Abstract
Synthetic peptide immunogens that mimic the conformation of a target epitope of pathological relevance offer the possibility to precisely control the immune response specificity. Here, we performed conformational analyses using a panel of peptides in order to investigate the key parameters controlling their conformation upon integration into liposomal bilayers. These revealed that the peptide lipidation pattern, the lipid anchor chain length, and the liposome surface charge all significantly alter peptide conformation. Peptide aggregation could also be modulated post-liposome assembly by the addition of distinct small molecule β-sheet breakers. Immunization of both mice and monkeys with a model liposomal vaccine containing β-sheet aggregated lipopeptide (Palm1–15) induced polyclonal IgG antibodies that specifically recognized β-sheet multimers over monomer or non-pathological native protein. The rational design of liposome-bound peptide immunogens with defined conformation opens up the possibility to generate vaccines against a range of protein misfolding diseases, such as Alzheimer disease.
Keywords: Amyloid, Liposomes, Neurodegeneration, Peptide Conformation, Protein Folding, Protein Misfolding, Vaccines
Introduction
The role of immunogen conformation in the specificity and efficacy of the humoral immune response has been recognized in a number of recent therapeutic and prophylactic vaccines (1–4). Conformation-specific antibodies generated by immunization with conformationally restricted immunogens are also important tools for disease diagnosis as biochemical probes for establishing protein structure-function relationships (5, 6) and in affinity purification (7, 8). Development of such peptide immunogens, however, has thus far been hindered by the lack of general design strategies to control the conformation of a given peptide sequence. In addition, most naked peptides have low inherent immunogenicity. Although design of peptide templates for the generation of antibodies that selectively recognize protein epitopes with an α-helical conformation has been reported (2, 4, 9, 10), general strategies to design short peptide fragments as immunogens for targeting other protein conformations are lacking.
Protein aggregation resulting from aberrant folding is characterized by the formation of proteinaceous deposits called amyloid, which exhibit β-sheet structure (11). Misfolding is now associated with over 20 human diseases, including Creutzfeldt-Jakob disease, Alzheimer disease (AD),2 Huntington disease, and Type II diabetes mellitus (12). Vaccines that can elicit antibodies against only the pathological β-sheet multimers of these targets would thus be considered advantageous from an efficacy perspective but also particularly from a safety perspective, where the non-folded protein has physiological relevance. In AD, β-amyloid peptides (Aβ) Aβ(1–40) and Aβ(1–42) have been associated with disease progression over several decades of research, and the β-sheet aggregates of Aβ are one of the major pathological hallmarks of AD (13). Although the molecular pathology of AD has not yet been elucidated, a large variety of soluble and insoluble β-sheet Aβ(1–40/42) oligomers with different degrees of polymerization have been identified in vitro or in AD brain as synaptotoxic and likely causal agents of cognitive impairment (14–18).
The discovery that active vaccination of transgenic mice overexpressing human amyloid precursor protein (APP) with Aβ-derived peptides could be an effective means to modify disease progression has opened up new vistas in AD therapy (19–21). The importance of immunogen aggregation state upon immune response specificity has been demonstrated by the generation of antibodies with binding specificities for fibrillar (22, 23) as well as a range of soluble forms of Aβ(1–40/42) (24–26). Control of peptide aggregation state is, however, precarious due to the stability issues associated with oligomers held together by non-covalent bonds. As a result of this, peptide immunogens are frequently used as heterogeneous mixtures of different secondary and quaternary conformations. On the other hand, vaccines that make use of covalently stabilized peptide multimers have hitherto been designed using de novo strategies that are not readily adaptable to target different oligomeric species.
We have previously reported that the tetrapalmitoylated β-amyloid 1–15 peptide (Palm1–15) embedded into liposomes along with monophosphoryl lipid A (MPLA), to generate liposomal vaccine ACI-24, could elicit an immune response that restored the cognitive impairment of APP transgenic mice (27). This truncated Aβ peptide sequence was chosen because it retains the immunodominant B-cell epitope of Aβ(1–42) but lacks the T-cell epitope (27). This T-cell epitope was associated with meningoencephalitis in a phase IIa study by ELAN Pharmaceuticals (22, 28). Surprisingly, we found that liposomal Palm1–15 (ACI-24) adopted a predominant β-sheet conformation although the N-terminal region of Aβ(1–40/42) is thought to be unstructured in vitro (29). Moreover, introduction of a PEG spacer between peptide and lipid anchor removed the peptide from the liposome surface and gave rise to a liposomal peptide with a random coil conformation. When tested in APP transgenic mice, only ACI-24 restored the cognitive deficit, suggesting a link between peptide immunogen conformation and in vivo efficacy (27).
In this study, we describe experiments aimed at understanding better the β-sheet conformation and establishing a structure-conformation relationship for liposomal Palm1–15 (ACI-24). In addition, we investigated whether the formation of a defined conformation is dependent upon the amino acid sequence. To this end, various peptides were lipidated and integrated into liposomes. To evaluate the utility of such constructs to generate conformation-specific antibodies in vivo, mice and monkeys were immunized with ACI-24, and the resulting antibodies were analyzed for conformation-specific binding to distinct conformations of Aβ(1–42) peptide as well as native protein.
EXPERIMENTAL PROCEDURES
CD Spectroscopy
CD spectra were acquired on a Jasco-815 spectropolarimeter (Japan) with a 0.1 cm path length quartz cuvette at 23 °C. Measurements were made with a 1.0 nm bandwidth and 0.5 nm resolution. A scan speed of 50 nm/min was employed with response time of 1 s. Solvent control spectra (four scans) were averaged and subtracted from the average of eight scans of each sample spectra. All spectra ([θ]obs, degrees) were smoothed after being converted to mean residue molar ellipticity ([θ], degrees cm2 dmol−1) with the equation, [θ] = [θ]obs × (MRW/10lc), where MRW is the mean residue molecular weight (molecular weight/number of residues), l is the optical path length (cm), and c is the concentration (g/cm3). For analysis, liposomal suspensions were diluted 6-fold in PBS to give final peptide concentrations between 10 and 40 μm. Spectra of the corresponding empty liposomes lacking peptide were subtracted.
Thioflavin T (ThT) Fluorescence
Fluorescence emission spectra were measured at 25 °C on a Tecan M200 spectrofluorimeter with an excitation of 440 nm. Liposomes were diluted 8-fold in PBS to give a final peptide concentration of 15 μm, and then ThT was added at different concentrations. Data were fitted using non-linear regression analysis (SigmaPlot version 10.0) assuming single-site binding saturation in order to calculate apparent Kd and maximal binding Bmax. For disaggregation experiments, see the supplemental “Results.”
Magic Angle Spinning (MAS)-NMR Spectroscopy
Experiments were conducted using a 3.2-mm triple-resonance (1H, 13C, 15N) MAS probe (Bruker Biospin) at a static magnetic field of 16.4 T, corresponding to 700-MHz proton resonance frequency. For analysis, the amino acids Ala-2, Ser-8, and Gly-9 of Palm1–15 were uniformly labeled with 13C and 15N. This peptide Palm1–15(ASG) was then incorporated into liposomes composed of phospholipids (1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dimyristoyl-sn-glycero-3-(phosphor-rac-(1-glycerol)) (DMPG), or 1,2-dimyristoyl-3-trimethylammonium-propane (DMTAP)), cholesterol, and MPLA, giving a final peptide concentration of 121 μm. Spectra were acquired and processed as described under supplemental “Methods.”
Vaccine Preparation for Immunization
ACI-24 was prepared using the lipids DMPC, DMPG, cholesterol, and MPLA (all from Avanti Polar Lipids) at a molar ratio of 9:1:7:0.06 respectively. Palm1–15 was added to give a peptide/phospholipid ratio of 1:100 and at a final concentration of 121 μm (see supplemental “Methods” for details). Vaccine was characterized by cryo-EM and HPLC for peptide, MPLA, and lipid content (data not shown).
