Abstract
Two-pore domain potassium (K2P) channels play a key role in setting the membrane potential of excitable cells. Despite their role as putative targets for drugs and general anesthetics, little is known about the structure and the drug binding site of K2P channels. We describe A1899 as a potent and highly selective blocker of the K2P channel TASK-1. As A1899 acts as an open-channel blocker and binds to residues forming the wall of the central cavity, the drug was used to further our understanding of the channel pore. Using alanine mutagenesis screens, we have identified residues in both pore loops, the M2 and M4 segments, and the halothane response element to form the drug binding site of TASK-1. Our experimental data were used to validate a K2P open-pore homology model of TASK-1, providing structural insights for future rational design of drugs targeting K2P channels.
Keywords: Drug Design, Ion Channels, Molecular Dynamics, Potassium Channels, Protein Conformation, ion Channel Blocker
Introduction
The acid-sensitive K2P4 channel TASK-1 (TWIK-related acid-sensitive K+) gives rise to instantaneous and non-inactivating potassium currents, which can be blocked by extracellular acidification (1). Five TASK channel family members have been described. The closest relatives of the TASK-1 channel are TASK-3 and TASK-5 (2, 3). The TASK-4 and TASK-2 channels show less homology and are functionally different as they are primarily blocked by protons in the physiological pH range (4, 5). Previous studies revealed that TASK channels are key regulators for many physiological processes in cells of the cardiovascular system (6), the central nervous system (7), and the adrenal gland (8). In addition, they are also involved in peripheral chemosensation (9) and the regulation of the immune system (10). TASK channels are the molecular targets of volatile and local anesthetics (11, 12). Although studies of TASK channel knock-out mice have advanced our knowledge about the physiological function of these channels, a selective TASK channel blocker would be very valuable to perform future in vivo experiments. In the present study, we describe a highly potent blocker of the TASK-1 channel, A1899, active in the one-digit nanomolar range. As suggested by the high affinity, we found that A1899 is selective for TASK-1 channels. Although anandamide has been reported as a physiological blocker of TASK-1 channels (13), this lipid does not combine the high affinity and specificity as A1899.
K2P channels have a transmembrane topology that is unique among potassium channels. One subunit contains four transmembrane domains and two pore-forming domains. The functional channel is formed by a dimer. Although other potassium channels are tetramers with a pore formed by four identical transmembrane domains, the domains and symmetry of the K2P channel pore have long been a matter of speculation. The different topology of K2P channels does not allow a direct transfer of data from the crystal structures of other potassium channels without experimental proof of validity. The pore-forming domains and the rotation of these domains can be validated by an open-channel pore blocker as the amino acids of the binding site should face the water-filled pore cavity. We have identified the binding site of A1899 in the open pore of TASK-1 channels. In good agreement with recently published data (14), our results suggest that the M2 and M4 segments of two TASK subunits form the walls of the inner pore. Using A1899, we have confirmed the orientation of the M2 and M4 segments and provide a model of the mammalian K2P channel TASK-1 in the open state.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
Mutations in the human TASK-1 (KCNK3, NM_002246) cDNA were introduced with the QuikChange site-directed mutagenesis kit (Stratagene) according to the instructions of the manufacturer.
Oocyte Preparation, cRNA Synthesis, and cRNA Injection
Oocytes were obtained from anesthetized Xenopus laevis frogs and incubated in OR2 solution containing in mm: 82.5 NaCl, 2 KCl, 1 MgCl2, 5 HEPES (pH 7.5) substituted with 2 mg/ml collagenase II (Sigma) to remove residual connective tissue. Subsequently, oocytes were stored at 18 °C in ND96 solution supplemented with 50 mg/liter gentamycin, 274 mg/liter sodium pyruvate, and 88 mg/liter theophylline. Human TASK-1 channels were subcloned into a pSGEM or pBF1 vector, and cDNA was linearized with NheI or MluI, respectively. cRNA was synthesized with mMESSAGE mMACHINE kit (Ambion). The quality of cRNA was tested using gel electrophoresis. Oocytes were each injected with 50 nl of cRNA.
