Abstract
Transcription regulation by histone modifications is a major contributing factor to the structural and functional diversity in biology. These modifications are encrypted as histone codes or histone languages and function to establish and maintain heritable epigenetic codes that define the identity and the fate of the cell. Despite recent advances revealing numerous histone modifications associated with transcription regulation, how such modifications dictate the process of transcription is not fully understood. Here we describe spatial and temporal analyses of the histone modifications that are introduced during estrogen receptor α (ERα)-activated transcription. We demonstrated that aborting RNA polymerase II caused a disruption of the histone modifications that are associated with transcription elongation but had a minimal effect on modifications deposited during transcription initiation. We also found that the histone H3S10 phosphorylation mark is catalyzed by mitogen- and stress-activated protein kinase 1 (MSK1) and is recognized by a 14-3-3ζ/14-3-3ϵ heterodimer through its interaction with H3K4 trimethyltransferase SMYD3 and the p52 subunit of TFIIH. We showed that H3S10 phosphorylation is a prerequisite for H3K4 trimethylation. In addition, we demonstrated that SET8/PR-Set7/KMT5A is required for ERα-regulated transcription and its catalyzed H4K20 monomethylation is implicated in both transcription initiation and elongation. Our experiments provide a relatively comprehensive analysis of histone modifications associated with ERα-regulated transcription and define the biological meaning of several key components of the histone code that governs ERα-regulated transcription.
Keywords: Breast Cancer, Chromatin Histone Modification, Estrogen, Gene Regulation, Transcription
Introduction
Histones are basic proteins that organize genomic DNA into a hierarchical chromatin structure (1). Histones undergo a plethora of post-translational modifications, including phosphorylation, acetylation, methylation, ubiquitination, sumoylation, and ADP-ribosylation, which occur in their flexible N- and C-terminal tails or within their globular folds in the nucleosome core (2). Acting individually or in combination, these modifications, in conjunction with DNA methylation, are believed to encipher inheritable epigenetic programs that encode distinct nucleosome functions such as gene transcription, X-chromosome inactivation, heterochromatin formation, mitosis, and DNA repair and replication (2–4). Mechanistically, these functions are mediated either directly by altering nucleosome interactions within chromatin or indirectly by recruiting effector proteins that possess characteristic modules that recognize specific histone modifications in a sequence-dependent manner (5, 6). The underlying basis of these epigenetic codes resides in the substrate specificity of the enzymes that catalyze the numerous covalent modifications, as well as the enzymes that remove these marks to alter the modifications.
Given that chromatin is the physiological template for all DNA-mediated processes, it is not surprising that histone modifications represent an essential component in controlling the structure and/or function of the chromatin fiber, with different modifications yielding distinct functional consequences. Indeed, recent studies have shown that site-specific histone modifications correlate well with particular biological functions such as gene transcription (7, 8). For instance, histone H3 lysine 9 acetylation (H3K9ac),2 H3 serine 10 phosphorylation (H3S10ph), and H3 lysine 4 trimethylation (H3K4me3) are believed to be associated with transcriptional activation (9). Conversely, H3K9me3 and hypoacetylation of H3 and H4 have been shown to be correlated with transcriptional repression. As stated above, ultimately, the functions of histone modifications are manifested by the recognition of histone code or histone language by a particular cellular machinery such as the transcription apparatus (10).
The gene transcription cycle begins with the binding of activators/coactivators on gene promoters. This event leads to the recruitment of general transcription factors. RNA polymerase II (pol II), together with general transcription factors including TFIIH, is positioned at the core promoter to form a preinitiation complex and carry out transcription (11–13). Transcription can be divided into three distinct yet closely coupled phases: initiation, elongation, and termination. Given the chromatinized nature of the genome organization, it is not surprising that the majority of the coactivators identified are chromatin modifiers. Indeed, a large number of coactivators that have been implicated in estrogen receptor α (ERα)-regulated transcription, including SRC-1, CBP/p300, pCAF, G9a, PRMT1, CARM1, and MLL2, all possess chromatin-remodeling capabilities (14–22).
Although abundant chromatin modifiers and histone modifications have been described in transcription regulation, including transcription activation by ERα, how the distinct transcription processes are dictated by histone modifications is not fully understood. Here we present our analysis of different histone modifications that are associated with distinct processes of ERα-regulated transcription as well as our identification of the biological effector proteins of several key components of the histone code that governs ERα-regulated transcription.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagents
Antibodies for SMYD3, TFIIH, MSK1, ERα, and 14-3-3ζ/ϵ were purchased from Santa Cruz Biotechnology. Histone peptides were from Millipore, and antibodies for specific histone modifications were from Abcam. 17β-Estradiol, α-amanitin for 14-3-3ζ/ϵ siRNAs, and phosphorylation kinase inhibitors were purchased from Sigma-Aldrich. Control siRNA and siRNAs for MSK1, SMYD3, and SET8 were synthesized by Shanghai GeneChem Inc. (Shanghai, China). The sequences used for siRNA were: control, 5′-UUCUCCGAACGUGUCACGU-3′; MSK1, 5′-GCTAAAGACCTCCTTCAGCGTCTTT-3′; SET8, 5′-UGUAACUGAGUUCUCUUCCTT-3′; and SMYD3, 5′-CCGCGUCGCCAAAUACUGUAG-3′.
Cell Culture and Reporter Assay
MCF-7 cells and T-47D were from American Type Culture Collection (Manassas, VA). Cells were maintained in DMEM or RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). Transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Luciferase activity was measured using a Dual-Luciferase (Promega, Madison, WI) according to the manufacturer's protocol. Each experiment was performed in triplicate.
GST Pulldown Assay
GST fusion constructs were expressed in BL21 Escherichia coli cells, and crude bacterial lysates were prepared by sonication in TEDGN (50 mm Tris-HCl, pH 7.4, 1.5 mm EDTA, 1 mm dithiothreitol, 10% (v/v) glycerol, 0.4 m NaCl) in the presence of the protease inhibitor mixture. The in vitro transcription and translation experiments were done with rabbit reticulocyte lysate (TnT Systems, Promega) according to the manufacturer's recommendation. In GST pulldown assays, about 10 μg of the appropriate GST fusion proteins was mixed with 5–8 μl of the in vitro transcribed/translated products and incubated in binding buffer (75 mm NaCl, 50 mm HEPES, pH 7.9) at room temperature for 30 min in the presence of the protease inhibitor mixture. The binding reaction was then added to 30 μl of glutathione-Sepharose beads and mixed at 4 °C for 2 h. The beads were washed three times with binding buffer, resuspended in 30 μl of 2× SDS-PAGE loading buffer, and resolved on 10% gels. Immunoreactive bands were visualized using Western blotting.
ChIP and Re-ChIP Assay
ChIP experiments were performed according to the procedure described previously (24–27). Re-ChIPs were done essentially the same as the primary immunoprecipitations. Beads eluted from the first immunoprecipitation were incubated with 10 mm DTT at 37 °C for 30 min and diluted 1:50 in dilution buffer (1% Triton X-100, 2 mm EDTA, 150 mm NaCl, 20 mm Tris-HCl, pH 8.1) followed by reimmunoprecipitation with the second antibodies. ChIP primers of TFF1 were as follows: primer a, 5′-CACCCCGTGAGCCACTGT-3′ (forward) and 5′-CTGCAGAAGTGATTCATAGTGAGAGAT-3′ (reverse); primer b, 5′-TGCGACAAAGACAAAGCG-3′ (forward) and 5′-CCGTGGTGAGGGAGGAT-3′ (reverse); primer c, 5′-GAAAGATGCAAAGTCCACAAACC-3′ (forward) and 5′-TGTCCAGTGAGGCGGATATAAA-3′ (reverse); and primer d, 5′-CCCCTCACCCCTGTAG-3′ (forward) and 5′-GCTCTGGGACTAATCACC-3′ (reverse).