Animal Care and Treatment
All treatments were carried out in accordance to local regulations. C57BL/6 adult mice (10 females/group) received subcutaneous injections of ACI-24 on three occasions with a 2-week interval between each immunization (at days 0, 14, and 28). Blood samples were collected, and plasma prepared at day 35. Cynomolgus monkeys were immunized four times at 3-week intervals. 8.5 months after the last immunization, monkeys were reimmunized subcutaneously in the leg area with two doses of ACI-24 at days 2 and 24.
Size Exclusion Chromatography (SEC) Aβ(1–42) Monomer and Oligomer Preparations
Aβ(1–42) used for SEC-HPLC fractionation was synthesized and purified by Dr. James I. Elliott (Yale University (New Haven, CT). Aβ(1–42) powder (1 mg) was equilibrated to room temperature for 6 min and dissolved in 50 μl of DMSO, followed by 800 μl of doubly distilled H2O and 10 μl of 2 m Tris-HCl, pH 7.4, and mixed by vortex. The Aβ preparation was incubated for 3–4 min at room temperature and centrifuged for 10 min at 6,000 × g at 4 °C. After centrifugation, the supernatant was immediately injected onto the HPLC and run over two columns, TSK-GEL G4000PWXL (Tosoh Bioscience) and Superose 6 10/300 GL (GE Healthcare), with a flow rate of 0.3 ml/min. Thirty fractions of 1 ml were collected and stored at 4 °C until use immediately after the fractionation was completed. The concentrations of Aβ monomer- and oligomer-enriched SEC-HPLC fractions were measured by absorbance at 280 nm in a glass cuvette.
Conformational Specificity of IgG Antibodies
ELISA plates were coated for 2 h at 4 °C with SEC Monomer and Oligomer Aβ(1–42) preparations diluted in PBS to obtain 10 μg/ml concentrations. After coating, washing with PBS, 0.05% Tween 20 was performed, followed by blocking with 1% BSA for 1 h at 37 °C. Serial dilutions of sera were added to the plates and incubated at 37 °C for 2 h. After washing, plates were incubated with alkaline phosphatase (AP)-conjugated anti-mouse IgG antibody (Jackson Immunoresearch) for 2 h at 37 °C. After final washing, plates were incubated with AP substrate para-nitrophenylphosphate for 2.5 h at room temperature and read at 405 nm using an ELISA plate reader. Results are expressed by reference to serial dilutions of monoclonal antibody 6E10 (Covance).
Human Post-mortem Tissues
Prepared cryosections of human Alzheimer brain (cortex) tissue were used as a positive control tissue, whereas normal human brain (cortex) tissue was used as a negative control. The panel of human tissues was obtained from Tissue Solutions Ltd., and analyses were performed by Covance Laboratories Ltd. (Harrogate, UK).
Serum and Antibodies
Mouse monoclonal antibody against β-amyloid (anti-human β-amyloid, M0872, Dako) was used as a positive control antibody. Monkey serum was used at 1:2000 dilution. Serum of one animal immunized with PBS was used as a negative control. A goat anti-monkey IgG (1:100 dilution) and a biotinylated rabbit anti-goat IgG were used to detect primary non-primate antibody.
Immunohistochemistry
∼5-μm-thick cryostat sections were prepared and fixed in acetone followed by 10% neutral buffered formalin. After washing, sections were incubated with Dako REAL serum blocking solution, Avidin solution, and Biotin solutions. Cryosections were incubated overnight with diluted sera and incubated with goat anti-monkey IgG, followed by biotinylated rabbit anti-goat. VECTASTAIN ABC/HRP and 3,3′-diaminobenzidine tetrahydrochloride were used to detect antibody complexes. After staining, sections were washed and incubated with 0.5% copper sulfate solution, counterstained in hematoxylin, and mounted. All reagents were diluted in antibody diluents, and all incubation steps were preceded by washing in PBS and were performed at room temperature.
Statistical Evaluation
The data were analyzed using the non-parametric Wilcoxon matched pair ranks. A value of p < 0.05 was considered to be statistically significant.
RESULTS
In order to investigate the structure-conformation relationship of liposomal tetrapalmitoylated peptides, a range of peptides were synthesized (Table 1), which differ in peptide sequence, length, and charge. The biophysical properties of these peptides either alone as “native” tetra-acetylated peptides or as tetrapalmitoylated peptides embedded into liposome bilayers were analyzed by various techniques.
TABLE 1.
Different peptide sequences synthesized bearing N- and C-terminal palmitoyl or acetyl chains
| Name | Peptide | Linker |
|---|---|---|
| Palm1–15 | H-K(X)-K(X)-DAEFRHDSGYEVHHQ-K(X)-K(X)-OH | X = Palmitoyl |
| Acetyl1–15 | X = Acetyl | |
| Palm15–1 | H-K(X)-K(X)-QHHVEYGSDHRFEAD-K(X)-K(X)-OH | X = Palmitoyl |
| Acetyl15–1 | X = Acetyl | |
| scPalm15 | H-K(X)-K(X)-GHEAYHSVERFDDQH-K(X)-K(X)-OH | X = Palmitoyl |
| scAcetyl15 | X = Acetyl | |
| Palm1–9 | H-K(X)-K(X)-DAEFRHDSG-K(X)-K(X)-OH | X = Palmitoyl |
| Acetyl1–9 | X = Acetyl | |
| Palm1–5 | H-K(X)-K(X)-DAEFR-K(X)-K(X)-OH | X = Palmitoyl |
| Acetyl1–5 | X = Acetyl | |
| Palm1–15(D7K) | H-K(X)-K(X)-DAEFRHKSGYEVHHQ-K(X)-K(X)-OH | X = Palmitoyl |
| Acetyl1–15(D7K) | X = Acetyl | |
| Palm1–15(E3A,D7K) | H-K(X)-K(X)-DAAFRHKSGYEVHHQ-K(X)-K(X)-OH | X = Palmitoyl |
| Acetyl1–15(E3A,D7K) | X = Acetyl | |
| Palm1–15(E3K,D7K) | H-K(X)-K(X)-DAKFRHKSGYEVHHQ-K(X)-K(X)-OH | X = Palmitoyl |
| Acetyl1–15(E3K,D7K) | X = Acetyl | |
| Palm1–15(E3K,D7K,E11K) | H-K(X)-K(X)-DAKFRHKSGYKVHHQ-K(X)-K(X)-OH | X = Palmitoyl |
| Acetyl1–15(E3K,D7K,E11K) | X = Acetyl |
Palm1–15 Forms β-Sheet Aggregates Similar to Aβ
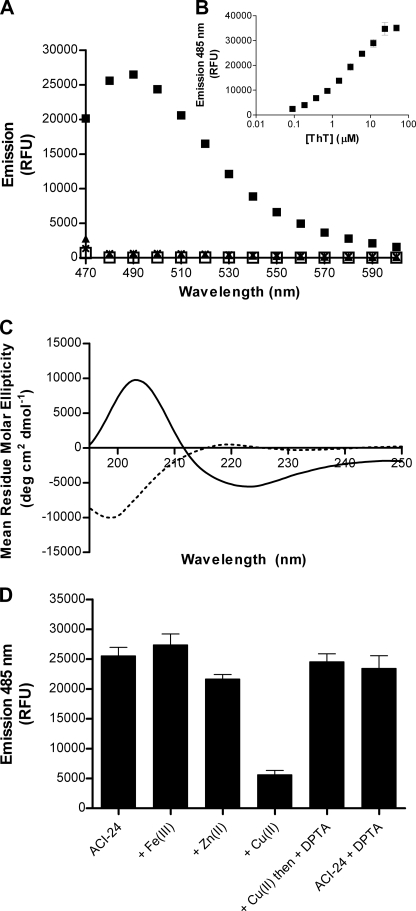
Previously, we reported that Palm1–15 (Table 1) adopts a β-sheet conformation within the lipid bilayer of ACI-24 (27). To explore if this conformation induces aggregation, we measured fluorescence of ThT upon the addition to ACI-24. ThT is a small benzothiazole dye that exhibits a strong red shift in its emission spectrum upon binding to aggregated β-sheet peptides (30, 31). As such, it has been widely used to detect and characterize the aggregation of β-amyloid peptides into both oligomers (32) and fibers (33). The addition of ThT to ACI-24 gave rise to a strong fluorescence emission at 485 nm, characteristic of the emission spectra for β-sheet aggregated amyloid (Fig. 1A). No emission above background was observed over the same range in the presence of liposomes lacking peptide (“empty” liposomes), confirming that the liposome matrix did not cause interference. Because Palm1–15 peptide is highly insoluble in PBS, an acetylated and PBS-soluble version, “Acetyl1–15” peptide was synthesized and analyzed; however, this showed no emission at 485 nm in the presence of ThT (Fig. 1A). For Palm1–15 in liposomes, the fluorescence response value at saturation Bmax and the apparent Kd value of 2.4 μm were estimated by fitting the fluorescence emission at 485 nm upon the addition of ThT at different concentrations to Palm1–15 (ACI-24) (Fig. 1B). This apparent Kd value is consistent with that reported for the interaction of ThT with Aβ(1–40) (11 μm) (34). ThT data were in agreement with the CD spectra, which show that the liposomal Palm1–15 adopts a β-sheet secondary structure, whereas “native” Acetyl1–15 peptide alone is unstructured in PBS solution (Fig. 1C). Thus, within the liposomal formulation of ACI-24, Palm1–15 forms β-sheet aggregates that are tinctorially similar to β-amyloid. Based upon liposomes of 100-nm diameter with 5-nm thick bilayers and standard lipid headgroup areas, we calculate that, on average, each liposome contains ∼335 peptides, assuming a random distribution of peptide between liposomes covering a minor portion of the surface.