Two-electrode Voltage Clamp Recordings
All two-electrode voltage clamp recordings were performed at room temperature (20–22 °C) with a TurboTEC 10CD (npi) amplifier and a Digidata 1200 Series (Axon Instruments) as analog/digital converter. Micropipettes were made from borosilicate glass capillaries GB 150TF-8P (Science Products) and pulled with a DMZ-Universal Puller (Zeitz). Recording pipettes had a resistance of 0.5–1.5 megaohms and were filled with 3 m KCl solution. Recording solution ND96 contained in mm: 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, 5 HEPES (pH 7.5). Block was analyzed with voltage steps from a holding potential of −80 mV. A first test pulse to 0 mV of 1-s duration was followed by a repolarizing step to −80 mV for 1 s directly followed by another 1-s test pulse to +40 mV. The sweep time interval was 10 s. For recordings with symmetrical potassium concentrations, the ND96 solution was modified; NaCl was reduced to 2 mm, and KCl concentration was increased to 96 mm. Data were acquired with Clampex 10 (Molecular Devices) and analyzed with Clampfit 10 (Molecular Devices) and Origin 7 (OriginLab Corp.).
Molecular Modeling
Based on the crystal structures of KcsA and KvAP (PDB IDs 1K4C and 1ORQ, respectively), homology models of TASK-1 were created using the Modeler 7 version 7 software. For ligand docking predictions, GOLD (version 4.0), AutoDock, and AutoDockTools were used to propose binding conformations for A1899 within the central cavity of TASK-1. Molecular dynamics (MD) simulations were performed with the Gromacs software. Detailed information on molecular modeling is provided in the supplemental Experimental Procedures, which includes details about the homology modeling, ligand docking experiments, and MD simulations.
Statistical Analyses
All values are expressed as means ± S.E. For all oocyte and CHO electrophysiological experiments, 3–12 cells were studied (n = 3–12). Error bars in Figs. 1 and 3–5 represent S.E. values. Significance was assessed using two-tailed Student's t test. Asterisks indicate significance: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 1.
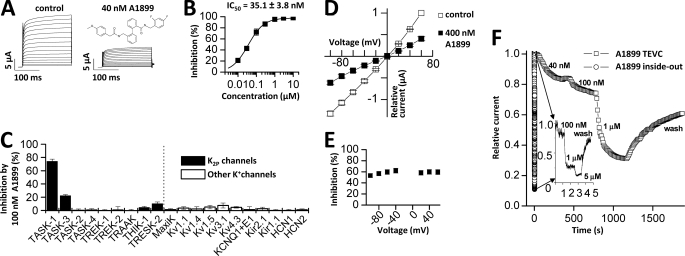
A1899 selectively blocks human TASK-1 channels expressed in Xenopus oocytes. A, human TASK-1 channels recorded with a voltage step protocol (200-ms steps from −70 mV to +70 mV with an increment of 10 mV; holding potential, −80 mV) before (left) and after (right) application of 40 nm A1899 (see inset for chemical structure of A1899). B, dose-response curve of A1899 on human TASK-1. Block was analyzed at the end of the test pulse to +40 mV. C, test for sensitivity of different potassium channels to 100 nm A1899 after heterologous expression in Xenopus oocytes. D, I-V relationships for TASK-1 in bath solution with high potassium concentration before (□) and after (■) application of 400 nm A1899. E, the percentage of inhibition by 400 nm A1899 from D plotted against the applied voltage. F, application of different concentrations of A1899 to TASK-3 channels recorded in intact whole oocytes (□) and in inside-out macropatches (○). The inset shows the recording of inside-out macropatches with a higher temporal resolution. TEVC, two-electrode voltage clamp.
FIGURE 3.
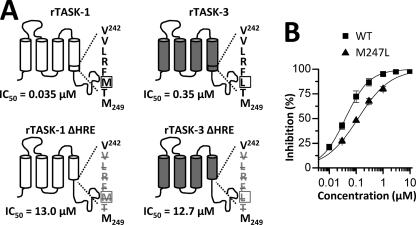
The HRE influences A1899 binding. A, the sequence of the HREs of TASK-1 and TASK-3 differ in one amino acid (VLRF(M/L)T) (indicated by a box). In both channels, in-frame deletion of the HRE (bottom) leads to a massive increase in IC50 values. rTASK, rat TASK. B, dose-response curves of A1899 on human TASK-1 WT (■) and an M247L (▴) mutant.
FIGURE 4.
