Abstract
HBR1 (hemoglobin response gene 1) is an essential gene in Candida albicans that positively regulates mating type locus MTLα gene expression and thereby regulates cell type-specific developmental genes. Hbr1p contains a phosphate-binding loop (P-loop), a highly conserved motif characteristic of ATP- and GTP-binding proteins. Recombinant Hbr1p was isolated in an oligomeric state that specifically bound ATP with Kd ∼2 μm. ATP but not ADP, AMP, GTP, or dATP specifically protected Hbr1p from proteolysis by trypsin. Site-directed mutagenesis of the highly conserved P-loop lysine (K22Q) and the less conserved glycine (G19S) decreased the binding affinity for soluble ATP and ATP immobilized through its γ-phosphate. ATP bound somewhat more avidly than ATPγS to wild type and mutant Hbr1p. Although Hbr1p exhibits sequence motifs characteristic of adenylate kinases, and adenylate kinase and ATPase activities have been reported for the apparent human ortholog of Hbr1p, assays for adenylate kinase activity, autophosphorylation, and ATPase activity proved negative. Overexpression of wild type but not the mutant forms of Hbr1p restored MTlα2 expression in an HBR1/hbr1 mutant, indicating that ATP binding to the P-loop is necessary for this function of Hbr1p.
Keywords: ATP, Fungi, Nucleotide, Site-directed Mutagenesis, Yeast, Candida albicans, P-loop, Adenylate Kinase, Nucleotide-binding Site
Introduction
Candida albicans typically resides as a commensal in the human gastrointestinal tract but becomes an opportunistic pathogen in immunocompromised hosts. Infections can spread by invasive colonization of host organs and vascular dissemination (1). Adaptation to each host organ may require changes in phenotype that are triggered by specific host environmental cues. For example, white to opaque cell type switching has been observed at some sites of infection (2, 3). Opaque phase cells, although vulnerable to host defenses, are necessary for efficient mating in C. albicans (4). However, suppression of this switching during vascular dissemination may help C. albicans elude host defenses. HBR1 (hemoglobin response gene 1) is a gene that represses white opaque switching and may play a key role to understanding C. albicans survival in a host.
HBR1 was identified based on its differential induction in cells cultured with hemoglobin and discovered to positively regulate MTLα genes (5). MTLα genes, in turn, control white opaque switching (4). HBR1 shows haplo-insufficiency for MTLα gene expression, indicating sensitivity of the MTL signaling pathway to levels of Hbr1p (5).
HBR1 is an essential gene in C. albicans (5), but its function in mating is not sufficient to explain its requirement during vegetative growth. In fact, the mating genes in C. albicans are not essential for viability (6, 7), suggesting that Hbr1p plays additional roles in vegetative growth regulation. HBR1 gene expression is maximal during early exponential growth, attenuated during the diauxic transition, and weak in the stationary phase (5).
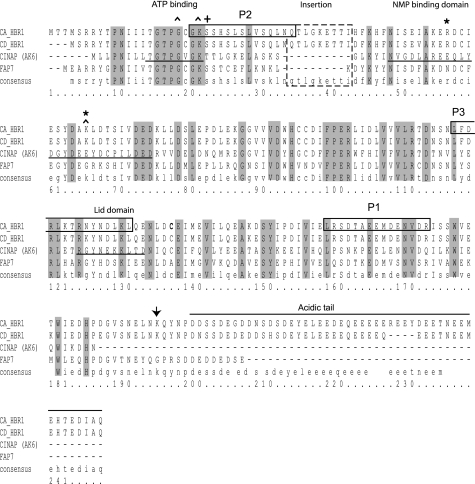
Hbr1p may serve a regulatory role at the level of protein-protein interactions. The yeast ortholog Fap7 is also an essential gene (8) and was initially shown to interact with Krr1p, the product of a gene required for ribosomal RNA processing (9). FAP7 heterozygosis also leads to an 18S rRNA processing defect (10) that can be recapitulated with the K20R mutation of its predicted phosphate-binding loop (P-loop)3 (11). In addition, a G19S mutation prevented oxidative stress-induced activation of a Gal4-Pos9 hybrid transcription factor, indicating an important functional role for this motif (8).
Hbr1p from C. albicans and Candida dubliniensis shares 68% amino acid sequence conservation with Fap7p and more than 40% identity with members of a group of nuclear-localized adenylate kinases (AK-6 family) (Fig. 1). This identity includes a predicted P-loop motif (12) that is characteristic of ATP- and GTP-binding proteins (13, 14). The P-loop pattern in Hbr1p (GTPGCGKS) most closely resembles that of the archaeal derived AK sequence (GXPGXGK(T/S)) (15). The absence of a terminal glycine that follows the invariable lysine, which is a unique feature of the archaeal AK family (15), is also a distinguishing feature of the AK-6 group from higher eukaryotes (16–18) (Fig. 1).
FIGURE 1.
Hbr1p amino acid sequence, predicted functional regions, and mutation sites. The 28.8-kDa amino acid sequence of Hbr1p from C. albicans is shown aligned with that of C. dubliniensis, the potential human ortholog AD-004/CINAP (coilin-interacting nuclear ATPase protein, AK-6), and the S. cerevisiae ortholog, Fap7p. The shaded areas indicate regions of identity between the four proteins. Mutations are indicated by carets and were designed to replace the consensus (K22Q) and nonconsensus (G19S) residues in the P-loop motif. A K66R mutation in the NMP-binding domain was designed to disrupt a predicted SUMO acceptor site (37). The designations for the ATP and NMP binding and the lid domains are taken from the crystal structure of CINAP (16) and are indicated by underlining. The three peptides used as epitopes for antibody production are indicated by solid boxed areas, and a Candida-specific insertion is indicated by a dashed boxed region. The lack of a terminal P-loop glycine that is a defining characteristic of the nuclear-localized group of adenylate kinase enzymes is indicated by a (+). The acidic carboxyl terminus contributes to the overall net charge of the molecule (−19.4 to −47).
The defining member of the nuclear-localized AK group is human AD-004 (CINAP; AK-6), and its crystal structure has been determined (16). The positions of the P-loop, the NMP-binding domain, and the lid domain are indicated in the Hbr1p primary sequence (Fig. 1). Both ATPase and AK activities have been demonstrated for AD-004 (16, 19). Nuclear localization of AD-004 may depend upon its interaction with coilin, a major protein component of Cajal bodies (19, 20). These bodies are extranucleosomal maturation sites of snRNAs, small nucleolar RNAs, and histone mRNA 3′ ends (21).
Despite major similarities, the primary amino acid sequence of Hbr1p differs in two aspects from these potential orthologs: (i) a 9-amino acid insertion between the P-loop and predicted NMP-binding domain and (ii) a highly negatively charged carboxyl terminus (Fig. 1). We examine here the nucleotide binding specificity and potential enzymatic activities of Hbr1p. We also report effects of site-directed mutations in the P-loop on ligand binding to Hbr1p and functional effects of Hbr1 on MTL gene expression.
EXPERIMENTAL PROCEDURES
Construction of HBR1 Expression Vectors
Thrombin-His6-tagged versions of Hbr1p were constructed in two steps. A clone containing the entire HBR1 open reading frame (5) was amplified by PCR using PFX polymerase (Invitrogen) and the following primers (5′ to 3′): GGTACCTATGACAACCATGTCCAAG to introduce a KpnI cloning site at the 5′ end of the gene and a 3′ primer containing coding sequences for a thrombin cleavage site followed by a His6 tag sequence TAATGGTGATGGTGATGATGACCAGCAGCAGAACCTCTAGGAACCAATTGTGCAATATCTTCTGTATGC (1.65 kDa). The purified PCR fragment was cloned into pCR4-Blunt TOPO (Invitrogen) using the manufacturer's standard protocol to generate plasmid pR1-THis. Second, the sequence cloned in pR1-THis was changed to standard codon usage by converting the leucine CUG codon at position 27 to one that would encode serine in standard usage, CCT (22). Primers GGAAATCATCTCATTCCTCATGTTTAGTTTCTCAACTC and GAGTTGAGAAACTAAACTAGAGGAATGAGATGATTTCC were used with the QuikChange II XL system (Stratagene, La Jolla, CA) to generate a codon-corrected version of the tagged HBR1 gene in plasmid pR1-THis-ls.