FIGURE 1.
Conformational analyses and metal binding properties of liposomal Palm1–15 peptide ACI-24 composed of Palm1–15 (121 μm), DMPC (10.9 mm), DMPG (1.21 mm), cholesterol (8.51 mm), and MPLA (77 μm). A, fluorescence emission of liposomal formulation of Palm1–15 peptide (ACI-24) (□) and upon the addition of ThT to ACI-24 (■), ACI-24-Empty (▴), and Acetyl1–15 peptide (×). Samples were analyzed in PBS at a peptide concentration of 15 μm with excitation at 440 nm. B, ThT binding isotherm for liposomal peptide Palm1–15 (ACI-24). C, CD spectra of lipidated peptide Palm1–15 embedded in liposomes (solid line) and of acetylated “native” peptide Acetyl1–15 (dotted line) in PBS. Spectra were recorded after 7-fold dilution for liposomal Palm1–15 in PBS with subtraction of a spectrum of the corresponding empty liposomes lacking peptide. D, fluorescence emission of ThT in the presence of liposomal vaccine ACI-24 upon the addition of different metal ions. ThT was added to ACI-24 alone or to ACI-24 (final concentration 15 μm) preincubated with different metals (final concentration 20 μm) prior to the addition of metal chelator diethylenetriamine pentaacetic acid (DPTA) (final concentration 200 μm). As a control, diethylenetriamine pentaacetic acid was added to liposomal Palm1–15 in the absence of Cu(II), resulting in no change to the ThT emission. Data are expressed as average ± S.D. (error bars) (n = 3). RFU, relative fluorescence units.
In order to further examine the similarities between Palm1–15 and Aβ(1–42) aggregates and to better understand the mechanism of β-sheet assembly upon incorporation into lipid bilayers, we investigated whether preformed β-sheet aggregates of liposomal Palm1–15 could be disaggregated by Cu(II), as reported for Aβ(1–42) fibers (35). To this end, ACI-24 was incubated with CuCl2 for 2 h and then analyzed by ThT. Likewise, the effect of adding ZnCl2 and FeCl3 was investigated. Fig. 1D shows that at near stoichiometric concentrations of metal/peptide, the ThT fluorescence is decreased by ∼75% upon the addition of Cu(II), whereas only ∼12% reduction was observed with Zn(II) and none with Fe(III). A shorter incubation time of just 5 min revealed a similar reduction upon the addition of Cu(II), whereas an EC50 value for Cu(II) disaggregation of ∼3 μm was determined. When the metal chelator diethylenetriamine pentaacetic acid was added to the metal-liposome solution instead of PBS alone, analysis by ThT fluorescence revealed an increase in fluorescence, similar to that found prior to the addition of Cu(II).
These data reveal that Palm1–15 is disaggregated by Cu(II) and can reassemble within the lipid bilayer post-liposome formation to form aggregates comparable with those formed prior to Cu(II)-induced disassembly. The ability of Cu(II) to disaggregate Palm1–15 β-sheets thus further highlights the similarities between Palm1–15 aggregates and Aβ(1–42) aggregates.
N- and C-terminal Dipalmitoylated Peptides Not Related to Aβ Form β-Sheet Aggregates in Liposome Bilayers
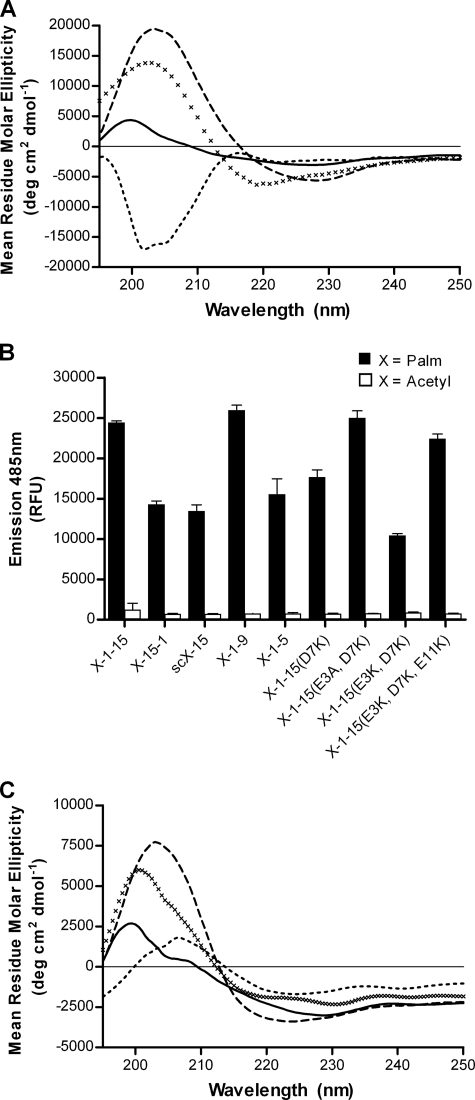
In order to determine whether the formation of β-sheet aggregates of liposomal Palm1–15 was related to the peptide sequence in Aβ1–15, the unrelated reverse sequence Palm15–1 and scrambled sequence scPalm15 were synthesized (Table 1). These peptides were then incorporated into liposomes as for ACI-24 and analyzed by CD spectroscopy. As can be seen in Fig. 2A, both liposomal peptides Palm15–1 and scPalm15 show minimal mean residue molar ellipticities at ∼220 nm. These spectra are similar to that found for liposomal Palm1–15 and characteristic of a β-sheet conformation. In contrast, the tetra-acetylated peptides Acetyl15–1 and scAcetyl15, corresponding to the sequences from Palm15–1 and scPalm15, were synthesized and upon analysis by CD were found to adopt random coil conformations (supplemental Fig. S1). When analyzed by ThT fluorescence, positive ThT emission signals were observed, whereas the corresponding acetylated peptides Acetyl15–1 and scAcetyl15 alone (native peptides) showed no fluorescence (Fig. 2B). Thus, the aggregation observed for Palm1–15 is not unique to the Aβ1–15 amino acid sequence but also occurs for other unrelated sequences upon peptide palmitoylation and incorporation into liposomes. Slight differences in fluorescence intensity were nevertheless observed between liposomal peptides Palm1–15, Palm15–1, and scPalm15 (Fig. 2B) despite similar binding affinities for ThT (supplemental Table S1 and Fig. S3), suggesting that the peptide sequence has only a minor influence on the extent of β-sheet aggregation.
FIGURE 2.