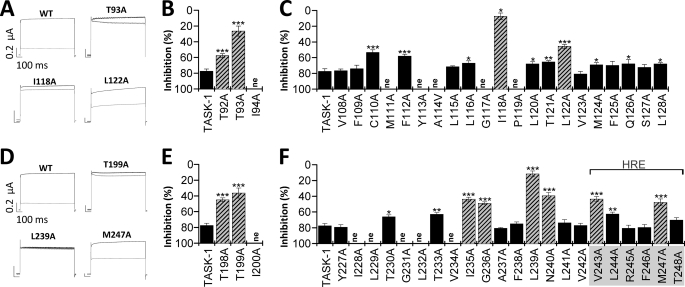
The A1899 binding site includes residues of the M2 and M4 segment and the selectivity filter. A–F, mutant TASK-1 channels were expressed in Xenopus oocytes, and block by 400 nm A1899 was analyzed. A and D, sample current traces for wild-type TASK-1 and mutated channels before (black) and after (gray) application of 400 nm A1899. B, C, E, and F, mutant TASK-1 channels with a reduced sensitivity to A1899 indicate residues as part of the binding site for A1899 (illustrated as striated bars). Threonine residues of the P1 and P2 pore loops are part of the A1899 binding site. In the M2 and M4 segment, residues of the binding site are located at a distance of 3–4 amino acids.
FIGURE 5.
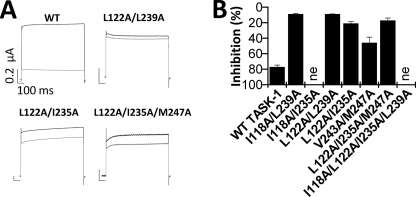
TASK-1 channels with multiple mutations at the drug binding site show a strong reduction of A1899 sensitivity. A, sample traces of TASK-1 wild-type, double, and triple mutant channels before (black) and after (gray) application of 400 nm A1899. B, inhibition of TASK-1 channels with multiple mutations by 400 nm A1899.
RESULTS
A1899 Is a Selective Blocker of TASK-1 Channels
We have tested the affinity of A1899 for TASK-1 channels expressed in Xenopus oocytes. Application of 40 nm A1899 led to a pronounced inhibition of TASK-1 currents (Fig. 1A). In oocytes, TASK-1 channels were blocked with an IC50 of 35.1 ± 3.8 nm, analyzed at +40 mV (Fig. 1B). The IC50 of A1899 for TASK-1 channels expressed in CHO cells was only 7 nm (Table 1), which is consistent with the notion that IC50 values determined in oocytes are mostly higher than those determined in mammalian cells. The IC50 for TASK-3 expressed in CHO cells was 70 nm (Table 1). Thus, TASK-3, the closest relative of TASK-1, binds A1899 with a 10-fold lower affinity. To further probe the selectivity of A1899, we expressed a set of different potassium channels in Xenopus oocytes and quantified the extent of current inhibition by 100 nm A1899 (Fig. 1C). Under these conditions, TASK-1 currents were blocked by 74.2 ± 3.4%. In contrast, the closest relative, TASK-3, was only blocked by 22.2 ± 1.6%. All the other channels tested showed only little (<11%) or no block (Fig. 1C). The IC50 values determined for several potassium channels (Table 1) indicate that A1899 is a specific TASK-1 blocker.
TABLE 1.
IC50 values of A1899 for different human potassium channels
Values were determined in Xenopus oocytes (n = 3–12) or when indicated in CHO cells (n = 3–12). TRAAK, TWIK-related arachidonic-acid-stimulated K+ channel.