Three additional mutations were introduced into pR1-THis-ls. The first single amino acid mutant G19S (Fig. 1B) occurs in a consensus region of the P-loop in Hbr1p orthologs (5) and AK (13) but not in the consensus region of a generalized P-loop motif (13). The second P-loop mutant K22Q occurs in a universal P-loop consensus region (13) that is believed to have a primary role in catalysis (23). A third mutant K66R occurs at a predicted small ubiquitin-related modifier (SUMO) acceptor site. The primer sequences (5′ to 3′) and the modification sites are as follows: G19S, CAGGTACACCTAGTTGTGGGAAATCATCTC and GAGATGATTTCCCACAACTAGGTGTACCTG; K22Q, CACCTGGGTGTGGGCAATCATCTCATTCCC and GGGAATGAGATGATTGCCCACACCCAGGTG; and SUMO, GATGCCAGGTTAGATACTTCGATTGTAGACG and CGTCTACAATCGAAGTATCTAACCTGGCATC. Protein expression vectors were constructed by releasing the tagged and modified sequences using KpnI and EcoRI and cloning into the yeast expression vector pYES2 (Invitrogen) cut at the same sites to create plasmids pYES-R1-THis-G19S, pYES-R1-THis-K22Q, and pYES-R1-THis-K66Q. All of the gene modifications were verified by sequence analysis.
Vectors for expression of mutated HBR1 genes were generated by amplifying the HBR1 gene with Platinum PFX DNA polymerase (Invitrogen) using primers CACCATGACAACCATGTCAAGAAGATATACACC and GAGCTGCAGGCTATTGTGCAATATCTTCTG to directionally clone into vector pENTR/d-TOPO (Invitrogen). The mutations were carried out by site-directed mutagenesis using the primers described above to generate G19S and K22Q mutations in the HBR1 gene. All of the gene modifications were verified by sequence analysis. A Gateway destination vector (Invitrogen) was constructed in plasmid pCaEXP-RFA by blunt end cloning of reading frame cassette A (Invitrogen) into the blunted BamHI site of pCaEXP (24) that had been partially digested with NdeI to release the RP10 gene. The RF cassette was sequenced to obtain a clone of the correct orientation that placed the attR1 lambda recombination site nearest the 3′ end of the MET3 promoter. The HBR1 wild type (WT) and mutated genes were transferred to pCaEXP-RFA using LR Clonase following the instructions of the manufacturer (Invitrogen). Selected constructs were sequenced to ensure that no mutations had occurred during manipulations and then introduced into the genome of C. albicans strain BWP17 (25) by digesting at the unique ClaI site in the HBR1 gene and transformation by the lithium acetate method. Ura+ colonies were selected for PCR analysis to ensure correct incorporation of the gene into the BWP17 genome. The presence of the integrated construct was confirmed using primers PN52 in the MET3 promoter (CAAGTATACGTAATCTCCCC) and PN55 in the HBR1 gene (CCTGCAGATATCTGTCAGGAATATAGCTGTC) located 60 bases 3′ of the ClaI site (data not shown).
Protein Expression
Transformation and protein expression followed standard protocols (26–28) and the manufacturer's instructions. Saccharomyces cerevisiae InvSC1 cells (Invitrogen) transformed with pYES-R1-THis-HBR1 were cultured at 30 °C and grown to OD650 nm ≅ 4.0 in 2% glucose YNB −URA medium (26). The cells were then washed and reinoculated to OD650 nm = 0.4 in 2% galactose YNB −URA medium, and protein expression was induced for 20–24 h (OD650 nm ≅ 2.0). The cells were centrifuged, and the protein extract was generated by cell lysis with 0.5-mm glass beads (29) (BioSpec Products, Inc., Bartlesville, OK) in extraction buffer that contained 50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm MgCl2, and the following protease inhibitors: 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (MP Biomedicals, Inc., Aurora, OH), 10 μm leupeptin, 20 μm pepstatin A, and 1 μm E-64 (Sigma).
Protein Purification
Hbr1p was purified from cell extracts by performing two sequential nickel affinity purification columns (2× nickel) according to the manufacturer's protocol (Amersham Biosciences). The optimized imidazole elution concentration for His-tagged Hbr1p was determined to be 150 mm. After the first purification, the Hbr1p sample was either dialyzed or diluted to 15 mm imidazole buffer and reapplied to the nickel column. After the second purification, the Hbr1p sample was dialyzed with a 12,000 molecular weight cutoff membrane (Spectrum Laboratories, Inc., Rancho Dominguez, CA) in dialysis buffer: 150 mm NaCl, 1 mm MgCl2, and 50 mm Tris-HCl, pH 7.5. For specified assays, Hbr1p was affinity-purified with cobalt, copper, or zinc substituting for nickel as described.
Gel Electrophoresis
Protein extracts were prepared in LDS sample buffer containing 50 mm DTT and heated to 95 °C before sample loading. Electrophoresis of protein samples in 12% Bis-Tris NuPAGE lithium dodecyl sulfate polyacrylamide gels was performed in 3-(N-morpholino)-propanesulfonic acid buffer according to the manufacturer's protocol (Invitrogen). The protein bands were visualized with Coomassie Brilliant Blue R-250 (Invitrogen).
Western Blot
Western blotting was performed according to a general protocol for semidry systems (30). For anti-His Western blots, 1:1500 primary (Amersham Biosciences) and 1:5000 goat anti-mouse HRP secondary antibody (KPL, Gaithersburg, MD) dilutions were used. For anti-peptide Western blots, 1:1000 primary (αP1, P2, or P3) and 1:5000 goat anti-rabbit HRP secondary antibody (KPL) dilutions were used. Reacting secondary antibodies were detected by SuperSignal enhanced chemiluminescent substrate (Pierce).
Polyclonal Antibodies
Rabbit antisera recognizing three peptide sequences of Hbr1p were prepared by immunizing rabbits with the following sequences cross-linked to BSA using glutaraldehyde: P1, 160LRSDTAEEMDENVDR174; P2, 21GKSSHSLSLVSQLNQ35; and P3, 118LFDRLKTRNYNDL130.
Mass Spectrometry (LC/MS)
One- and two-dimensional gel electrophoresis (30) was performed using 2× nickel affinity-purified Hbr1p. Discrete protein bands were excised and digested using trypsin, and standard LC/MS preparation, procedure, and analysis were performed (31, 32) using a S. cerevisiae database containing the C. albicans Hbr1p sequence.
Ion Exchange Chromatography
Ion exchange chromatography was performed according to a general protocol (33). 2× nickel affinity-purified Hbr1p samples were diluted to [NaCl] of ≤50 mm and applied to DEAE fast flow Sepharose columns (Amersham Biosciences). The columns were rinsed and equilibrated with 10 mm NaCl in one of the following buffers: 50 mm Tris-HCl, pH 7.5; 50 mm phosphate, pH 6.0; 50 mm histidine, pH 5.0; and 50 mm acetate, pH 4.0. Step (100 mm NaCl/step) or continuous NaCl gradients (10 mm to 1 m NaCl) were used for ion exchange elution. Eluted fractions were collected and monitored by gel electrophoresis with either R-250 stain or anti-His Western blotting.
Thrombin Cleavage
2× nickel affinity-purified Hbr1p was incubated with a thrombin-agarose resin for 2 h according to the manufacturer's protocol (Sigma). Cleaved Hbr1p was recovered in the supernatant after centrifugation and verified by gel electrophoresis with R-250 staining. Generally, the assays were performed with the His tag intact because the presence of the tag did not influence the activity.