Conformational analyses of various N- and C-terminal dipalmitoylated peptides of different primary sequence. A, CD spectra of liposomal peptides Palm15–1 (solid line), scPalm15 (dashed line), Palm1–9 (×), and Palm1–5 (dotted line). B, ThT fluorescence emission upon the addition to liposomes embedded with different sequences of N- and C-dipalmitoylated peptides (filled bars) and corresponding “native” acetylated peptides (open bars) in the absence of liposomes. Results are given as average ± S.D. (error bars) (n = 3). C, CD spectra of liposomal peptides Palm1–15(D7K) (solid line), Palm1–15(E3A,D7K) (dashed line), Palm1–15(E3K,D7K) (×), and Palm1–15(E3K,D7K,E11K) (dotted line). RFU, relative fluorescence units.
Short N- and C-terminal Dipalmitoylated Peptides Form β-Sheet Aggregates in Liposomes
Because the binding epitope within Aβ(1–42) of sera taken from mice immunized with ACI-24 was found to be in the region Aβ1–9 (27), we next decided to investigate the effect of peptide length on conformation. To this end, the truncated peptides Palm1–9 and Palm1–5 were synthesized (Table 1). Upon incorporation into liposomes, peptide Palm1–9 was found to form β-sheet aggregates by CD (Fig. 2A) similar to liposomal peptide Palm1–15 (Fig. 1A). On the contrary, lipopeptide Palm1–5 was found to give a CD spectrum consistent with a mixed proportion of extended β-sheet and random coil conformations. This was supported by ThT binding data showing lower ThT fluorescence intensity (Fig. 2B) and a lower Bmax value for liposomal peptide Palm1–5 compared with Palm1–15 or Palm1–9 (supplemental Table S2 and Fig S3). In summary, even short tetrapalmitoylated peptides generate β-sheet aggregates within the liposome bilayers, but with a minimal length of at least 5 amino acids.
N- and C-terminal Dipalmitoylated Peptides Form β-Sheet Aggregates in Liposome Bilayers Independent of Peptide Charge
To examine whether the preference for β-sheet formation would still arise for peptides with different net charges at physiological pH, we decided to synthesize a range of Aβ1–15 mutants (Palm1–15(D7K), Palm1–15(E3A,D7K), Palm1–15(E3K,D7K), and Palm1–15(E3K,D7K,E11K)), wherein different numbers of acidic amino acids within Aβ1–15 were replaced by either basic Lys or neutral Ala residues (Table 1). In this way, it was possible to create a series of peptides with very different isoelectric points ranging from 5.2 to 10.0 (supplemental Table S1), such that they carry net negative, neutral, and positive charges at physiological pH. These different peptides were then incorporated into liposomes carrying a negative surface charge as for ACI-24. CD spectroscopic analyses of these liposomal peptides revealed characteristic minima at ∼220 nm (Fig. 2C) indicative of β-sheet secondary structure. These findings were supported by ThT analyses, which showed that all liposomal tetrapalmitoylated peptides gave rise to positive ThT signals, whereas the corresponding acetylated “native” peptides did not (Fig. 2B, open bars). All liposomal peptides, Palm1–15(D7K), Palm1–15(E3A,D7K), Palm1–15(E3K,D7K), and Palm1–15(E3K,D7K,E11K), were found to have similar binding affinities for ThT as well as similar Bmax values, except for mutant Palm1–15(E3K,D7K) having a slightly lower Bmax value. Together, these data demonstrate that the β-sheet aggregation of N- and C- dipalmitoylated peptides embedded into anionic liposomes occurs for a range of peptide sequences independent of their electrostatic charge. In order to evaluate whether peptide-liposome interactions could explain the variations to the THT signals observed, binding analyses between the corresponding acetylated peptides of identical length with anionic liposomes were carried out using surface plasmon resonance (supplemental Fig. S2). No correlation was observed between peptide affinity for liposomes and aggregation of the corresponding palmitoylated peptide. Combined, these results suggest that specific interactions between peptide side chains and the liposome surface do not play a significant role in modulating the aggregation of N- and C-terminal dipalmitoylated liposomal peptides.
Liposome Surface Charge Modulates Peptide Aggregation
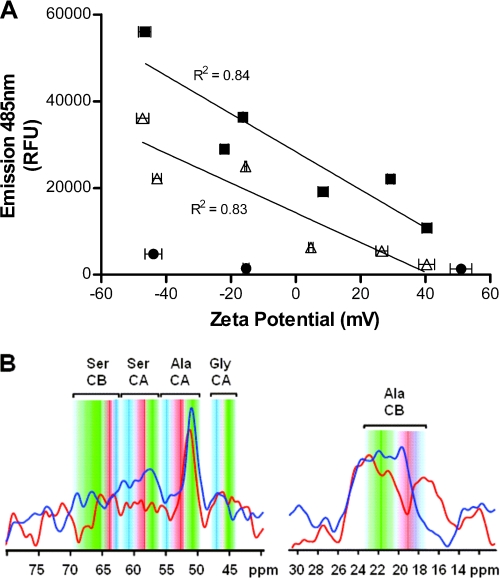
Next a series of liposomes were prepared using different molar ratios of the phospholipids DMPC and DMPG as well as the cationic phospholipid DMTAP (supplemental Table S3, formulations A–L), in order to explore whether the liposome surface charge could influence the conformation of anionic Palm1–15 (pI = 5.2) or cationic mutant Palm1–15(E3K,D7K,E11K) (pI = 10.0). Cholesterol and MPLA content was maintained at a cholesterol/phospholipid molar ratio of 7:10 and cholesterol/MPLA molar ratio of 7:0.06, respectively. Liposomes lacking peptide (formulations M, N, and O) were prepared as negative controls. Analysis by ThT fluorescence revealed much lower fluorescence (Bmax) for liposomes containing either Palm1–15 or mutant Palm1–15(E3K,D7K,E11K) when the liposomes displayed greater positive ζ potential (Fig. 3A). For liposomes lacking peptide, only minimal fluorescence was observed. Apparent binding constants (Kd values) were determined in order to determine whether the lower ThT fluorescence in the presence of cationic liposomes was due to lower affinity resulting from electrostatic repulsion between ThT and positively charged liposomes (supplemental Table S3 and Fig. S4). The resulting binding curves revealed similar apparent Kd values for different liposome formulations composed of the same peptide (supplemental Table S3, formulations A–F and G–L), indicating that charge does not weaken the ThT binding affinity. The lower affinity of ThT for liposomes containing cationic peptide Palm1–15(E3K,D7K,E11K) compared with liposomal Palm1–15 may explain the slightly lower ThT fluorescence observed for the liposome formulations containing Palm1–15(E3K,D7K,E11K) (Fig. 3A). In order to confirm these observations, MAS-NMR spectroscopy was performed using Palm1–15 uniformly labeled with 13C/15N at Ala-2, Ser-8, and Gly-9 (Palm1–15(ASG)). When Palm1–15(ASG) was incorporated into anionic liposomes as for ACI-24, 13C double quantum experiments revealed a significant population of extended backbone conformations, as shown by distinctive chemical shifts (36) of the Cα position of Ser-8 and the Cβ position of Ala-2 (Fig. 3B). Such backbone folds are typically found in amyloids forming intermolecular β-sheets during aggregation. On the other hand, when Palm1–15(ASG) was incorporated into cationic liposomes, a significant decrease in signal intensity was observed for the Cα of Ser-8, indicative of an increase in mobility. In addition, an upfield shift in the signal of Ala-2 Cβ was observed, along with a significant reduction in signal intensity consistent with a decrease in the proportion of peptide in β-strand conformation. This result further suggests that the intensity of the ThT signal is due to differences in peptide structure and aggregation and not due to differences in binding affinities for liposomes. According to our experiments, β-strand formation of N- and C-terminal dipalmitoylated peptides is favored upon incorporation into anionic rather than cationic liposomes, independent of the peptide charge.
FIGURE 3.
