Abstract
CD81 is a tetraspanin protein that is involved in several essential cellular functions, as well as in the hepatitis C virus (HCV) infection. CD81 interacts with a high stoichiometry with its partner proteins EWI-2, EWI-2wint, and EWI-F. These latter proteins modify the functions of CD81 and can thereby potentially inhibit infection or modulate cell migration. Here, we characterized the cleavage of EWI-2 leading to the production of EWI-2wint, which has been shown to inhibit HCV infection. We determined the regions of EWI-2/EWI-2wint and CD81 that are important for their interaction and their functionality. More precisely, we identified a glycine zipper motif in the transmembrane domain of EWI-2/EWI-2wint that is essential for the interaction with CD81. In addition, we found that palmitoylation on two juxtamembranous cysteines in the cytosolic tail of EWI-2/EWI-2wint is required for their interaction with CD81 as well as with CD9, another tetraspanin. Thus, we have shown that palmitoylation of a tetraspanin partner protein can influence the interaction with a tetraspanin. We therefore propose that palmitoylation not only of tetraspanins, but also of their partner proteins is important in regulating the composition of complexes in tetraspanin networks. Finally, we identified the regions in CD81 that are necessary for its functionality in HCV entry and we demonstrated that EWI-2wint needs to interact with CD81 to exert its inhibitory effect on HCV infection.
Keywords: Flavi Viruses, Glycosylation, Membrane Proteins, Protein Acylation, Protein-Protein Interactions, CD81, EWI-2/EWI-2wint Proteins, Hepatitis C Virus Entry, Palmitoylation, Tetraspanin-enriched Microdomains
Introduction
Tetraspanins comprise a family of evolutionary highly conserved proteins that all contain four transmembrane domains for which they are named, as well as a small and a large extracellular loop. They are ubiquitously expressed proteins that are involved in many cellular functions such as adhesion, migration, co-stimulation, signal transduction, and cell differentiation. Tetraspanins also play an important role in infection by several pathogens (reviewed in Ref. 1). However, their specific function in all these processes remains to be elucidated. Tetraspanins have the special feature to interact with each other and other transmembrane proteins, forming extended cholesterol-rich complexes on the cell surface, called tetraspanin-enriched microdomains (TEMs)6 (2, 3). In these domains, tetraspanins form primary complexes with a limited number of proteins called tetraspanin partners. These primary interactions are direct, highly specific, and occur at high stoichiometry. Tetraspanin molecules can form different partnerships in different cell types. The precise mechanisms of interaction within TEMs remain unknown. However, in addition to specific domains, cellular lipids such as cholesterol, glycosphingolipids, and palmitic acid seem to play an important role in the interaction of tetraspanins with each other and therefore in building the tetraspanin network (1).
In this study, we focused on tetraspanin CD81, which was initially characterized as important for B cell proliferation (4), but has since been found in many different cell lines, contributing to a variety of different cellular processes (5). Importantly, CD81 plays a major role in several infectious diseases. Indeed, CD81 is an essential entry factor for the hepatitis C virus (HCV) (6), it is necessary for infection with the malaria parasite Plasmodium (7) and plays a role in Listeria monocytogenes infection (8).
HCV infection is a global public health problem affecting over 130 million individuals worldwide. Its symptoms include chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (9). HCV encodes two envelope glycoproteins, E1 and E2, that interact to form a E1E2 heterodimer that is present at the surface of HCV particles and is therefore the obvious candidate ligand for cellular receptors. The use of surrogate models and HCV grown in cell culture (HCVcc) provided numerous evidence that CD81 plays a key role in the early steps of the HCV life cycle, likely during a post-attachment step. The CD81 large extracellular loop (LEL) is the critical region for the interaction with the E2 glycoprotein and for virus entry. Antibodies directed against CD81, as well as a soluble form of the CD81 LEL, are able to inhibit in vitro and in vivo HCV entry into hepatocytes. Moreover, the absence of CD81 expression or CD81 down-regulation using siRNA in hepatoma cells abolishes HCV infection (reviewed in Ref. 10).
In most cell lines, CD81 is associated in a high stoichiometry with EWI-F (also called CD9P-1, FPRP, or CD315) and EWI-2 (also called PGRL, IgSF8, or CD316) (11–15). Both are members of the EWI family, a small Ig-domain family whose members have a single transmembrane domain and several extracellular Ig domains with a conserved EWI motif, as well as a very short cytosolic tail (14). Recently, we have identified EWI-2wint, a cleavage product of EWI-2 in which the first of the 4 extracellular Ig domains is cleaved (16). This shorter protein still interacts with CD81 and CD9 and can be found, along with EWI-2, in most cell lines expressing EWI-2. Importantly, in contrast to full-length EWI-2, EWI-2wint inhibits the binding of HCV envelope glycoprotein 2 (E2) to CD81, thus inhibiting viral entry (16). To date, it still remains unclear how EWI-2wint inhibits the binding of E2 to CD81 and which cellular protease is responsible for the cleavage of EWI-2.
The direct interaction between tetraspanins and their partner proteins results in the modulation of their functions. CD81 functions in cellular processes and infectious diseases can therefore be affected by the association with the proteins EWI-F, EWI-2, or EWI-2wint. For example, EWI-2 has been shown to modulate cellular migration (17–19). As already described, EWI-2wint has an inhibitory effect on HCV infection by inhibiting the interaction between CD81 and HCV E2 (16). Very recently, it was also shown that overexpression of EWI-F inhibits Plasmodium infection, whereas its silencing increases infection efficiency (20).
Because of the ability of EWI-2 to inhibit cell invasion, migration, and tumor growth (21) and EWI-2wint to inhibit HCV entry (16), these two molecules have therapeutic interests. The characterization of the biogenesis of these molecules, as well as the mechanism leading to their interaction with CD81, is therefore essential. In addition, such a mechanism could be part of a more general mechanism of tetraspanin regulation. In our study, we performed biochemical and functional characterization of the interaction between EWI-2/EWI-2wint and CD81.
EXPERIMENTAL PROCEDURES
Antibodies
5A6 (anti-CD81 kindly provided by S. Levy (4)), TS82b (anti-CD82 (12)), and SYB-I (anti-CD9 (3)) mAbs were used in this study. M2 anti-FLAG mAb was from Sigma. Anti-HA mAbs (HA11 and 3F10) were from Covance and Roche Applied Science, respectively. Anti-NS5 was from AUSTRAL Biologicals. PE-labeled goat anti-mouse and PE-labeled 5A6 were from BD Pharmingen. FITC-labeled rabbit anti-mouse F(ab′)2 and irrelevant mouse IgG1 were from DAKO. Goat anti-mouse or anti-rat immunoglobulins conjugated to peroxidase were from Sigma and Jackson ImmunoResearch, respectively.
Plasmids
pcDNA3.1 plasmids expressing CD81 (11), CD81Plm− (22), EWI-2FLAG (16), and EWI-2FLAGFur (16) have been described previously. All other plasmids were generated by PCR-based mutagenesis with the aforementioned plasmids. Briefly, truncated forms of EWI-2 contain its signal peptide sequence followed by the EWI-2 sequence starting at residues Asp122 (Ig2–4) or Asp270 (Ig3–4) and a C-terminal FLAG tag sequence. In the ΔIg4 construct, the amino acid sequence between Arg425 and Ala518 was deleted from the C-terminal FLAG-tagged EWI-2 construct. Glycosylation mutants were based on a C-terminal HA-tagged EWI-2 (EWI-2HA). In these mutants, the potentially glycosylated Asn in Ig domains one, three, or four were replaced by Gln, either only in Ig1 (N1), in Ig1 and Ig3 (N1–2), or all (N1–2-3). Cloning details and oligonucleotide sequences are available upon request.
Cells and Cell Transfection
Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and supplemented with 100 nm nonessential amino acids (Invitrogen). Huh-7, CHO, and HEK-293T cells were from ATCC. CHO FD11 and CHO FD11 overexpressing furin were kindly provided by S. H. Leppla (23). Huh-7w7 (CD81−) were described previously (24). Cell transfections were performed as described in Ref. (16). For stably transfected Huh-7 and Huh-7w7 cells, cells were electroporated using the Gene Pulser apparatus (Bio-Rad) and neomycin was added 48 h post-transfection at 600 μg/ml. Huh-7 cell clones expressing EWI-2/EWI-2wint mutants were isolated using selection cylinders and maintained in neomycin medium.
Detection of Cell Surface-biotinylated Proteins
Cells were biotinylated as described in Ref. 16 and lysed in lysis buffer containing 1% Brij-97 in PBS with 2 mm EDTA (PBS/Brij/EDTA) and protease inhibitors (Complete, Roche Applied Science). Lysates were precleared for 2 h at 4 °C with protein A-Sepharose (GE Healthcare), then incubated for 2 h at 4 °C with specific mAbs immobilized onto protein A-Sepharose beads. Complexes were eluted with non-reducing Laemmli buffer, resolved by SDS-PAGE, transferred to a nitrocellulose membrane (GE Healthcare), and immunoblotted with peroxidase-conjugated streptavidin (Vector). All experiments were done at least three times and representative blots are shown.
Immunoblotting
Immunoprecipitated proteins or cell lysates were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with 3F10 anti-HA, 5A6 anti-CD81, or TS82b anti-CD82 antibodies followed by peroxidase-conjugated secondary antibodies.