RT-PCR and Real-time PCR Assay
Total cellular RNAs were isolated with the TRIzol reagent (Invitrogen) and used for first strand cDNA synthesis with the Reverse Transcription System (Promega, A3500). Quantitation of all gene transcripts was done by qPCR using Power SYBR Green PCR Master Mix and an ABI PRISM 7300 sequence detection system (Applied Biosystems, Foster City, CA) with the expression of GAPDH as the internal control. Experiments were repeated between two and four times with different inductions. The primer pairs used were as follows: TFF1, 5′-CAGGAGGTGGCTGGTTATG-3′ (forward) and 5′-GCACAGGGCAGGGTTGTT-3′ (reverse); MSK1, 5′-GAGCATACAAGGACAGAACGACA-3′ (forward) and 5′-TAGAACACCCAAACTCCACCAG-3′ (reverse); GAPDH, 5′-CCCACTCCTCCACCTTTGAC-3′ (forward) and 5′-CATACCAGGAAATGAGCTTGACAA-3′ (reverse); SMYD3, 5′-TTCCCGATATCAACATCTACCAG-3′ (forward) and 5′-AGTGTGTGACCTCAATAAGGCAT-3′ (reverse); SET8, 5′-TCAAAGACGCCAGGAAAG-3′ (forward) and 5′-TGGAATCACAAGATGAGGGT-3′ (reverse); 14-3-3ϵ, 5′-TTGGGTGTTAGCTTGAGGTG-3′ (forward) and 5′-GAGGAGTCGGCAAGAATGAG-3′ (reverse); 14-3-3ζ, 5′-GGCTAGTGATTGGAGGAAACC-3′ (forward) and 5′-TCATATCGCTCTGCCTGCTC-3′ (reverse); CATD, 5′-T CCAGACATCCTCTCTGGAA-3′ (forward) and 5′-GGAGCGGAGGGTCCATTC-3′ (reverse); EBAG9, 5′-GATGCACCCACCAGTGTAAAGA-3′ (forward) and 5′-AGTCAGGTTCCAGTTGTTCCAAAG-3′ (reverse); and GRIP1, 5′-AAGCCTTTGCCAGATTCAG-3′ (forward) and 5′-CAACGAGAGTGCCATCAGAC-3′ (reverse).
Histone Peptide Pulldown and Mass Spectrometry Analysis Assay
Nuclear extracts from MCF-7 cells were prepared using a standard high salt extraction protocol. The salt concentration was diluted in the extract to 150 mm by dialyzing against buffer D1 (150 mm KCl, 0.2 mm EDTA, 1 mm PMSF, 20% glycerol, 0.1% Triton X-100, 20 mm Tris-HCl, pH 7.9). Protease inhibitors and phosphatases were added at every step of extract preparation. Triton X-100 was added to the nuclear extracts to a final concentration of 0.1%, and the solution was centrifuged at 16,000 × g for 10 min at 4 °C to remove any precipitate that may have formed. The nuclear extract was precleared with avidin beads by adding 80 μl of 50% immobilized avidin slurry and prewashed with buffer D1. Beads were incubated for 1 h at 4 °C with rotation, and supernatants were collected by centrifugation at low speed. For each peptide, 40 μl of 50% peptide-bound slurry was added and incubated with rotation for 3 h or overnight at 4 °C. Beads were washed eight times with 1 ml of buffer D2 (300 mm KCl, 0.2 mm EDTA, 1 mm PMSF, 20% glycerol, 0.1% Triton X-100, 20 mm Tris-HCl, pH 7.9). The final wash was performed with low-HEPES buffer. Beads were eluted twice for 10 min at room temperature with 100 mm glycine, pH 2.8; 1–2 equivalents of the bead volume were used for each elution. The supernatant was transferred to a fresh tube. Eluates from each pulldown were analyzed by SDS-PAGE, and proteins were visualized by silver staining using a mass spectrometry-compatible protocol. Gels were handled with caution to prevent contamination with keratin. Protein bands present in the pulldown gel lane after 17β-estradiol (E2) stimulation were excised, and entrapped proteins were identified by mass spectrometry.
RESULTS
Histone Modifications That Are Introduced on Gene TFF1 Activated by ERα
ERα-regulated transcription and ERα-mediated pathophysiology are a primary research focus in our laboratory (24, 26, 28–31). To gain a comprehensive understanding of the histone modifications associated with ERα-regulated gene transcription, we first verified the temporal pattern of the recruitment of ERα on the promoter of TFF1 (Trefoil factor 1), a well characterized ERα target gene, in the estrogen-responsive breast carcinoma cell line MCF-7 by quantitative chromatin immunoprecipitation (qChIP). As shown in Fig. 1A, the recruitment of ERα on the TFF1 promoter was detected at 15 min after E2 stimulation; it peaked at 45 min and decreased to the initial levels by around 90 min. Examination of TFF1 mRNA expression by real-time RT-PCR indicated that TFF1 mRNA accumulation started at 30 min and reached a maximum 6 h after E2 induction (Fig. 1B). These results are consistent with previous reports by us and others (24, 28, 29, 32, 33), and they validate our system for the analysis of histone modifications.
FIGURE 1.
Histone modifications associated with TFF1 transcription activated by ERα. A, recruitment of ERα on the TFF1 gene promoter. MCF-7 cells were grown in the absence of estrogen for 3 days and then treated with 100 nm E2 for the indicated times. The cells were collected then, and qChIP experiments were performed with a primer described under “Experimental Procedures.” Results are expressed relative to the values of the negative control (ethanol). Each bar represents the mean ± S.D. for triplicate measurements. B, accumulation of TFF1 mRNA after E2 stimulation. MCF-7 cells were grown in the absence of estrogen for 3 days and then treated with 100 nm E2 for the indicated times. Total RNAs were extracted, and the expression of TFF1 mRNA was measured by real-time RT-PCR. Results are expressed relative to the values of the negative control (ethanol). Each bar represents the mean ± S.D. for triplicate measurements. C, schematic representation of ChIP primer designing for analysis of TFF1 for histone modifications. D, histone modifications induced during E2 stimulation. MCF-7 cells were grown in the absence of estrogen for 3 days and then treated with E2 for the indicated times. The cells were then collected, and ChIP or qChIP experiments were performed with specific antibodies against the indicated histone modifications and primers. Each point represents the mean ± S.D. for triplicate measurements.
We then designed one set of primers that covers the upstream regulatory region as well as three sets that cover different coding regions of TFF1 (Fig. 1C) to analyze histone modifications associated with different regions of the TFF1 and representing different transcription phases in ERα-regulated transcription. We chose 10 different histone modifications including H2BK5ac, H2BK5me1, H3K9ac, H3S10ph, H3K4me3, H3K36me2, H3K36me3, H3K79me2, H4K5ac, and H4K20me1 on the basis of a recommendation from the Alliance for the Human Epigenome and Disease (AHEAD) (34). In addition, H3K9ac, H3S10ph, H3K4me3, H3K36me2, H3K36me3, H3K79me2, and H4K5ac were shown to function during transcription activation, and H2BK5ac, H2BK5me1, and H4K20me1 were shown to distribute to transcriptionally active regions (35–39). Spatial and temporal alterations in the levels of these histone modifications on the TFF1 gene in MCF-7 cells under treatment with E2 were analyzed by ChIP or qChIP. The results show that the increase in H2BK5ac appeared to be transient and occurred at 30 min after E2 induction, mainly at the TFF1 promoter (Fig. 1D). Likewise, the level of H2BK5me1 was found to be elevated at the TFF1 promoter in response to E2 stimulation (Fig. 1D). In response to E2 stimulation, the level of H3K9ac was increased at the TFF1 promoter around 30 min after E2 induction; however, deposition of this modification was found to be decreased at the coding region of TFF1, and H3K9ac was found to be increased at the TFF1 promoter in T-47D cells at 45 min after E2 stimulation (Fig. 1D). The results of these experiments showed that increases in the level of H3S10 phosphorylation were detected mainly in the TFF1 promoter; the increase was seen at 15 min after E2 stimulation, achieved a maximum at 45 min, and returned to the basal level at 60 min (Fig. 1D). The same pattern was observed in the T-47D cell line (Fig. 1D). During 90 min of E2 induction, there was a pronounced increase in H3K4me3 near the 5′-end of the coding region; no changes in H3K4me3 were found at the promoter of the TFF1 gene (Fig. 1D). A steady increase in H3K36me2 was detected at the promoter region after 120 min of E2 induction. More pronounced increases in this modification were observed near the 3′-end of the coding regions, whereas the change in H3K36me2 near the 5′-end of the coding region was minimal (Fig. 1D). In addition, H3K36me3 increased about 3-fold at the promoter and 20-fold in the middle of the coding region after 30 min of E2 stimulation (Fig. 1D). The level of H3K79me2 increased at the promoter as well as at the 5′-end of the coding region of the TFF1 gene under E2 induction (Fig. 1D). H4K5ac, on the other hand, was enriched at the promoter as well as at the coding region of TFF1 during 60 min of E2 induction, although the dynamics of change in different regions appeared to be different (Fig. 1D). In the case of H4K20me1, it was elevated at the promoter by 11-fold at 120 min after E2 induction, and it also increased downstream of the coding regions, especially in the 3′-end of the coding region, where it was increased about 50-fold at 90 min (Fig. 1D). Collectively, these results indicate that E2 induced strong increases in the levels of H2BK5ac, H2BK5me1, H3K9ac, H3S10ph, H3K36me2, H3K36me3, H3K79me2, H4K5ac, and H4K20me1 on the promoter of TFF1, suggesting that these particular modifications might be involved in transcription initiation. On the other hand, significant elevations in H3K4me3, H3K36me2, H3K36me3, H3K79me2, H4K5ac, and H4K20me1 and decreases in H3K9ac were detected after E2 induction in the various regions of the TFF1 coding sequence, suggesting that these modifications might be involved in transcription elongation.
