highly conserved basic helix-loop-helix and Per/ARNT/Sim (PAS) domains involved in DNA binding and protein-protein interactions, respectively. The PAS domain may also contribute to HIF target gene specificity and may serve as a target for small molecules that disrupt specific HIF complexes (6, 7).
The levels of HIF-α subunits increase during hypoxia due to impaired modifications of two proline residues (8, 9) situated within the oxygen-dependent degradation domain (10), part of a larger domain known as the N-terminal activation domain (NTAD) located in the midportion of HIF-α proteins. These two proline residues are otherwise selectively hydroxylated under normoxic conditions by oxygen-dependent prolyl hydroxylases (8, 9, 11) and subsequently target the HIF-α proteins for proteasomal degradation by the von Hippel-Lindau (VHL) ubiquitin-protein ligase complex (12–16).
A second oxygen-dependent hydroxylation by asparaginyl hydroxylases (17, 18) targets an asparagine residue within the C-terminal activation domain (CTAD) of HIF-α under normoxic conditions, thereby blocking recruitment of the coactivators p300 and CBP to the C terminus and decreasing HIF transactivation capacity (18). Mutation of the modified proline and asparagine residues results in oxygen-insensitive, “constitutively active” HIF-α proteins.
Despite the overall sequence conservation in some regions, each HIF-α protein has distinct physiological roles (19, 20) that are in part conferred by target gene selectivity. The divergent portion of HIF-α that we refer to as the unique region, located in the C terminus between the NTAD/oxygen-dependent degradation domain and CTAD (21), likely participates in HIF isoform selective signaling (22). Indeed, several factors have been identified that bind in a HIF-α protein-selective manner or that mediate their action through the unique region of either HIF-1α (23) or HIF-2α (24–26).
Sirt1, a deacetylase initially identified in aging studies of lower eukaryotes (27) and implicated in diverse physiological processes in mammals, augments HIF-2 signaling during hypoxia in cell culture as well as mouse models (21). Augmentation of HIF-2 signaling by Sirt1 requires an intact deacetylase activity of Sirt1 and is conferred by deacetylating specific lysine residues located in the HIF-2α unique region that are acetylated. Absolute levels of HIF-2α acetylation increase during hypoxia, although acetylated HIF-2α is only evident if Sirt1 activity is inhibited through pharmacological or genetic means.
Despite the increase in HIF-2α protein levels during hypoxia, Sirt1 action is so efficient that acetylated HIF-2α is normally undetectable unless Sirt1 deacetylase activity is inhibited. How Sirt1 action is maintained or increased during hypoxia is unclear. In this study, we asked if Sirt1 expression was itself altered during hypoxia. We noted that Sirt1 gene expression in cells as well as in mice increased during acute hypoxia and determined that HIF signaling was primarily responsible for the increased Sirt1 expression observed early after the onset of hypoxia.
EXPERIMENTAL PROCEDURES
Reporter and Expression Plasmids
The human SIRT1 promoter region (−354/+54) was isolated by PCR amplification of human genomic DNA and inserted into the firefly luciferase reporter plasmid pGL3-Basic (Promega, Madison, WI). We introduced mutations of HIF-responsive element 5 (HRE5) or flanking ETS-binding sites (EBS) into the (−354/+54) SIRT1 promoter using PCR-based site-directed mutagenesis (QuikChange II, Stratagene) or extension PCR mutagenesis. We previously described the oxygen-independent (PPN) HIF-1α, PPN HIF-2α, wild type (WT) vsv-g:SIRT1, and deacetylase mutant vsv-g:SIRT1 expression vectors (21).
siRNA Knockdown
The day before transfection, we trypsinized and plated 2.0 × 105 Hep3B cells on each well of a 6-well plate in 2 ml of antibiotic-free complete DMEM. Using DharmaFECT1 (catalog no. T-2001-03, Thermo Fisher Scientific (Lafayette, CO)), we transfected non-targeting control (catalog no. D-001810–10-20, Thermo Fisher Scientific), HIF-1α (catalog no. L-004018-00-0005, Thermo Fisher Scientific), or HIF-2α (catalog no. L-004814-00-0005, Thermo Fisher Scientific) siRNA. After 24 or 48 h, cells were harvested for mRNA or protein analysis, respectively.
Immunoblotting and Cell Fractionation
To prepare nuclear extracts from Hep3B or HT1080 cells, we used NE-PER® nuclear and cytoplasmic extraction reagents (catalog no. 78833, Pierce). To prepare whole cell extracts from Hep3B or HT1080 cells, we used cytoBuster protein extraction reagent (catalog no. 71009, Novagen (Gibbstown, NJ)). Equivalent amounts of whole cell extracts or nuclear extracts in SDS-PAGE sample buffer were analyzed by immunoblotting using anti-human Sirt1 (1:1,000 dilution; catalog no. 07131, Upstate Biotechnology, Inc.), anti-human HIF-1α (1:1,000 dilution; catalog no. 610959, BD Biosciences), anti-human HIF-2α (1:1,000 dilution; catalog no. NB100-132, Novus Biologicals (Littleton, CO)), anti-HA (1:5,000 dilution; catalog no. H9658, Sigma), anti-vsv-g antibodies (1:5,000 dilution; catalog no. ab18612, Abcam (Cambridge, MA)), anti-α-tubulin (1:5,000 dilution; catalog no. T5168, Sigma), or anti-TATA-binding protein (1:1,000 dilution; catalog no. sc-204, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)) antibodies.
Detection of Exogenous HIF-2α Acetylation
Hep3B or HT1080 cells were transfected with expression vectors encoding N-terminal S-peptide (SP) epitope-tagged and C-terminal hemagglutinin A (HA) epitope-tagged PPN HIF-1α (SP:PPN HIF-1α:HA) or PPN HIF-2α (SP:PPN HIF-2α:HA) as described previously (21). Twenty-four hours after transfection, cells were exposed to hypoxia for the indicated time. Eight hours before the harvest time, cells were treated with 5 μm sirtinol plus 10 mm nicotinamide. After harvesting, SP:PPN HIF-1α:HA, and SP:PPN HIF-2α:HA were purified for immunoblot analyses of acetylated lysine residues or the HA epitope as described previously (21).
Cell Culture and Transfections
We used the indicated amounts of reporter plasmids (30 ng/well) and expression plasmids (100 ng/well) in transient transfection analyses. We maintained human renal clear cell carcinoma 786-0 (catalog no. CRL-1932, ATCC), human fibrosarcoma HT1080 (catalog no. CCL-121, ATCC), and human hepatocellular carcinoma Hep3B (catalog no. HB-8064, ATCC) cells in complete media (DMEM, 4.5 g/liter glucose, 4 mm glutamate (catalog no. SH30022, HyClone, Logan, UT), 10% fetal bovine serum (FBS; catalog no. S10650H, Atlanta Biologicals (Lawrenceville, GA)) with penicillin (100 units/ml)/streptomycin (100 μg/ml) (catalog no. 15140-148, Invitrogen)) in a 5% CO2, 95% N2 incubator. We transfected Hep3B or HT1080 cells in 48-well plates (catalog no. 3548, CoStar, Corning Glass) at 50–60% confluence using Lipofectamine 2000 (catalog no. 11668-019, Invitrogen) and made up the balance with pIRES. At 24 h post-transfection, we harvested cells for luciferase assays. For hypoxia treatments, we transferred cells to a humidified environmental chamber (Coy Laboratory Products, Inc., Grass Lake, MI), replaced culture media with deoxygenated media, and maintained the cells under hypoxic (1% O2, 5% CO2, 94% N2) conditions for the specified periods. We prepared cell extracts within the chamber.