Metabolic Labeling
For the kinetics of EWI-2wint production, EWI-2FLAG expressing CHO cells were starved in Met-Cys-free medium for 30 min and pulse-labeled for 10 min with 35S-Protein Labeling Mix (100 mCi/ml, PerkinElmer Life Sciences). After the indicated time of chase, cells were lysed with PBS, 1% Triton X-100, 2 mm EDTA and the lysates were used for immunoprecipitation with the M2 anti-FLAG mAb.
To assess palmitic acid incorporation, transfected CHO cells were incubated in serum-free medium containing sodium pyruvate for 3 h and labeled 4 h at 37 °C with 300 mCi/ml of [9,10-3H]palmitic acid (PerkinElmer Life Sciences) in serum-free medium containing sodium pyruvate with 2% fatty acid-free BSA (Sigma). The cells were then lysed in PBS/Triton X-100/EDTA and used for immunoprecipitations with M2 anti-FLAG, 5A6 anti-CD81 or TS82b anti-CD82 mAbs.
Endoglycosidase Digestions
Digestions with endo-β-N-acetylglucosaminidase H (Endo H) and peptide N-glycosidase F (PNGase F) (both from New England Biolabs) were carried out as previously described (25). Digestions with peptide N-glycosidase, neuraminidase, glycopeptide-N-acetylgalactosaminidase (O-Glycanase) of unlabeled proteins were performed as previously described (12).
Confocal Microscopy
Huh-7 cells stably expressing wild-type (wt) EWI-2/EWI-2wint proteins or palmitoylation mutant (wtPlm−) were grown on 12-mm glass coverslips and fixed with paraformaldehyde. After blocking with goat serum, cells were incubated with anti-CD81 1.3.3.22 (mouse IgG1, Santa Cruz Biotechnology) and anti-HA 3F10 (rat IgG1, Roche Applied Science) primary antibodies, and then with Alexa 488-conjugated goat anti-mouse IgG and Alexa 555-conjugated goat anti-rat IgG (Invitrogen) secondary antibodies. Image acquisition was performed with an LSM710 confocal microscope (Zeiss) using a ×63 oil immersion objective with a 1.4 numerical aperture. Signals were sequentially collected using single fluorescence excitation and acquisition settings to avoid crossover. Images were assembled and analyzed by using Zen software (Zeiss).
HCVcc Infection
HCVcc used in this study were based on the JFH1 strain (26), kindly provided by T. Wakita (National Institute of Infectious Diseases, Tokyo, Japan), and contained cell culture-adaptive mutations CS and N6 (27). HCVcc stocks were produced as previously reported (27).
For the HCVcc infection assay, cells grown in 24-well plates were incubated with HCVcc (multiplicity of infection = 1) for 2 h at 37 °C, washed, and incubated for an additional 40 h at 37 °C. Infections were scored by flow cytometry.
Flow Cytometry Analysis
For single NS5 staining, infected cells were permeabilized with PBS, 2% BSA, 0.05% saponin. After rinsing with PBS, 2% BSA, cells were incubated 1 h at 4 °C with anti-NS5 mAb (2F6/G11). After rinsing with washing solution, cells were incubated with PE-labeled goat anti-mouse for 45 min at 4 °C. Cells were washed, detached with PBS, 2 mm EDTA, and fixed with formalin solution (formaldehyde 4%, Sigma).
For double staining, NS5-stained cells were incubated with FITC-labeled rabbit anti-mouse F(ab′)2 immunoglobulins. Cells were then incubated with irrelevant mouse IgG1 for 30 min, washed, and incubated 1 h at 4 °C with PE-labeled 5A6 anti-CD81 mAb. Cells were washed, detached with PBS, 2 mm EDTA and fixed with formalin solution. Cells stained only with the secondary antibodies or PE-labeled isotype control were used as negative controls. Labeled cells were analyzed using a FACS Beckman EPICS-XL MCL.
For CD81 staining, cells were detached with PBS, 2 mm EDTA and incubated 1 h at 4 °C with 5A6. After rinsing with washing solution, cells were incubated with PE-labeled goat anti-mouse for 45 min at 4 °C. Cells were washed and fixed with formalin solution.
RESULTS
Cleavage and Glycosylation of EWI-2
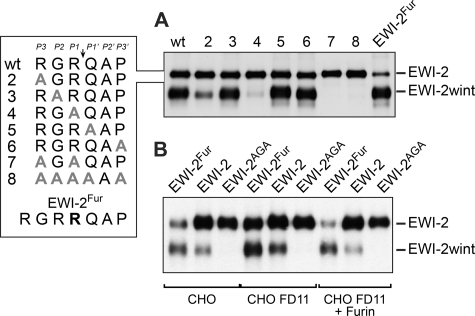
To better understand the production of EWI-2wint, we first analyzed the processing of EWI-2, a ∼70 kDa protein. It is cleaved between the two N-terminal Ig domains in the sequence RGRQAP, where Q is the first amino acid of EWI-2wint, a ∼55 kDa protein (16) (Fig. 1A). To identify the amino acid sequence recognized by the protease, a mutagenesis analysis was performed in which every residue in this sequence was sequentially replaced by an alanine. The mutant proteins were expressed in CHO cells (a cell line that naturally cleaves EWI-2) and analyzed for their cleavage. As shown in Fig. 1A, mutation of the two arginine residues abolished the cleavage of EWI-2, whereas the replacement of all other amino acids did not affect the cleavage, indicating that RXR likely corresponds to the minimal site recognized by the protease involved in the production of EWI-2wint.
FIGURE 1.
Cleavage of EWI-2 leading to EWI-2wint production. A, wild type FLAG-tagged EWI-2 protein (wt) and proteins with mutations in the putative cleavage site (see box) were expressed in CHO cells. Cells were surface biotinylated and the proteins were precipitated with the M2 anti-FLAG antibody. Proteins were separated by SDS-PAGE under nonreducing conditions and detected using HRP-streptavidin. Expression of EWI-2 (wt) in CHO cells leads to the production of EWI-2 and EWI-2wint. B, wild type FLAG-tagged EWI-2 protein (EWI-2) or EWI-2 containing a furin cleavage consensus site (EWI-2Fur) or EWI-2 in which the RXR consensus site was mutated (EWI-2AGA, mutant 7 in A) were expressed either in regular CHO cells or CHO cells that were deficient for furin expression (CHO FD11), or CHO FD11 cells that overexpressed furin (CHO FD11 + Furin). Proteins were surface biotinylated, immunoprecipitated with M2 anti-FLAG antibody, separated by SDS-PAGE under nonreducing conditions, transferred to a nitrocellulose membrane, and detected using HRP-streptavidin.
Insertion of an additional arginine in the sequence to generate a furin cleavage consensus site (RGRR, EWI-2Fur) led to a higher degree of cleavage of EWI-2 (Fig. 1A). This fact and the polybasic cleavage preference suggested that furin is the protease that cleaves EWI-2, even though the sequence does not correspond to the preferred furin cleavage sequence. To address this point, we tested cleavage of EWI-2 and EWI-2Fur in furin-deficient CHO cells (CHO FD11 (23)). We used EWI-2 in which the RXR consensus site was mutated (EWI-2AGA, mutant 7 in Fig. 1A) as a control. CHO FD11 cells were still able to cleave EWI-2 and EWI-2Fur, which both contain the original cleavage site (Fig. 1B), suggesting that furin is unlikely to be involved in EWI-2wint production. Concurring with this, overexpression of furin in CHO FD11 cells led to an increased cleavage of EWI-2Fur, whereas the cleavage of EWI-2 was not affected (Fig. 1B, CHO FD11 + Furin).
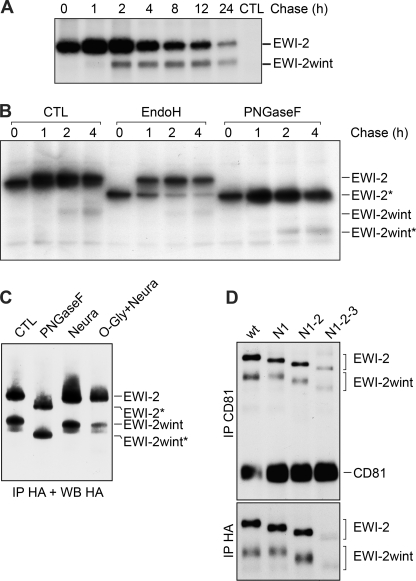
We next performed pulse-chase experiments to analyze the kinetics of the cleavage of EWI-2. These experiments showed that EWI-2wint starts to appear after 1 h and accumulates over time for at least 24 h (Fig. 2A). Glycosylation predictions indicated that EWI-2/EWI-2wint might be N-glycosylated at three/two sites and possibly be O-glycosylated as well. To analyze the N-glycosylation status of EWI-2/EWI-2wint over time, we performed digestion with Endo H and PNGase F in pulse-chase experiments (Fig. 2B). PNGase F hydrolyzes nearly all types of N-glycan chains from glycoproteins, whereas Endo H removes the chitobiose core of high-mannose and some hybrid forms of N-linked sugars, excluding the complex forms (28). Resistance to digestion with Endo H is indicative that glycoproteins have moved from the ER to at least the medial and trans Golgi, where complex sugars are added. As shown in Fig. 2B, EWI-2 was sensitive to PNGase F digestion, whereas it became partially resistant to Endo H after 1 h of chase, with the majority of the protein being resistant to Endo H deglycosylation after 4 h. Interestingly, EWI-2wint was resistant to Endo H deglycosylation from the beginning, indicating that it carries only mature N-glycans and that it is produced after glycan maturation of EWI-2. Together, these results suggest that the cleavage of EWI-2 to produce EWI-2wint likely occurs in a post-ER compartment.