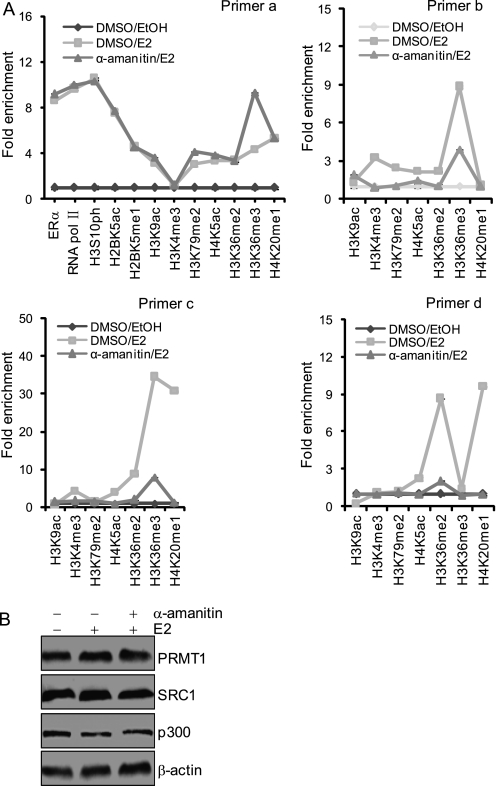
Aborting RNA Polymerase II Affects the Histone Modifications Associated with Elongation but Not Initiation
To further support the above observations and to investigate how the different processes of ERα-activated transcription are dictated by distinct histone modifications, we treated MCF-7 cells with E2 in the presence of α-amanitin, which interacts with the bridge helix in RNA pol II and interferes with the RNA and DNA translocation that is needed to empty the site for the next round of RNA synthesis (40–43). We found that α-amanitin did not significantly affect the recruitment of ERα and pol II and the occurrence of the histone modifications, including H2BK5ac, H2BK5me1, H3K9ac, H3S10ph, H3K36me2, H3K36me3, H3K79me2, H4K5ac, and H4K20me1 on the TFF1 promoter (Fig. 2A). However, the levels of H3K9ac, H3K4me3, H3K36me2, H3K36me3, H3K79me2, H4K5ac, and H4K20me1, which occur in the coding region of TFF1, were all reduced in cells treated with α-amanitin (Fig. 2A). We sought to ascertain whether the α-amanitin sensitivity of the modifications H3K9ac, H3K4me3, H3K36me2, H3K36me3, H3K79me2, H4K5ac, and H4K20me1 was a nonspecific consequence of a general block in the transcription of genes encoding for histone-modifying factors. Thus we examined the expression of several key proteins including SRC-1, p300, and PRMT1 that possess either histone acetyltransferase (HAT) or methyltransferase (HMT) activity and are implicated in ERα-regulated gene transcription (44–46) by Western blotting and found that the abundance of these proteins was not affected by treatment with α-amanitin (Fig. 2B). These results indicate that aborting pol II affected the histone modifications that are associated with elongation but not initiation, supporting the notion that different transcription processes are governed by distinct histone codes.
FIGURE 2.
Aborting RNA polymerase II affects the histone modifications associated with elongation but not with initiation. A, recruitment patterns of ERα and RNA pol II and the deposition of different histone modifications on the TFF1 gene. MCF-7 cells were grown in the absence of estrogen for 3 days and then treated with α-amanitin for 1 h before E2 stimulation. Cells were induced with E2 for 45 min to determine the changes of histone modifications at the promoter but for 1 h for the changes in the coding region. The cells were then collected for qChIP experiments with the indicated primers. Each point represents the mean ± S.D. for triplicate measurements. B, the expression of SRC-1, p300, and PRMT1 was determined by Western blotting in the MCF-7 cells described in A. DMSO, dimethyl sulfoxide.
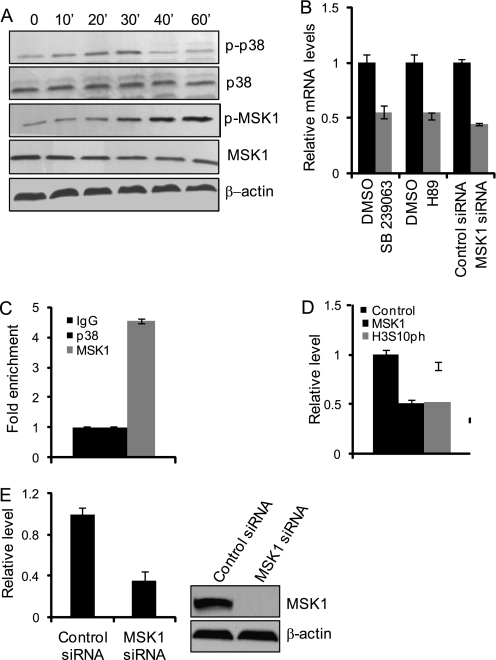
H3S10ph Is Deposited by MSK1 in Response to the Activation of MAPK Signaling by E2
In the above ChIP experiment, we found several histone modifications associated with transcription initiation, and our experiments indicated that the increase in the level of H3S10ph was restricted to the TFF1 promoter. In addition, our experiments showed that H3S10ph occurred at 15 min after E2 stimulation, peaked at 45 min, and decreased to the basal level at 60 min; these changes were rapid and significant (Fig. 1D). These observations support an argument that phosphorylation of H3S10 is an early event in transcriptional activation by ERα.
To further understand the role of H3S10 phosphorylation in ERα-regulated gene transcription, we first set out to identify the kinase that is responsible for the deposit of this modification. In this regard, it is important to note that activation of the MAPK signaling pathway by various mitogens and extracellular growth stimulatory factors is known to be an early event in their cellular functions (35, 47) and that cross-talk between ERα and the MAPK signaling pathway have been well documented (48–50). Examination of the activation of MAPK pathway kinases in response to E2 stimulation revealed that MAPK p38, as well as its immediate downstream targets, mitogen- and stress-activated protein kinase 1 (MSK1), were activated in response to E2 treatment of MCF-7 cells (Fig. 3A). Consistent with this observation, treatment of MCF-7 cells with SB239063 or H89, potent inhibitors of p38 and MSK1, respectively (51–54), or knockdown of the expression of MSK1 with its specific siRNA, led to marked reductions in E2-induced TFF1 expression as measured by real-time RT-PCR (Fig. 3B), Moreover, ChIP experiments detected the recruitment of MSK1 but not p38 on the TFF1 promoter in MCF-7 cells (Fig. 3C). However, treatment of MCF-7 cells with MSK1 siRNA resulted in a pronounced reduction in the recruitment of MSK1 and in the level of H3S10ph on the TFF1 promoter (Fig. 3D). MSK1 RNAi efficiency is shown in Fig. 3E. Collectively, these results strongly suggest that the MAPK kinase signaling pathway is activated in ERα-regulated transcription and that activated MSK1 is responsible for catalyzing the H3S10ph mark.
FIGURE 3.