Chromatin Immunoprecipitation Assays in Cells
We seeded Hep3B cells at 2 × 106 (150-mm plates) 48 h prior to use, next exposed the cells to normoxia or hypoxia, and then harvested for whole cell protein or RNA. EPO induction after hypoxia exposure was confirmed by real-time RT-PCR. Chromatin immunoprecipitation (ChIP) assays were carried out using the ChIP-ITTM Express Magnetic assay kit (catalog no. 53009, Active Motif). The antisera used were normal mouse IgG (1–2 μg/ml; catalog no. 2027, Santa Cruz Biotechnology, Inc.), normal rabbit IgG (1–2 mg/ml; catalog no. NI01, EMD Chemicals, Inc., Gibbstown, NJ), anti-human EPAS1 antiserum (2 mg/ml; NB 100-132, Novus Biologicals), and mouse anti-human HIF-1α (2 mg/ml; catalog no. 610958, BD Biosciences). After ChIP, the precipitated genomic DNA was analyzed by quantitative PCR using an Applied Biosystems ABI Prism 7000 thermocycler (Applied Biosystems; Foster City, CA) and Power SYBR Green Master Mix (catalog no. 4367659, Applied Biosystems) using the following human SIRT1 promoter primers: 5′-AGCAAGGAGCAGAAAAAGGAGCAAAAGAGGAG-3′ (forward) and 5′-TCTTCCAACTGCCTCTCTGGCCCTCCTCCC-3′ (reverse). The captured genomic DNA was normalized to input material and compared between the normoxic and hypoxic treatment samples.
Chromatin Immunoprecipitation Assays in Mice
CD1 mice were exposed to 6% oxygen for 2 h and then euthanized for harvest of liver samples inside a hypoxia chamber. We minced 30 mg of fresh tissue to 1–3 mm3, transferred tissue into 10 ml of PBS plus protease inhibitors/g of tissue, added formaldehyde (final concentration 1%), and rotated tubes at room temperature for 15 min. We stopped cross-linking with fresh glycine (final concentration of 0.125 m). After 5 min at room temperature, the tissue was pelleted at low speed centrifugation at 4°C, washed once with cold PBS plus protease inhibitors, and then repelleted. We resuspended the washed pellet in 1 ml of PBS on ice and ground the tissue using a micro-tissue grinder on ice. Cells were pelleted by microcentrifugation at 4 °C. ChIP was carried out using the ChIP-ITTM Express Magnetic assay kit (catalog no. 53009, Active Motif). After ChIP, the precipitated genomic DNA was analyzed for the presence of a mouse Sirt1 amplicon by quantitative PCR using an Applied Biosystems ABI Prism 7000 thermocycler (Applied Biosystems, Foster City, CA), Power SYBR Green Master Mix (catalog no. 4367659, Applied Biosystems), and the following primers: 5′-GGCAACAGGCCCCGAGGGCTGGCTTGGGCA-3′ (mouse Sirt1 promoter forward) and 5′-TCTTCCAACTGCCTCTCTGGCCCTCCGCCC-3′ (mouse Sirt1 promoter reverse). The captured genomic DNA was normalized to input material and compared between the normoxic and hypoxic treatment samples, with the results of a representative experiment shown.
Real Time RT-PCR Analyses
For cells, total RNA was extracted using GenEluteTM mammalian total RNA kit (catalog no. RTN70-1KT, Sigma). For mouse liver, total RNA was isolated from about one-eighth of the mouse liver (right lobule) and extracted using the FastRNA Pro Green kit (catalog no. 6045, MP Biomedicals (Solon, OH)). 1 μg of total RNA was reverse transcribed with oligo(dT) primers using a Moloney murine leukemia virus reverse transcriptase kit (catalog no. 28025-013, Invitrogen). Real-time quantitative PCR was performed on an Applied Biosystems ABI Prism 7000 thermocycler using Power SYBR Green Master Mix following the manufacturer's protocol and one-tenth of total cDNA with the following pairs primers: CYCLOPHILIN B (forward), 5′-ATGTGGTTTTCGGCAAAGTTCTA-3′; CYCLOPHILIN B (reverse), 5′-GGCTTGTCCCGGCTGTCT-3′, SIRT1 (forward), 5′-GCAGGTTGCGGGAATCCAA-3′; SIRT1 (reverse), 5′-GGCAAGATGCTGTTGCAAA-3′; PGK1 (forward), 5′-TTAAAGGGAAGCGGGTCGTTA-3′; PGK1 (reverse), 5′-TCCATTGTCCAAGCAGAATTTGA-3′; EPO (forward), 5′-GAGGCCGAGAATATCACGACGGG-3′; and EPO (reverse), 5′-TGCCCGACCTCCATCCTCTTCCAG-3′ (human primers) or using cyclophilin B (forward), 5′-ATGTGGTTTTCGGCAAAGTTCTA-3′; cyclophilin B (reverse), 5′-GGCTTGTCCCGGCTGTCT-3′; Sirt1 (forward), 5′-GCAGGTTGCAGGAATCCAA-3′; and Sirt1 (reverse), 5′-GGCAAGATGCTGTTGCAAA-3′ (mouse primers). We expressed the results of three independent experiments, each measured in triplicate, as 2−(EPO number of cycles − cyclophilin number of cycles). Levels for genes of interest were normalized to cyclophilin B mRNA.
Mouse Hypoxia Experiments
All mice were housed under standard 12 h/12 h light/dark conditions and fed ad lib with standard chow. We established C57BL/6J HIF-1α heterozygous (28) × 129S6/SvEvTac HIF-2α (29, 30) heterozygous mating pairs for generation of F1 hybrid (129S6/SvEvTac:C57BL/6J) wild type, HIF-1α heterozygous, HIF-2α heterozygous, and HIF-1α/HIF-2α compound heterozygous mice. For hypoxic experiments, we placed age- and gender-matched wild type CD1 or wild type, HIF-1α, HIF-2α, and HIF-1α/HIF-2α compound heterozygous F1 hybrid mice in a hypoxia chamber with flow by air supply and subjected them to normoxic (21% oxygen) or continuous hypoxic (6% oxygen) treatment for 2 h. Mice were then euthanized, and tissues were collected and snap-frozen in liquid nitrogen for subsequent extraction of total RNA. Real-time RT-PCR determinations were made for genes of interest as described.
Mouse Adenoviral Experiments
We transduced adult CD1 wild type female mice (Charles River Laboratories, Wilmington, MA) with PPN HIF-1α:HA, PPN HIF-2α:HA, or control (GFP) adenovirus as described (21). Hepatic Sirt1 and Cyclophilin mRNA levels were determined by real-time RT-PCR analyses.