Effect of liposome surface charge upon conformation of N- and C-terminal dipalmitoylated peptides integrated into liposomal bilayers. A, effect of liposome surface charge upon conformation of Palm1–15 peptide (■, liposome formulations A–F; see supplemental Table S3), and cationic mutant Palm1–15(E3K,D7K,E11K) (△, liposome formulations G–L) measured by ThT fluorescence. Liposomes are composed of peptide, phospholipids (different proportions of DMPC, DMPG, and/or DMTAP), cholesterol, and MPLA in a molar ratio of 0.1:10:7:0.06, respectively. Empty liposomes lacking peptide (●, liposome formulations M–O) were used as negative controls. ζ potential values are given as average ± S.D. (n = 3). B, 13C-13C double quantum MAS-NMR spectra of Palm1–15(ASG) peptide uniformly (13C/15N) labeled at positions Ala-2, Ser-8, and Gly-9 and incorporated into anionic liposomes (DMPC/DMPG/cholesterol/MPLA (molar ratio 9:1:7:0.06)) (dark blue line) or cationic liposomes (DMTAP/cholesterol/MPLA (molar ratio 10:7:0.06)) (dark red line). Conformation-dependent chemical shift ranges (36) compatible with α-helix (blue), random coil (red), and β-sheet (green) backbone structure are annotated. RFU, relative fluorescence units. CA and CB indicate the α- and β-Z carbon atoms of the corresponding amino acids.
Peptide Lipidation Pattern and Lipid Anchor Chain Length Modulate Peptide Aggregation
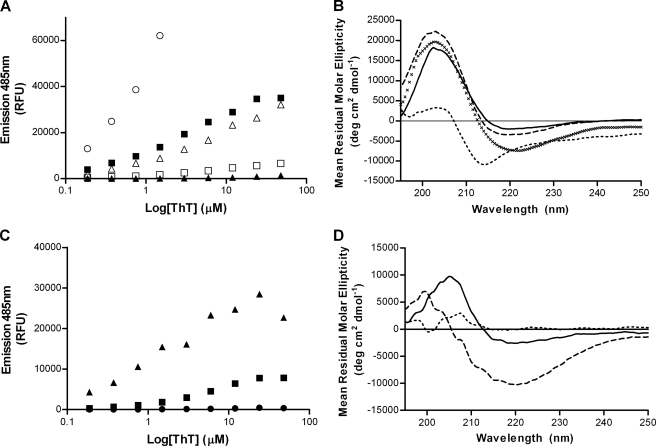
Next we decided to investigate the effect of the palmitoyl chains on the conformation of Palm1–15 peptide embedded within liposomal bilayers. A series of palmitoylated peptides were synthesized (Table 2) in which both the number and position of lipid anchors were varied. These peptides were incorporated into liposomes, and their conformations were analyzed using both ThT fluorescence and CD spectroscopy. Liposomal peptide Palm1–15(4C) containing four palmitic chains on the peptide C terminus showed a much higher ThT fluorescence emission (Bmax) than peptide Palm1–15 (ACI-24) or peptide Palm1–15(2C) (Fig. 4A and supplemental Table S4), indicating that the C-terminal tetrapalmitoylated peptide has a greater tendency to form β-sheet aggregates. Importantly, the apparent Kd values for ThT binding to Palm1–15 or to Palm1–15(4C) were similar (supplemental Table S4 and Fig. S5). This shows that the increase in Bmax for Palm1–15(4C) compared with Palm1–15 (2N2C) cannot be explained simply by a difference in the binding affinity of ThT. Palm1–15(2C) peptide gave rise to a similar fluorescence signal as Palm1–15 (2N2C), whereas the peptide Palm1–15(1N1C) containing one palmitic chain on both the N and C termini was only very weakly fluorescent upon the addition of ThT (Fig. 4A). The latter showed significantly lower binding affinity to ThT, consistent with a non-aggregated conformation (supplemental Table S4 and Fig. S5). Similarly, C-terminally monopalmitoylated liposomal peptide Palm1–15(1C) showed only a very low signal compared with liposomal peptide Palm1–15 (ACI-24).
TABLE 2.
Synthesized lipopeptides with different lipidation patterns
| Name | Peptide |
|---|---|
| Palm1–15(4C) | H-DAEFRHDSGYEVHHQ-K(Palm)-K(Palm)-K(Palm)-K(Palm)-OH |
| Palm1–15(2C) | H-DAEFRHDSGYEVHHQ-K(Palm)-K(Palm)-OH |
| Palm1–15(1N1C) | H-K(Palm)-DAEFRHDSGYEVHHQ-K(Palm)-OH |
| Palm1–15(1C) | H-DAEFRHDSGYEVHHQ-K(Palm)-OH |
| Dodecyl1–15 | H-K(Dodecanoyl)-K(Dodecanoyl)-DAEFRHDSGYEVHHQ-K(Dodecanoyl)-K(Dodecanoyl)-OH |
| Octyl1–15 | H-K(Octanoyl)-K(Octanoyl)-DAEFRHDSGYEVHHQ-K(Octanoyl)-K(Octanoyl)-OH |
| Butyl1–15 | H-K(Butanoyl)-K(Butanoyl)-DAEFRHDSGYEVHHQ-K(Butanoyl)-K(Butanoyl)-OH |
FIGURE 4.
Secondary and quaternary structural analysis of liposomal lipopeptides. A, ThT fluorescence upon the addition to liposomal peptide constructs Palm1–15(4C) (○), Palm1–15 (■), Palm1–15(2C) (▴), Palm1–15(1N1C) (△), and Palm1–15(1C) (□). B, CD spectra of liposomal peptide constructs Palm(4C) (solid line), Palm1–15(2C) (×), Palm1–15(1N1C) (dashed line), and Palm1–15(1C) (dotted line). C, ThT fluorescence upon the addition to liposomal peptide constructs Dodecyl1–15 (▴), Octyl1–15 (■), and Butyl1–15 (●). D, CD spectra of liposomal peptide constructs Dodecyl1–15 (dashed line), Octyl1–15 (solid line), and Butyl1–15 (dotted line). RFU, relative fluorescence units.
CD spectroscopic analyses of these liposomes were performed to measure the peptide secondary structure. Liposomal peptides Palm1–15(4C) and Palm1–15(2C) showed classical β-sheet CD spectra with minimum absorption at ∼220 nm and crossing the x axis at ∼214 nm, similar to ACI-24 (Fig. 4B). Interestingly, the spectrum of dipalmitoylated Aβ1–15 peptide Palm1–15(1N1C) showed an extended β-strand conformation by CD but did not show any fluorescence in the presence of ThT. This highlights the need to measure both peptide secondary structure (by CD) and peptide quaternary structure (by ThT fluorescence). It therefore appears that peptide Palm1–15(1N1C) adopts an extended β-strand conformation but is insufficiently aggregated to enable Thioflavin T to bind. In contrast, monopalmitoylated peptide Palm1–15(1C) shows a distinct CD spectrum with a minimum at ∼215 nm and crossing the x axis at ∼207 nm, consistent with a mixture of random coil and β-sheet conformations.
Next we investigated the effect of replacing the palmitoyl anchors with shorter homologs. To this end, a series of liposomes were prepared containing either dodecanoyl, octanoyl, or butanoyl lipid chains replacing those of palmitoyl in the Pal1–15 peptide (Table 2). ThT fluorescence measurements showed a substantial decrease in fluorescence upon reducing the peptide lipid anchor chain length from dodecanoyl (C12) to octanoyl (C8) and to butanoyl (C4), whereupon no fluorescence could be observed (Fig. 4C). Analysis of peptide secondary structure by CD spectroscopy likewise revealed that the liposomal vaccine containing octanoyl lipid anchor chains had reduced ellipticity at 220 nm compared with the longer dodecanoyl analog. Likewise, the peptide containing butanoyl chains did not show any negative ellipticity at 220 nm, consistent with a lack of secondary structure.
In summary, these results show that both the conformation and extent of β-sheet aggregation of palmitoylated peptides are strongly dependent upon the number of lipid chains at the peptide termini as well as the lipid anchor chain length, suggesting that hydrophobic association of peptide lipid anchor chains is a key driver of peptide aggregation.