| Channel | IC50 |
|---|---|
| μm | |
| TASK-1 | 0.035 ± 0.003 |
| TASK-3 | 0.318 ± 0.030 |
| TASK-1 CHO | 0.007 ± 0.001 |
| TASK-3 CHO | 0.070 ± 0.009 |
| TASK-2 | 12.0 ± 2.2 |
| TASK-4 | 8.1 ± 2.3 |
| TREK-1 | 23.8 ± 1.8 |
| TREK-2 | 8.4 ± 1.1 |
| TRAAK | >20 |
| THIK-1 | 2.2 ± 0.2 |
| TRESK | 0.9 ± 0.1 |
| Kv1.1 | 2.7 ± 0.3 |
Subsequently, we recorded TASK-1 currents in the presence of an increased extracellular K+ concentration ([K+]in = [K+]out). Under these conditions, it was possible to test for a potential voltage dependence of block. We found that 400 nm A1899 blocked inward and outward currents to the same extent, indicating that the block of A1899 is not voltage-dependent (Fig. 1, D and E). However, the extent of block at +40 mV elicited by 400 nm A1899 in a bath solution with a high concentration of extracellular potassium was only 59.7% in comparison with about 90% in ND96 (Fig. 1, B and D), reflecting an almost 10-fold increase in IC50 of A1899. A reduced blocker affinity in bath solutions with elevated extracellular potassium concentrations is typical for an open-channel block mechanism (15, 16). Most open-channel blockers act from the intracellular side by binding in the water-filled central cavity (15, 17). To assess whether A1899 blocks TASK channels in the central cavity, we tested the time course of inhibition by A1899 using whole-cell and inside-out macropatch recordings from Xenopus oocytes. For these experiments, we tested inhibition of TASK-3 channels as these give rise to large macroscopic currents, necessary for inside-out macropatch recordings. In whole-cell recordings, the block of TASK-3 channels by A1899 developed very slowly. When the blocker was applied from the cytosolic phase during inside-out macropatch experiments, the block was faster by several orders of magnitude (Fig. 1F). These experiments suggest that A1899 acts as an open-channel blocker of TASK channels, presumably by binding to the inner cavity. If A1899 acts as an open-channel blocker, then the domains and the respective residues that interact with A1899 should form the walls of the central cavity. Hence, we continued to identify the drug binding site of A1899.
A1899 Interacts with the M2 and M4 Transmembrane Segments of TASK-1 Channels
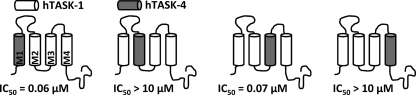
As a first step toward identification of the drug binding site and the pore-forming transmembrane domains, we constructed chimeric channels between the drug-sensitive TASK-1 and the drug-insensitive TASK-4. We replaced one of the TASK-1 transmembrane segments at a time by a TASK-4 transmembrane domain (Fig. 2), expressed the chimeric channels in oocytes, and determined the IC50 values of A1899 (Fig. 2). Replacing the M1 or M3 segment of TASK-1 with TASK-4 did not cause a major reduction in the binding affinity of A1899. The IC50 values were 0.06 μm for the M1 exchange and 0.07 μm for the M3 exchange (Fig. 2), whereas the wild-type IC50 was 0.035 μm. In contrast, replacing the M2 or M4 transmembrane segments of TASK-1 increased the IC50 values of A1899 by several orders of magnitude (IC50 > 10 μm) (Fig. 2). We conclude from these findings that the M2 and M4 segments of TASK-1 are pore-forming domains that are crucial for the binding of A1899 and the subsequent block of the channel.
FIGURE 2.
M2 and M4 segments of TASK-1 are part of the binding site for A1899. Four different chimeras, with TASK-4 transmembrane segments introduced into a TASK-1 background replacing M1, M2, M3, or M4, were analyzed for their IC50 of block by A1899. hTASK, human TASK.
The Halothane Response Element Is Part of the A1899 Binding Site
Volatile anesthetics activate TASK-1 and TASK-3 channels via the halothane response element (HRE). Therefore, we tested A1899 on wild-type TASK-1 and a mutant TASK-1 channel with a deletion of the HRE sequence, 243VLRFMT248 (Fig. 3A, left column). The in-frame deletion of the HRE in TASK-1 led to a drastic reduction of the A1899 affinity, and the IC50 increased 371-fold from 35 nm to 13 μm. The decrease in A1899 sensitivity of the ΔHRE channel mutant strongly suggests an involvement of the halothane response element in binding of the drug. A similar in-frame deletion of the HRE in TASK-3 channels also led to a drastic reduction in drug affinity (Fig. 3A, right column). The IC50 value increased 36-fold from 350 nm to 12.7 μm.