FIGURE 2.
Kinetics of production and glycosylation of EWI-2/EWI-2wint. A, CHO cells expressing C-terminal FLAG-tagged EWI-2 were pulse-labeled for 10 min with 35S-Protein Labeling Mix. After the indicated time of chase, cells were lysed in PBS, 1% Triton X-100 and proteins were precipitated with M2 antibody, separated by SDS-PAGE under nonreducing conditions, and detected by autoradiography. B, cells were labeled and lysed as described in A, after immunoprecipitation (IP) the proteins were either left untreated (CTL), treated with Endo H to remove high-mannose type glycans or treated with PNGase F to remove all N-linked glycosylations. Deglycosylated proteins are indicated by an asterisk. C, C-terminal HA-tagged EWI-2 proteins were expressed in CHO cells and precipitated using HA11 antibody. After precipitation, the samples were either left untreated (CTL), treated with PNGase F, or treated with neuraminidase (Neura) or O-glycanase and neuraminidase (O-Gly + Neura). Digested samples were resolved by SDS-PAGE and immunoblotted with the 3F10 anti-HA antibody. Deglycosylated proteins are indicated by an asterisk. D, expression of the glycosylation mutants of EWI-2 in which the potentially glycosylated Asn in Ig-domains one, three, or four were replaced by Gln, either only in Ig1 (N1), in Ig1 and Ig3 (N1–2), or all (N1–2-3). HA-tagged EWI-2 and glycosylation mutants were co-expressed with CD81 in CHO cells. Surface proteins were biotinylated and precipitated using HA11 anti-HA or 5A6 anti-CD81 antibodies. Proteins were detected using HRP-streptavidin.
Further analyses using neuraminidase and O-glycanase digestions suggested that EWI-2/EWI-2wint are only N-glycosylated, but not O-glycosylated (Fig. 2C). In addition, successive mutations of the potential N-glycosylation sites showed that EWI-2 and EWI-2wint proteins are glycosylated on their three and two N-glycosylation sites, respectively (Fig. 2D). Interestingly, the absence of N-glycan on EWI-2 did not affect EWI-2wint production. However, it should be noted that the completely unglycosylated proteins (N1-2-3) were expressed at lower levels, likely due to protein instability. Glycosylation mutants were next tested for their interaction with CD81 in co-immunoprecipitation experiments. In this study, all experiments using co-precipitation of EWI-2/EWI-2wint with CD81 were performed after lysis with a detergent (1% Brij 97, 2 mm EDTA) that keeps primary complexes intact (CD81/EWI-2/EWI-2wint), but disrupts tetraspanin-tetraspanin interactions (11). As shown in Fig. 2D, all glycosylation mutants of EWI-2/EWI-2wint co-precipitated with CD81. Altogether, these results indicate that N-glycosylation of EWI-2/EWI-2wint does not play a major role in the interaction with CD81 nor in the cleavage of EWI-2 to EWI-2wint.
Palmitoylation of Juxtamembranous Cysteines and the Presence of a Glycine-zipper Motif in TM Domain of EWI-2/EWI-2wint Are Essential for the Interaction with CD81
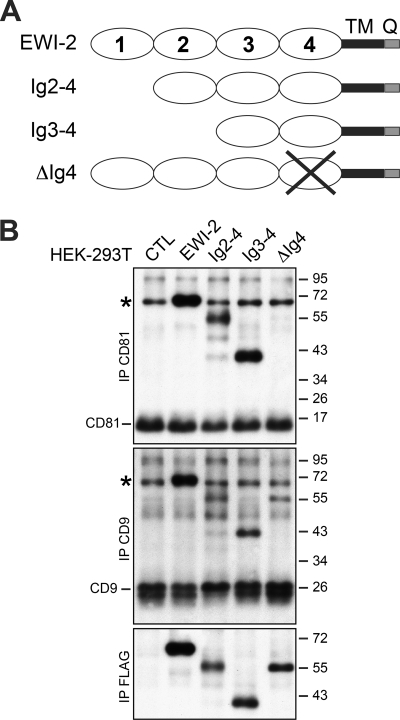
Because we identified EWI-2wint, a shorter form of EWI-2, through direct interaction with CD81 (16), we next tried to map domain(s) required for this interaction. Toward this aim, we first constructed a set of mutants consecutively shortened by one of the extracellular Ig domains (Fig. 3A). However, the shortest form containing only the most membrane-proximal Ig4-domain could not be expressed. We therefore made an additional mutant in which the Ig4 domain was deleted (ΔIg4). Truncated proteins were equally expressed as controlled by direct immunoprecipitation with the anti-FLAG mAb (Fig. 3B, IP FLAG). Interestingly, the co-precipitation of Ig2-4 and Ig3-4 mutant proteins with CD81 was similar to that of the full-length protein, whereas the ΔIg4 construct failed to interact with CD81 (Fig. 3B, IP CD81), indicating that the membrane-proximal Ig4 domain is necessary for the interaction of EWI-2/EWI-2wint with CD81. Interestingly, the ΔIg4 construct interacted with CD9, another tetraspanin that directly interacts with EWI-2/EWI-2wint (Fig. 3B, IP CD9), indicating that the overall conformation of the ΔIg4 protein is conserved and the interaction of EWI-2 with tetraspanins CD81 and CD9 involves domains that are different. It should be noted that this experiment was performed in HEK-293T cells that express endogenous EWI-2 (asterisk in Fig. 3B) but in which EWI-2 cleavage does not occur, as previously described (16).
FIGURE 3.
Characterization of extracellular deleted variants of EWI-2 protein. A, schematic representation of the extracellular deleted variants of FLAG-tagged EWI-2. B, deleted EWI-2 proteins were expressed in HEK-293T cells. Surface-biotinylated proteins were immunoprecipitated with anti-CD81, anti-CD9, or anti-FLAG mAbs. Proteins were separated by SDS-PAGE under nonreducing conditions, transferred to a nitrocellulose membrane, and detected using HRP-streptavidin. EWI-2, CD81, and CD9 proteins are endogenously expressed in HEK-293T cells. The cleavage of EWI-2 does not occur in HEK-293T cells. Endogenous EWI-2 is indicated by an asterisk.
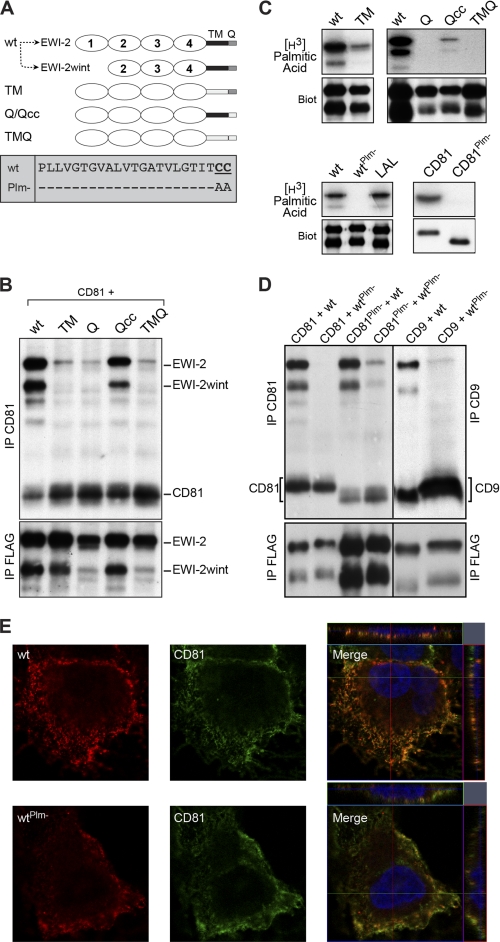
Next, we generated chimeric EWI-2 proteins in which either the transmembrane (TM) domain, or the cytosolic tail (Q), or both (TMQ) were replaced by the corresponding domains of the human MHC class II molecule (MHCII; HLA-DBQ1), which are exactly the same length (Fig. 4A). Although the Q and TMQ chimeras were slightly less expressed, domain exchanges in chimeras did not significantly affect their cell surface expression, as controlled by direct immunoprecipitation of biotinylated cell lysates (Fig. 4B, IP FLAG). As shown by co-immunoprecipitation experiments, only the wild type EWI-2/EWI-2wint interacted with CD81 (Fig. 4B, IP CD81), suggesting that both TM and Q domains may play a role in the interaction with CD81. However, EWI-2/EWI-2wint contain two juxtamembranous cysteine residues in their Q domain that are potential palmitoylation sites. We therefore constructed an additional chimera with the cytosolic tail of MHCII, but which still contained two cysteine residues (Qcc) (Fig. 4A). Interestingly, the presence of these residues restored the interaction with CD81 (Fig. 4B, IP CD81), as compared with the Q chimera. To verify that EWI-2/EWI-2wint and the chimeras containing the juxtamembranous cysteines were indeed palmitoylated, proteins were metabolically labeled with [3H]palmitate (Fig. 4C). Our results clearly showed that EWI-2/EWI-2wint, as well as the TM and Qcc chimeras, incorporated [3H]palmitate, whereas the Q and TMQ chimeras did not. Altogether, our results indicate that in addition to the Ig4 domain, the TM domain of EWI-2/EWI-2wint and palmitoylation on their juxtamembranous cysteine residues are likely important for the interaction with CD81.