H3S10ph at the TFF1 promoter is deposited by MSK1 in response to p38-MAPK signaling. A, activation of p38 and MSK1 by E2. MCF-7 cells were grown in the absence of estrogen for 3 days and then treated with 100 nm E2 for the indicated times. Whole cellular proteins were prepared for Western blotting analysis with the indicated antibodies. B, the level of TFF1 mRNA was measured by real-time RT-PCR in MCF-7 cells treated with SB239063, H89, or MSK1 siRNA. Each bar represents the mean ± S.D. for triplicate measurements. DMSO, dimethyl sulfoxide. C, recruitment of MSK1 on the TFF1 promoter. MCF-7 cells were grown in the absence of estrogen for 3 days and then treated with 100 nm E2 for 30 min. The cells were then collected, and qChIP experiments were performed with primer a as described under “Experimental Procedures.” Each bar represents the mean ± S.D. for triplicate measurements. D, the presence of phosphorylated MSK1 and the change in H3S10ph at the TFF1 promoter in MCF-7 cells treated with MSK1 siRNA as measured by qChIP assays. Each bar represents the mean ± S.D. for triplicate measurements. E, efficiency of MSK1 RNAi. MCF-7 cells were transfected with 50 nm of nonsilencing siRNA and MSK1 siRNA. The expression levels of the MSK1 mRNA and protein were determined by real-time qPCR and Western blotting, respectively.
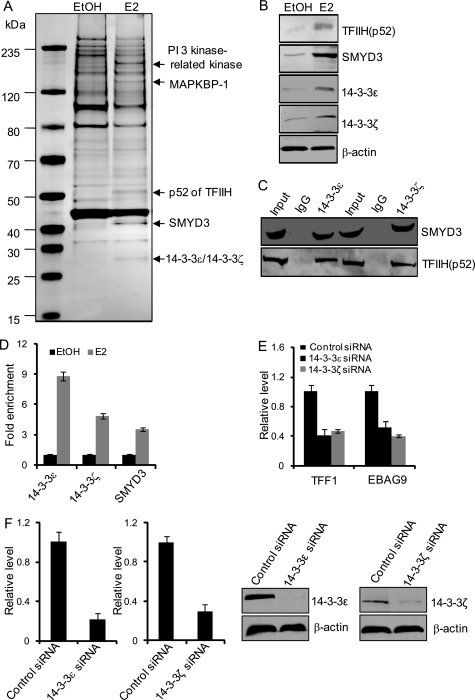
The 14-3-3ζ/ϵ Heterodimer Recognizes the H3S10ph Mark in ERα-regulated Transcription
To further understand the functional role of H3S10 phosphorylation in ERα-regulated gene transcription, we next performed histone peptide pulldown assays to identify the downstream effector proteins for H3S10ph. In these experiments, synthesized histone H3 peptides with H3S10ph were incubated with cellular extracts from MCF-7 cells that were cultured in the presence or absence of E2. Peptide-bound materials were then resolved on SDS-PAGE, and the protein bands were subjected to mass spectrometry analysis. These experiments identified MAPKBP1 (MAPK-binding protein 1), PP1 (serine/threonine protein phosphatase 1), SMYD3 (SET and MYND domain containing 3), the epsilon (ϵ) and zeta (ζ) isoforms of the 14-3-3 proteins, and the p52 subunit of the general transcription factor TFIIH in extracts of E2-treated MCF-7 cells that were associated with H3S10ph peptides (Fig. 4A). The association of these proteins with the H3S10ph peptide was further confirmed by probing the peptide-bound materials by Western blotting with antibodies against SMYD3, 14-3-3ζ, 14-3-3ϵ, and p52 of THIIH (Fig. 4B).
FIGURE 4.
H3S10ph is recognized by 14-3-3ζ and 14-3-3ϵ in ERα-activated transcription. A, mass spectrometry analysis of H3S10ph-associated proteins after E2 stimulation. MCF-7 cells were deprived of E2 for 3 days and then treated with E2 for 45 min. Nuclear extracts from these cells were prepared and subjected to affinity purification with H3S10ph peptide that was immobilized on avidin beads. The bound materials were resolved on SDS-PAGE and silver-stained, and the bands were retrieved and analyzed by mass spectrometry. B, Western blotting analysis of the peptide-bound materials for the abundance of the indicated proteins. C, physical interaction between 14-3-3 proteins and p52 or SMYD3. Cellular nuclear extracts from MCF-7 cells were prepared and immunoprecipitated with antibodies against 14-3-3 proteins. The immunoprecipitated materials were subjected to Western blotting analysis with antibodies against p52 or SMYD3. D, recruitment of 14-3-3 proteins and SMYD3 on the TFF1 promoter. MCF-7 cells were grown in the absence of estrogen for 3 days and then treated with 100 nm E2 for 30 min. The cells were then collected, and qChIP experiments were performed with antibodies against the indicated proteins. Each bar represents the mean ± S.D. for triplicate measurements. E, the levels of TFF1 and EBAG9 mRNA were measured by real-time RT-PCR in MCF-7 cells treated with 14-3-3ϵ/ζ siRNAs. Each bar represents the mean ± S.D. for triplicate measurements. F, efficiencies for 14-3-3ϵ/ζ RNAi. MCF-7 cells were transfected with 50 nm of nonsilencing siRNA and 14-3-3ϵ/ζ siRNA. The expression levels of the 14-3-3ϵ/ζ mRNA and protein were determined by real-time qPCR and Western blotting, respectively.
The 14-3-3 proteins are a family of highly conserved, acidic, regulatory molecules found in all eukaryotes (55). These proteins have been documented to interact with diverse cellular proteins by means of their ability to specifically bind to phosphoserine/threonine and by virtue of their capacity to form either homo- or heterodimers among their seven family members: β, γ, ϵ, σ, ζ, τ, and η (55). Indeed, 14-3-3 γ, ϵ, and ζ have been reported to recognize the H3S10ph mark in c-fos and c-jun mononucleosomes upon gene activation (56). Our experiments suggest that, in ERα-regulated gene transcription, a 14-3-3ζ/ϵ heterodimer recognizes H3S10 phosphorylation.
The identification of MAPKBP1, SMYD3, and the TFIIH p52, which are associated with H3S10ph under E2 treatment, is interesting. First, although its exact biological function has not been characterized, it is known that MAPKBP1 is involved in MAPK signaling, and we showed that this signaling pathway is activated in ERα-regulated transcription. Secondly, SMYD3 has been identified as a histone methyltransferase that catalyzes the addition of H3K4me3 (57), and it was recently reported that SMYD3 functions as a coactivator of ERα and potentiates ERα activity in response to ligand (44). Third, TFIIH is a component of the basal transcriptional machinery that is essential for transcription initiation. However, none of the three (MAPKBP1, SMYD3, or TFIIH p52) has been shown to possess phosphoserine/threonine binding capacity. Therefore, it is likely that the association of these proteins with H3S10ph is indirect, possibly through their interactions with the 14-3-3ζ/ϵ heterodimer. To investigate this hypothesis, co-immunoprecipitation experiments were performed in MCF-7 cells with antibodies against 14-3-3ζ or 14-3-3ϵ, and the immunoprecipitates were then immunoblotted with antibodies against p52 or SMYD3. These experiments revealed that p52 and SMYD3 were indeed associated with 14-3-3ζ and 14-3-3ϵ in vivo (Fig. 4C). In addition, ChIP assays revealed E2-induced recruitment of 14-3-3ζ, 14-3-3ϵ, and SMYD3 on the promoters of TFF1 in MCF-7 cells (Fig. 4D). To determine whether 14–3-3ϵ/ζϵ heterodimer is involved in ERα-regulated transcription, MCF-7 cells were treated with siRNAs specific for 14-3-3ϵ and 14-3-3ζ. Real-time RT-PCR analysis indicated that knockdown of 14-3-3ϵ or 14-3-3ζ resulted in a significant decrease in E2-induced expression of ERα target genes including TFF1 and EBAG9 (Fig. 4E). Efficiencies for 14-3-3ϵ/ζ RNAi were shown in Fig. 4F. Collectively, our experiments suggest a scenario in the initiation phase of ERα-regulated gene transcription in which H3S10ph is catalyzed by MSK1 and recognized by a 14-3-3ζ/ϵ heterodimer, which in turn leads to the recruitment of additional histone-modifying enzymes such as SMYD3 and the binding of components of the transcription apparatus such as p52 of the TFIIH. If this interpretation is correct, it means that the H3S10ph mark might encode multiple messages and that this histone language could be translated into at least the action of additional histone-modifying enzymes and the recruitment of basal transcription machinery.