The M2 and M4 segments that we have identified as part of the A1899 binding site share about 90% sequence homology between TASK-1 and TASK-3. In each of the two segments, there are only two amino acids that are not identical (supplemental Fig. S1). One of these four residues is located in the HRE. TASK-1 has a methionine at position 247, whereas TASK-3 has a leucine residue at the same position (Fig. 3A). We introduced an M247L mutation in the TASK-1 channel, resulting in an HRE with the sequence of TASK-3. The M247L TASK-1 mutant was only blocked by 63.7 ± 4.2% by 400 nm A1899. The IC50 of this mutant expressed in oocytes increased from 35 nm for wild-type TASK-1 to 117 nm (Fig. 3B), reflecting a 3.3-fold increase in IC50 by converting the amino acid sequence to that of the TASK-3 HRE. The experiments indicate that the HRE is part of the A1899 drug binding site and that the sequence variations in the TASK-1 and TASK-3 halothane response elements might contribute to the different drug affinities of TASK-1 and TASK-3.
A1899 Is a Pore Blocker Binding to Threonine Residues of the Signature Sequence
Our data described above indicate that A1899 blocks TASK-1 channels from the intracellular side and that the M2 and M4 segments including the HRE are involved in A1899 binding. For different Na+, Ca2+, and K+ channels, it has been described that threonine residues of the signature sequence directly face into the central cavity and interact with open-channel blockers (18–21). If A1899 is in fact binding in the central cavity formed by M2 and M4 segments, the drug could interact with the corresponding threonine residues of the signature sequence to plug the channel pore. The positions of the analogous threonine residues (bold) are 92TTIGYG97 in the P1 loop and 198TTIGFG203 in the P2 loop. Therefore, we tested the affinity of the Thr93 and Thr199 alanine mutations of TASK-1 for 400 nm A1899. Although wild-type TASK-1 channels were blocked by 78 ± 5%, the T93A and T199A mutants were blocked by only 26.4 ± 6.4 and 36.4 ± 6.4%, respectively (Fig. 4, A–B and D–E). The strong reduction of block resulting from either mutation strongly suggests that A1899 interacts with the two threonine residues of the signature sequence and that A1899 deeply protrudes into the central cavity of the TASK-1 channel to induce a plugging of the pore.
Identification of the A1899 Binding Site by Alanine Mutagenesis Screening
As our data indicate that A1899 is an open-channel blocker of TASK-1 and that the M2 and M4 segments, the pore loops, and the halothane response elements are involved in the binding of A1899, we performed a systematic alanine mutagenesis screen of these domains to identify the A1899 binding site. TASK-1 channels with single mutations to alanine (or alanine to valine) were expressed in Xenopus oocytes, and block by 400 nm A1899 was measured (Fig. 4). The rationale was that mutations that remove or strongly reduce block should be part of the A1899 binding site (Fig. 4, striated columns). The following residues were identified by the alanine scan: Thr92 and Thr93 in the P1 region (Fig. 4B), Ile118 and Leu122 of the M2 segment (Fig. 4C), Thr198 and Thr199 of the P2 region (Fig. 4E), Ile235, Gly236, Leu239, and Asn240 of the M4 segment (Fig. 4F), and Val243 and Met247 of the HRE (Fig. 4F). The alanine scan revealed a helical pattern for the A1889 binding site in the M2 and M4 segment, meaning that substitutions by alanine that disturbed block were located in distances of about 3–4 amino acids. Consequently, the amino acids identified as parts of the binding site all point to the same direction of the respective α-helical transmembrane segment. As the identified residues should interact with the drug in the central cavity, our data can be used to validate the rotation of the M2 and M4 segment in a pore homology model.
To confirm the residues identified by our alanine scan, we introduced multiple mutations of the drug binding site simultaneously in the TASK-1 channels and tested these channels for their A1899 sensitivity (Fig. 5). Fig. 5A shows currents of the L122A/I235A and L122A/L239A double mutations and the L122A/I235A/M247A triple mutation, which all showed little or almost no block by 400 nm A1899. As expected, other TASK-1 constructs harboring multiple mutations at the A1899 binding site also showed a strong reduction in block (Fig. 5B). Summarizing, we have identified the residues of the A1899 binding site and conclude that the M2 and M4 segments build the pore of TASK-1 channels and that these amino acids forming the binding site of A1899 line the inner face of the pore.