Gel Filtration
Hbr1p was further purified by size exclusion chromatography (34). A 250-μl sample of 2× nickel affinity-purified Hbr1p (between 10 and 25 μm) was loaded into a liquid chromatography work station (Dionex, Sunnyvale, CA) that controlled a Superdex-75 column (Amersham Biosciences). The column was equilibrated with a buffer containing 50 mm Tris-HCl, pH 7.5, and 500 mm NaCl, and with a flow rate of 0.7 ml/min. Sample elution was monitored by absorbance at 280 nm. Gel filtration standards (Bio-Rad) were used to calibrate the column. For renatured Hbr1p, a 2× nickel affinity purification was performed in 8 m urea. A ∼20 μm WT Hbr1p sample was loaded into the column as above and renatured by exchanging with buffer on the column.
ATP Affinity Chromatography
Binding and elution of Hbr1p to ATP immobilized on a polyacrylamide resin (ATP-Binders Resin; Novagen, San Diego, CA) was performed according to the manufacturer's instructions with the following modifications. 2× nickel affinity-purified Hbr1p samples containing ∼10 μm each of WT and mutant protein (G19S, K22Q, and K66R) were incubated for 2.5 h at 4 °C with a pretreated ATP resin. After Hbr1p incubation, the ATP resin was washed with buffer containing three potential ATP competitors: ADP, AMP, and NADH. The resin elution was performed using three fractions containing ATP. Each eluted fraction represented three column volumes of 20 mm ATP buffer incubated for 5 min at room temperature. Western blotting with anti-His antibody was used to visualize Hbr1p in the washed and eluted fractions. ImageQuant 5.0 software (Molecular Dynamics, Sunnyvale, CA) was used to quantify Western blot band intensities. The experiments were performed in duplicate for Hbr1p mutants and triplicate for WT Hbr1p.
Equilibrium Dialysis
Dialysis was carried out in paired 100 μl of volume dialysis chambers fitted with 3500 molecular weight cutoff membranes (Spectrum Laboratories, Inc.) and equilibrated with dialysis buffer (150 mm NaCl, 1 mm MgCl2, and 50 mm Tris-HCl, pH 7.5). One side of the dialysis membrane contained 100 μl of 10 μm Hbr1p (WT, G19S, K22Q, or K66R). Both sides of dialysis membrane contained varying concentrations (0–300 μm) of nonradioactive ATPγS (Roche Applied Science) and fixed concentrations of trace radiolabeled [γ-35S]ATP (≤0.625 nm) (MP Biomedicals, Irvine, CA). The chambers were equilibrated at room temperature by rotation mixing for 48 h, and the radioactivity in each chamber was quantified by liquid scintillation counting. Each assay was performed and counted in triplicate.
Equilibrium Filtration
10 μm Hbr1p (WT, G19S, K22Q, and K66R) samples were incubated in dialysis buffer (above) with a fixed amount of trace radiolabeled [γ-32P]ATP (≤1 nm) (MP Biomedicals) and varied concentrations of nonradioactive ATP (Sigma) at room temperature for 1 h. Bound Hbr1p was centrifuged through a micro-Sephadex G-50 column (Amersham Biosciences), and radioactivity was quantified by liquid scintillation counting. The samples were prepared in triplicate and counted in duplicate.
Ligand Binding Analysis
The raw data (counts/min versus [ATPγS]added) represent the amount of [γ-35S]ATP bound to Hbr1p. Assuming that the ligands [γ-35S]ATP and ATPγS bind with equal affinity to Hbr1p, the fraction of [γ-35S]ATP bound equals the fraction of ATPγS bound. Because the fraction of nonlabeled ATPγS added is known, we calculated the concentration of bound ATPγS and free ATPγS. In this format, bound versus free, the data were fit to a single site homogenous equilibrium-binding isotherm. The equilibrium dissociation constant and maximum bound concentrations are fitted variables. The fit parameter for maximum bound concentration of [Hbr1p:ATPγS] varied from 7.5 to 9.4 μm, which is in good agreement with the known concentration of 10 μm Hbr1p measured by BCA protein assay (Pierce).
Treatment of the data for equilibrium filtration was the same as above except that the ligands used were [γ-32P]ATP and ATP. The fit parameter for maximum bound concentration of [Hbr1p: ATP] varied from 0.5 to 1.2 μm, which is below the measured concentration of Hbr1p (10 μm). This may indicate that a large (∼90%) fraction of Hbr1p remains on the G-50 column. The analysis relied on the assumption that the fraction of eluted Hbr1p represented the equilibrium before the filtration. The error in fitting these data is larger than for equilibrium dialysis but still appears to fit well to a single site homogenous model.
Trypsin Digestion
5 μm samples of Hbr1p (WT, G19S, K22Q, and K66R) were each incubated at room temperature for 1 h with 0.25 μm tosylphenylalanyl chloromethyl ketone-treated trypsin (Cooper Biomedical, Malvern, PA) in dialysis buffer either alone or with 20 mm ATP. For nucleotide comparison, WT Hbr1p was used as above with incubation buffers containing 20 mm each of the nucleotides ATP, ADP, AMP, GTP, or dATP. Gel electrophoresis and Coomassie staining monitored the extent of proteolysis after trypsin digestion.
Circular Dichroism
A 10 μm sample of WT Hbr1p was dialyzed into a 50 mm potassium phosphate buffer (pH 7.3) containing 1 mm MgCl2 and incubated at room temperature with and without ATP (200 μm) for 1 h. CD measurements were made using a Jasco J810 spectropolarimeter at 0 °C at 20 nm/min, using a time constant of 2 s. A cuvette pathlength of 0.1 cm was used to minimize ATP absorbance in the far UV range. A circular dichroism fitting program was used to fit the spectra to the Fasman algorithm (35). The spectral interpretation estimated the percentage secondary structure of α-helix, β-sheet, and random coil. Other algorithms used to fit the CD spectra gave similar secondary structure predictions (35).
ATPase assay
1 μm samples of WT Hbr1p or 30 μg/ml 2× nickel affinity-purified S. cerevisiae extract (blank) were incubated for 1.5 h at room temperature in 150 mm NaCl, 50 mm Tris, pH 7.5, 1 mm MgCl2, and either ∼1 nm [γ-32P]ATP or ∼1 nm [γ-35S]ATP. The Hbr1p samples contained protein purified at each stage of three sequential methods: 2× nickel affinity chromatography, ion exchange chromatography, and gel filtration. Peak fractions eluting from the ion exchange column at 500 mm NaCl in 50 mm pH 7.5 Tris buffer were collected. Hbr1p eluted from the gel filtration column at 15 min. Protein samples were normalized to 1 μm in a 50-μl reaction volume. Separation of nucleotide components was accomplished by poly(ethyleneimine)-cellulose TLC (EMD Chemicals, Gibbstown, NJ) using 0.5 m LiCl and 1 m formic acid as the mobile phase (36). Retention factor (Rf) values of radiolabeled [γ-32P]ATP or free 32P were measured by exposure of TLC plates to BioMax XAR film (Eastman Kodak Co.).
Adenylate Kinase Activity Assay
Samples containing 1 μm WT Hbr1p or 30 μg/ml C. albicans extract were each incubated for 20 min at 37 °C using 50 mm Tris (pH 7.5), 0.5 mg/ml BSA, 5 mm dithiothreitol, 5 mm MgCl2, 1 nm [γ-32P]ATP, and 0.4 mm NMP (either AMP, TMP, CMP, UMP, or GMP). The samples were loaded and dried onto poly(ethyleneimine)-cellulose TLC plates and eluted using 1 m LiCl (36). Rf values of radiolabeled [γ-32P]ATP, [β-32P]NDP, or free 32P were visualized by autoradiography.
Autophosphorylation Assay
5 μm WT Hbr1p was incubated for 1 min in dialysis buffer with 1 nm [γ-32P]ATP, 1× phosphatase inhibitors (phosphatase inhibitor mixture set II; Calbiochem, San Diego, CA), and varying concentrations (0–10 mm) of nonlabeled ATP. Gel electrophoresis and autoradiography was used to assess phosphate transfer to Hbr1p.