H3S10ph Is Required for H3K4me3
The interaction between H3S10ph and SMYD3 suggests that there might be a functional connection between H3S10 phosphorylation and H3K4 methylation. To test this hypothesis, MCF-7 cells were deprived of estrogen for 3 days and then treated with E2 in the presence or absence of the MSK1 inhibitor H89. qChIP experiments with anti-SMYD3 or anti-H3K4me3 showed that the recruitment of SMYD3 and the deposition of H3K4me3 in the TFF1 gene promoter and coding region were greatly reduced in MCF-7 cells that had been treated with H89 (Fig. 5A). Similar results were also obtained when MSK1 was knocked down in MCF-7 cells (Fig. 5B). However, knockdown of the expression of SMYD3 did not have any significant effect on the recruitment of MSK1 and the phosphorylation of H3S10 (Fig. 5C). Efficiency for SMYD3 RNAi is shown in Fig. 5D. Given the interaction of SMYD3 with H3S10ph, these results suggest that the recruitment of SMYD3, and thus the methylation of H3K4, is dependent on the recruitment of MSK1 and thus the phosphorylation of H3S10ph. Consistent with this proposition, it appeared from our profiling of the histone modifications that the occurrence of H3S10ph precedes the deposition of H3K4me3 in response to E2 stimulation.
FIGURE 5.
H3S10ph is required for H3K4me3. A, recruitment of SMYD3 and the change in H3K4me3 on TFF1 in MCF-7 cells treated with H89. MCF-7 cells grown in the absence of estrogen were treated with H89 for 1 h before the addition of E2. qChIP assays were performed with antibodies against the indicated proteins and with the indicated primers. Each bar represents the mean ± S.D. for triplicate measurements. DMSO, dimethyl sulfoxide. B, recruitment of SMYD3 and the change in H3K4me3 on TFF1 in MCF-7 cells treated with MSK1 siRNA. MCF-7 cells were transfected with MSK1 siRNA and grown in the absence of estrogen for 72 h prior to treatment with E2. qChIP assays were performed with antibodies against the indicated proteins and with the indicated primers. Each bar represents the mean ± S.D. for triplicate measurements. C, recruitment of MSK1 and the change in H3S10ph on the TFF1 promoter in MCF-7 cells treated with SMYD3 siRNA. MCF-7 cells were transfected with SMYD3 siRNA and grown in the absence of estrogen for 72 h prior to treatment with E2. qChIP or ChIP assays were performed with antibodies against MSK1 or H3S10ph. Each bar represents the mean ± S.D. for triplicate measurements. D, efficiency for SMYD3 RNAi. MCF-7 cells were transfected with 50 nm of nonsilencing siRNA and SMYD3 siRNA. The expression levels of the SMYD3 mRNA and protein were determined by real-time qPCR and Western blotting, respectively.
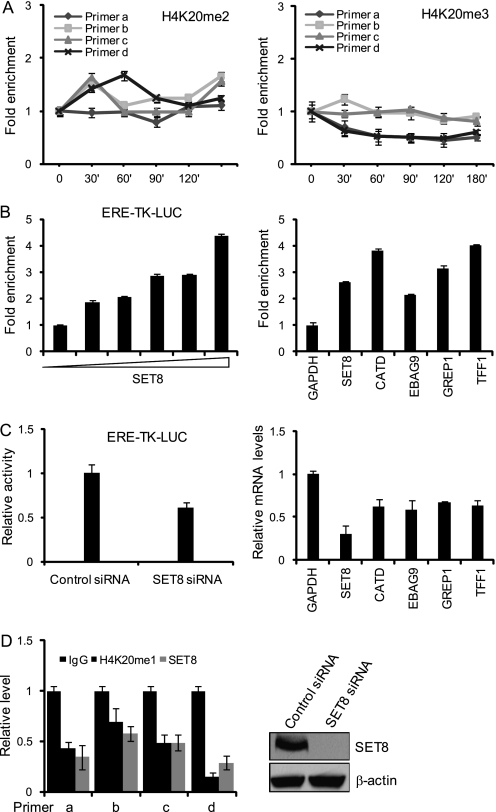
H4K20me1 Is Deposited by SET8/PR-Set7/KMT5A and Is Involved in both Initiation and Elongation of ERα-regulated Transcription
In our experiment, we found several histone modifications related to transcription elongation. Among these modifications, changes of H4K20me1 at the promoter and coding region of TFF1 were the most significant, and its role in transcription has been obscure thus far. Although H4K20 methylation was one of the first post-translationally modified histone residues to be discovered (58), the biological importance of this histone modification is only now beginning to emerge. H4K20me1 has been implicated in both transcriptional activation and gene silencing. In support of its role in transcription activation, recent profiling of histone methylations in the human genome shows a strong correlation between H4K20me1 and gene activation in regions downstream from the transcription start site (59). However, a role for this histone modification in ERα-regulated transcription has not been described. Our experiments indicate that H4K20me1 modification shows a differential pattern of distribution on the various regions of the TFF1 gene upon activation by ERα. At the TFF1 promoter, H4K20me1 was found to be increased 11-fold at 120 min, and it maintained a high level for a long time (Fig. 1D). In the immediate downstream region of the transcription start site of TFF1, there were no significant increases in H4K20me1 for as long as 120 min after E2 stimulation. However, a drastic increase in the level of H4K20me1 was detected further downstream in the gene body of TFF1 in response to E2 treatment (Fig. 1D). There were no significant changes in H4K20me2 in ERα-activated transcription, whereas the level of H4K20me3 actually decreased during this process (Fig. 6A).
FIGURE 6.
H4K20me1 is deposited by SET8/PR-Set7/KMT5A. A, qChIP measurement of the changes in H4K20me2 and H4K20me3 on TFF1 in MCF-7 cells treated with E2. Each point represents the mean ± S.D. for triplicate measurements. B, MCF-7 cells were co-transfected with the estrogen response element (ERE)-Luc reporter and SET8. Luciferase activity was measured by a Dual reporter kit, and the mRNA expression of the indicated endogenous ERα target genes was determined by real-time RT-PCR. Each bar represents the mean ± S.D. for triplicate measurements. C, MCF-7 cells were co-transfected with the ERE-Luc reporter and SET8 siRNA. Luciferase activity was measured by a Dual reporter kit. MCF-7 cells were treated with SET8 RNAi prior to stimulation with E2 for 6 h. The mRNA expression of TFF1 and EBAG9 was then determined by real-time RT-PCR. Each bar represents the mean ± S.D. for triplicate measurements. D, MCF-7 cells were treated with SET8 RNAi and qChIP was performed to determine the levels of H4K20me1. Each bar represents the mean ± S.D. for triplicate measurements. The efficiency of SET8 RNAi was determined by Western blotting.
SET8/PR-Set7/KMT5A is the only known enzyme that specifically catalyzes the addition of H4K20me1 (60, 61). To investigate whether SET8 was also responsible for the addition of the H4K20me1 mark in ERα-regulated transcription, we first tested whether SET8 was involved ERα-regulated transcription. To this end, MCF-7 cells were co-transfected with a SET8 expression construct and an estrogen response element-driven luciferase reporter. Reporter activity assays indicated that overexpression of SET8 potentiated ERα-regulated transcription in a dose-dependent fashion (Fig. 6B, left). In addition, real-time RT-PCR analysis indicated that overexpression of SET8 resulted in a significant increase in E2-induced expression of ERα target genes including TFF1, CATD, GRIP1, and EBAG9 (Fig. 6B, right). On the other hand, knockdown of SET8 expression impaired E2-stimulated reporter activity and the expression of endogenous genes (Fig. 6C). These experiments strongly support an argument that SET8 is involved in ERα-regulated transcription.
To test the hypothesis that SET8 is responsible for the deposition of H4K20me1 in ERα-regulated transcription, the expression of SET8 was knocked down in MCF-7 cells and the change in H4K20me1 on the TFF1 gene was examined by ChIP experiments. The results of these experiments indicated that E2-induced enrichment of H4K20me1 was almost abrogated in SET8-depleted MCF-7 cells (Fig. 6D), strongly supporting the argument that H4K20me1 is written by SET8 in ERα-activated transcription. The efficiency of SET8 RNAi is shown in Fig. 6D.