Open-state Pore Homology Model of TASK-1 Validated by the A1899 Drug Binding Site
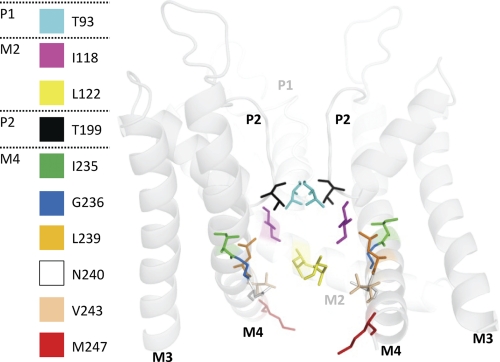
Based on the crystal structure of KvAP, we created an open-state homology model of TASK-1 using the M2 and M4 segments as the pore-forming helices. The model is in good agreement with our experimental data, expecting that residues of the A1899 binding site are facing into the central cavity (Fig. 6). In the model, the identified residues Thr93 (cyan) and Thr199 (black) of the first and second pore loop, Ile118 (magenta) and Leu122 (yellow) of the M2 segment, Ile235/Gly236 (green/blue), Leu239/Asn240 (orange/white) of the M4 segment, and Val243 (sand) and Met247 (red) of the HRE element are facing into the central cavity (Fig. 6) or are, in the case of Met247, accessible on the way into the pore (Fig. 6). Thus, the A1899 binding site data validate our in silico TASK-1 pore homology model.
FIGURE 6.
Open-state pore homology model of TASK-1 based on the KvAP structure. The figure illustrates residues identified as pore-facing using A1899. For better display, the backbone of half a TASK-1 subunit (M1, P1 and M2) is hidden, whereas the identified residues are still depicted. The backbone is shown transparent for a better view on the residues of the drug binding site.
Confirmation of the A1899 Binding Site by Docking of the Drug to the Open Pore
A proportional ligand molecule was used to perform docking experiments. Dockings of the ligand molecule into the open state of TASK-1 yielded optimal binding scores. When bound, the drug clearly blocks the channel pore and impedes passage of K+ ions. In this conformation, the difluorobenzene end of the molecule lies closest to the selectivity filter, with the carbonyl oxygen of the amide linkage suggested to accept hydrogen bonds from residues of the signature sequence, Thr93 and Thr199 (Fig. 7, dotted lines). The majority of the other ligand-protein contacts are hydrophobic in nature, with Ile118, Leu122, Leu232 (the latter mutant did not express in oocytes), Ile235, and Leu239 all within 4 Å of the first three rings of the molecule. The fourth ring was found to be localized lower in the cavity toward the cytosolic entrance. At this position, the ring forms contacts with Leu122 and Val243, whereas the attached methoxy moiety could potentially form hydrogen bonds with Asn240. No interaction of the drug was predicted for Met247, which was located too low in the M4 segment. For a table listing the nature of the drug molecule interactions, see supplemental Fig. S2. It is noteworthy that the majority of the sites predicted by the docking experiments to the open state are in good agreement with our electrophysiological data as the model mostly highlights the same positions as the mutagenic screen (Figs. 4 and 6). Thus, the docking experiments reconfirmed the validity of the open-pore homology model and the data obtained by the alanine scanning approach.
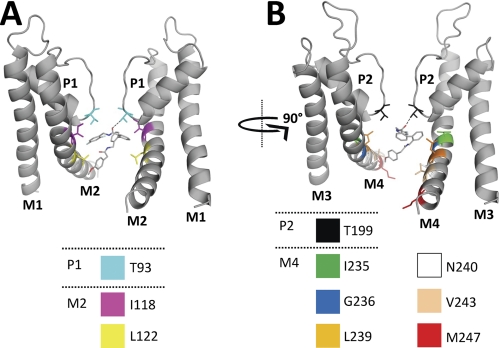
FIGURE 7.
Docking of A1899 to an open-state pore homology model of TASK-1. The binding mode of A1899 in the pore of TASK-1 as predicted by docking using the software GOLD is shown. All amino acids predicted to interact with the drug molecule are illustrated. For a better display of A1899 binding, the TASK-1 channel is shown in two orientations (rotated by 90°). The left panel shows the two M1, P1, and M2 segments, and the right panel shows the two M3, P2, and M4 segments. H-bonds are depicted as dotted lines between A1899 and the threonine residues of the selectivity filter.