FIGURE 4.
Transmembrane domain and palmitoylation of EWI-2/EWI-2wint are important for the interaction with CD81. A, schematic representation of the different EWI-2 chimeric proteins and palmitoylation mutant. Expression of EWI-2 (wt) in CHO cells leads to the production of EWI-2 and EWI-2wint. B, FLAG-tagged chimeras of EWI-2 containing the transmembrane domain (TM) and/or the cytosolic tail (Q) of MHC II were co-expressed with CD81 in CHO cells. Surface-biotinylated proteins were lysed in PBS/Brij97/EDTA, immunoprecipitated, separated by SDS-PAGE under nonreducing conditions, and detected using HRP-streptavidin. C, metabolic labeling of EWI-2/EWI-2wint and CD81 mutants using [3H]palmitic acid, followed by IP. Expression of each protein was controlled by cell surface biotinylation (Biot). D, wild type proteins and palmitoylation-deficient mutants were co-expressed in CHO cells, biotinylated, lysed in PBS/Brij97/EDTA, and immunoprecipitated. Proteins were resolved by SDS-PAGE under nonreducing conditions and detected using HRP-streptavidin. In all experiments, anti-CD81, anti-CD9, and anti-FLAG IPs were performed with 5A6, SYB-I, and M2 antibodies, respectively. E, Huh-7 cells stably expressing either wild type EWI-2/EWI-2wint (wt) or palmitoylation mutant (wtPlm-) were labeled with antibodies to CD81 or HA epitope present at the N terminus of the EWI-2/EWI-2wint construct. Cells were imaged with an LSM710 confocal microscope using a ×63 oil immersion objective with a 1.4 numerical aperture. Representative confocal images are shown. Confocal sections of the Z-stack along the indicated lines are shown on the top and the right of merged images.
To further confirm the importance of palmitoylation in the interaction between EWI-2/EWI-2wint and CD81, we made a mutant of EWI-2/EWI-2wint in which the two cysteine residues were replaced by alanines (Fig. 4A, box wtPlm-). In addition, we used a CD81 mutant lacking all potential palmitoylation sites (CD81Plm-) (22). As shown by [3H]palmitate incorporation, both mutants were no longer palmitoylated (Fig. 4C), in contrast to the wild type proteins. It should be noted that EWI-2wint incorporated less [3H]palmitic acid than EWI-2 (Fig. 4C), indicating that cleavage of the first Ig domain may regulate EWI-2 palmitoylation. Cell surface biotinylation showed that wild type and palmitoylation mutant proteins were expressed at equal levels at the plasma membrane indicating that the absence of palmitoylation did not affect transport to the cell surface (Fig. 4C, Biot). Co-immunoprecipitation experiments showed that wild type EWI-2/EWI-2wint interacted equally well with CD81 as with CD81Plm- (Fig. 4D), suggesting that palmitoylation of CD81 is not necessary for the interaction with its partners. In contrast, the unpalmitoylated EWI-2/EWI-2wint failed to co-precipitate with CD81 and was found only in trace amounts in complexes with CD81Plm- (Fig. 4D). In addition, unpalmitoylated EWI-2 proteins also failed to interact with CD9 (Fig. 4D), another tetraspanin that directly interacts with EWI-2/EWI-2wint. Elution of immunoprecipitated proteins in the presence of reducing agents showed similar migration profiles (data not shown) indicating that the absence of co-immunoprecipitation of unpalmitoylated EWI-2/EWI-2wint with CD81 or CD9 was not due to the formation of covalent high molecular mass complexes. In addition, the absence of palmitoylation on EWI-2/EWI-2wint did not affect their co-localization at the plasma membrane with CD81, as controlled by confocal microscopy (Fig. 4E). Thus, we showed that palmitoylation of EWI-2/EWI-2wint is critical for the interaction with tetraspanins.
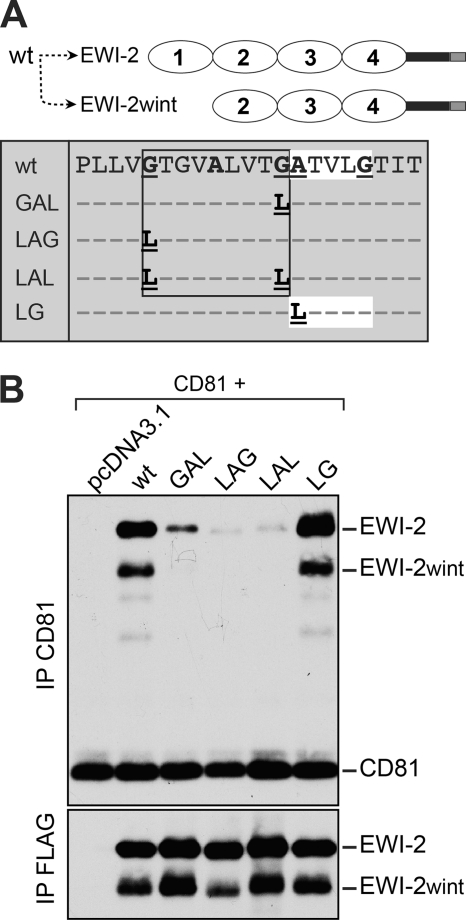
Next, we performed a more detailed analysis of the TM domain of EWI-2/EWI-2wint, in which we mapped a glycine-zipper motif (GXXXAXXXG, where X is any amino acid) (29) and a single (A/G)XXXG motif (Fig. 5A, box). Simple glycine-rich motifs have been shown to be involved in direct packing between glycine faces in TM helices (30). Glycine-zipper motifs ((G,A,S)XXXGXXX(G,S,T)) have been described as motifs important for oligomerization via TM domains, which can also be found in asymmetric packings (29). To test the importance of these motifs, we generated different proteins in which glycine-rich motifs were mutated by substitution of glycine/alanine for leucine residues (Fig. 5A, box). Loss of either one (LAG, GAL) or both (LAL) of the glycines in the glycine-zipper motif dramatically reduced the interaction with CD81 (Fig. 5B), without affecting palmitoylation (Fig. 4C, LAL), whereas mutation in the AXXXG motif (LG) did not have any effect (Fig. 5B). Combined together, these results demonstrate that the GXXXAXXXG motif in the TM domain of EWI-2/EWI-2wint plays a major role in hetero-oligomerization with CD81.
FIGURE 5.
A glycine-zipper motif in the TM domain of EWI-2/EWI-2wint is essential for the interaction with CD81. A, schematic representation of the different EWI-2 mutants. Expression of EWI-2 (wt) in CHO cells leads to the production of EWI-2 and EWI-2wint. Residues in the glycine-zipper or glycine-rich motif of EWI-2 were replaced by leucines. FLAG-tagged proteins were co-expressed with CD81 in CHO cells, biotinylated, lysed in PBS/Brij97/EDTA, and immunoprecipitated with 5A6 or M2. Proteins were resolved by SDS-PAGE under nonreducing conditions and detected using HRP-streptavidin.