Statistical Analyses
Statistical analyses were performed using Microsoft Excel (Microsoft Corp., Redmond, WA) and StatPlus:mac LE (AnalystSoft, Inc.) by Student's t test or by analysis of variance as indicated. One-tailed analyses were performed because we anticipated that decreases or increases in HIF signaling would blunt or augment Sirt1 expression, respectively. p values less than 0.10 were deemed statistically significant.
RESULTS
Sirt1 Expression Increases during Hypoxia
The activity of Sirt1 can be controlled by transcriptional as well as by post-translational mechanisms. Recently, we determined that Sirt1 selectively augmented HIF-2α signaling during hypoxia. We surmised that the participation of Sirt1 in HIF-2 signaling during hypoxia reflects a physiologically relevant interaction of two crucial stress-responsive signaling pathways. However, the mechanism by which Sirt1 activity is maintained or even increased during hypoxia was unknown. We hypothesized that Sirt1 regulation during hypoxia could be conferred by changes in Sirt1 gene expression.
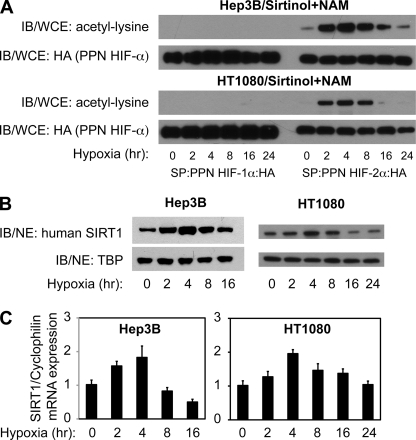
We first performed a time course analysis of acetylation during hypoxia in Hep3B cells, a model hypoxia-responsive cell line, as well as in HT1080 cells (Fig. 1A), which have recently been used in Sirt1/HIF studies (31). Ectopic HIF-1α and HIF-2α exhibited marked differences in the ability to undergo acetylation during hypoxia in both cell lines. HIF-2α, but not HIF-1α, was efficiently acetylated within 2 h, peaked by 4–8 h, and returned to base-line values by 24 h following hypoxia exposure.
FIGURE 1.
Sirt1 expression increases during hypoxia. A, acetylation time course of ectopic HIF-1α and HIF-2α expressed in Hep3B or HT1080 cells exposed to hypoxia. The top panel for each cell line shows immunoblotting (IB) of acetyl-lysine residues in whole cell extract (WCE) after SP affinity purification of ectopic HIF-1α and HIF-2α from cells exposed to hypoxia (1% oxygen) for the indicated period of time. The bottom panel shows the same membrane reprobed for HA epitope tag. B, immunoblot analyses of human SIRT1 or TATA-binding protein (TBP) in nuclear extract (NE) from Hep3B or HT1080 cells exposed to hypoxia (1% oxygen) for the indicated period of time. C, SIRT1 mRNA levels as measured by real-time RT-PCR analyses of total RNA obtained from Hep3B or HT1080 cells exposed to hypoxia (1% oxygen) for the indicated period of time. SIRT1 mRNA levels were normalized to cyclophilin B (cyclophilin) mRNA levels for each sample. The data represent the average of three independent samples obtained at each time point with each sample measured in triplicates. Error bars, S.E. p = 0.00002 for Hep3B samples and p = 0.006 for HT1080 samples with comparisons made by one-way analysis of variance.
We next examined SIRT1 protein levels during hypoxia in Hep3B and in HT1080 cells (Fig. 1B). Immunoblotting of SIRT1 revealed a time-dependent increase in protein levels for both cell lines with the peak of expression at 4 h following hypoxia exposure. To determine whether the observed changes in SIRT1 protein levels during hypoxia were due to changes in SIRT1 gene expression, we measured steady-state SIRT1 mRNA levels by real-time RT-PCR (Fig. 1C) in parallel samples as assessed in Fig. 1B. SIRT1 mRNA levels increased in a time-dependent manner during hypoxia for both Hep3B and HT1080 cells and peaked at the same time point (4 h) as observed for the peak of SIRT1 protein expression.
HIF-α Members Activate and Are Recruited to the Sirt1 Proximal Promoter
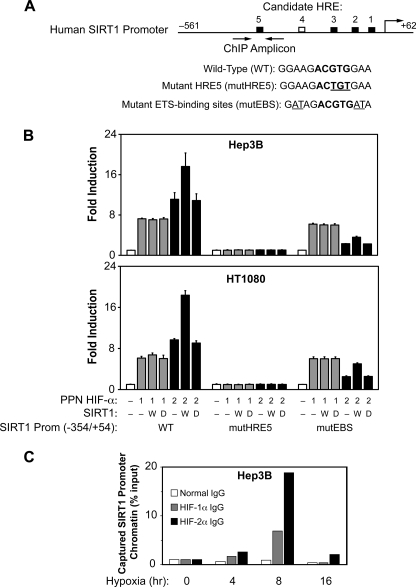
Because SIRT1 gene expression increased during hypoxia, we hypothesized that HIF members might directly participate in the increase in SIRT1 gene expression during hypoxia. The human proximal SIRT1 promoter region contains several candidate HIF-responsive elements (HREs) conserved between the mouse and human promoters that could potentially serve as functional transcriptional regulatory elements (Fig. 2A). One of these candidate HREs, HRE5, also is flanked by candidate ETS-binding sites (EBS), which have previously been shown to confer HIF-2-selective activation (32–34). To test whether the proximal SIRT1 promoter would respond to HIF activation, we isolated the human SIRT1 proximal promoter and fused it to firefly luciferase coding sequences for use in transient transfection assays with Hep3B and HT1080 cells.
FIGURE 2.
HIF activates the Sirt1 promoter. A, schematic representation of the human SIRT1 proximal promoter region. The HREs conserved between mouse and human in sequence and relative position are shown by black boxes, whereas an HRE unique to the human SIRT1 promoter is indicated as a white box. The sequences for HRE5 and surrounding EBS are shown in the parental WT construct as well as for the site-directed mutants in HRE5 (mutHRE) or in the flanking EBS (mutEBS). The nucleotide substitutions in the core HRE (ACGTG) or in the core EBS (GGA) sequences are underlined. B, transient transfection assays of the isolated human SIRT1 proximal promoter in Hep3B or HT1080 cells. Transfections were performed with the indicated isolated SIRT1 proximal promoter reporter and expression plasmids encoding no protein (−) (basal values) or oxygen-insensitive (PPN) HIF-1α or PPN HIF-2α along with control (−), wild type (W), or deacetylase mutant (D) SIRT1 expression plasmids. The data represent the average of three independent experiments with each experiment performed in triplicate. Error bars, S.E. For Hep3B cells, p = 0.048 for PPN HIF-2α versus PPN HIF-2α + wild type Sirt1 with wild type SIRT1 promoter, and p = 0.001 for PPN HIF-2α versus PPN HIF-2α + wild type Sirt1 with mutEBS SIRT1 promoter. For HT1080 cells, p = 0.0004 for PPN HIF-2α versus PPN HIF-2α + wild type Sirt1 with wild type SIRT1 promoter, and p = 0.0003 for PPN HIF-2α versus PPN HIF-2α + wild type Sirt1 with mutEBS SIRT1 promoter. All comparisons were made by one-tailed t test. C, ChIP assays of HIF protein binding to the SIRT1 proximal promoter region. Binding of HIF-1α and HIF-2α to the SIRT1 proximal promoter region in Hep3B cells during hypoxia as assessed by ChIP assays is shown with a PCR amplicon centered on HRE5. The cell extracts from each time point were subjected to ChIP assays using normal, anti-human HIF-1α or anti-human HIF-2α IgG.