To further support this proposition, co-immunoprecipitation was carried out in MCF-7 cells. Immunoprecipitation with antibodies against ERα followed by immunoblotting with antibodies against SET8 revealed that ERα interacts with SET8 in vivo (Fig. 7A). In addition, GST pulldown experiments with GST-fused SET8 and in vitro transcribed/translated ERα demonstrated that ERα is capable of interacting with SET8 in vitro (Fig. 7A). Furthermore, ChIP assays found that SET8 was indeed recruited to the TFF1 promoter under E2 induction (Fig. 7B). Collectively, these data suggest that SET8 is recruited by ERα to its target gene promoters and that it is involved in ERα-regulated transcription.
FIGURE 7.
H4K20me1 is involved in both the initiation and elongation of ERα-regulated transcription. A, physical interaction between ERα and SET8. Lysates from MCF-7 and were co-immunoprecipitated with antibodies against ERα, and the immunoprecipitates were then immunoblotted with antibodies against SET8 (upper panel). GST pulldown experiments were performed with GST-fused SET8 and in vitro transcribed/translated ERα (lower panel). B, ChIP analysis of the recruitment of SET8 on the TFF1 promoter in MCF-7 cells transfected with SET8 and treated with E2 for 45 min. C, co-immunoprecipitation with MCF-7 cellular lysates using antibodies against differentially phosphorylated pol II. The immunoprecipitates were then immunoblotted with antibodies against SET8. D, ChIP/Re-ChIP analysis of the co-existence of Ser-2-phosphorylated RNA polymerase II, ERα, and SET8 on the TFF1 gene. Soluble chromatin was prepared from MCF-7 cells treated with E2 for 45 min and divided into three aliquots. Each aliquot was first immunoprecipitated with specific antibodies against IgG, ERα, or Ser-2-phosphorylated RNA polymerase II followed by reimmunoprecipitation with antibodies against SET8. Each bar represents the mean ± S.D. for triplicate measurements.
The observation that H4K20me1 modification occurred both in the promoter and coding region of TFF1 suggests that H4K20me1 might be involved in both the initiation and elongation of ERα-regulated transcription. In this regard, it is important to note that it is believed that the distinct transcription phases are coupled by pol II by means of the differential phosphorylation of the C-terminal domain of its largest subunit. This subunit functions in the recruitment of different histone modification enzymes and protein factors to fulfill corresponding transcriptional activity (62, 63). Therefore, to support both the observation that H4K20me1 also occurs in the coding region and the argument that it is involved in transcription elongation, co-immunoprecipitation experiments were performed to test whether SET8 was associated with either the Ser-2- or Ser-5-phosphorylated forms of pol II. Using antibodies that distinguish different forms of phosphorylated pol II, we found that SET8 was co-immunoprecipitated with Ser-2-phosphorylated pol II but not with Ser-5-phosphorylated pol II (Fig. 7C), suggesting that SET8 is associated with the transcription-competent form of pol II in vivo.
Next, ChIP/Re-ChIP assays were performed, and the existence of ERα, SET8, and pol II on the different regions of TFF1 was examined. In these experiments, soluble chromatins obtained from MCF-7 cells were first immunoprecipitated with antibodies against SET8 followed by reimmunoprecipitation with antibodies against ERα or Ser-2-phosphorylated pol II (Fig. 7D). We found that ERα and SET8 co-existed only at the promoter of TFF1, whereas SET8 and phosphorylated pol II co-occupied only the coding region of this gene. Taken together, these experiments suggest that SET8 is able to interact with hyperphosphorylated and transcription-competent pol II and that H4K20me1 might carry an important message for transcription elongation or other hyperphosphorylated pol II-associated events.
DISCUSSION
Eukaryotic transcription is a dynamic process that is regulated at multiple levels and by a variety of mechanisms. Intricate interactions between RNA polymerase II and its chromatin environment are essential for proper transcription progression through initiation, elongation, and termination (64, 65). Although chromatin remodeling has been widely investigated in ERα-regulated transcription, and various histone-modifying factors, including histone acetyltransferases p300/CBP and GCN5/pCAF (15, 22) and histone methyltransferases CARM1, PRMT1, G9a, RIZ1, NSD1, MLL2, and SMYD3 have been implicated in this process, how histone modifications dictate ERα-regulated transcription is still not fully understood. We have reported here the spatial and temporal analyses of histone modifications that are introduced in ERα-activated TFF1 transcription. We have shown that these modifications appear to exhibit a bimodal pattern and occur in two phases. The first phase facilitates the initiation of transcription and is characterized by a rapid increase in H2BK5ac, H3K9ac, H4K5ac, H3S10ph, H2BK5me, H3K36me2, H3K36me3, H3K79me2, and H4K20me1 at the TFF1 gene promoter. The second phase, which takes place during transcription elongation, is concomitant with active transcription and is characterized by increases in H3K4me3, H3K36me2, H3K36me3, H3K79me2, H4K5ac, and H4K20me1 and decrease in H3K9ac in the coding region of the gene. In support of the notion that different processes in ERα-activated transcription are dictated by distinct histone codes, our experiments showed that pol II inhibitor α-amanitin drastically affected the occurrence of modifications in the second phase but had a limited effect on modifications in the first phase.
Phosphorylation of H3S10 is associated with transcriptional activation in different organisms and also with chromosome condensation during mitosis (66, 67). Interestingly, it appears that H3S10 phosphorylation is catalyzed by different kinases in various systems in response to distinct stimuli or under different cellular micromilieu. In mammalian cells, for example, stimulation with growth factors induces rapid phosphorylation of H3S10 at c-fos and c-jun promoters by MSK1, MSK2, and RSK2 kinases (67–69). Also, in response to inflammatory cytokines, IκB kinase α (IKKα) phosphorylates H3S10 at NF-κB-responsive promoters (70, 71), whereas in c-Myc-mediated transcriptional activation and oncogenic transformation, PIM1 is responsible for phosphorylation of H3S10 (72). Our experiments showed that E2 treatment led to rapid and transient activation of the MAPK signaling pathway and that MSK1 in this pathway was responsible for the phosphorylation of H3S10. It is unlikely that the H3S10ph mark added by different kinases itself carries different messages. However, it is possible that different kinases could be responsible for H3S10 phosphorylation in different cells, which could lead to the recruitment of different 14-3-3 isoforms based on the different contexts of promoters and promoter-associated proteins and the availability of the 14-3-3 protein isoforms in the particular cell type. Thus, H3S10ph could be interpreted in different ways. In this sense, it is interesting to note that our experiments identified ζ and ϵ isoforms, but not other isoforms, of 14-3-3 proteins that are associated with E2-triggered H3S10ph. Based on their characteristic phosphoserine/threonine binding ability and their functional mode as a homodimer or heterodimer (55), we propose that a 14-3-3ζ/14-3-3ϵ heterodimer recognizes the H3S10ph in ERα-activated transcription, although we cannot exclude the possibility that a 14-3-3ζ/14-3-3ϵ heterodimer, a 14-3-3ζ/14-3-3ζ homodimer, or/and a 14-3-3ϵ/14-3-3ϵ homodimer is in effect on a different allele of a particular ERα target gene or on different ERα target genes in MCF-7 cells.
There are seven family members in the 14-3-3 family (55). It is currently unknown whether the H3S10ph added by different kinases is recognized by the same set of 14-3-3 proteins. At least in c-fos and c-jun activation, three members of the 14-3-3 family (γ, ϵ, and ζ) have been identified as recognizing the H3S10ph mark (56). However, it is important to note that the transcription of c-fos and c-jun might be activated by a variety of stimuli, and it is possible that the identification of 14-3-3 γ, ϵ, and ζ was a reflection of c-fos and c-jun activation by multiple environmental cues. Overall, it remains likely that the H3S10ph added by different kinases is recognized by different 14-3-3 species, which in turn could be determined by the cellular abundance and availability of these proteins in a particular cell type. Alternatively, different members of the 14-3-3 family might be differentially activated by different signaling pathways to function in H3S10ph recognition. In any case, the same H3S10ph mark could be recognized in different ways through its differential association with 14-3-3 proteins.
We also found that H3S10ph is associated with the p52 subunit of TFIIH. This protein has not been reported as able to recognize phosphoserine/threonine. Therefore, we propose that the interaction between H3S10ph and the p52 subunit of TFIIH is indirect, possibly through the interaction of p52 with 14-3-3ζ/14-3-3ϵ. Indeed, we showed that p52 is physically associated with 14-3-3ζ/14-3-3ϵ in vivo. TFIIH comprises nine polypeptides and is involved in three of the most important functions of cells: DNA repair, cell cycle control, and transcription. During transcription initiation, the ATP-dependent helicase activity of TFIIH is essential for the formation of an open DNA complex. After establishment of the open complex, the kinase activity of TFIIH phosphorylates the fifth residue of the heptapeptide repeat present in the C-terminal domain of the largest subunit of pol II, enabling transcription (73–75).