Mutations in EWI-2/EWI-2wint That Cause Defective Interaction with CD81 Also Affect the Inhibitory Effect of EWI-2wint on HCV Infection
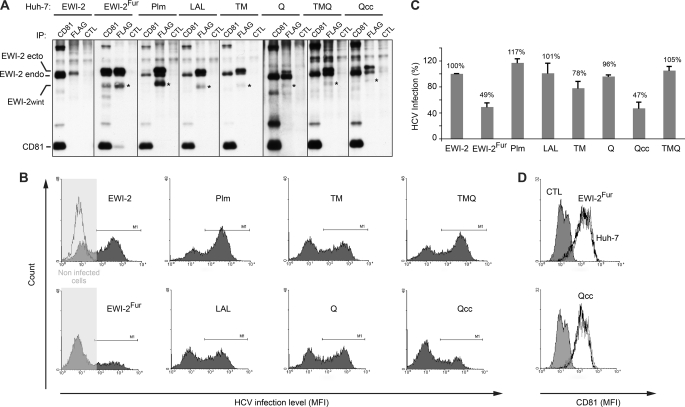
Because we found that the ectopic expression of EWI-2wint in Huh-7 target cells inhibits HCV infection (16), we then sought to determine whether mutations in EWI-2/EWI-2wint that abolish interaction with CD81 also affect the inhibitory effect of EWI-2wint on HCV infection. We generated Huh-7 cellular clones expressing either the palmitoylation mutant (Plm) or the double mutant of the glycine-zipper (LAL) or chimeric EWI-2 proteins (TM, Q, TMQ, and Qcc). Due to technical difficulties, we could not generate clones expressing the ΔIg4 protein. It should be noted that EWI-2 is endogenously expressed in Huh-7 cells (Fig. 6A, EWI-2 endo) but its cleavage naturally does not occur in Huh-7 cells preventing EWI-2wint production, as previously described (16). Mutated and chimeric EWI-2 proteins were therefore modified to express an additional arginine directly downstream of the RGR cleavage site generating a RGRR furin cleavage consensus site, as previously described (Fig. 1 and Ref. 16). We also generated cellular clones expressing unmodified full-length EWI-2 protein (Fig. 6A, EWI-2), which does not have any effect on HCV infection (16). Cellular clones expressing wild type EWI-2 with the furin site (producing EWI-2wint) were generated as positive controls of inhibition of HCV infection (Fig. 6A, EWI-2Fur). Several clones expressing every EWI-2 protein were generated. Protein expression in each clone was controlled by flow cytometry (data not shown) and immunoprecipitation of cell biotinylated lysate (Fig. 6A). A representative clone of every EWI-2 protein is presented in Fig. 6. Although at slightly different levels, each of the ectopic EWI-2 proteins was correctly expressed in selected clones, as controlled by direct immunoprecipitation with the anti-FLAG mAb. As shown by co-immunoprecipitation with CD81, only EWI-2, EWI-2Fur, and Qcc related proteins interacted with CD81 (Fig. 6A), as described above in CHO cells (Figs. 4 and 5). Cellular clones were next infected with HCVcc, which represent the most relevant tool to study the HCV life cycle. HCV infection levels were evaluated by flow cytometry using a mAb directed against the HCV non-structural protein 5 (NS5). A representative histogram of every clone is shown in Fig. 6B. Histograms are characterized by two peaks, one corresponding to the non-infected cells (left) and the other corresponding to the infected cells (right). In Fig. 6C, results are presented as relative percentages to full-length EWI-2 expressing cells and reported as the mean ± S.D. of three independent experiments. As expected, the proportion of infected cells was reduced in EWI-2wint producing cells (Fig. 6, B and C, EWI-2Fur) when compared with full-length EWI-2 expressing cells (Fig. 6, B and C, EWI-2). Interestingly, the proportion of infected cells was similarly reduced in cells expressing the Qcc protein, which still interacts with CD81. In contrast, infection levels of cells expressing ectopic EWI-2 proteins that no longer interact with CD81 (Plm, LAL, TM, Q, TMQ) were not significantly affected (Fig. 6, B and C). It should be noted that inhibition of HCV infection in cells expressing either EWI-2Fur or Qcc proteins was not due to a reduction of the CD81 expression level, as controlled by anti-CD81 flow cytometry (Fig. 6D). In addition, the absence of inhibition of HCV infection in cells expressing TM, Q, and TMQ chimeras was not due to the low expression level of EWI-2wint in these cells because cells expressing the Qcc chimera expressed equal amounts of EWI-2wint (Fig. 6A, asterisk).
FIGURE 6.
Mutations in EWI-2/EWI-2wint that abolish interaction with CD81 also affect the inhibitory effect of EWI-2wint on HCV infection. A, Huh-7 cell clones expressing the different FLAG-tagged mutants of EWI-2/EWI-2wint were biotinylated, lysed in PBS/Brij97/EDTA, immunoprecipitated, and detected using HRP-streptavidin. CD81 and FLAG IP were performed with 5A6 and M2 mAbs, respectively. CTL IP were performed with irrelevant immunoglobulins. Endogenous (EWI-2 endo) and ectopic (EWI-2 ecto) EWI-2 proteins are indicated. Asterisks show bands corresponding to EWI-2wint protein. B and C, HCVcc infection levels in Huh-7 cells expressing EWI-2/EWI-2wint proteins. Infected cells were stained using anti-NS5 mAb (2F6/G11) and secondary antibodies were conjugated with PE. Noninfected cells were used as negative controls. A representative histogram of every clone is shown in B. Histograms are characterized by two peaks, one corresponding to the noninfected cells (left) and the other corresponding to the infected cells (right). In C, results are presented as relative percentages to full-length EWI-2 expressing cells and reported as the mean ± S.D. of three independent experiments. D, CD81 expression on cells expressing EWI-2Fur or Qcc proteins. Cells were stained using 5A6 and secondary antibodies conjugated with PE. CTL corresponds to cells that were stained only with secondary antibodies. CD81 expression on cells expressing EWI-2Fur or Qcc EWI-2/EWI-2wint proteins (black line) was compared with that of parental Huh-7 cells (gray line).
Altogether, our results indicate that mutations in EWI-2/EWI-2wint that abolish their interaction with CD81 also affect the inhibitory effect of EWI-2wint on HCV infection. Therefore, EWI-2wint needs to physically interact with CD81 to exert its inhibitory effect on the HCV life cycle.
Interaction with EWI-2 and EWI-2wint Depends on CD81 TM Domains 3 and 4 and the Extracellular Domains
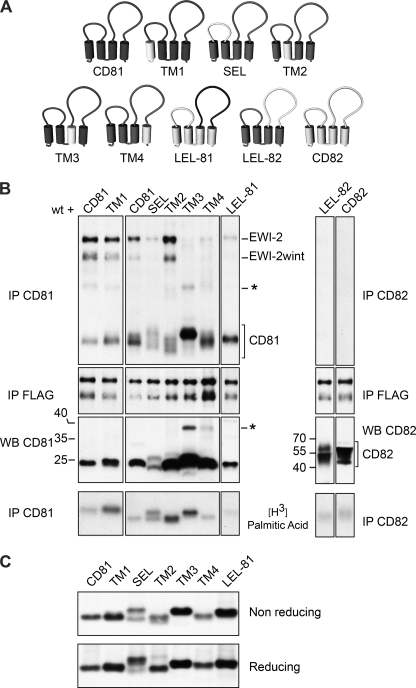
After showing that palmitoylation of CD81 is not essential for interaction with its partner, we wanted to find the essential components of CD81 for this interaction. CD81, like other tetraspanins, is composed of a small (SEL) and a large (LEL) extracellular loop, four TM domains (TM1–4), and three cytosolic domains (Fig. 7A). To identify domains involved in EWI-2/EWI-2wint interaction, we constructed a set of chimeras in which we successively exchanged domains of CD81 with corresponding domains of CD82 (Fig. 7A), another tetraspanin that does not form strong enough complexes with EWI-2 to withstand our lysis conditions (Fig. 7B, IP CD82). As shown in Fig. 7B, all chimeras were correctly expressed as seen by immunoprecipitation of cell surface-biotinylated lysates (IP CD81) and immunoblotting (WB CD81/CD82), and they were all palmitoylated ([3H]palmitic acid). In our experiments, CD82 and LEL-82 chimeras were only very weakly labeled after surface biotinylation, but expression of these proteins was clearly visible after immunoblotting (Fig. 7B, WB CD82). EWI-2 and EWI-2wint were equally expressed in all samples as seen by direct co-immunoprecipitation (Fig. 7B, IP FLAG). Co-immunoprecipiation experiments (Fig. 7B, IP CD81) showed that replacement of the first two TM domains (TM1 and TM2) of CD81 did not affect the interaction with EWI-2/EWI-2wint, whereas exchange of the SEL led to a decreased interaction. However, this chimera showed a diffuse migration pattern with two bands under nonreducing conditions (Fig. 7B, SEL) and only one band under reducing conditions (Fig. 7C, SEL). This was likely due to an altered conformation of CD81 because it has been shown that CD81 SEL helps stabilize the LEL active conformation (31) and might therefore influence the interaction of CD81 with its partners. Substitution of TM3 or TM4 with corresponding domains of CD82 dramatically reduced the interaction of CD81 with its partners (Fig. 7B). It should be noted that the TM3 chimera and to a lesser extent, the TM4 chimera showed an additional band in anti-CD81 immunoprecipitations, which was also detected in CD81 immunoblotting (Fig. 7B, asterisk). This indicates that these proteins have an increased tendency to oligomerize under our experimental conditions, suggesting a potential role for TM3 and TM4 domains in the oligomerization of CD82. Replacement of the LEL of CD81 by that of CD82 also abolished the interaction with EWI-2/EWI-2wint (Fig. 7, LEL-82). In contrast, replacement of the LEL of CD82 by that of CD81 led to some co-precipitation of EWI-2/EWI-2wint (Fig. 7, LEL-81), as compared with CD82. In conclusion, our results show the importance of the extracellular, TM3 and TM4 domains of CD81 for its interaction with EWI-2/EWI-2wint proteins.
FIGURE 7.
Motifs in CD81 important for its interaction with EWI-2/EWI-2wint. A, schematic representation of the different CD81/CD82 chimeras. B, EWI-2/EWI-2wint (wt) and chimeric CD81 proteins were co-expressed in CHO cells, surface biotinylated, labeled with [3H]palmitic acid, or directly lysed in PBS/Brij97/EDTA. IP were performed with 5A6, TS82b, or M2 antibodies. Proteins were detected using streptavidin-HRP, autoradiography, or immunoblotting. Oligomers of CD81 are indicated by an asterisk. C, chimeric CD81 proteins were expressed in CHO cells, biotinylated, lysed in PBS/Brij97/EDTA, and precipitated with 5A6. Proteins were resolved by SDS-PAGE under reducing (bottom) or nonreducing (top) conditions and detected using streptavidin-HRP.