To evaluate the effect of HIF signaling on the isolated SIRT1 promoter, we overexpressed oxygen-independent (PPN) mutant forms of HIF-1α or HIF-2α that are not modified by the oxygen-dependent prolyl or asparaginyl hydroxylases (Fig. 2B). These constructs allow for HIF signaling during normoxia, thereby avoiding signaling induced by other hypoxia-activated regulators. Both PPN HIF-1α and PPN HIF-2α increased activity of the isolated SIRT1 promoter reporter in both Hep3B and HT1080 cells (Fig. 2B). Reporter activity was more pronounced with PPN HIF-2α compared with PPN HIF-1α overexpression, despite equivalent or even greater levels of PPN HIF-1α (see Fig. 1A, 0 h hypoxia time point). Moreover, SIRT1 co-expression augmented HIF-2α, but not HIF-1α, stimulation of the SIRT1 proximal promoter reporter.
HIF signaling is primarily mediated through binding of HIF heterodimers to HREs in regulatory regions of HIF target genes, although additional cis-elements may influence responsiveness of a given HRE to either HIF-1 or HIF-2. We hypothesized that HRE5 in the proximal SIRT1 promoter region might mediate HIF signaling, given its location in a typical enhancer position and the presence of flanking EBS that are important for HIF-2 signaling. To specifically evaluate the contribution of HRE5 to HIF signaling, we generated a point mutation of HRE5 in the (−354/+54) SIRT1 promoter and observed a profound effect on SIRT1 promoter activity such that it was rendered unresponsive to either PPN HIF-1α or PPN HIF-2α (Fig. 2B). These data indicate that HRE5 is probably a functional and crucial HRE conferring HIF-mediated induction of SIRT1 gene expression during hypoxia.
To determine whether the EBS that flank HRE5 contribute to HIF isoform activation of the SIRT1 promoter, we constructed and tested a SIRT1 promoter point mutant that lacked functional flanking EBS. As seen (Fig. 2B), ablation of the flanking EBS blunts fold-induction by PPN HIF-2α, but not by PPN HIF-1α. However, the EBS mutations do not prevent augmentation of PPN HIF-2α signaling by SIRT1. Similar to our results with other HIF-responsive regulatory regions in HEK293 and Hep3B cells (21), we did not observe an effect of SIRT1 on induction by PPN HIF-1α of isolated SIRT1 promoter activity in either Hep3B or HT1080 cells.
We next asked whether HIF-1α or HIF-2α are recruited to the endogenous SIRT1 promoter during hypoxia. We performed ChIP assays using extracts from Hep3B cells exposed to various periods of hypoxia with a PCR amplicon that centered on HRE5 of the SIRT1 promoter region (Fig. 2C). As shown, endogenous HIF-1α and HIF-2α are recruited to the endogenous SIRT1 proximal promoter region during hypoxia (Fig. 2C).
HIF-1α and HIF-2α Contribute to Sirt1 Induction during Hypoxia in Cells
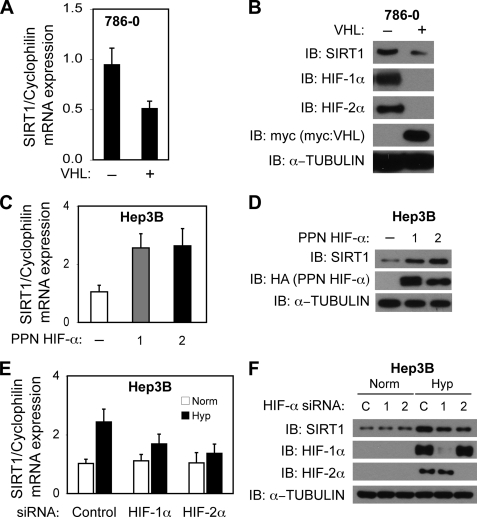
Because exogenous PPN HIF-1α as well as PPN HIF-2α increased activity of the isolated SIRT1 promoter in transient transfection assays, we asked if pathological states involving up-regulation of HIF-α members would result in increases in SIRT1 mRNA and SIRT1 protein levels. We began with examination of Sirt1 expression in 786-0 cells, derived from VHL tumors in which the inactivation of the VHL protein results in constitutive HIF-α expression, after transfection with control or VHL-encoding expression plasmids. In the absence of functional VHL protein, HIF-1α or HIF-2α proteins were stabilized under normoxic conditions in 786-0 cells (Fig. 3B). Introduction of a functional VHL protein resulted in the expected decrease in HIF-1α and HIF-2α proteins and was accompanied by a reduction of SIRT1 mRNA (Fig. 3A) and SIRT1 protein (Fig. 3B) levels.
FIGURE 3.
HIF activates endogenous Sirt1 gene expression in cells. A, SIRT1 mRNA levels in 786-0 cells transfected with a control or VHL-encoding expression plasmid. SIRT1 mRNA levels were analyzed and are presented as in Fig. 1A. p = 0.037 by one-tailed t test. B, immunoblot (IB) analyses for SIRT1, HIF-1α, and HIF-2α in 786-0 cells treated in parallel as in A. C, SIRT1 mRNA levels in Hep3B cells after transfection with expression plasmids encoding control (−), or oxygen-insensitive (PPN) HIF-1α or HIF-2α. p = 0.024 for control versus PPN HIF-1α, and p = 0.034 for control versus PPN HIF-2α. All comparisons were made by one-tailed t test. D, immunoblot analyses for SIRT1, HIF-1α, and HIF-2α in Hep3B cells treated in parallel as in C. E, SIRT1 mRNA levels in Hep3B cells after transfection with control, HIF-1α, or HIF-2α siRNA. p = 0.123 for control versus HIF-1α siRNA and p = 0.059 for control versus HIF-2α siRNA hypoxia samples. All comparisons were made by one-tailed t test. F, immunoblot analyses for SIRT1, HIF-1α, and, HIF-2α in Hep3B cells treated in parallel as in E. The data represent the average of three independent experiments with each experiment performed in triplicate. Error bars, S.E.
Because VHL loss may affect other signaling pathways besides HIF, we asked if HIF signaling alone, as conferred by exogenous PPN HIF-1α or PPN HIF-2α overexpression, could affect SIRT1 protein levels in Hep3B cells. Exogenous PPN HIF-1α as well as PPN HIF-2α significantly increased expression of endogenous SIRT1 mRNA (Fig. 3C) and SIRT1 protein (Fig. 3D) levels in Hep3B cells. Expression of exogenous PPN HIF-2α resulted in an increase in Sirt1 gene expression similar to the increase observed with exogenous PPN HIF-1α, although ectopic expression of exogenous PPN HIF-2α was less than exogenous PPN HIF-1α.