Distinct Small Molecules Modify Peptide Aggregation Post-liposome Assembly
Because we observed disaggregation of the β-sheet aggregates of liposomal Palm1–15 with Cu(II), we decided also to explore whether the addition of small molecules that have been reported to disassemble Aβ aggregates could also modify the aggregation state of Palm1–15 embedded within the liposomal bilayer. For this, tannic acid, rosmarinic acid, and Methylene Blue were incubated with ACI-24, and the ThT fluorescence emission was monitored. A decrease of fluorescence was observed upon the addition of all compounds, with the most efficient disaggregation observed upon the addition of Methylene Blue (supplemental Fig. S6). When added to the β-sheet aggregates of the unrelated peptide sequence scPalm15, a similar fluorescence decrease was observed (supplemental Fig. S6), showing that β-sheet disaggregation occurs independent of peptide sequence and in a manner similar to that of Aβ(1–40/42).
Peptide Aggregation Determines Immune Response Specificity
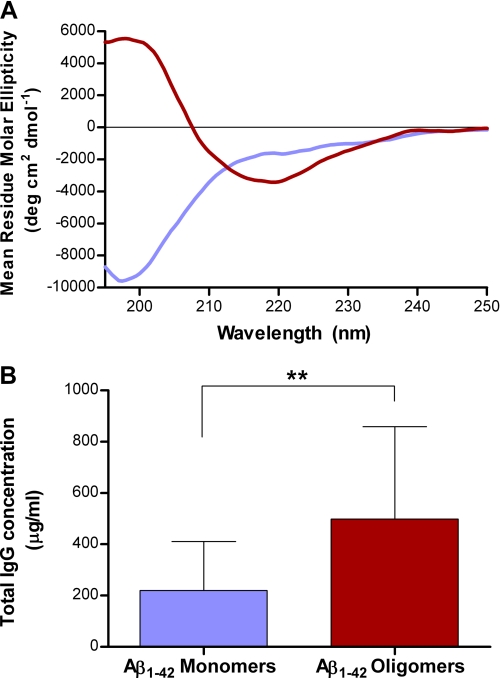
In order to study the relationship between the aggregation state of liposomal peptide immunogens and the binding specificity of the corresponding polyclonal antibodies generated in vivo, the liposomal vaccine ACI-24 containing Palm1–15 peptide was prepared, characterized (see supplemental material), and injected into mice, and the IgG titers were determined by ELISA. To analyze the conformational specificity of the polyclonal antibody response, Aβ(1–42) was allowed to partially aggregate and was subsequently fractionated by SEC (see supplemental Fig. S7) to give conformationally pure monomeric random coil and oligomeric β-sheet fractions (Fig. 5A). The plasma taken from individual mice immunized with ACI-24 was analyzed for polyclonal binding to the different fractions of Aβ. As can be seen in Fig. 5B, ACI-24 elicits a significantly higher (p = 0.002, Wilcoxon matched pair ranks) IgG response against the Aβ(1–42) oligomeric β-sheet, compared with the unstructured Aβ(1–42) monomer fraction.
FIGURE 5.
Conformation-specific binding of anti-ACI-24 polyclonal antibodies to Aβ(1–42). A, CD spectra of SEC purified oligomer (red) and monomer fractions (blue) of Aβ(1–42). B, recognition of distinct Aβ(1–42) conformations by polyclonal plasma taken from WT mice after immunization with liposomal vaccine ACI-24. Monomeric (random coil) and oligomeric (β-sheet) Aβ(1–42) species were coated onto an ELISA plate, and binding of plasma was quantified relative to control IgG antibody 6E10. Data are expressed in mean ± S.D. (error bars) (n = 10). **, p < 0.01 by non-parametric Wilcoxon matched pairs ranks.
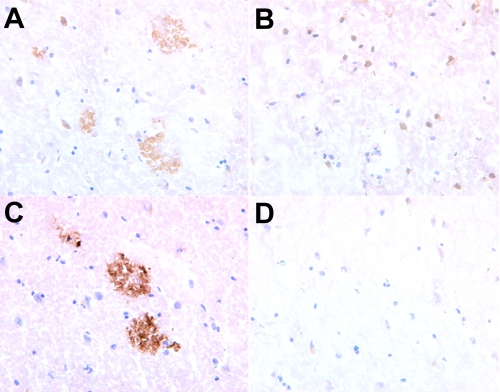
To assess whether antibodies against ACI-24 would show conformation-specific binding to the pathological β-sheet Aβ plaques, over the non-folded native healthy APP protein, ACI-24 was injected into monkeys that share 100% APP sequence homology with human APP. The binding properties of the antibodies raised against ACI-24 were then assessed using histologically prepared cryostat sections taken from human healthy and AD brain. Fig. 6 shows clearly that serum taken from the ACI-24-immunized monkey stains AD human brain, whereas no cross-reactivity is observed to healthy non-AD brain. Thus, Palm1–15 peptide aggregation within the liposomal vaccine is critical for determining the conformational specificity of the immune response.
FIGURE 6.
Staining of pathological misfolded β-sheet plaques in human AD brain. Shown is staining of human AD brain (A) and healthy human brain (B), with sera (diluted 1:2000) taken from a monkey immunized with ACI-24. Shown is staining of human AD brain with positive control monoclonal antibody (Dako mouse mAb) (C) or with sera from a monkey immunized with PBS (D). Brain slices are magnified 40-fold.
DISCUSSION
Peptide N- or C-terminal lipidation is a widely used approach to embed peptides into liposome bilayers. We have previously reported that incorporating a range of peptides conjugated at both the N and C termini with two palmitoyl chains into liposomes composed of phospholipids (DMPC and DMPG), cholesterol, and MPLA rapidly induced high IgG titers in mice (21, 27, 37). When we incorporated the N- and C-terminal dipalmitoylated peptide Palm1–15 into liposomes, we observed that the peptide adopted a β-sheet conformation, whereas the non-lipidated form was found to exist in a non-structured random coil conformation. In this report, we show that the Palm1–15 peptide forms aggregates on the liposome surface that exhibit ThT fluorescence similar to aggregates of Aβ, implying that the two species are morphologically similar.
To better understand the properties and mechanism of formation of the β-sheet aggregates formed by liposomal peptide Palm1–15, we investigated whether preformed aggregates could undergo disassembly within the lipid bilayers of ACI-24. We report that Cu(II) can rapidly disaggregate liposomal Palm1–15 aggregates in a similar manner to Aβ(1–42) fibers (35), whereas Zn(II) or Fe(III) show little effect, consistent with the high affinity coordination of copper to the N-terminal amino acids (38). Presumably, copper disrupts peptide-liposome or peptide-peptide stabilizing interactions such that the hydrophobic interactions between palmitic chains are insufficient to maintain β-sheet formation. Reaggregation of Palm1–15 upon the addition of a metal chelating agent shows that peptide aggregation occurs within the lipid membrane and implies that the peptide aggregates are formed as a single thermodynamically stable structure.
Structure-conformation studies show that a range of peptides of unrelated primary sequence also form β-sheet aggregates upon integration into liposomes of identical composition with only a minor influence of amino acid composition upon aggregation. Therefore, β-sheet aggregation can be induced by lipidation of unstructured peptides and incorporation into liposomes. Previously, Chiti and Dobson (39) demonstrated that many polypeptide sequences can form amyloid-like aggregates when suitably forcing conditions are applied. In our liposome systems, we have succeeded in generating amyloid from unstructured peptide sequences even under physiological conditions. We find that induction of peptide misfolding into β-sheets still occurs for a 9-mer peptide, Palm1–9. This is even shorter than the minimum length (12-mer) found to be necessary to enable β-sheet conformation in a model peptide (KIGAKI)2 (40) and highlights the additional negative free folding energy brought by peptide tetrapalmitoylation.