A1899 Docking to a Closed-state Homology Model Supports a Binding to the Open State
Next, we compared the docking of A1899 to the open state (Fig. 7) with dockings to a closed-state model of TASK-1, which was generated on the basis of the KcsA structure (supplemental Fig. S3). Using the software GOLD, binding to the closed state was only possible if the M2 side chain and Phe125 were rotated so that this residue pointed up, toward the selectivity filter. This facilitates a strong binding mode that is in good agreement with the binding observed in the open-state model. In addition, the drug adopts the same orientation as in the open state. However, in this case, Phe125 would strongly interact with the drug, which is in disagreement with our experimental data (supplemental Fig. S3). Using the docking software Autodock, the drug does not fit into the binding site, despite the displacement of Phe125 (as done for GOLD). In addition, using Autodock, enabling a more flexible binding site does not appear to improve the results. The experiments suggest that A1899 does not fit into a closed channel pore and that the channels have to open to allow an optimal binding of A1899.
MD Simulations of A1899 in the Open Pore of TASK-1 Channels
To test the binding of the drug molecule in more detail, MD simulations were performed to assess the dynamic interactions of the open-channel complex. Throughout all four of the 30-ns simulations, the complex remained intact, with the protein stabilizing to a typical Cα root mean square deviation of between 3 and 3.5 Å. The percentage of contacts, for each protein side chain within 4 Å of the drug molecule, was calculated at 10-ps intervals for all four simulations. Most contacts were observed for Thr92 and Thr93 of P1, Ile118 and Leu122 of M2, Thr198 and Thr199 of P2, and Ile235, Gly236, Leu239, and Asn240 of M4 (Fig. 8). Thus, overall, the majority of contacts remained the same as described for the original docking (Fig. 7 and supplemental Fig. S2). As a putative false positive, the MD simulations predicted an interaction with the residue Gln126, a residue that was not tested positive by our alanine scan. Similar to the initial docking experiments, the MD simulations did not predict an interaction with Phe125. The MD simulations (Fig. 8) and the original docking (supplemental Fig. S2) predict an interaction with Leu232. Unfortunately, introducing mutations at Leu232 led to non-functional channels. The MD simulations indicate a hydrogen bonding between drug and the side chains of Thr93 and Thr199 of the selectivity filter and Asn240 of the inner transmembrane helices. In addition to the H-bonding with Thr93 and Thr199, the uppermost carbonyl oxygen of the drug also appears to coordinate the S4 K+ ion. Moreover, in two simulations tested, the ions starting at the S3 site move down to S4 to interact with the ligand (Fig. 8). It is noteworthy that open-channel blockers have been previously reported to interact with the K+ ion at site 4 of the selectivity filter (20–23). At depolarized potentials, when TASK-1 is open, the K+ ion seems to favor the occupancy at site 4 (22), which in turn appears to favor binding of the open-channel blocker A1899.
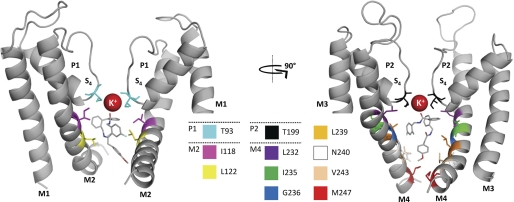
FIGURE 8.
Molecular dynamics simulations validate the A1899 binding site. For a better display of A1899 binding, the TASK-1 channel is shown in two orientations (rotated by 90°). The left panel shows the two M1, P1, and M2 segments, and the right panel shows the two M3, P2, and M4 segments. All amino acids predicted to have multiple contacts with the drug molecule are illustrated. In addition, A1899 interacts with the potassium ion at S4 of the selectivity filter.
DISCUSSION
In recent years, K2P channels have been considered to represent promising drug targets. However, little is known about the structure of these channels, and in addition, there is a lack of specific blockers. We have identified A1899 as the most potent synthetic K2P channel blocker currently known. After showing that A1899 acts from the cytosolic phase as a pore blocker, we used a combined approach of chimeras and alanine scanning to identify the first drug binding site of a K2P channel. K2P channels have a unique transmembrane topology. Crystal structures of ion channels with different transmembrane topologies, including for example KcsA (24), rKv1.2 (25), and KirBac1.1 (26), suggest that the transmembrane segments following the selectivity filter are pore-forming inner helices. Our data support the idea that the M2 and M4 segments of K2P channels form the wall of the inner cavity. In addition, the interaction between the drug located in the central cavity and the amino acids of the M2 and M4 segments was assessed experimentally to provide data analyzing the rotation of the inner helices.