RESULTS
Hbr1 Protein Expression
C. albicans Hbr1-His6 was expressed in S. cerevisiae and purified by two sequential nickel affinity chromatographic steps. Denaturing and reducing gel electrophoresis of the recombinant protein preparation revealed a major component with an apparent molecular mass of ∼42 kDa and several other smaller prominent bands (Fig. 2A, lane 1, regions A–E). Because the calculated molecular mass of the His-tagged protein is 31 kDa, we used Western blot analysis to identify proteins containing the His epitope. Reactive bands greater than ∼31 kDa were identified, suggesting that the smaller species may be carboxyl-terminal truncations of the Hbr1p-His6 fusion protein (Fig. 2A, lane 2).
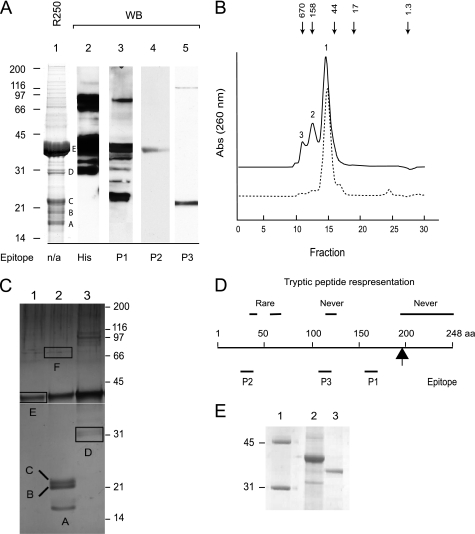
FIGURE 2.
Analysis of Hbr1p. A, Hbr1p-His6 protein was purified twice on a nickel affinity column. The samples were prepared by heating to 95 °C for 10 min in LDS buffer containing 50 mm DTT and electrophoresed under reducing conditions in a 12% acrylamide gel. Lane 1, Coomassie Blue R-250-stained Hbr1p-His6 preparation. Protein bands referred to in the text are labeled. Lane 2, Western blot (WB) of Hbr1p-His6 using His6 antibody. Lanes 3–5, Western blot using polyclonal anti-Hbr1p peptide antibodies generated as indicated in D. B, isocratic elution of purified recombinant Hbr1p-His6 from a Superdex-75 column with 50 mm Tris-HCl, pH 7.5, and 500 mm NaCl as the mobile phase. Solid line, Hbr1p-His6 preparation; dotted line, combined fractions 1–3 renatured after urea denaturation. Arrows represent the elution positions of molecular weight standards: thyroglobulin (bovine), 670,000; γ-globulin (bovine), 158,000; ovalbumin (chicken), 44,000; myoglobin (horse), 17,000; vitamin B12, 1,350. C, analysis of peak fractions from B on 12% LDS-PAGE as above. The gel regions excised for MS analysis are indicated (regions A–F). D, positions of peptide epitopes used for antibody generation and peptide representation from MS analysis. The arrow indicates the terminal basic residue. E, anomalous migration of Hbr1p. An Hbr1p-His6 preparation was digested with thrombin to release the His tag and electrophoresed using 12% reducing LDS-PAGE as above. Lane 1, MW markers; lane 2, Hbr1p-His6; lane 3, Hbr1p.
Gel filtration chromatography of this affinity-purified Hbr1p-His6 preparation revealed three major species (Fig. 2B, solid line). Analysis of each by LDS-PAGE identified six major protein bands similar in size to those of the affinity-purified preparation (Fig. 2C, regions A–F). These individual protein bands were excised, digested, and analyzed using LC/MS. The mass spectra were analyzed using a S. cerevisiae protein database with appended C. albicans Hbr1p data.
Tryptic peptides from Hbr1p-His6 extending to the terminal basic residue (Lys196) were identified in the mass spectra from all six gel slices (Fig. 2C). Peptides distal to Lys196 would not be identified in this analysis because these His fusion peptides would be filtered. The peptide sequence 122LKTRNYNDLK131 was never identified and, interestingly, was part of the peptide used to generate antibody P3 (Fig. 2D). This fragment also contains a high probability Type II nonconsensus sumoylation site (37). 40ETTIHFK46 and 56ERDCIESYDAK66 were only rarely identified, and both contain lower probability sumoylation sites (37) (Fig. 2D).
Rabbit polyclonal antibodies were generated to three peptide sequences that are most likely on surface-exposed regions of Hbr1p (Fig. 2D). The most carboxyl-terminal P1 epitope (residues 160–174) recognized the primary Hbr1p-His6 band at ∼42 and ∼22 kDa but did not recognize the 20- or 17-kDa fragments (Fig. 2A, lane 3). This is consistent with progressive Hbr1p truncations from the carboxyl terminus deleting the acidic region (22 kDa) to epitope P1 (20 kDa) and to epitope P3 (17 kDa). These smaller fragments were not detected with the anti-His antibody, which is consistent with this interpretation (Fig. 2A, lane 2). The P-loop epitope antibody P2 detected only the primary ∼42-kDa Hbr1p band (Fig. 2A, lane 4).
The anti-P3 antibody (residues 118–130) bound only to a 20–22-kDa fragment and not the primary 42-kDa Hbr1p-His6 band (Fig. 2A, lane 5). This epitope may be masked by partial renaturation upon transfer to the membrane, and cleavage of Hbr1p may selectively expose this internal epitope in the 22-kDa fragment.
Hbr1p-His6 renatured after urea treatment exhibited only the major 65-kDa peak (Fig. 2B, dotted line). Samples from the three major peaks of Hbr1p monitored by gel electrophoresis showed the same molecular mass (∼42 kDa), as did the single band for Hbr1p renatured from urea, suggesting that the three peaks represent different oligomerization states of Hbr1p (data not shown). Western blots using anti-His antibody confirmed that all peaks are Hbr1p-His6 (data not shown). The smallest mass estimated by gel filtration (65 kDa) is larger than the observed mass by gel electrophoresis (42 kDa) and approximately twice as large as the theoretical mass of Hbr1p-His6 (31 kDa), suggesting that this may be a dimer.
The preceding results demonstrated anomalous behavior of Hbr1p during electrophoresis. Hbr1p is a highly acidic protein with a net charge of −47 and a theoretical pI of 3.9. These values change only slightly when the His6 tag is included in the calculation (−47 and 4.12, respectively). Highly acidic proteins can exhibit electrophoretic mobilities differing from that expected for a completely unfolded globular protein (38). In most cases, protein mobility is decreased, resulting in an overestimation of the true mass, which may be explained by decreased detergent binding. Untagged Hbr1p also migrated more slowly than expected, indicating that the His tag is not the reason for the anomalous migration (Fig. 2E).
Hbr1p Binds to Immobilized ATP
As a first step to analyze the functional significance of the P-loop of Hbr1p, we used quantitative affinity chromatography (39) with an acrylamide-ATP resin. Hbr1p-His6 was quantitatively bound and could be completely released using a 1000-fold molar excess of ATP in three successive column washes (Fig. 3A). No Hbr1p-His6 release was detected in column washes containing the same excess of ADP, AMP, or NADH after prebinding in the presence of ATP (data not shown).
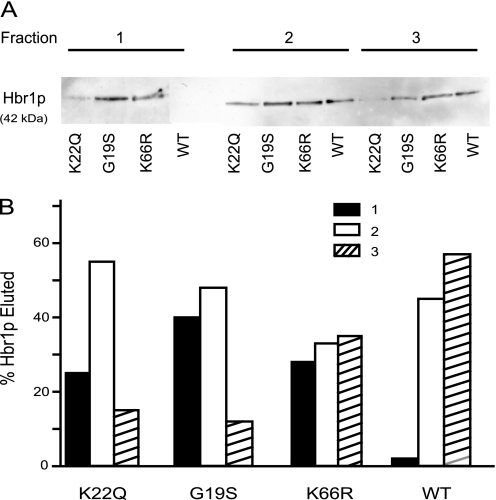
FIGURE 3.
Binding of WT and mutant Hbr1p to ATP immobilized on polyacrylamide resin. A, Western blot analysis using an anti-His antibody. Three fractions were sequentially collected by elution with 20 mm ATP. B, relative band intensities from above quantified by digital image analysis (ImageQuant).