We next asked whether knockdown of endogenous HIF-α subunits affects endogenous Sirt1 induction during hypoxia. Following introduction of siRNA recognizing either HIF-1α or HIF-2α, Hep3B cells were exposed to normoxia or hypoxia, and total RNA was prepared. Knockdown of HIF-1α trended toward lower induction, and knockdown of HIF-2α significantly blunted induction of SIRT1 mRNA during hypoxia but had no effect in cells maintained under normoxic conditions (Fig. 3E). These reductions in SIRT1 mRNA levels were not a general inhibitory effect of HIF knockdown; the HIF-1 target gene PGK1 was only reduced after HIF-1α knockdown, whereas the HIF-2 preferential target gene EPO was predominantly affected after HIF-2α (supplemental Fig. S1, A and B). The reduction in SIRT1 mRNA levels resulting from HIF-1α or HIF-2α knockdown was accompanied by a blunting of the increase in SIRT1 protein levels during hypoxia, but did not affect SIRT1 protein levels during normoxia (Fig. 3F).
HIF Members Regulate Sirt1 Gene Expression in Mice
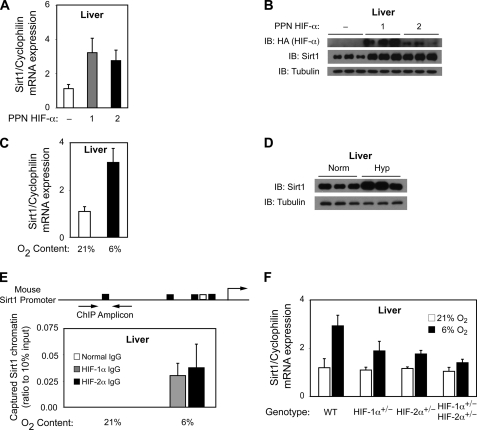
We have previously shown that ectopic expression of PPN HIF-1α or PPN HIF-2α in the livers of mice results in activation of HIF-1 or HIF-2 target genes. We now asked if Sirt1 levels differed in mice expressing ectopic PPN HIF-1α or PPN HIF-2α. Similar to results obtained in Hep3B cells (Fig. 3, C and D), ectopic PPN HIF-1α or PPN HIF-2α expression in mouse livers significantly increased endogenous liver Sirt1 mRNA (Fig. 4A) and Sirt1 protein (Fig. 4B) levels. Furthermore, and as seen in Hep3B cells, ectopic PPN HIF-1α resulted in an increase in Sirt1 protein levels similar to that observed with ectopic PPN HIF-2α, despite the lower levels of the latter compared with the former ectopic HIF-α protein.
FIGURE 4.
HIF activates endogenous Sirt1 gene expression in mice. A, CD1 mice transduced with control (−) adenovirus or adenovirus encoding PPN HIF-1α:HA or PPN HIF-2α:HA. Livers were isolated 7 days following transduction for determination of Sirt1 and cyclophilin B levels from total RNA. p = 0.022 for control versus PPN HIF-1α, and p = 0.019 for control versus PPN HIF-2α. All comparisons were made by one-tailed t test. n = 5 mice per group. Each sample was measured in triplicate. Error bars, S.E. B, immunoblot (IB) analyses of Sirt1 and α-tubulin expression in livers isolated from three independent mice for each group examined in A. C, CD1 mice exposed to room air (21% oxygen) or continuous hypoxia (6% oxygen) for 2 h followed by harvesting of livers for total RNA preparation. Sirt1 and cyclophilin B mRNA levels were determined as in A. p = 0.004 for normoxia versus hypoxia by one-tailed t test. n = 6 mice per group. Each sample was measured in triplicate. Error bars, S.E. D, immunoblot analyses of Sirt1 and α-tubulin expression in livers isolated from three independent mice for each group examined in C. E, schematic representation of the mouse Sirt1 proximal promoter region as well as the amplicon centered on HRE5 and used in ChIP assays of HIF binding to the Sirt1 proximal promoter region. Binding of HIF-1α and HIF-2α to the Sirt1 proximal promoter region, as assessed by ChIP assays, in mice maintained under normoxia or exposed to hypoxia is shown. No signal was evident with control IgG under either condition. n = 3 mice per group. Each sample was measured in triplicate. Error bars, S.E. F, mice with intact (WT) HIF levels or haploinsufficient for HIF-1α (HIF-1α+/−), HIF-2α (HIF-2α+/−), or both HIF-2α and HIF-2α (HIF-1α+/−/HIF-2α+/−) were exposed to room air (21% oxygen) or continuous hypoxia (6% oxygen) for 2 h followed by harvesting of livers for total RNA preparation. Sirt1 and cyclophilin B mRNA levels were determined as in A. p = 0.055 for WT versus HIF-1α+/−, p = 0.015 for WT versus HIF-2α+/−, and p = 0.004 for WT versus HIF-1α+/−/HIF-2α+/−. All comparisons were for hypoxia samples and were made by one-tailed t test. n = 6 mice per group. The data represent the average of three independent experiments with each experiment performed in triplicates. Error bars, S.E.
Because Sirt1 gene expression increases in cells during hypoxia, we next asked whether endogenous Sirt1 mRNA and Sirt1 protein levels were altered in livers of mice exposed to hypoxia. Hepatic Sirt1 mRNA (Fig. 4C) and protein (Fig. 4D) levels increased during hypoxia exposure relative to mice maintained under normoxic conditions. This was accompanied by recruitment of endogenous HIF-1α and HIF-2α to the Sirt1 promoter region in liver after hypoxia exposure, as revealed by in vivo chromatin immunoprecipitation assays (Fig. 4E). The importance of HIF members for induction of Sirt1 gene expression was demonstrated by the blunted induction of Sirt1 mRNA in mice that were haploinsufficient for HIF-1α, HIF-2α, or both HIF-1α and HIF-2α (Fig. 4F).
Impairment of HIF Signaling Affects Sirt1 Activity
Sirt1 gene expression increases during hypoxia, but the increase is blunted by reduction in either HIF-1α or HIF-2α protein levels. We have previously shown that Sirt1 deacetylates HIF-2α. We next asked if a reduction in Sirt1 levels conferred by HIF-1α would affect the acetylation status of HIF-2α.
We first confirmed that knockdown of HIF-1α resulted in decreased SIRT1 levels. Reduction of SIRT1 protein was evident when HIF-1α was knocked down (Fig. 5A, lane 2) and was unaffected by inhibition of SIRT1 activity mediated by the pharmacological inhibitors sirtinol and nicotinamide (Fig. 5A, lane 4). Ectopic expression of wild type (Fig. 5A, lane 6) or deacetylase-inactive (Fig. 5A, lane 8) SIRT1 did not affect HIF-1α knockdown.