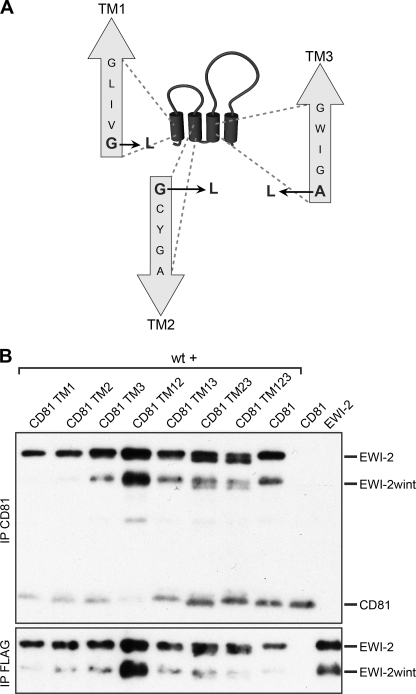
We identified three glycine-rich motifs in TM1, TM2, and TM3 domains of CD81 (Fig. 8A). Because we found that a glycine-zipper motif in the TM domain of EWI-2/EWI-2wint is essential for the interaction with CD81, we next assessed the role of glycine-rich motifs in TM domains of CD81 in the interaction with EWI-2/EWI-2wint. For this purpose, we generated different CD81 proteins in which glycine-rich motifs were mutated by substitution of glycine/alanine for leucine residues (Fig. 8A). Glycine motifs were mutated separately (CD81 TM1, TM2, and TM3) or in combination with another motif (CD81 TM12, TM13, TM23). We also generated a CD81 protein in which all three glycine-rich motifs were mutated (CD81 TM123). As shown in Fig. 8B, the mutation of glycine-rich motifs did not lead to any drastic effects on the interaction with EWI-2/EWI-2wint, indicating that these motifs in TM domains of CD81 do not play a major role in the interaction with EWI-2/EWI-2wint. It should be noted that mutation of polar residues (NEE) in TM domains of CD81 also did not affect its interaction with EWI-2/EWI-2wint (data not shown).
FIGURE 8.
Mutation of glycine-rich motifs in transmembrane domains of CD81. A, schematic representation of mutations of glycine-rich motifs in TM1, TM2, and TM3 of CD81. The first amino acid (G/A), in each motif was replaced by a leucine residue (L). B, mutated CD81 proteins were co-expressed with wild-type EWI-2/EWI-2wint proteins (wt), biotinylated, lysed in PBS/Brij97/EDTA, and immunoprecipitated with 5A6 or M2 antibodies. Proteins were separated by SDS-PAGE under nonreducing conditions and detected using HRP-streptavidin.
Extracellular, TM3 and TM4 Domains in CD81 Are Necessary for CD81 Functionality in HCV Entry
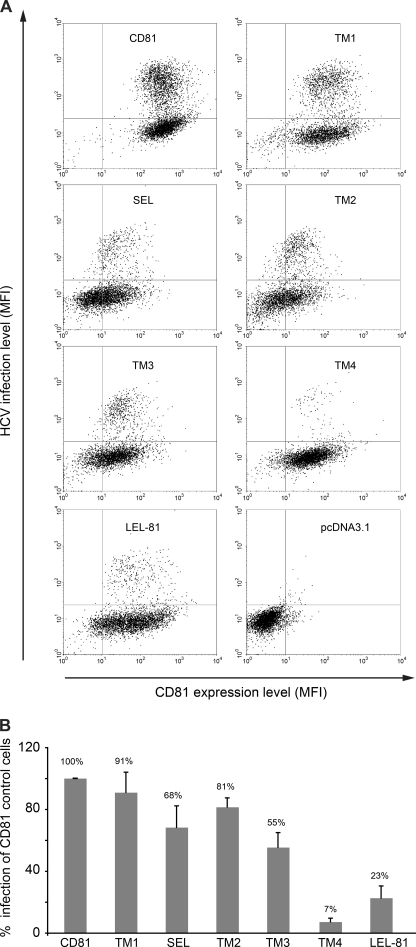
The HCV entry stage is a multistep process involving several cellular factors including CD81, which plays a major role during a post-attachment step (reviewed in Ref. 10). Recently, we described a Huh-7 cell line (Huh-7w7) that has lost CD81 expression and is therefore resistant to HCV infection (24). Huh-7w7 cells can be infected when CD81 is ectopically expressed. We therefore took advantage of these cells to analyze the capacity of the different CD81 chimeras to restore permissivity of Huh-7w7 cells to HCVcc. Because CD81 LEL is the critical region for interaction with the HCV E2 envelope protein and virus entry, we generated Huh-7w7 cell lines stably expressing only chimeras containing CD81 LEL (Fig. 9). Protein expression in each cell line was controlled by flow cytometry (Fig. 9A) and immunoblotting (data not shown). Cell lines were next infected with HCVcc particles and infection levels were evaluated by flow cytometry. Because CD81 is essential for HCV infection, infection levels were evaluated only for CD81 positive cells. A representative double staining dot plot of each cell line is shown in Fig. 9A. Huh-7w7 cells transfected with the pcDNA3.1 empty vector were used as a negative control. In Fig. 9B, results are presented as relative percentages to full-length CD81 expressing cells and reported as the mean ± S.D. of three independent experiments. Huh-7w7 cells expressing TM1 or TM2 chimeras were infected at the same level as control cells. In contrast, cells expressing SEL or TM3 chimeras showed reduced infection levels. To a higher extent, cells engineered to express TM4 or LEL-81 chimeras were almost resistant to HCVcc infection (Fig. 9). It should be noted that the TM4 and LEL-81 chimeras were more cell surface expressed than the TM2 chimera that was functional in HCV entry. This indicates that differences in infectivity were not due to the expression levels of chimeras. As an additional control, we generated Huh-7w7 cells expressing LEL-82, which as expected was completely resistant to HCV infection despite good expression of the chimeric protein (data not shown). Together, these results indicate that the CD81 sequence of the extracellular loops, TM3 and TM4, are necessary for CD81 function in HCV entry.
FIGURE 9.
Functionality of CD81/CD82 chimeras in HCV infection. A, Huh-7w7 cells stably expressing CD81/CD82 chimeras were infected with HCVcc and infection levels were evaluated by flow cytometry. Cells were double stained by using anti-CD81 and anti-NS5 mAbs, as described under “Experimental Procedures.” A representative double staining dot plot of each cell line is shown in A. Huh-7w7 cells transfected with the pcDNA3.1 empty vector were used as negative controls. In B results are presented as relative percentages to full-length CD81 expressing cells and reported as the mean ± S.D. of three independent experiments.
DISCUSSION
Here, for the first time, we characterized the essential requirements for the functionality and the interaction of CD81 with two of its major partners: EWI-2 and EWI-2wint.
In our study, we showed that the membrane-proximal Ig4 and TM domains of EWI-2/EWI-2wint are essential for the interaction with CD81. More precisely, a glycine-zipper motif in the TM domain, and palmitoylation on two juxtamembranous cysteines are essential for this process. These results concur with recent findings, which demonstrate the necessity of TM regions for the interaction of EWI-F, another protein of the EWI family, which also interacts with CD81 (20, 32). Interestingly, EWI-F, like EWI-2/EWI-2wint, displays a glycine-zipper motif in its TM domain that may potentially mediate the CD81/EWI-F interaction. Our results stand in contrast to a previous report that showed mouse EWI-2 lacking membrane distal Ig1-Ig2 domains were no longer able to co-precipitate CD81 (13), which might be due to differences in co-precipitation approaches. The finding that palmitoylation of EWI-2/EWI-2wint is important for the interaction shed light on the loss of interaction between CD81 and EWI-2 containing the cytoplasmic tail of CD2 (33), as this sequence cannot be palmitoylated.
Analysis of requirements for the interaction also showed that there is an interaction between CD81 TM3, TM4, and EWI-2/EWI-2wint TM domains, which might be strengthened by the additional extracellular interactions. The structure of the LEL of CD81 is known through x-ray defraction (34) and the complete molecule has been modeled (35). Based on this model, TM3 and TM4 domains have a common face and the TM domain of EWI-2 might interact on this side of the molecule bringing the Ig4 extracellular domain in close contact with the LEL of CD81, both domain exchanges decreased the CD81/EWI-2/EWI-2wint interaction. In the context of the development of new antiviral molecules and therapeutic strategies against HCV, the interacting extracellular domains constitute a good basis for the design of peptides or miniproteins interacting with CD81 and leading to an inhibition of HCV infection.
We showed that interaction with the Ig4 domain is specific for CD81. Our results, taken together with a published report on the interaction of EWI-2 with CD9 (11), indicate that the interaction between EWI-2 and its direct tetraspanin partners might be similar. Both tetraspanins require TM3 and LEL regions to fully interact with EWI-2. In contrast, only the TM4 of CD81 is required for the CD81-EWI-F interaction (20) and it has been shown that deletion of the most membrane proximal Ig domain of EWI-F does not affect interaction with CD81 and CD9 (32). Until now, there are no clear data regarding the role that palmitoylation might play in this interaction, as the sequence used to replace the EWI-F cytosolic tail in this report (32) still contains a cysteine that could be palmitoylated (based on CSS-Palm 2.0, a palmitoylation site prediction software (36)).