Hbr1p mutations altering the P-loop consensus (G19S and K22Q) or the consensus putative SUMO attachment sites (K66R) (Fig. 1) were examined. The three purified Hbr1p His6 mutants were quantitatively retained on the resin. However, each eluted earlier than Hbr1p-His6, indicating decreasing affinities in the order WT > K66R > K22Q > G19S (Fig. 3A). Quantification of the Western blots using densitometry is shown in Fig. 3B. Interestingly, the putative SUMO attachment site mutant (K66R) was also destabilized for binding, suggesting a long range interaction with the P-loop or a conformational change that influences ATP binding.
Binding of ATPγS and ATP to Hbr1p
Equilibrium dialysis was used to assess the affinity of ATPγS for binding to WT and mutant Hbr1p proteins. ATPγS bound saturably to WT Hbr1p, and the data could be fit using a single site homogenous binding model with one binding site per Hbr1p (Fig. 4A). The dissociation constants obtained (±S.D.) are listed in Table 1. The K22Q mutation was the most destabilizing and had the highest Kd value for ATPγS, whereas the mutations G19S and K66R had less but still significant effects on ATPγS binding to Hbr1p.
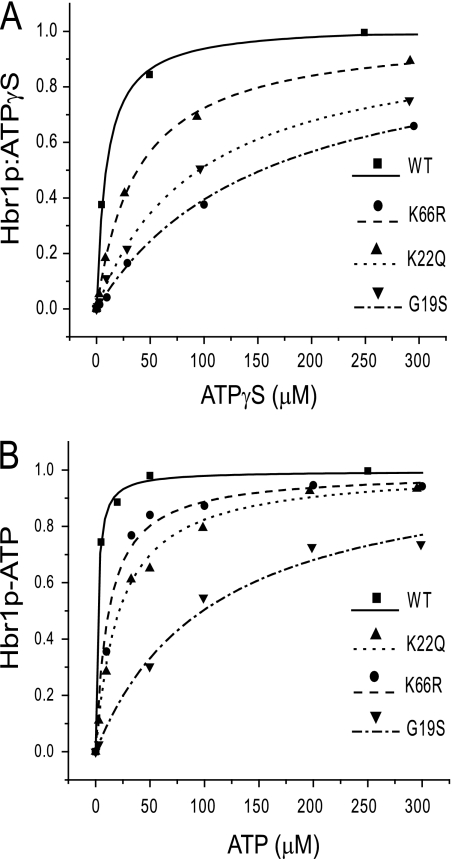
FIGURE 4.
Equilibrium binding of ATP to WT and mutant Hbr1p-His6. A, equilibrium dialysis of Hbr1p and radiolabeled [γ-35S]ATP displaced with nonlabeled ATPγS is represented as the concentration of ATPγS bound to Hbr1p versus free ATPγS. The data points are represented by symbols, and the calculated single site homogenous binding isotherms are represented as lines. B, equilibrated mixtures of Hbr1p-His6, radiolabeled [γ-32P]ATP, and competing amounts of nonlabeled ATP were column-filtered to separate free ATP from bound. The equilibrium isotherm is represented as the concentration of ATP bound Hbr1p-His6 versus free ATP using symbols for data and lines for fits.
TABLE 1.
Dissociation constants of Hbr1 protein with ATPγS or ATP
| Protein | ATPγS |
ATP |
||
|---|---|---|---|---|
| Kd | Kd/Kd(WT) | Kd | Kd/Kd(WT) | |
| μm | μm | |||
| WT | 7 ± 2 | 1 | 1.5 ± 0.5 | 1 |
| G19S | 155 ± 33 | 22.1 | 104 ± 60 | 69.3 |
| K22Q | 40 ± 2 | 5.71 | 20 ± 8 | 13.3 |
| K66R | 96 ± 17 | 13.7 | 17 ± 6 | 11.3 |
Because a minor phosphatase contaminant in Hbr1p precluded equilibrium dialysis using ATP (see below), equilibrium filtration was used to measure ATP binding to Hbr1p WT and the mutants. WT Hbr1p had the greatest affinity for ATP, and the relative affinities of the mutants followed that of ATPγS, indicating that each of the mutations destabilize ATP binding (Fig. 4B and summarized in Table 1).
WT Hbr1p displayed a 5-fold greater affinity for ATP than ATPγS. The calculated dissociation constants displayed in Table 1 are also listed as ratios of dissociation constants normalized to WT (Kd(mutant)/Kd(WT)). The similar binding ratio of the K66R mutant for ATP versus ATPγS shows that this mutant has the same ligand selectivity as WT Hbr1p despite an overall lower binding affinity. The difference in the ratios for the G19S and K22Q mutants indicates that they differ in their ligand selectivity from WT Hbr1p (Table 1).
ATP Protects Hbr1p from Trypsin Digestion
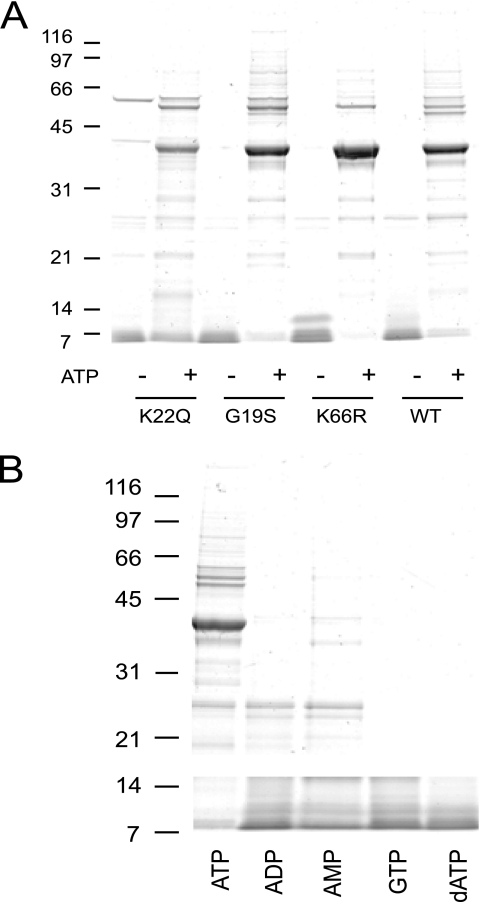
The above analysis indicates that ATP binds at the Hbr1p P-loop but also that a distal mutation (K66R) has an effect on ATP binding affinity. This suggested that ATP binding may alter the conformation of Hbr1p, which could be reflected in altered protease sensitivity. Preliminary experiments demonstrated that a 20:1 molar ratio of Hbr1p-His6:trypsin was the minimal ratio in the absence of ATP to achieve complete loss of intact WT and mutant proteins within the time scale of these experiments (Fig. 5A and data not shown). In contrast, proteolysis of both WT and mutant Hbr1p was prevented in the presence of a saturating concentration of ATP (20 mm; Fig. 5A).
FIGURE 5.
ATP binding confers resistance of Hbr1p-His6 to proteolysis by trypsin. Gel electrophoresis was used to monitor levels of intact Hbr1p-His6 after 1 h of trypsin incubation at 20:1 Hbr1p-His6:trypsin molar ratio. Hbr1p-His6 WT and mutants with and without preincubation with 20 mm ATP (A) or 20 mm of the indicated nucleotides (B). Migration of molecular mass markers is indicated (kDa).
To examine the nucleotide binding selectivity, ATP, ADP, AMP, GTP, or dATP was preincubated with WT Hbr1p-His6 before adding trypsin. Of these, only ATP protected WT Hbr1p-His6 from digestion (Fig. 5B). These results indicate that ATP selectively binds and stabilizes Hbr1p and is probably its natural ligand.