The role of peptide charge in determining the ability to form β-sheet aggregates upon liposomes was examined both by generating a series of mutated peptides and by directly monitoring the binding interactions of the non-lipidated “native” peptides with liposomes. Both approaches revealed that the amino acid side chains have little effect upon aggregation despite the fact that the peptide sequences were chosen in order to have very different isoelectric points. This suggests that it is peptide backbone-liposome interactions that play an important role in determining the peptide secondary and quaternary structure. This model is supported by the modulation of peptide aggregation we observe by modification of the liposome surface potential with anionic or cationic lipids. In accordance with our finding, anionic lipid surfaces were also reported to greatly enhance the fibrillization rate of both Aβ(1–42) (41) and human islet amyloid peptide (42). The decreased aggregation we observe either in the absence of DMPG or in the presence of DMTAP implies that it is the net surface potential that is responsible for aggregation.
Conformational changes of mono- or dilipidated peptides upon interaction with lipid surfaces have been reported, including unordered to β-sheet (43) or unordered to β-turn (44). Our study attempted to extend these observations to determine the rules that govern the conformational transitions of lipopeptides anchored on lipid bilayers. We find that the number and position of peptide lipidation sites have a dramatic influence upon β-sheet aggregation. Interestingly, the total number of palmitoyl chains at one end of Palm1–15 was found to correlate well with the extent of β-sheet aggregation, whereas the total number of lipidation sites poorly correlated. Peptide self-assembly therefore appears to be driven by hydrophobic interactions between palmitoyl chains. This observation is supported by the changes to both peptide secondary and quaternary structure upon reducing the chain length of the lipid anchor attached to the peptide. Increased hydrophobicity of Aβ(1–42) compared with Aβ(1–40) has also been considered to be the factor responsible for the increased aggregation rate of Aβ(1–42) (45). Likewise, cholesterol adducts of Aβ(1–42) (46) and α-synuclein (47) have been found to display accelerated aggregation propensities.
An attractive alternative strategy to control peptide conformation post-liposome assembly would be via the addition of distinct small molecules. We find that known small molecule β-sheet breakers can disaggregate the β-sheets of liposomal peptides independent of peptide sequence. This shows that β-sheet disaggregation can occur not only in solution phase but also within the two-dimensional plane at the solid-liquid interface. This observation is pertinent following the identification of Aβ(1–40/42) dimers within lipid rafts of neuronal membranes, which were correlated with memory impairment (48). In this context, liposome-bound peptide systems such as those described here may also prove useful as models to screen for therapeutic agents capable of interfering with the aggregation of peptides associated to cell membranes.
The relevance of lipopeptide aggregation state to the conformational specificity of the immune response is demonstrated in this report by analyzing the Aβ binding properties of plasma taken from mice or monkeys immunized with aggregated liposomal Palm1–15 (ACI-24). Polyclonal antibodies in plasma from mice immunized with ACI-24 show high conformation specific binding to Aβ(1–42) in the synaptotoxic oligomeric β-sheet conformation, whereas the monomeric Aβ(1–42) is only poorly recognized. The preferential recognition of the N-terminal region within Aβ(1–42) β-sheet aggregates by antibodies was previously reported to be critical for restoring the memory capacity in a transgenic mouse model of AD (27). The selective recognition of the oligomeric β-sheet conformation of Aβ(1–42) oligomers over monomers described in this report also implies that the N-terminal region of Aβ(1–42) may be capable of adopting a more structured conformation than has previously been considered. This observation is consistent with recent data showing that even a single N-terminal point mutation (Tyr1) to Aβ(1–42) is sufficient to alter peptide assembly kinetics and aggregate size distribution (49).
The selective recognition of the pathological misfolded conformation of Aβ plaques in human AD brain over non-AD brain that we observe further highlights the conformational specificity of the antibody response. Moreover, it can be expected that this selective recognition will increase the efficacy of the antibody response by circumventing loss of therapeutic antibodies that cross-bind to peripheral healthy APP protein or to Aβ(1–40) monomers circulating in peripheral blood. Sera taken from humans that were vaccinated with preaggregated full-length Aβ(1–42) was reported by others to be specific for binding human AD brain over healthy brain (22). However, in contrast to our findings, these antibodies were not able to recognize soluble oligomeric forms of Aβ(1–42) that are associated with toxicity. In a related context, conformation-specific antibodies, such as those described here, also hold promise as tools for disease diagnosis. Notably, antibodies that can discriminate different protein folds, such as those of prion protein, are under investigation for use in detection of transmissible spongiform encephalopathies (50).
Induction of β-sheet “misfolded” peptide conformations under physiological conditions using a range of unstructured peptide sequences as described here raises important questions concerning the molecular requirements for amyloid formation. We find that the forces that drive the self-assembly of the liposomal lipopeptides can be attributed to a combination of hydrophobic interactions between palmitoyl anchors and interactions between peptide backbone and liposome surface. Development of strategies described in this report to modulate these interactions thus extends the repertoire of available methods to alter the aggregation of lipopeptide immunogens within liposome bilayers (27).
In this study, ACI-24 was chosen as a model system to demonstrate that peptide conformation within liposome bilayers can be translated into a conformation-specific immune response in vivo. The ability to control peptide conformation by modulation of the peptide lipidation pattern and spacer and liposome composition or via co-administration with small molecules implies that this technology may also be applied to the rational design of other liposomal vaccines. Selective targeting of the pathological conformation of a target protein, as described here, can be expected to contribute to the generation of safer and more efficacious therapies for a range of human disease, in particular those based on misfolded proteins, such as AD.
Acknowledgment
We thank Professor K. Kostarelos (University of London) for helpful discussions.
Solid-state NMR studies were supported by Netherlands Organization for Scientific Research Grant 700.26.121. D. R. is a member of the Board of Directors and Scientific Advisory Board of AC Immune SA.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods” and “Results,” including Tables S1–S4 and Figs. S1–S7.
- AD
- Alzheimer disease
- Aβ
- β-amyloid protein
- APP
- amyloid precursor protein
- DMPC
- 1, 2-dimyristoyl-sn-glycero-3-phosphocholine
- DMPG
- 1,2-dimyristoyl-sn-glycero-3-(phospho-rac-(1-glycerol))
- DMTAP
- 1, 2-dimyristoyl-3-trimethylammonium-propane
- MAS
- magic angle spinning
- MPLA
- monophosphoryl lipid A
- ThT
- thioflavin T
- SEC
- size exclusion chromatography.