EWI family members have a propensity for oligomerization. EWI-101/CD101 has been described as a disulfide-bonded homodimeric polypeptide (37) and it has been recently demonstrated that EWI-F forms oligomers at the cell surface that are directly associated with tetraspanins CD9 and CD81 (32). Interestingly, EWI-2 has also been observed to form protein complexes containing at least two EWI-2 molecules (32). EWI-2wint oligomerizes with EWI-2 as well. It should be noted that all domain exchanges and mutations in the EWI-2 protein described in this study did not affect oligomerization of EWI-2wint with EWI-2. We therefore cannot exclude the possibility that EWI-2wint needs EWI-2 to interact with CD81.
Most, if not all, tetraspanins are palmitoylated and the palmitoylation plays a role in heterologous tetraspanin-tetraspanin interaction and therefore TEM assembly. In accordance with previous studies (11, 39), we found that palmitoylation of CD81 and CD9 is not involved in their interaction with EWI-2/EWI-2wint. The absence of palmitoylation might even favor interactions with their partners, an observation made with an unpalmitoylated CD9 mutant for which an enhanced EWI-2 association has been observed (39) and is also visible in our experiments (Fig. 4). In contrast, we showed that the interaction between a tetraspanin and a partner molecule can be dependent on the palmitoylation of the partner protein. Indeed, palmitoylation of EWI-2/EWI-2wint is important for interaction with CD81, as well as CD9, showing that the effect of this post-translational modification is a more general feature. Palmitoylation increases the hydrophobicity of proteins and contributes to their membrane anchoring. Palmitoylation also influences the trafficking of some transmembrane proteins; it affects aggregation, endocytosis, recycling, protein stability, transport from the ER to the plasma membrane, and association with lipid rafts (40, 41). Through its reversibility, palmitoylation also provides mechanisms for regulation of functional activities of transmembrane proteins. Thus, palmitoylation of EWI-2/EWI-2wint may regulate interaction with CD81 and CD9 tetraspanins for a number of possible reasons. Palmitoylation may control homo- and hetero-oligomerization of EWI-2 and EWI-2wint, which might be required for their interaction with CD81. Alternatively, palmitoylation may contribute to the enrichment/stabilization of EWI-2/EWI-2wint in lipid microdomains favorable to the interaction with tetraspanins. Although EWI-2Plm is recognized by 8A12, a conformation-sensitive anti-EWI-2 monoclonal antibody (11), we cannot exclude the possibility that the absence of palmitoylation alters the conformation of Ig domains of EWI-2/EWI-2wint, as it has been described for MHC class I molecules (42).
So far, there are only two reports regarding palmitoylation of the tetraspanin partner protein (43, 44). In the case of tetraspanin CD151 and integrin α6β4, it has been shown that palmitoylation occurs early in biosynthesis and undergoes little subsequent turnover (44). Depalmitoylation of both CD151 and its interactor α6β4 has no effect on the direct association of those two molecules, but weakens the interaction with other tetraspanins and the TEMs in total. In the case of synaptotagmin VII (Syt VII), a Ca2+ sensor that regulates lysosome exocytosis and plasma membrane repair, mutation of palmitoylation sites disrupts Syt VII targeting to lysosomes. Flannery et al. (43) demonstrated that palmitoylation-dependent association with the lysosomal tetraspanin CD63 is a requirement for the intracellular trafficking of Syt VII from the Golgi complex to lysosomes. Generally, palmitoylation of tetraspanins is thought to stabilize the tetraspanin web and to be relatively stable; nevertheless, it has been shown that palmitoylation of CD81 can be modulated by oxidative stress and that non-palmitoylated CD81 could then interact with intracellular signaling protein 14-3-3 (45). The fact that the interaction of EWI-2/EWI-2wint with CD81 and CD9 tetraspanins depends on the palmitoylation of EWI-2/EWI-2wint suggests that these interactions can be regulated by depalmitoylation, thus allowing the cell to regulate functions of the tetraspanins as well as signaling through dissociation of tetraspanin partner complexes. Together these results suggest that regulation of protein-protein interactions through palmitoylation is a general principle used in TEMs.
Although its exact role is still unclear, CD81 is essential for the HCV life cycle. HCV is a hepatotropic pathogen of significant importance to public health. It has a very narrow host range due to its specificity for the human homologues of some of its receptors (46). It also has a very narrow cell tropism in humans, infecting almost exclusively liver cells. This cell tropism might be partly due to the fact that most cells express EWI-2, which is subsequently cleaved to produce EWI-2wint, which in turn inhibits the interaction of CD81 with the viral E2 glycoprotein. In hepatocytes, this cleavage is notably absent, enabling the binding of E2 to CD81 and infection. We found that cleavage of EWI-2 is dependent on two basic residues (RXR, where X is any amino acid) and occurs after N-glycan maturation, suggesting that it takes place in a post-ER compartment. These results might now help us to identify the protease responsible for the cleavage, thus leading to a better understanding of the cellular tropism of HCV. Such a protease might either be lacking in liver cells or accessibility of EWI-2 to protease(s) may be modulated by other components of the TEMs that vary in different cell types (1).
EWI-2 has been shown to be involved in cellular processes such as cellular migration. It has been found that its expression reduces migration on fibronectin and laminin (18, 19), whereas its silencing increases migration toward the chemokine SDF-1α (17). As already described, EWI-2wint has a functional effect on HCV infection through inhibition of the interaction between CD81 and HCV E2 (16). Interestingly, we found in this study that mutations in EWI-2/EWI-2wint that abolish their interaction with CD81 also affect the inhibitory effect of EWI-2wint on HCV infection. Therefore, the inhibitory effect of EWI-2wint on HCV infection depends on the interaction of EWI-2/EWI-2wint with CD81.
The LEL of CD81 is the major HCV E2 glycoprotein binding site on CD81 (6, 47) and characterization of chimeric proteins between CD81 and CD9, its closest counterpart, confirmed that the LEL sequence is the region of CD81 defining HCV entry (48). In our study, we generated chimeras between CD81 and CD82, a tetraspanin that interacts with different proteins than CD81 or CD9. Interestingly, we found that expression of CD81/CD82 chimeras, which interact less or no longer with EWI-2, only partially restored susceptibility to HCV infection. In particular, cells expressing TM3, TM4, and LEL-81 chimeras showed highly reduced infection levels. Although chimeras are recognized by 5A6, a conformation-sensitive anti-CD81 monoclonal antibody (4), and behave similarly (except for SEL) under reducing and nonreducing conditions, we cannot exclude that domain exchanges lead to conformational changes in LEL structure incompatible with the viral entry. Further experiments with a soluble form of E2 will be necessary to determine whether domain exchanges lead to a direct disruption of CD81-E2 binding. However, our results indicate that HCV infection levels might correlate with interaction levels of CD81 with its partners. EWI-2 is likely not directly involved in HCV life cycle because its silencing has no effect on infection (16). In contrast, the tight junction protein Claudin-1 forms functional complexes with CD81, which play a major role in the HCV entry process (38, 49–52). Domain exchanges in CD81/CD82 chimeras might affect CD81·Claudin-1 complex formation. It has been shown that homoclustered CD9 shifts toward heteroclusters upon EWI-2 and EWI-F expression (39), indicating that partner proteins modulate tetraspanin oligomerization. We therefore propose that TM3, TM4, and LEL-81 chimeras are not functional in HCV entry because in the absence of interaction with their partners, these proteins may have a higher tendency to homo-oligomerize. In accordance with this, we showed that TM3 and TM4 chimeras have an increased tendency to oligomerize under our experimental conditions. As already suggested (24), homoclustered CD81 might be less competent for HCV entry.
Acknowledgments
We thank Sophana Ung, André Pillez, Clare Whitehead, Alison Liewen, and Johanna Jarmer for technical assistance. We are grateful to S. Levy and S. H. Leppla for providing us with reagents. We thank the Microscopy-Imaging-Cytometry Platform of the Lille Pasteur Campus for access to the instruments and Elisabeth Werkmeister for help with image acquisition.
This work was supported in part by the “Agence Nationale de Recherches sur le Sida et les hépatites virales” (ANRS) and the “Fondation pour la Recherche Médicale.”
- TEMs
- tetraspanin-enriched microdomains
- HCV
- hepatitis C virus
- E2
- HCV envelope glycoprotein 2
- IP
- immunoprecipitation
- TM
- transmembrane
- Q
- cytosolic tail
- Endo H
- endo-β-N-acetylglucosaminidase H
- O-glycanase
- glycopeptide-N-acetylgalactosaminidase
- Syn VII
- synaptotagmin VII
- PNGase F
- peptide N-glycosidase F
- LEL
- large extracellular loop
- SEL
- small extracellular loop
- HCVcc
- HCV grown in cell culture
- PE
- phosphatidylethanolamine.