CD Measurements of Hbr1p
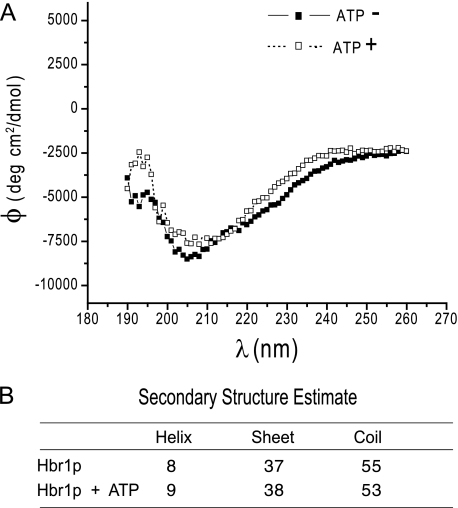
Protection of Hbr1p-His6 from proteolysis by ATP suggested a possible ligand-induced change in secondary structure. As shown in Fig. 6A, the far-UV CD spectrum of Hbr1-His6 did not change significantly in the presence of a saturating concentration of ATP. The minimum near 200 nm and the low ellipticity at 220 nm indicate a low overall content of ordered secondary structure in Hbr1p (Fig. 6B). This is one characteristic of intrinsically unstructured proteins (40). This contrasts with the apparent human ortholog AD-004 (CINAP; AK-6), which contains 58% α-helix and 18% β-sheet and has a highly structured core as determined from the crystal structure (16).
FIGURE 6.
Hbr1p secondary structure does not alter when equilibrated with ATP. A, circular dichroism spectra of Hbr1p before and after exposure to ATP show little change in the ellipticity of the optical signal. B, the Fasman algorithm was used to estimate helix, β-sheet, and random coil secondary structure for the measured data (35).
Hbr1p Lacks ATPase Activity
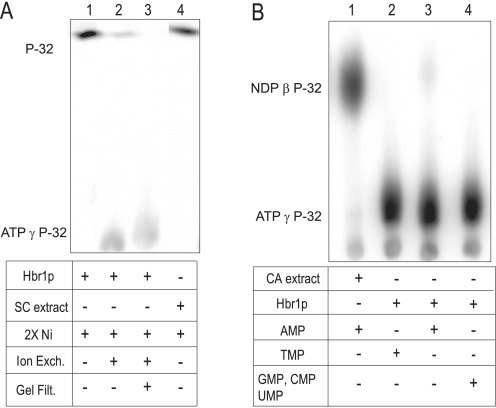
AD-004 was reported to possess ATPase activity, which is a necessary for its role as an adenylate kinase (AK) (19). We assessed this activity in Hbr1p using poly(ethyleneimine)-cellulose thin layer chromatography (36). Incubation of Hbr1-His6 that was purified by two sequential nickel affinity chromatographic steps (2× nickel) exhibited complete conversion of [γ-32P]ATP to orthophosphate (Fig. 7A, lane 1). However, the ATPase activity of Hbr1p dramatically decreased with further protein purification steps. Ion exchange chromatography on DEAE Sepharose resulted in decreased orthophosphate release (Fig. 7A, lane 2), and a further gel filtration step resulted in a protein preparation essentially lacking detectable ATPase activity (Fig. 7A, lane 3). Furthermore, a control experiment using an equivalent molar concentration of extract from S. cerevisiae lacking an HBR1 expression vector and mock purified by 2× nickel affinity showed identical ATPase activity eluting in the equivalent Hbr1p fractions (Fig. 7A, lane 4). Therefore, the ATPase activity probably originates from a contaminant that co-purifies with Hbr1p and not Hbr1p itself. No degradation of [γ-35S]ATP was detected by this method, validating its use for the long incubation times required for the equilibrium dialysis studies.
FIGURE 7.
Purified Hbr1p lacks ATPase and adenylate kinase activities. A, ATPase activity was measured by formation of radiolabeled 32P from the hydrolysis of [γ-32P]ATP. Hbr1p was sequentially purified by 2× nickel affinity chromatography (2× Ni), ion exchange, and gel filtration and added to the reaction mixture as indicated. The products were separated by TLC and detected by autoradiography. A control assay used 2× nickel affinity-purified S. cerevisiae extract, which contained no Hbr1p. B, adenylate kinase activity was measured by formation of NDP β-32P from phospho-transfer of [γ-32P]ATP to the indicated unlabeled NMP.
Hbr1p Lacks Adenylate Kinase Activity
Apparent orthologs of Hbr1p including human AD-004 (16), Drosophila melanogaster DAK6 (18), and Caenorhabditis elegans ADLP (17) were all reported to possess AK activity: 2ADP ↔ ATP + AMP. We therefore tested Hbr1p-His6 for this activity by measuring the transfer of [32P]orthophosphate from [γ-32P]ATP to AMP. The reaction mixture was analyzed by thin layer chromatography to detect [β-32P]ADP formation. Using the 2× nickel Hbr1p-His6 protein preparation, only minimal [β-32P]ADP was formed (Fig. 7B, lane 3). Similarly, no NDP β-32P formation was observed using other NMP substrates (Fig. 7B, lanes 2 and 4). However, a C. albicans extract at equivalent protein concentration showed almost quantitative conversion of [γ-32P]ATP to [β-32P]ADP (Fig. 7B, lane 1). Therefore, the slight [β-32P]ADP formation in the Hbr1p-His6 protein preparation may be due to the presence of a contaminant rather than Hbr1p.
Several attempts were made to optimize assay conditions to promote AK activity, such as changing buffer conditions (pH 3.2 to 7.9), monovalent ionic strength (5–500 mm NaCl or KCl), cations (Mg2+, Mn2+, Zn2+, Fe3+), temperature (20–70 °C), reducing agents (2-mercaptoethanol, dithiothreitol, or tributylphosphine), denaturant (0.06–1.0 m urea), and polyamines (1 μm to 5 mm spermine, spermidine, or putrescine). The greatest rate of [β-32P]ADP formation observed was calculated to be <1 nmol min−1 mg−1 at 30 °C in 30 mm HEPES, pH 7.0, 5 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol, 1 mm AMP, 1 nm [γ-32P]ATP, and 2 mm spermine. This specific activity is 3 orders of magnitude less than reported for AD-004 (16) and 6 orders of magnitude less than for an Escherichia coli AK (23). Consistent with these results, Juhnke et al. (8) were unable to complement uridylate kinase (URA6Δ) and adenylate kinase (ADK1Δ) deletion mutants with Fap7p in S. cerevisiae.
No Evidence for Hbr1p Autophosphorylation
Hbr1p was incubated with radiolabeled [γ-32P]ATP in the presence of phosphatase inhibitors and then monitored by gel electrophoresis and autoradiography for phosphorylation of the ∼42-kDa Hbr1p band (see “Experimental Procedures”). No significant autophosphorylation was detected (data not shown).
P-loop Mutations Result in the Deregulation of Hbr1p Target Genes
The initial genetic studies of HBR1 indicated that the gene is haplo-insufficient, and the 50% decrease in its mRNA level associated with deletion of one allele abrogated MTLα gene expression (5). The absence of the MTLα2 gene product also promoted secondary effects such as CAG1 derepression caused by the absence of the A1/α2 repressor (41). We studied the effects of P-loop mutations on the expression of the MTLα2 and CAG1 genes in C. albicans cells to determine whether ATP binding is required for this function. WT and G19S or K22Q mutants of Hbr1p were expressed from a MET3 promoter contained on a plasmid integrated into the HBR1 locus of a WT C. albicans strain. We measured steady-state transcription of the MTLα2 and CAG genes in the presence (repressing) and absence of 2.5 mm methionine (inducing conditions).
The HBR1 heterozygous strain CAMP 43 displayed HBR1 levels approximately half that of the parental strain (Table 2) in agreement with previous results (5). As predicted by the regulatory model for the mating locus (5, 41), this decrease in Hbr1p level was sufficient to cause CAG1 derepression caused by the absence of MTLα2 protein (Table 2). Expression of WT HBR1 from the MET3 promoter (−Met) restored MTLα2 expression levels and effectively decreased CAG1 levels, although not to the levels seen in the parental strain (Table 2). This lack of complete repression of CAG1 may be due to the high levels of Hbr1p, which resulted in an almost 6-fold increase in MTLα2 levels (Table 2). The effectiveness of the Mtla1/α2 repressor complex is apparently compromised in this strain. S. cerevisiae cells overexpressing Matα2 display abnormal bud site selection and morphology (42). Therefore, off-target effects of Mtlα2 overexpression may be expected.