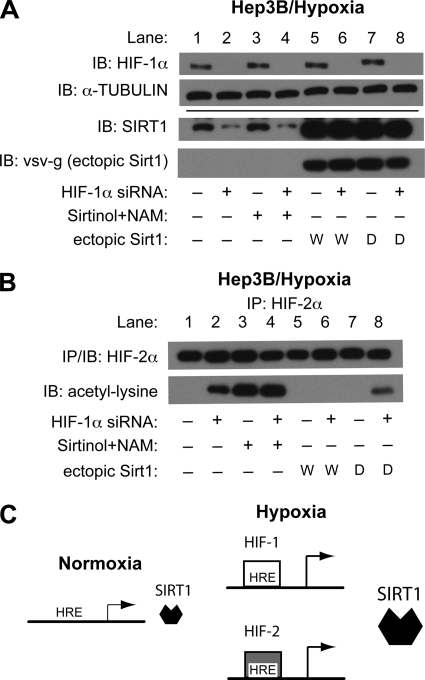
FIGURE 5.
HIF deficiency alters Sirt1 function during hypoxia. A, endogenous HIF-1α, SIRT1, and α-tubulin as well as ectopic SIRT1 (vsv-g) protein levels were determined by immunoblot (IB) analyses in Hep3B cells after transfection with control (−) or HIF-1α (+) siRNA in the absence or presence of ectopic wild type (W) or deacetylase-defective (D) Sirt1 expression. Cells were exposed to hypoxia for 6 h beginning 48 h after siRNA transfection. Samples were harvested in the absence or presence of the Sirt1 inhibitors sirtinol and nicotinamide (NAM). B, endogenous HIF-2α was immunoprecipitated from samples prepared as in A and subjected to immunoblot analysis with antibody recognizing HIF-1α or acetylated lysine. C, a model for Sirt1 activation during acute hypoxia. Upon hypoxia exposure, HIF-1α and HIF-2α stabilize and bind to the HREs in the proximal Sirt1 promoter region, resulting in increased Sirt1 gene expression.
We examined whether a reduction in endogenous Sirt1 protein levels, as a result of HIF-1α knockdown, affected acetylation of endogenous HIF-2α during hypoxia. Acetylated HIF-2α is normally undetectable without Sirt1 inhibition (Fig. 5B, lane 1). However, after knockdown of HIF-1α, acetylated HIF-2α was readily detectable in the absence of the pharmacological inhibitors (Fig. 5B, lane 2). This was not due to an overall increase in acetylated HIF-2α levels, because the addition of Sirt1 pharmacological inhibitors resulted in similar levels of acetylated HIF-2α in the absence (Fig. 5B, lane 3) or presence (Fig. 5B, lane 4) of HIF-1α knockdown. Moreover, acetylated HIF-2α, which was detectable after HIF-1α knockdown, was deacetylated by ectopic wild type Sirt1 (Fig. 5B, lane 6) but not by deacetylase mutant (Fig. 5B, lane 8) Sirt1.
DISCUSSION
Why is it important to understand how Sirt1 action is maintained during hypoxia? Studies of Sirt1 activation to date have primarily focused on post-translational mechanisms of regulation. These mechanisms include changes in pyridine nucleotide levels or ratios that affect Sirt1 enzymatic activity, Sirt1 phosphorylation or other kinase-induced effects (35–40), or Sirt1 subcellular localization (41–44). Changes in pyridine nucleotide levels or ratios are perhaps the best studied mechanism for regulating Sirt1 function (45). However, this mechanism is unlikely to be responsible for activation of Sirt1 during hypoxia.
During ischemia, the change in redox state, which mirrors the pyridine nucleotide ratio, does not favor Sirt1 activation. Similarly, the alterations in pyridine nucleotide levels during hypoxia are opposite that associated with increased Sirt1 enzymatic activity. Although most hypoxia/pyridine nucleotide studies examined total NAD+ or NADH levels rather than free NAD+ or NADH levels or ratios, which are the relevant molecular species for NAD+/NADH-regulated enzymes such as Sirt1, it is nevertheless unlikely that a major change in direction of the cellular redox state during hypoxia would occur if free NAD+/free NADH ratios were calculated. Therefore, an alternative mechanism for activation of Sirt1 during hypoxia, besides changes in redox state and pyridine nucleotide levels/ratios, must exist.
Transcription factors that participate in hypoxia-dependent regulation and that may be regulated by Sirt1 are attractive candidates for controlling Sirt1 activity during hypoxia. Sirt1 and HIF are critical regulators of physiological processes in mammals, including several affected by hypoxia or ischemia (46), such as metabolic adaptation (47–49), angiogenesis (50–55), and oxidative stress defense mechanisms (56). A logical expectation is that feedback mechanisms may exist for transcription factors whose target genes encode genetic regulators, which in turn act upon the transcription factor itself. This is the case for FoxO, a transcription factor whose activity is also augmented by Sirt1 deacetylation (57) and which acts as a positive regulator of Sirt1 gene expression (58).
Based upon our data obtained from cell culture and animal models, we suggest that Sirt1 induction during acute hypoxia is mediated through binding of HIF complexes to, and subsequent activation of, the proximal Sirt1 promoter (Fig. 5C). An increase in either HIF-1 or HIF-2 signaling is sufficient to increase Sirt1 gene expression, whereas a decrease in either HIF-1 or HIF-2 signaling blunts the induction of Sirt1 gene expression during acute hypoxia. Finally, although Sirt1 acts selectively with HIF-2α to augment HIF-2 signaling, Sirt1 does not augment HIF-1 induction of isolated HIF-responsive regulatory regions from the Epo enhancer, VEGF promoter, Sod2 promoter, and, as now reported, the Sirt1 proximal promoter in Hep3B cells (21), raising the possibility of positive feedback regulation.
As with other HIF-regulated regulatory regions, we only observed a selective and stimulatory action of Sirt1 on HIF-2, and not HIF-1, transactivation of the Sirt1 promoter. The link between Sirt1 and HIF-2 pathways involves Sirt1-dependent deacetylation of acetylated HIF-2α; we have previously found significant acetylation of HIF-2α, and not HIF-1α, using either exogenous or endogenous HIF-α proteins in HEK293 and Hep3B cells (21). Determining how acetylation of HIF-2α, by a currently undetermined acetyltransferase, and Sirt1-mediated deacetylation act to increase HIF-2 signaling will require additional studies.
Whether Sirt1 plays a role in HIF-1 signaling during acute hypoxia is less clear. We have only found a functional and positive role for Sirt1 in HIF-2-dependent, and not HIF-1-dependent, induction of endogenous target genes during acute hypoxia in Hep3B cells and in mice (21). In the current study, we extended our evaluation of HIF acetylation in a more prolonged hypoxia time course in both Hep3B cells and HT1080 cells, the cell line used in the study that reported HIF-1α acetylation (31). In contrast to the results of Lim et al. (31), we found no significant acetylation of ectopic HIF-1α in either Hep3B or HT1080 cells during hypoxia.
The reasons for the discrepancy between our results and those of Lim et al. (31) is unclear, although we note that Lim et al. (31) evaluated HIF-1α acetylation sites induced by ectopic PCAF overexpression rather than by hypoxia. Because it is probably more robust, acetylation induced by exogenous PCAF may allow for easily detectable amounts of acetylated HIF-1α. However, this raises the question of whether low level acetylation of HIF-1α during hypoxia, if present, is physiologically relevant. Irrespective of whether or not low level acetylation of HIF-1α occurs during hypoxia, we have not observed a functional role for Sirt1 in HIF-1 signaling in cells or in mice exposed to acute hypoxia (21). Additional experiments will be needed to clarify if Sirt1 plays a role in HIF-1 signaling during chronic hypoxia.