REFERENCES
- 1. Charrin S., le Naour F., Silvie O., Milhiet P. E., Boucheix C., Rubinstein E. (2009) Biochem. J. 420, 133–154 [DOI] [PubMed] [Google Scholar]
- 2. Berditchevski F., Odintsova E., Sawada S., Gilbert E. (2002) J. Biol. Chem. 277, 36991–37000 [DOI] [PubMed] [Google Scholar]
- 3. Rubinstein E., Le Naour F., Lagaudrière-Gesbert C., Billard M., Conjeaud H., Boucheix C. (1996) Eur. J. Immunol. 26, 2657–2665 [DOI] [PubMed] [Google Scholar]
- 4. Oren R., Takahashi S., Doss C., Levy R., Levy S. (1990) Mol. Cell. Biol. 10, 4007–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Levy S., Shoham T. (2005) Nat. Rev. Immunol. 5, 136–148 [DOI] [PubMed] [Google Scholar]
- 6. Pileri P., Uematsu Y., Campagnoli S., Galli G., Falugi F., Petracca R., Weiner A. J., Houghton M., Rosa D., Grandi G., Abrignani S. (1998) Science 282, 938–941 [DOI] [PubMed] [Google Scholar]
- 7. Silvie O., Rubinstein E., Franetich J. F., Prenant M., Belnoue E., Rénia L., Hannoun L., Eling W., Levy S., Boucheix C., Mazier D. (2003) Nat. Med. 9, 93–96 [DOI] [PubMed] [Google Scholar]
- 8. Tham T. N., Gouin E., Rubinstein E., Boucheix C., Cossart P., Pizarro-Cerda J. (2010) Infect. Immun. 78, 204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lemon S. M., Walker C., Alter M. J., Yi M. (2007) in Fields Virology (Knipe D. M., Howley P. M. eds) Fifth Ed., pp. 1253–1304, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 10. Dubuisson J., Helle F., Cocquerel L. (2008) Cell Microbiol. 10, 821–827 [DOI] [PubMed] [Google Scholar]
- 11. Charrin S., Le Naour F., Labas V., Billard M., Le Caer J. P., Emile J. F., Petit M. A., Boucheix C., Rubinstein E. (2003) Biochem. J. 373, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Charrin S., Le Naour F., Oualid M., Billard M., Faure G., Hanash S. M., Boucheix C., Rubinstein E. (2001) J. Biol. Chem. 276, 14329–14337 [DOI] [PubMed] [Google Scholar]
- 13. Clark K. L., Zeng Z., Langford A. L., Bowen S. M., Todd S. C. (2001) J. Immunol. 167, 5115–5121 [DOI] [PubMed] [Google Scholar]
- 14. Stipp C. S., Kolesnikova T. V., Hemler M. E. (2001) J. Biol. Chem. 276, 40545–40554 [DOI] [PubMed] [Google Scholar]
- 15. Stipp C. S., Orlicky D., Hemler M. E. (2001) J. Biol. Chem. 276, 4853–4862 [DOI] [PubMed] [Google Scholar]
- 16. Rocha-Perugini V., Montpellier C., Delgrange D., Wychowski C., Helle F., Pillez A., Drobecq H., Le Naour F., Charrin S., Levy S., Rubinstein E., Dubuisson J., Cocquerel L. (2008) PLoS ONE 3, e1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sala-Valdés M., Ursa A., Charrin S., Rubinstein E., Hemler M. E., Sánchez-Madrid F., Yáñez-Mó M. (2006) J. Biol. Chem. 281, 19665–19675 [DOI] [PubMed] [Google Scholar]
- 18. Stipp C. S., Kolesnikova T. V., Hemler M. E. (2003) J. Cell Biol. 163, 1167–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang X. A., Lane W. S., Charrin S., Rubinstein E., Liu L. (2003) Cancer Res. 63, 2665–2674 [PubMed] [Google Scholar]
- 20. Charrin S., Yalaoui S., Bartosch B., Cocquerel L., Franetich J. F., Boucheix C., Mazier D., Rubinstein E., Silvie O. (2009) J. Biol. Chem. 284, 31572–31578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kolesnikova T. V., Kazarov A. R., Lemieux M. E., Lafleur M. A., Kesari S., Kung A. L., Hemler M. E. (2009) Neoplasia 11, 77–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Silvie O., Charrin S., Billard M., Franetich J. F., Clark K. L., van Gemert G. J., Sauerwein R. W., Dautry F., Boucheix C., Mazier D., Rubinstein E. (2006) J. Cell Sci. 119, 1992–2002 [DOI] [PubMed] [Google Scholar]
- 23. Gordon V. M., Klimpel K. R., Arora N., Henderson M. A., Leppla S. H. (1995) Infect. Immun. 63, 82–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rocha-Perugini V., Lavie M., Delgrange D., Canton J., Pillez A., Potel J., Lecoeur C., Rubinstein E., Dubuisson J., Wychowski C., Cocquerel L. (2009) BMC Microbiol. 9, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cocquerel L., Meunier J. C., Pillez A., Wychowski C., Dubuisson J. (1998) J. Virol. 72, 2183–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Kräusslich H. G., Mizokami M., Bartenschlager R., Liang T. J. (2005) Nat. Med. 11, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delgrange D., Pillez A., Castelain S., Cocquerel L., Rouillé Y., Dubuisson J., Wakita T., Duverlie G., Wychowski C. (2007) J. Gen. Virol. 88, 2495–2503 [DOI] [PubMed] [Google Scholar]
- 28. Robbins P. W., Trimble R. B., Wirth D. F., Hering C., Maley F., Maley G. F., Das R., Gibson B. W., Royal N., Biemann K. (1984) J. Biol. Chem. 259, 7577–7583 [PubMed] [Google Scholar]
- 29. Kim S., Jeon T. J., Oberai A., Yang D., Schmidt J. J., Bowie J. U. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14278–14283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Russ W. P., Engelman D. M. (2000) J. Mol. Biol. 296, 911–919 [DOI] [PubMed] [Google Scholar]
- 31. Yalaoui S., Zougbédé S., Charrin S., Silvie O., Arduise C., Farhati K., Boucheix C., Mazier D., Rubinstein E., Froissard P. (2008) PLoS Pathog. 4, e1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. André M., Chambrion C., Charrin S., Soave S., Chaker J., Boucheix C., Rubinstein E., Le Naour F. (2009) J. Proteomics 73, 93–102 [DOI] [PubMed] [Google Scholar]
- 33. Kolesnikova T. V., Stipp C. S., Rao R. M., Lane W. S., Luscinskas F. W., Hemler M. E. (2004) Blood 103, 3013–3019 [DOI] [PubMed] [Google Scholar]
- 34. Kitadokoro K., Bordo D., Galli G., Petracca R., Falugi F., Abrignani S., Grandi G., Bolognesi M. (2001) EMBO J. 20, 12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seigneuret M. (2006) Biophys. J. 90, 212–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2008) Protein Eng. Des. Sel. 21, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bagot M., Martinel I., Charue D., Weill F., Boulland M. L., Wechsler J., Freeman G. J., Bensussan A., Boumsell L. (1997) Tissue Antigens 50, 439–448 [DOI] [PubMed] [Google Scholar]
- 38. Mee C. J., Harris H. J., Farquhar M. J., Wilson G., Reynolds G., Davis C., van Ijzendoorn S. C., Balfe P., McKeating J. A. (2009) J. Virol. 83, 6211–6221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang X. H., Kovalenko O. V., Kolesnikova T. V., Andzelm M. M., Rubinstein E., Strominger J. L., Hemler M. E. (2006) J. Biol. Chem. 281, 12976–12985 [DOI] [PubMed] [Google Scholar]
- 40. Bijlmakers M. J., Marsh M. (2003) Trends Cell Biol. 13, 32–42 [DOI] [PubMed] [Google Scholar]
- 41. Linder M. E., Deschenes R. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 42. Gruda R., Achdout H., Stern-Ginossar N., Gazit R., Betser-Cohen G., Manaster I., Katz G., Gonen-Gross T., Tirosh B., Mandelboim O. (2007) J. Immunol. 179, 3655–3661 [DOI] [PubMed] [Google Scholar]
- 43. Flannery A. R., Czibener C., Andrews N. W. (2010) J. Cell Biol. 191, 599–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang X., Kovalenko O. V., Tang W., Claas C., Stipp C. S., Hemler M. E. (2004) J. Cell Biol. 167, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clark K. L., Oelke A., Johnson M. E., Eilert K. D., Simpson P. C., Todd S. C. (2004) J. Biol. Chem. 279, 19401–19406 [DOI] [PubMed] [Google Scholar]
- 46. Ploss A., Rice C. M. (2009) EMBO Rep. 10, 1220–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Flint M., Thomas J. M., Maidens C. M., Shotton C., Levy S., Barclay W. S., McKeating J. A. (1999) J. Virol. 73, 6782–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang J., Randall G., Higginbottom A., Monk P., Rice C. M., McKeating J. A. (2004) J. Virol. 78, 1448–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Coller K. E., Berger K. L., Heaton N. S., Cooper J. D., Yoon R., Randall G. (2009) PLoS Pathog. 5, e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harris H. J., Davis C., Mullins J. G., Hu K., Goodall M., Farquhar M. J., Mee C. J., McCaffrey K., Young S., Drummer H., Balfe P., McKeating J. A. (2010) J. Biol. Chem. 285, 21092–21102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harris H. J., Farquhar M. J., Mee C. J., Davis C., Reynolds G. M., Jennings A., Hu K., Yuan F., Deng H., Hubscher S. G., Han J. H., Balfe P., McKeating J. A. (2008) J. Virol. 82, 5007–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Krieger S. E., Zeisel M. B., Davis C., Thumann C., Harris H. J., Schnober E. K., Mee C., Soulier E., Royer C., Lambotin M., Grunert F., Dao Thi V. L., Dreux M., Cosset F. L., McKeating J. A., Schuster C., Baumert T. F. (2010) Hepatology 51, 1144–1157 [DOI] [PubMed] [Google Scholar]