We showed that H3S10ph is also associated with SMYD3. Similarly, we propose that this association might also be mediated by 14-3-3ζ/14-3-3ϵ. Significantly, we demonstrated that H3S10ph is functionally coupled with SMYD3-catalyzed H3K4me3, in which the addition of H3K4me3 appears to be dependent on the occurrence of H3S10ph. Interdependence among different histone modifications appears to be a predominant feature in the constitution of histone code and the dynamics of chromatin biology (76). This feature also functions in removing histone marks, as we demonstrated in our recent report that deacetylation and demethylation are coupled through the physical association of LSD1 and the NuRD complex (77). For H3S10ph, in addition to the coordination between H3S10ph and H3K4me3, it has been reported that H3S10ph cooperates with H3K9ac, H4K16ac, and other acetylation modifications (78), whereas H3S10ph antagonizes H3K9 methylation (79). It is conceivable that other histone modifications, including others studied in this report, also occur in a coordinated fashion. The discovery of the sequence of events and the mechanisms underlying the deposition of various histone modifications is the key to deciphering the histone code hypothesis.
As mentioned previously, although H4K20 methylation was among the first histone modifications identified (58), the biological function of this histone modification is not understood. H4K20me1 has been implicated in both transcriptional activation and gene silencing. It has also been demonstrated that H4K20me1 displays two distinct patterns, a peak close to the promoter and a further enrichment across the gene body region, and that it is required for the transcription activation of certain genes (23, 80). Our results indicate that H4K20me1 is correlated with both the initiation and elongation of transcription.
SET8 is the only known enzyme that catalyzes the addition of H4K20me1. Indeed, we have demonstrated that SET8 is required for ERα-mediated transcription and is responsible for the deposition of H4K20me1. Interestingly, we found that SET8 is associated with both ERα and a phosphorylated form of pol II in vivo. In light of its occurrence in both the initiation and elongation of transcription, a logical theme emerges in which SET8 is recruited first by ERα at the gene promoter, and once transcription-competent pol II is transformed, SET8 is delivered by ERα to pol II to function in transcription elongation. Future studies will be needed to investigate the validity of this scheme. In addition, exactly how H4K20me1 might impact transcription elongation is currently unknown. Additional experiments are warranted to address this issue.
Because of technical limitations, our experiments involved a limited number of histone modifications and involved only one ERα target gene. It will be important in future investigations to determine the scope of the studied histone modifications and the variety of other histone modifications involved in ERα-regulated transcription. Nonetheless, our current study provides a relatively comprehensive analysis of histone modifications introduced in ERα-regulated transcription. We have defined the enzymes, effector proteins, and meaning of several key components of the histone code governing TFF1 transcription regulated by ERα.
This work was supported by Grants 30830032 and 30921062 (to Y. S.) from the National Natural Science Foundation of China and Grants 2011CB504204 and 2007CB914503 (973 Program) (to Y. S.) from the Ministry of Science and Technology of China.
- H3K9ac
- histone H3 lysine 9 acetylation
- H3S10ph
- histone H3 serine 10 phosphorylation
- H3K4me3
- histone H3 lysine 4 trimethylation
- pol II
- polymerase II
- E2
- 17β-estradiol
- ERα
- estrogen receptor α
- MSK1
- mitogen- and stress-activated protein kinase 1
- MAPKBP1
- MAPK-binding protein 1
- qPCR
- quantitative PCR
- qChIP
- quantitative chromatin immunoprecipitation.
REFERENCES
- 1. Luger K., Hansen J. C. (2005) Curr. Opin. Struct. Biol. 15, 188–196 [DOI] [PubMed] [Google Scholar]
- 2. Peterson C. L., Laniel M. A. (2004) Curr. Biol. 14, R546–551 [DOI] [PubMed] [Google Scholar]
- 3. Bhaumik S. R., Smith E., Shilatifard A. (2007) Nat. Struct. Mol. Biol. 14, 1008–1016 [DOI] [PubMed] [Google Scholar]
- 4. Ho L., Crabtree G. R. (2010) Nature 463, 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Couture J. F., Trievel R. C. (2006) Curr. Opin. Struct. Biol. 16, 753–760 [DOI] [PubMed] [Google Scholar]
- 6. Martin C., Zhang Y. (2005) Nat. Rev. Mol. Cell Biol. 6, 838–849 [DOI] [PubMed] [Google Scholar]
- 7. Berger S. L. (2007) Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 8. Li B., Carey M., Workman J. L. (2007) Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 9. Tshuikina M., Nilsson K., Oberg F. (2008) Gene 410, 259–267 [DOI] [PubMed] [Google Scholar]
- 10. Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 11. Hong S. W., Hong S. M., Yoo J. W., Lee Y. C., Kim S., Lis J. T., Lee D. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14276–14280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. (2000) Cell 103, 667–678 [DOI] [PubMed] [Google Scholar]
- 13. Orphanides G., Lagrange T., Reinberg D. (1996) Genes Dev. 10, 2657–2683 [DOI] [PubMed] [Google Scholar]
- 14. Leo C., Chen J. D. (2000) Gene 245, 1–11 [DOI] [PubMed] [Google Scholar]
- 15. Das C., Lucia M. S., Hansen K. C., Tyler J. K. (2009) Nature 459, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson B. J., Tremblay A. M., Deblois G., Sylvain-Drolet G., Giguère V. (2010) Mol. Endocrinol. 24, 1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Le Romancer M., Treilleux I., Leconte N., Robin-Lespinasse Y., Sentis S., Bouchekioua-Bouzaghou K., Goddard S., Gobert-Gosse S., Corbo L. (2008) Mol. Cell 31, 212–221 [DOI] [PubMed] [Google Scholar]
- 18. Qi C., Chang J., Zhu Y., Yeldandi A. V., Rao S. M., Zhu Y. J. (2002) J. Biol. Chem. 277, 28624–28630 [DOI] [PubMed] [Google Scholar]
- 19. Carascossa S., Dudek P., Cenni B., Briand P. A., Picard D. (2010) Genes Dev. 24, 708–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. (1999) Science 284, 2174–2177 [DOI] [PubMed] [Google Scholar]
- 21. Lee D. Y., Northrop J. P., Kuo M. H., Stallcup M. R. (2006) J. Biol. Chem. 281, 8476–8485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mo R., Rao S. M., Zhu Y. J. (2006) J. Biol. Chem. 281, 15714–15720 [DOI] [PubMed] [Google Scholar]
- 23. Wakabayashi K., Okamura M., Tsutsumi S., Nishikawa N. S., Tanaka T., Sakakibara I., Kitakami J., Ihara S., Hashimoto Y., Hamakubo T., Kodama T., Aburatani H., Sakai J. (2009) Mol. Cell. Biol. 29, 3544–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. (2000) Cell 103, 843–852 [DOI] [PubMed] [Google Scholar]
- 25. Sun X., Zhang H., Wang D., Ma D., Shen Y., Shang Y. (2004) J. Biol. Chem. 279, 32839–32847 [DOI] [PubMed] [Google Scholar]
- 26. Wu H., Chen Y., Liang J., Shi B., Wu G., Zhang Y., Wang D., Li R., Yi X., Zhang H., Sun L., Shang Y. (2005) Nature 438, 981–987 [DOI] [PubMed] [Google Scholar]
- 27. Wu H., Sun L., Zhang Y., Chen Y., Shi B., Li R., Wang Y., Liang J., Fan D., Wu G., Wang D., Li S., Shang Y. (2006) J. Biol. Chem. 281, 21848–21856 [DOI] [PubMed] [Google Scholar]