Our data suggest a functional role for both HIF-1 and HIF-2 in the control of Sirt1 gene expression during acute hypoxia that is mediated by transcriptional activation of the Sirt1 locus. Moreover, impeding HIF-1 signaling has functional consequences upon HIF-2 acetylation, which is evident after HIF-1α knockdown in the absence of Sirt1 inhibitors. The reduction in Sirt1 protein levels, the increase in HIF-2α acetylation due to reduced Sirt1 levels, and the lack of compensatory changes in Sirt1 gene expression by HIF-2α after HIF-1α knockdown indicate that impeding HIF-1 signaling will probably directly affect Sirt1 as well as HIF-2 signaling, thereby intricately linking Sirt1 and HIF signaling. Indeed, the blunting of EPO gene expression in Hep3B cells after HIF-1α knockdown, albeit a more muted effect compared with that resulting from HIF-2α knockdown, might reflect loss of Sirt1 augmentation of HIF-2 signaling in addition to, or as opposed to, a reduction in HIF-1-mediated induction of EPO directly.
The sensitivity of Sirt1 gene expression to HIF-α gene dosage indicates an exquisite reliance upon HIF signaling, one that results in an increase in Sirt1 activity during initial hypoxia exposure. Similar to other HIF-2-responsive regulatory regions (21), HIF-2 activation of the isolated Sirt1 promoter is augmented by Sirt1. HIF-2 activation of the Sirt1 promoter is probably influenced by other trans-acting factors, besides Sirt1, because mutations in the candidate EBS that flank HRE5 selectively affect HIF-2-mediated, and not HIF-1-mediated, induction of the isolated Sirt1 promoter. The spatial relationship of the HRE and flanking EBS are consistent with that found in other HIF-2-selective HIF target genes (32–34). Definitive proof of a role for Ets family members in Sirt1 regulation will require further studies.
With continued hypoxic exposure beyond 8–12 h in cell culture, Sirt1 mRNA and protein levels begin to decline. The biphasic response of Sirt1 to hypoxia suggests a favorable role for Sirt1 in the acute phase of hypoxia and a neutral, or even deleterious, role for Sirt1 in the latter phases of hypoxia. Although the complexities of Sirt1 regulation during hypoxia have not been completely elucidated, redox-dependent mechanisms, which repress Sirt1 gene expression in a HIF-independent manner, may be relevant in the chronic hypoxia state (59).
Although we cannot exclude post-translational mechanisms as contributions to Sirt1 activation during hypoxia, our results suggest that Sirt1 activity is maintained, and even increased, during acute hypoxia through mass action effects mediated by HIF-dependent increases in Sirt1 gene expression. HIF and Sirt1 activity can also be increased in hypoxia-independent manners, including by oxidative stress, and thus Sirt1/HIF interplay may have broader relevance. Finally, given the augmentation of HIF-2 signaling by Sirt1 and now the role of HIF-2 in induction of Sirt1 gene expression, we propose that a feedback system linking Sirt1 activation with HIF-2 signaling is active during acute hypoxia and probably other environmental stress states. Additional studies will be needed to confirm the existence of such a system and to better understand the relevance of Sirt1/HIF signaling to mammalian physiological and pathophysiological states.
Acknowledgments
We thank A. Das and E. Ballard for technical assistance, M. Alexander for suggestions for in vivo chromatin immunoprecipitation assays, Ko Uyeda for insightful comments regarding measurements of cellular redox state and their current limitations, Rick Bruick for the VHL expression plasmid, and Randall Johnson for HIF-1α founder mice.
This work was supported by the American Heart Association and the Department of Veterans Affairs.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- HIF
- hypoxia-inducible factor
- ARNT
- aryl hydrocarbon receptor nuclear translocator
- HRE
- HIF-response element
- PAS
- Per/ARNT/Sim
- VHL
- von Hippel-Lindau
- EBS
- ETS-binding site(s)
- SP
- S-peptide
- NTAD
- N-terminal activation domain
- CTAD
- C-terminal activation domain
- PPN
- triple substitution mutant for two NTAD proline residues and one CTAD asparagine residue in the HIF-α protein that results in an oxygen-independent, “constitutively active” protein.
REFERENCES
- 1. Semenza G. L. (2000) Genes Dev. 14, 1983–1991 [PubMed] [Google Scholar]
- 2. Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tian H., McKnight S. L., Russell D. W. (1997) Genes Dev. 11, 72–82 [DOI] [PubMed] [Google Scholar]
- 4. Ema M., Taya S., Yokotani N., Sogawa K., Matsuda Y., Fujii-Kuriyama Y. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4273–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gu Y. Z., Moran S. M., Hogenesch J. B., Wartman L., Bradfield C. A. (1998) Gene Expr. 7, 205–213 [PMC free article] [PubMed] [Google Scholar]
- 6. Zelzer E., Wappner P., Shilo B. Z. (1997) Genes Dev. 11, 2079–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang J., Zhang L., Erbel P. J., Gardner K. H., Ding K., Garcia J. A., Bruick R. K. (2005) J. Biol. Chem. 280, 36047–36054 [DOI] [PubMed] [Google Scholar]
- 8. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 9. Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 10. Huang L. E., Gu J., Schau M., Bunn H. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bruick R. K., McKnight S. L. (2001) Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 12. Cockman M. E., Masson N., Mole D. R., Jaakkola P., Chang G. W., Clifford S. C., Maher E. R., Pugh C. W., Ratcliffe P. J., Maxwell P. H. (2000) J. Biol. Chem. 275, 25733–25741 [DOI] [PubMed] [Google Scholar]
- 13. Kamura T., Sato S., Iwai K., Czyzyk-Krzeska M., Conaway R. C., Conaway J. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10430–10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. (1999) Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 15. Ohh M., Park C. W., Ivan M., Hoffman M. A., Kim T. Y., Huang L. E., Pavletich N., Chau V., Kaelin W. G. (2000) Nat. Cell Biol. 2, 423–427 [DOI] [PubMed] [Google Scholar]
- 16. Tanimoto K., Makino Y., Pereira T., Poellinger L. (2000) EMBO J. 19, 4298–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hewitson K. S., McNeill L. A., Riordan M. V., Tian Y. M., Bullock A. N., Welford R. W., Elkins J. M., Oldham N. J., Bhattacharya S., Gleadle J. M., Ratcliffe P. J., Pugh C. W., Schofield C. J. (2002) J. Biol. Chem. 277, 26351–26355 [DOI] [PubMed] [Google Scholar]