TABLE 2.
Effects of P-loop mutations on Hbr1p target gene expression
| Strain | HBR1 | CAG1 | MTLα2 |
|---|---|---|---|
| SC5314 | 1.0 ± .04a | 1.0 ± .02 | 1.0 ± .04 |
| CAMP43 | 0.4 ± .05 | 44.5 ± 2.0 | ND |
| WT − Met | 4.2 ± 0.9 | 8.4 ± .38 | 5.7 ± .34 |
| WT + Met | 0.81 ± .1 | 20 ± 1.1 | ND |
| G19S − Met | 1.6 ± .6 | 36.7 ± 4.5 | NDbc |
| G19S + Met | 0.23 ± .04 | 21.7 ± 1.6 | ND |
| K22Q − Met | 5.8 ± .6 | 70.5 ± 6.1 | ND |
| K22Q + Met | 0.95 ± .3 | 105.8 ± 6.5 | ND |
a Fold change (KSD, n = 4) as measured by quantitative RT-PCR. The transcription levels were normalized to 1.0 in the WT strain SC5314. All of the individual samples were adjusted to the levels of CDC36, which was used as an internal control. The mean cycle threshold values of CDC36 did not vary more than 5% between samples.
b G19S mutation in Fap7p causes a slow growth phenotype (8).
c ND, not detected. Ct > 37.
The G19S mutant strain in the absence of methionine displayed WT levels of Hbr1p and yet was unable to cause transactivation of MTLα2. As expected, decreasing Hbr1 levels further (+Met) did not affect MTLα2 mRNA levels and decreased CAG1 mRNA levels by ∼30% (Table 2). This indicates that the G19S mutation interferes with the ability of Hbr1 to activate MTLα2 gene expression. We did not further investigate the reasons for the secondary effects on CAG1 expression.
The K22Q strain, despite an almost 6-fold overexpression of Hbr1p, could not transactivate MTLα2. As expected, a further decrease did not result in any further change in MTLα2 levels (Table 1). Interestingly, CAG1 levels in this strain were more than twice that of the HBR1 deletion strain CAMP 43. This suggested that the K22Q mutation caused a dominant negative effect, perhaps by further interference with endogenous A1/α2 repressor complexes. Neither the G19S nor the K22Q P-loop mutant proteins were able to restore MTLα2 to detectable levels, indicating a lack of functionality.
Hbr1p P-loop Mutations Do Not Alter Growth Rate
The original S. cerevisiae mutant strain Fap7–1 was identified as an isolate that failed to activate a peroxide-sensitive promoter and possessed a slow growth phenotype (8). However, the growth rates of all the strains listed in Table 2 were similar, with doubling times of ∼80 min at 30 °C in YPD medium.
DISCUSSION
These studies demonstrate that the predicted P-loop of Hbr1p contributes to a high affinity binding site for ATP. Other NTPs, dATP, and nucleotides tested did not bind, indicating that ATP is a specific ligand for Hbr1p. Although the potential human ortholog of Hbr1p has been reported to possess NTPase and AK activities, we could not detect enzymatic activity of Hbr1p. As initially purified, Hbr1p contained some ATPase activity, but further purification of the protein removed this apparent contaminant. This is not consistent with Hbr1p being a functional ortholog of the AK family of nuclear enzymes (16–19). The S. cerevisiae ortholog Fap7p has also been localized in the yeast nucleus (8), but its proposed function in the final step of 18 S rRNA maturation is cytoplasmic (11).
The primary amino acid sequences of Hbr1p from C. albicans and C. dubliniensis are more than 40% identical with AD-004 (AK-6 in Fig. 1). This includes the absence of a terminal glycine in the P-loop consensus (Ser23 in Hbr1p) that is a unique feature of the new AK-6 group. On the other hand, Hbr1p is unique in having a nine-amino acid insertion between these two domains (Fig. 1). In the AD-004 crystal structure, this insertion corresponds precisely with a small loop connecting the first α helix and the second β sheet (16).
Overall, Hbr1p is a highly acidic protein with a significantly higher net charge (−47, pI = 3.9) than members of the AK-6 group (e.g. AD-004, −17.5, pI = 4.36). This is due to the acidic tail. When this is excluded from the calculation, Hbr1p is similar to AK-6 members (−19.4, pI = 4.32). Another difference from the AK-6 group is the presence of two basic residues and seven acidic residues in the NMP-binding domain instead of 10 acidic and no basic residues (Fig. 1). In the AD-004 crystal structure, these acidic residues form a negatively charged surface and are conserved in higher eukaryotes (16). The S. cerevisiae ortholog Fap7p is similar to Hbr1p in this regard (Fig. 1).
The anomalous electrophoretic behavior of Hbr1p may be explained by its high negative charge, which decreases LDS binding (38, 43, 44). Notably, AD-004 protein is less highly charged and lacks this anomaly (16). Gel filtration indicates that purified Hbr1p oligomerizes, and the predominant isoform may be a dimer. However, further studies are required to confirm this conclusion.
Hbr1p possesses five cysteine residues, two of which are conserved with the AK-6 family members and two with S. cerevisiae Fap7 (Fig. 1). The fifth residue occurs as a part of a doublet, distal to the NMP-binding domain (Fig. 1). Because reducing conditions did not alter the gel migration pattern of Hbr1p, intersubunit disulfide bonding is unlikely.
Circular dichroism measurements indicate that ATP binding does not significantly change the secondary structure of Hbr1p. On the other hand, the near-UV CD spectra minima near 200 nm and low 220 nm ellipticity indicate a general lack of ordered secondary structure. However, some change in Hbr1p structure must occur to explain its protection from proteolysis when bound to ATP. ATP binding may stabilize Hbr1p by inducing movements of secondary structure elements that do not significantly perturb the CD spectrum but are sufficient to mask trypsin-sensitive residues. For example, substrate-induced changes in the three-dimensional structure of shikimate kinase involve concerted movements of secondary structure rather than alterations in α-helix or β-sheet content (45).
Hbr1p meets several of the six criteria proposed for classification as a highly unstructured protein (40). These include its high negative charge and a lack of CD spectral signatures for an ordered secondary structure. Highly unstructured proteins typically adopt a rigid structure upon ligand binding. This is consistent with the increased trypsin resistance of Hbr1p following ATP binding. Further biophysical and biochemical characterization is necessary to determine whether Hbr1p is indeed an intrinsically disordered protein.
Although enzymatic activity of Hbr1p cannot be completely excluded, it clearly lacks the NTPase and NMP kinase activities that have been demonstrated in the related AK family. We propose that the main function of the ATP-binding site of Hbr1p is to sense ATP levels and thereby modulate downstream targets. Mutations that disrupt ATP binding to Hbr1p diminish or eliminate its ability to regulate expression of its known target genes in the mating pathway (Table 2). In the absence of an enzymatic activity, ATP binding could alter the affinity of Hbr1p for interaction with other regulatory proteins in the cell. Indeed, single amino acid mutations in the P-loop as well as in the NMD-binding domain of the S. cerevisiae ortholog Fap7p abrogate its ability to process 20 S rRNA and interact with ribosomal protein Rps14p (11).
The identities of ATP-sensitive targets of Hbr1p remain unknown, but studies of its yeast ortholog Fap7 and human AD-004 may provide clues. Depletion of AD-004 by siRNA decreased mammalian cell viability, presumably because of a misdirection of components needed for histone or small nuclear ribonucleoprotein processing in the Cajal body (20). However, a 50% depletion of Hbr1p does not affect cell viability. On the other hand, the rRNA processing activity of Fap7p may indicate that these proteins either play similar roles in RNA transit or act directly in rRNA maturation. Our ongoing studies support this hypothesis.