REFERENCES
- 1. Misumi S., Nakayama D., Kusaba M., Iiboshi T., Mukai R., Tachibana K., Nakasone T., Umeda M., Shibata H., Endo M., Takamune N., Shoji S. (2006) J. Immunol. 176, 463–471 [DOI] [PubMed] [Google Scholar]
- 2. Villard V., Agak G. W., Frank G., Jafarshad A., Servis C., Nébié I., Sirima S. B., Felger I., Arevalo-Herrera M., Herrera S., Heitz F., Bäcker V., Druilhe P., Kajava A. V., Corradin G. (2007) PLoS ONE 2, e645–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Su H. P., Golden J. W., Gittis A. G., Hooper J. W., Garboczi D. N. (2007) Virology 368, 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pimentel T. A., Yan Z., Jeffers S. A., Holmes K. V., Hodges R. S., Burkhard P. (2009) Chem. Biol. Drug Des. 73, 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gao J., Sidhu S. S., Wells J. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3071–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yokoyama T., Kimura K. M., Ushiki Y., Yamada S., Morooka A., Nakashiba T., Sassa T., Itohara S. (2001) J. Biol. Chem. 276, 11265–11271 [DOI] [PubMed] [Google Scholar]
- 7. Weaver-Feldhaus J. M., Miller K. D., Feldhaus M. J., Siegel R. W. (2005) Protein Eng. Des. Sel. 18, 527–536 [DOI] [PubMed] [Google Scholar]
- 8. Liebman H. A., Limentani S. A., Furie B. C., Furie B. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 3879–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fieser T. M., Tainer J. A., Geysen H. M., Houghten R. A., Lerner R. A. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 8568–8572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu S. M., Hodges R. S. (2002) J. Biol. Chem. 277, 23515–23524 [DOI] [PubMed] [Google Scholar]
- 11. Selkoe D. J. (2003) Nature 426, 900–904 [DOI] [PubMed] [Google Scholar]
- 12. Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C. M., Stefani M. (2002) Nature 416, 507–511 [DOI] [PubMed] [Google Scholar]
- 13. Roychaudhuri R., Yang M., Hoshi M. M., Teplow D. B. (2009) J. Biol. Chem. 284, 4749–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chimon S., Shaibat M. A., Jones C. R., Calero D. C., Aizezi B., Ishii Y. (2007) Nat. Struct. Mol. Biol. 14, 1157–1164 [DOI] [PubMed] [Google Scholar]
- 15. Cleary J. P., Walsh D. M., Hofmeister J. J., Shankar G. M., Kuskowski M. A., Selkoe D. J., Ashe K. H. (2005) Nat. Neurosci. 8, 79–84 [DOI] [PubMed] [Google Scholar]
- 16. Noguchi A., Matsumura S., Dezawa M., Tada M., Yanazawa M., Ito A., Akioka M., Kikuchi S., Sato M., Ideno S., Noda M., Fukunari A., Muramatsu S., Itokazu Y., Sato K., Takahashi H., Teplow D. B., Nabeshima Y., Kakita A., Imahori K., Hoshi M. (2009) J. Biol. Chem. 284, 32895–32905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ono K., Condron M. M., Teplow D. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14745–14750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shankar G. M., Li S., Mehta T. H., Garcia-Munoz A., Shepardson N. E., Smith I., Brett F. M., Farrell M. A., Rowan M. J., Lemere C. A., Regan C. M., Walsh D. M., Sabatini B. L., Selkoe D. J. (2008) Nat. Med. 14, 837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Janus C., Pearson J., McLaurin J., Mathews P. M., Jiang Y., Schmidt S. D., Chishti M. A., Horne P., Heslin D., French J., Mount H. T., Nixon R. A., Mercken M., Bergeron C., Fraser P. E., St George-Hyslop P., Westaway D. (2000) Nature 408, 979–982 [DOI] [PubMed] [Google Scholar]
- 20. Morgan D., Diamond D. M., Gottschall P. E., Ugen K. E., Dickey C., Hardy J., Duff K., Jantzen P., DiCarlo G., Wilcock D., Connor K., Hatcher J., Hope C., Gordon M., Arendash G. W. (2000) Nature 408, 982–985 [DOI] [PubMed] [Google Scholar]
- 21. Nicolau C., Greferath R., Balaban T. S., Lazarte J. E., Hopkins R. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2332–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hock C., Konietzko U., Papassotiropoulos A., Wollmer A., Streffer J., von Rotz R. C., Davey G., Moritz E., Nitsch R. M. (2002) Nat. Med. 8, 1270–1275 [DOI] [PubMed] [Google Scholar]
- 23. O'Nuallain B., Wetzel R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmechel A., Zentgraf H., Scheuermann S., Fritz G., Pipkorn R., Reed J., Beyreuther K., Bayer T. A., Multhaup G. (2003) J. Biol. Chem. 278, 35317–35324 [DOI] [PubMed] [Google Scholar]
- 25. Lambert M. P., Viola K. L., Chromy B. A., Chang L., Morgan T. E., Yu J., Venton D. L., Krafft G. A., Finch C. E., Klein W. L. (2001) J. Neurochem. 79, 595–605 [DOI] [PubMed] [Google Scholar]
- 26. Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003) Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 27. Muhs A., Hickman D. T., Pihlgren M., Chuard N., Giriens V., Meerschman C., van der Auwera I., van Leuven F., Sugawara M., Weingertner M. C., Bechinger B., Greferath R., Kolonko N., Nagel-Steger L., Riesner D., Brady R. O., Pfeifer A., Nicolau C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9810–9815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Orgogozo J. M., Gilman S., Dartigues J. F., Laurent B., Puel M., Kirby L. C., Jouanny P., Dubois B., Eisner L., Flitman S., Michel B. F., Boada M., Frank A., Hock C. (2003) Neurology 61, 46–54 [DOI] [PubMed] [Google Scholar]
- 29. Petkova A. T., Ishii Y., Balbach J. J., Antzutkin O. N., Leapman R. D., Delaglio F., Tycko R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16742–16747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. LeVine H., 3rd (1993) Protein Sci. 2, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khurana R., Coleman C., Ionescu-Zanetti C., Carter S. A., Krishna V., Grover R. K., Roy R., Singh S. (2005) J. Struct. Biol. 151, 229–238 [DOI] [PubMed] [Google Scholar]
- 32. Maezawa I., Hong H. S., Liu R., Wu C. Y., Cheng R. H., Kung M. P., Kung H. F., Lam K. S., Oddo S., Laferla F. M., Jin L. W. (2008) J. Neurochem. 104, 457–468 [DOI] [PubMed] [Google Scholar]
- 33. Groenning M. (2010) J. Chem. Biol. 3, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Levine H., 3rd (1995) Amyloid Int. J. Exp. Clin. Invest. 2, 1–6 [Google Scholar]
- 35. House E., Mold M., Collingwood J., Baldwin A., Goodwin S., Exley C. (2009) J. Alzheimers. Dis. 18, 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y., Jardetzky O. (2002) Protein. Sci. 11, 852–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tosi P. F., Radu D., Nicolau C. (1995) Biochem. Biophys. Res. Commun. 212, 494–500 [DOI] [PubMed] [Google Scholar]
- 38. Sarell C. J., Syme C. D., Rigby S. E., Viles J. H. (2009) Biochemistry 48, 4388–4402 [DOI] [PubMed] [Google Scholar]
- 39. Chiti F., Dobson C. M. (2009) Nat. Chem. Biol. 5, 15–22 [DOI] [PubMed] [Google Scholar]
- 40. Meier M., Seelig J. (2008) J. Am. Chem. Soc. 130, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 41. Bokvist M., Lindström F., Watts A., Gröbner G. (2004) J. Mol. Biol. 335, 1039–1049 [DOI] [PubMed] [Google Scholar]
- 42. Jha S., Sellin D., Seidel R., Winter R. (2009) J. Mol. Biol. 389, 907–920 [DOI] [PubMed] [Google Scholar]
- 43. Laczkó I., Hollósi M., Vass E., Tóth G. K. (1998) Biochem. Biophys. Res. Commun. 249, 213–217 [DOI] [PubMed] [Google Scholar]
- 44. Löwik D. W., Linhardt J. G., Adams P. J., van Hest J. C. (2003) Org. Biomol. Chem. 1, 1827–1829 [DOI] [PubMed] [Google Scholar]
- 45. Burdick D., Soreghan B., Kwon M., Kosmoski J., Knauer M., Henschen A., Yates J., Cotman C., Glabe C. (1992) J. Biol. Chem. 267, 546–554 [PubMed] [Google Scholar]
- 46. Zhang Q., Powers E. T., Nieva J., Huff M. E., Dendle M. A., Bieschke J., Glabe C. G., Eschenmoser A., Wentworth P., Jr., Lerner R. A., Kelly J. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4752–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bosco D. A., Fowler D. M., Zhang Q., Nieva J., Powers E. T., Wentworth P., Jr., Lerner R. A., Kelly J. W. (2006) Nat. Chem. Biol. 2, 249–253 [DOI] [PubMed] [Google Scholar]
- 48. Kawarabayashi T., Shoji M., Younkin L. H., Wen-Lang L., Dickson D. W., Murakami T., Matsubara E., Abe K., Ashe K. H., Younkin S. G. (2004) J. Neurosci. 24, 3801–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maji S. K., Ogorzalek Loo R. R., Inayathullah M., Spring S. M., Vollers S. S., Condron M. M., Bitan G., Loo J. A., Teplow D. B. (2009) J. Biol. Chem. 284, 23580–23591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paramithiotis E., Pinard M., Lawton T., LaBoissiere S., Leathers V. L., Zou W. Q., Estey L. A., Lamontagne J., Lehto M. T., Kondejewski L. H., Francoeur G. P., Papadopoulos M., Haghighat A., Spatz S. J., Head M., Will R., Ironside J., O'Rourke K., Tonelli Q., Ledebur H. C., Chakrabartty A., Cashman N. R. (2003) Nat. Med. 9, 893–899 [DOI] [PubMed] [Google Scholar]