- 18. Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu C. J., Wang L. Y., Chodosh L. A., Keith B., Simon M. C. (2003) Mol. Cell. Biol. 23, 9361–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Warnecke C., Zaborowska Z., Kurreck J., Erdmann V. A., Frei U., Wiesener M., Eckardt K. U. (2004) FASEB J. 18, 1462–1464 [DOI] [PubMed] [Google Scholar]
- 21. Dioum E. M., Chen R., Alexander M. S., Zhang Q., Hogg R. T., Gerard R. D., Garcia J. A. (2009) Science 324, 1289–1293 [DOI] [PubMed] [Google Scholar]
- 22. O'Rourke J. F., Tian Y. M., Ratcliffe P. J., Pugh C. W. (1999) J. Biol. Chem. 274, 2060–2071 [DOI] [PubMed] [Google Scholar]
- 23. Baek J. H., Mahon P. C., Oh J., Kelly B., Krishnamachary B., Pearson M., Chan D. A., Giaccia A. J., Semenza G. L. (2005) Mol. Cell 17, 503–512 [DOI] [PubMed] [Google Scholar]
- 24. Bracken C. P., Whitelaw M. L., Peet D. J. (2005) J. Biol. Chem. 280, 14240–14251 [DOI] [PubMed] [Google Scholar]
- 25. Chen L., Uchida K., Endler A., Shibasaki F. (2007) J. Biol. Chem. 282, 12707–12716 [DOI] [PubMed] [Google Scholar]
- 26. Elvert G., Kappel A., Heidenreich R., Englmeier U., Lanz S., Acker T., Rauter M., Plate K., Sieweke M., Breier G., Flamme I. (2003) J. Biol. Chem. 278, 7520–7530 [DOI] [PubMed] [Google Scholar]
- 27. Tissenbaum H. A., Guarente L. (2001) Nature 410, 227–230 [DOI] [PubMed] [Google Scholar]
- 28. Ryan H. E., Lo J., Johnson R. S. (1998) EMBO J. 17, 3005–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tian H., Hammer R. E., Matsumoto A. M., Russell D. W., McKnight S. L. (1998) Genes Dev. 12, 3320–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scortegagna M., Morris M. A., Oktay Y., Bennett M., Garcia J. A. (2003) Blood 102, 1634–1640 [DOI] [PubMed] [Google Scholar]
- 31. Lim J. H., Lee Y. M., Chun Y. S., Chen J., Kim J. E., Park J. W. (2010) Mol. Cell 38, 864–878 [DOI] [PubMed] [Google Scholar]
- 32. Aprelikova O., Wood M., Tackett S., Chandramouli G. V., Barrett J. C. (2006) Cancer Res. 66, 5641–5647 [DOI] [PubMed] [Google Scholar]
- 33. Le Bras A., Lionneton F., Mattot V., Lelièvre E., Caetano B., Spruyt N., Soncin F. (2007) Oncogene 26, 7480–7489 [DOI] [PubMed] [Google Scholar]
- 34. Ohradanova A., Gradin K., Barathova M., Zatovicova M., Holotnakova T., Kopacek J., Parkkila S., Poellinger L., Pastorekova S., Pastorek J. (2008) Br. J. Cancer 99, 1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kang H., Jung J. W., Kim M. K., Chung J. H. (2009) PloS One 4, e6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zschoernig B., Mahlknecht U. (2009) Biochem. Biophys. Res. Commun. 381, 372–377 [DOI] [PubMed] [Google Scholar]
- 37. Ford J., Ahmed S., Allison S., Jiang M., Milner J. (2008) Cell Cycle 7, 3091–3097 [DOI] [PubMed] [Google Scholar]
- 38. Nasrin N., Kaushik V. K., Fortier E., Wall D., Pearson K. J., de Cabo R., Bordone L. (2009) PloS One 4, e8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo X., Williams J. G., Schug T. T., Li X. (2010) J. Biol. Chem. 285, 13223–13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hisahara S., Chiba S., Matsumoto H., Tanno M., Yagi H., Shimohama S., Sato M., Horio Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15599–15604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sugino T., Maruyama M., Tanno M., Kuno A., Houkin K., Horio Y. (2010) FEBS Lett. 584, 2821–2826 [DOI] [PubMed] [Google Scholar]
- 43. Tanno M., Kuno A., Yano T., Miura T., Hisahara S., Ishikawa S., Shimamoto K., Horio Y. (2010) J. Biol. Chem. 285, 8375–8382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tanno M., Sakamoto J., Miura T., Shimamoto K., Horio Y. (2007) J. Biol. Chem. 282, 6823–6832 [DOI] [PubMed] [Google Scholar]
- 45. Denu J. M. (2003) Trends Biochem. Sci. 28, 41–48 [DOI] [PubMed] [Google Scholar]
- 46. Cai Z., Manalo D. J., Wei G., Rodriguez E. R., Fox-Talbot K., Lu H., Zweier J. L., Semenza G. L. (2003) Circulation 108, 79–85 [DOI] [PubMed] [Google Scholar]
- 47. Huang Y., Hickey R. P., Yeh J. L., Liu D., Dadak A., Young L. H., Johnson R. S., Giordano F. J. (2004) FASEB J. 18, 1138–1140 [DOI] [PubMed] [Google Scholar]
- 48. Seagroves T. N., Ryan H. E., Lu H., Wouters B. G., Knapp M., Thibault P., Laderoute K., Johnson R. S. (2001) Mol. Cell. Biol. 21, 3436–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oktay Y., Dioum E., Matsuzaki S., Ding K., Yan L. J., Haller R. G., Szweda L. I., Garcia J. A. (2007) J. Biol. Chem. 282, 11750–11756 [DOI] [PubMed] [Google Scholar]
- 50. Carmeliet P., Dor Y., Herbert J. M., Fukumura D., Brusselmans K., Dewerchin M., Neeman M., Bono F., Abramovitch R., Maxwell P., Koch C. J., Ratcliffe P., Moons L., Jain R. K., Collen D., Keshert E. (1998) Nature 394, 485–490 [DOI] [PubMed] [Google Scholar]
- 51. Duan L. J., Zhang-Benoit Y., Fong G. H. (2005) Circulation 111, 2227–2232 [DOI] [PubMed] [Google Scholar]
- 52. Peng J., Zhang L., Drysdale L., Fong G. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8386–8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Morita M., Ohneda O., Yamashita T., Takahashi S., Suzuki N., Nakajima O., Kawauchi S., Ema M., Shibahara S., Udono T., Tomita K., Tamai M., Sogawa K., Yamamoto M., Fujii-Kuriyama Y. (2003) EMBO J. 22, 1134–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ding K., Scortegagna M., Seaman R., Birch D. G., Garcia J. A. (2005) Invest. Ophthalmol. Vis. Sci. 46, 1010–1016 [DOI] [PubMed] [Google Scholar]
- 55. Dioum E. M., Clarke S. L., Ding K., Repa J. J., Garcia J. A. (2008) Invest. Ophthalmol. Vis. Sci. 49, 2714–2720 [DOI] [PubMed] [Google Scholar]
- 56. Scortegagna M., Ding K., Oktay Y., Gaur A., Thurmond F., Yan L. J., Marck B. T., Matsumoto A. M., Shelton J. M., Richardson J. A., Bennett M. J., Garcia J. A. (2003) Nat. Genet. 35, 331–340 [DOI] [PubMed] [Google Scholar]
- 57. Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 58. Nemoto S., Fergusson M. M., Finkel T. (2004) Science 306, 2105–2108 [DOI] [PubMed] [Google Scholar]
- 59. Zhang Q., Wang S. Y., Fleuriel C., Leprince D., Rocheleau J. V., Piston D. W., Goodman R. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 829–833 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]