Acknowledgments
We thank Dr. Mark Lowenthal for performing LC/MS experiments, the late Dr. Henry Krutzsch for peptide synthesis and antibody preparation, and Dr. Toshiaki Hara for performing CD experiments.
This work was supported, in whole or in part, by the National Institutes of Health through the National Cancer Institute Center for Cancer Research Intramural Research Program, ZIA SC 009173.
- P-loop
- phosphate binding loop
- AK
- adenylate kinase
- LDS
- lithium dodecyl sulfate
- MTLα
- mating type-like locus α
- SUMO
- small ubiquitin-related modifier
- NMP
- nucleoside monophosphate.
REFERENCES
- 1. Odds F. C. (1988) Candida and Candidosis, 2nd ed., pp. 206–228, Bailliere Tindall, London [Google Scholar]
- 2. Kvaal C., Lachke S. A., Srikantha T., Daniels K., McCoy J., Soll D. R. (1999) Infect. Immun. 67, 6652–6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Soll D. R. (1992) Clin. Microbiol. Rev. 5, 183–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller M. G., Johnson A. D. (2002) Cell 110, 293–302 [DOI] [PubMed] [Google Scholar]
- 5. Pendrak M. L., Yan S. S., Roberts D. D. (2004) Eukaryot. Cell 3, 764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hull C. M., Raisner R. M., Johnson A. D. (2000) Science 289, 307–310 [DOI] [PubMed] [Google Scholar]
- 7. Magee B. B., Magee P. T. (2000) Science 289, 310–313 [DOI] [PubMed] [Google Scholar]
- 8. Juhnke H., Charizanis C., Latifi F., Krems B., Entian K. D. (2000) Mol. Microbiol. 35, 936–948 [DOI] [PubMed] [Google Scholar]
- 9. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 10. Peng W. T., Robinson M. D., Mnaimneh S., Krogan N. J., Cagney G., Morris Q., Davierwala A. P., Grigull J., Yang X., Zhang W., Mitsakakis N., Ryan O. W., Datta N., Jojic V., Pal C., Canadien V., Richards D., Beattie B., Wu L. F., Altschuler S. J., Roweis S., Frey B. J., Emili A., Greenblatt J. F., Hughes T. R. (2003) Cell 113, 919–933 [DOI] [PubMed] [Google Scholar]
- 11. Granneman S., Nandineni M. R., Baserga S. J. (2005) Mol. Cell. Biol. 25, 10352–10364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saraste M., Sibbald P. R., Wittinghofer A. (1990) Trends Biochem. Sci. 15, 430–434 [DOI] [PubMed] [Google Scholar]
- 14. Ramakrishnan C., Dani V. S., Ramasarma T. (2002) Protein Eng. 15, 783–798 [DOI] [PubMed] [Google Scholar]
- 15. Leipe D. D., Koonin E. V., Aravind L. (2003) J. Mol. Biol. 333, 781–815 [DOI] [PubMed] [Google Scholar]
- 16. Ren H., Wang L., Bennett M., Liang Y., Zheng X., Lu F., Li L., Nan J., Luo M., Eriksson S., Zhang C., Su X. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhai R., Meng G., Zhao Y., Liu B., Zhang G., Zheng X. (2006) FEBS Lett. 580, 3811–3817 [DOI] [PubMed] [Google Scholar]
- 18. Meng G., Zhai R., Liu B., Zheng X. (2008) Biochemistry 73, 38–43 [DOI] [PubMed] [Google Scholar]
- 19. Santama N., Ogg S. C., Malekkou A., Zographos S. E., Weis K., Lamond A. I. (2005) J. Biol. Chem. 280, 36429–36441 [DOI] [PubMed] [Google Scholar]
- 20. Zhang J., Zhang F., Zheng X. (2010) Cell Mol. Life Sci. 67, 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cioce M., Lamond A. I. (2005) Annu. Rev. Cell Dev. Biol. 21, 105–131 [DOI] [PubMed] [Google Scholar]
- 22. Santos M. A., Tuite M. F. (1995) Nucleic Acids Res. 23, 1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reinstein J., Schlichting I., Wittinghofer A. (1990) Biochemistry 29, 7451–7459 [DOI] [PubMed] [Google Scholar]
- 24. Care R. S., Trevethick J., Binley K. M., Sudbery P. E. (1999) Mol. Microbiol. 34, 792–798 [DOI] [PubMed] [Google Scholar]
- 25. Wilson R. B., Davis D., Mitchell A. P. (1999) J. Bacteriol. 181, 1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sherman F. (1991) in Guide to Yeast Genetics and Molecular Biology (Guthrie C., Fink G. R. eds) pp. 3–21, Academic Press, San Diego [Google Scholar]
- 27. Adams A., Gottschling D. E., Kaiser C. A., Stearns T. (1998) Methods in Yeast Genetics, pp. 99–102, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 28. Giniger E., Varnum S. M., Ptashne M. (1985) Cell 40, 767–774 [DOI] [PubMed] [Google Scholar]
- 29. Dunn B., Wobbe C. R. (1992) Curr. Protoc. Mol. Biol. Chapter 13 Unit 13, 11–19 [DOI] [PubMed] [Google Scholar]
- 30. Adams L. D., Gallagher S. R. (2004) Curr. Protoc. Mol. Biol. Chapter 10 Unit 14, 11–23 [DOI] [PubMed] [Google Scholar]
- 31. Stone K. L., Williams K. R. (2004) Current Protoc. Protein. Sci. Chapter 11 Unit 13, 1–10 [DOI] [PubMed] [Google Scholar]
- 32. Moore R. E., Young M. K., Lee T. D. (2000) Current. Protoc. Protein. Sci. Chapter 16 Unit 10, 11–19 [DOI] [PubMed] [Google Scholar]
- 33. Williams A., Frasca V. (1998) Curr. Protoc. Mol. Biol. Chapter 10 Unit 10, 11–30 [DOI] [PubMed] [Google Scholar]
- 34. Begg G. E., Harper S. L., Speicher D. W. (1999) Current Protoc. Protein. Sci. Chapter 7 Unit 10, 11–15 [DOI] [PubMed] [Google Scholar]
- 35. Deléage G., Geourjon C. (1993) Comput. Appl. Biosci. 9, 197–199 [DOI] [PubMed] [Google Scholar]
- 36. Randerath K., Randerath E. (1964) J. Chromatogr. A 16, 111–125 [DOI] [PubMed] [Google Scholar]
- 37. Ren J., Gao X., Jin C., Zhu M., Wang X., Shaw A., Wen L., Yao X., Xue Y. (2009) Proteomics 9, 3409–3412 [DOI] [PubMed] [Google Scholar]
- 38. Alves V. S., Pimenta D. C., Sattlegger E., Castilho B. A. (2004) Biochem. Biophys. Res. Commun. 314, 229–234 [DOI] [PubMed] [Google Scholar]
- 39. Oda Y., Kasai K., Ishii S. (1981) J. Biochem. 89, 285–296 [DOI] [PubMed] [Google Scholar]
- 40. Uversky V. N. (2002) Eur. J. Biochem. 269, 2–12 [DOI] [PubMed] [Google Scholar]
- 41. Bennett R. J., Johnson A. D. (2005) Annu. Rev. Microbiol. 59, 233–255 [DOI] [PubMed] [Google Scholar]
- 42. Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S. G., Cyert M., Hughes T. R., Boone C., Andrews B. (2006) Mol. Cell 21, 319–330 [DOI] [PubMed] [Google Scholar]
- 43. Uversky V. N., Gillespie J. R., Fink A. L. (2000) Proteins 41, 415–427 [DOI] [PubMed] [Google Scholar]
- 44. García-Ortega L., De los Ríos V., Martínez-Ruiz A., Oñaderra M., Lacadena J., Martínez del Pozo A., Gavilanes J. G. (2005) Electrophoresis 26, 3407–3413 [DOI] [PubMed] [Google Scholar]
- 45. Krell T., Coggins J. R., Lapthorn A. J. (1998) J. Mol. Biol. 278, 983–997 [DOI] [PubMed] [Google Scholar]