- 28. Zhang H., Sun L., Liang J., Yu W., Zhang Y., Wang Y., Chen Y., Li R., Sun X., Shang Y. (2006) EMBO J. 25, 4223–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang H., Yi X., Sun X., Yin N., Shi B., Wu H., Wang D., Wu G., Shang Y. (2004) Genes Dev. 18, 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shang Y., Brown M. (2002) Science 295, 2465–2468 [DOI] [PubMed] [Google Scholar]
- 31. Zhang Y., Zhang H., Liang J., Yu W., Shang Y. (2007) EMBO J. 26, 2645–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reid G., Hübner M. R., Métivier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F. (2003) Mol. Cell 11, 695–707 [DOI] [PubMed] [Google Scholar]
- 33. Métivier R., Penot G., Hübner M. R., Reid G., Brand H., Kos M., Gannon F. (2003) Cell 115, 751–763 [DOI] [PubMed] [Google Scholar]
- 34. American Association for Cancer Research Human Epigenome Task Force and European Union Network of Excellence Scientific Advisory Board (2008) Nature 454, 711–71518685699 [Google Scholar]
- 35. Ivaldi M. S., Karam C. S., Corces V. G. (2007) Genes Dev. 21, 2818–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ke Q., Li Q., Ellen T. P., Sun H., Costa M. (2008) Carcinogenesis 29, 1276–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steger D. J., Lefterova M. I., Ying L., Stonestrom A. J., Schupp M., Zhuo D., Vakoc A. L., Kim J. E., Chen J., Lazar M. A., Blobel G. A., Vakoc C. R. (2008) Mol. Cell. Biol. 28, 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edmunds J. W., Mahadevan L. C., Clayton A. L. (2008) EMBO J. 27, 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee J. S., Shilatifard A. (2007) Mutat. Res. 618, 130–134 [DOI] [PubMed] [Google Scholar]
- 40. Kaplan C. D., Larsson K. M., Kornberg R. D. (2008) Mol. Cell 30, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rödicker F., Ossenbühl F., Michels D., Benecke B. J. (1999) Gene Expr. 8, 165–174 [PMC free article] [PubMed] [Google Scholar]
- 42. Kedinger C., Gniazdowski M., Mandel J. L., Jr., Gissinger F., Chambon P. (1970) Biochem. Biophys. Res. Commun. 38, 165–171 [DOI] [PubMed] [Google Scholar]
- 43. Lindell T. J., Weinberg F., Morris P. W., Roeder R. G., Rutter W. J. (1970) Science 170, 447–449 [DOI] [PubMed] [Google Scholar]
- 44. Kim H., Heo K., Kim J. H., Kim K., Choi J., An W. (2009) J. Biol. Chem. 284, 19867–19877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang N., vom Baur E., Garnier J. M., Lerouge T., Vonesch J. L., Lutz Y., Chambon P., Losson R. (1998) EMBO J. 17, 3398–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Glass C. K., Rosenfeld M. G. (2000) Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 47. Berndsen C. E., Tsubota T., Lindner S. E., Lee S., Holton J. M., Kaufman P. D., Keck J. L., Denu J. M. (2008) Nat. Struct. Mol. Biol. 15, 948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marino M., Galluzzo P., Ascenzi P. (2006) Curr. Genomics 7, 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seval Y., Cakmak H., Kayisli U. A., Arici A. (2006) J. Clin. Endocrinol. Metab. 91, 2349–2357 [DOI] [PubMed] [Google Scholar]
- 50. Frigo D. E., Basu A., Nierth-Simpson E. N., Weldon C. B., Dugan C. M., Elliott S., Collins-Burow B. M., Salvo V. A., Zhu Y., Melnik L. I., Lopez G. N., Kushner P. J., Curiel T. J., Rowan B. G., McLachlan J. A., Burow M. E. (2006) Mol. Endocrinol. 20, 971–983 [DOI] [PubMed] [Google Scholar]
- 51. Benvenisti-Zarom L., Chen-Roetling J., Regan R. F. (2006) Neurosci. Lett. 398, 230–234 [DOI] [PubMed] [Google Scholar]
- 52. Kim H. G., Lee K. W., Cho Y. Y., Kang N. J., Oh S. M., Bode A. M., Dong Z. (2008) Cancer Res. 68, 2538–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vermeulen L., De Wilde G., Van Damme P., Vanden Berghe W., Haegeman G. (2003) EMBO J. 22, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thomson S., Clayton A. L., Hazzalin C. A., Rose S., Barratt M. J., Mahadevan L. C. (1999) EMBO J. 18, 4779–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Morrison D. K. (2009) Trends Cell Biol. 19, 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Macdonald N., Welburn J. P., Noble M. E., Nguyen A., Yaffe M. B., Clynes D., Moggs J. G., Orphanides G., Thomson S., Edmunds J. W., Clayton A. L., Endicott J. A., Mahadevan L. C. (2005) Mol. Cell 20, 199–211 [DOI] [PubMed] [Google Scholar]
- 57. Hamamoto R., Furukawa Y., Morita M., Iimura Y., Silva F. P., Li M., Yagyu R., Nakamura Y. (2004) Nat. Cell Biol. 6, 731–740 [DOI] [PubMed] [Google Scholar]
- 58. Murray K. (1964) Biochemistry 3, 10–15 [DOI] [PubMed] [Google Scholar]
- 59. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 60. Couture J. F., Collazo E., Brunzelle J. S., Trievel R. C. (2005) Genes Dev. 19, 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xiao B., Jing C., Kelly G., Walker P. A., Muskett F. W., Frenkiel T. A., Martin S. R., Sarma K., Reinberg D., Gamblin S. J., Wilson J. R. (2005) Genes Dev. 19, 1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Egloff S., Murphy S. (2008) Trends Genet. 24, 280–288 [DOI] [PubMed] [Google Scholar]
- 63. Phatnani H. P., Greenleaf A. L. (2006) Genes Dev. 20, 2922–2936 [DOI] [PubMed] [Google Scholar]
- 64. Tang G. Q., Roy R., Bandwar R. P., Ha T., Patel S. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 22175–22180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hansen J. C., Wolffe A. P. (1992) Biochemistry 31, 7977–7988 [DOI] [PubMed] [Google Scholar]
- 66. Cheung P., Allis C. D., Sassone-Corsi P. (2000) Cell 103, 263–271 [DOI] [PubMed] [Google Scholar]
- 67. Nowak S. J., Corces V. G. (2004) Trends Genet. 20, 214–220 [DOI] [PubMed] [Google Scholar]
- 68. Sassone-Corsi P., Mizzen C. A., Cheung P., Crosio C., Monaco L., Jacquot S., Hanauer A., Allis C. D. (1999) Science 285, 886–891 [DOI] [PubMed] [Google Scholar]
- 69. Soloaga A., Thomson S., Wiggin G. R., Rampersaud N., Dyson M. H., Hazzalin C. A., Mahadevan L. C., Arthur J. S. (2003) EMBO J. 22, 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yamamoto Y., Verma U. N., Prajapati S., Kwak Y. T., Gaynor R. B. (2003) Nature 423, 655–659 [DOI] [PubMed] [Google Scholar]
- 71. Anest V., Hanson J. L., Cogswell P. C., Steinbrecher K. A., Strahl B. D., Baldwin A. S. (2003) Nature 423, 659–663 [DOI] [PubMed] [Google Scholar]
- 72. Zippo A., De Robertis A., Serafini R., Oliviero S. (2007) Nat. Cell Biol. 9, 932–944 [DOI] [PubMed] [Google Scholar]
- 73. Akhtar M. S., Heidemann M., Tietjen J. R., Zhang D. W., Chapman R. D., Eick D., Ansari A. Z. (2009) Mol. Cell 34, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fukuda A., Hisatake K. (2000) Tanpakushitsu Kakusan Koso 45, 1505–1512 [PubMed] [Google Scholar]
- 75. Svejstrup J. Q., Vichi P., Egly J. M. (1996) Trends Biochem. Sci. 21, 346–350 [PubMed] [Google Scholar]
- 76. Suganuma T., Workman J. L. (2008) Cell 135, 604–607 [DOI] [PubMed] [Google Scholar]
- 77. Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L., Li R., Li Y., Zhang Y., Li Q., Yi X., Shang Y. (2009) Cell 138, 660–672 [DOI] [PubMed] [Google Scholar]
- 78. Zippo A., Serafini R., Rocchigiani M., Pennacchini S., Krepelova A., Oliviero S. (2009) Cell 138, 1122–1136 [DOI] [PubMed] [Google Scholar]
- 79. Duan Q., Chen H., Costa M., Dai W. (2008) J. Biol. Chem. 283, 33585–33590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Karlic R., Chung H. R., Lasserre J., Vlahovicek K., Vingron M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2926–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]