While we were in the process of finalizing our study, Kollewe et al. (14) proposed a structure for the Drosophila K2P channel dORK. The group utilized long range electrostatic compensation of positively charged amino acids in the selectivity region by negatively charged residues introduced into the M2 or M4 segment to identify pore-facing amino acids. This approach was used before in Kir2.1 channels by Chatelain et al. (27), who showed that charge and distance between the two mutations are decisive for a rescue effect and that electrostatic rescue does not work for residues that are located in close proximity or in great distance from each other. Because of this limitation, Kollewe et al. (14) could only test amino acids in a restricted region of the lower M2 and M4 segment. They probed residues corresponding to 119–127 of the TASK-1 M2 segment and 233–241 of the TASK-1 M4 segment (supplemental Fig. S4), whereas our more comprehensive approach scanned the whole length of those segments: 108–128 of the M2 and 227–248 of the M4 segment. Strikingly, five of the eight amino acids of dORK (Gly134, Leu257, Val258, Met261, and Thr262), identified by Kollewe et al. (14) as pore-facing, were also identified by our alanine scan. These residues correspond to the TASK-1 residues Leu122 in M2 and Ile235, Gly236, Leu239, and Asn240 in M4 (supplemental Fig. S4). The data of dORK and TASK-1 can only be partially compared. In addition to the limited region of overlap, the mammalian TASK-1 channel shares only little sequence homology with dORK, and the structures of the two channels might differ as well. For example, we did not identify the M2 residues Thr121, Phe125, and Gln126 to be pore-facing as expected from the study with dORK (supplemental Fig. S4). In contrast, we identified Ile118, a residue that was not in the scanning range of the electrostatic interaction approach, as pore-facing (supplemental Fig. S4). In addition, we identified two residues in the late M4 segment (HRE), namely Val243 and Met247, which were also not within the spatial range for electrostatic interactions (supplemental Fig. S4).
The M2 and M4 segments of TASK-1 share low sequence homology (supplemental Fig. S4). Thus, not as expected from the primary sequence, the TASK-1 drug binding site has a remarkably high symmetry (Fig. 6). The primary binding site in the central cavity is formed by three rings of four identical residues. The first ring, located at the selectivity filter, is formed by four threonine residues, including two Thr93 residues of the P1 loop and two Thr199 residues of the P2 loop. The Ile118 of M2 together with Ile235 of M4 form a ring of four isoleucines, located underneath the selectivity filter in the central cavity. One helix turn below, there is a ring of four leucine residues formed by Leu122 of M2 and Leu239 of M4. In addition to these residues of a four-fold symmetrical binding site, we have identified Gly236 and Asn240 in the M4 segment and Val243 and Met247 in the HRE, which appear only twice in the binding site.
Interestingly, the HRE is a molecular prerequisite for the high-affinity block of A1899. This part of the channel is known to be the site of action for volatile anesthetics that activate TASK channels, and it has been described as a domain essential for the inhibition via Gq-coupled receptors. However, it was not possible to identify single amino acids responsible for the action of anesthetics or the G-protein coupling within this region. In contrast, our alanine scan points out that two residues of the HRE, Val243 and Met247, are responsible for the binding of A1899 to the HRE. Although the docking of A1899 to the pore homology model predicts an interaction of the HRE residue Val243 with the drug, Met247 seems to be too remote from the final binding site. Nevertheless, our mutagenesis data suggest that the difference in amino acid sequence at position 247 of the halothane response element contributes to the different drug affinities of TASK-1 and TASK-3. An explanation is that Met247 is important for a trapping of the drug in the central cavity. Our 30-ns MD simulations, however, showed no evidence for a trapping of the drug through gate closure at Met247, although this process likely requires MD simulations on a longer timescale. A more likely explanation lies within the fact that Met247 is located in the access pathway into the central cavity (Fig. 6). We suggest that position 247 is not part of the binding site but that it influences drug sensitivity because it regulates accessibility of the pore for A1899. The role of the HRE in high-affinity drug binding presented in this study suggests that the mechanism of synthetic modulators and endogenous mechanisms of TASK-1 regulation might be similar. Analogous actions of drug binding and endogenous G protein-coupled receptor modulation of TASK-1 might be the focus of future studies.
Our alanine scan does not suggest any impact of the F125A mutation on drug binding. The position Phe125 is analogous to Phe656 in human ERG channels, which is essential for the binding of most ERG blockers (19, 28). Former studies propose the corresponding amino acids in dORK (Phe137) and TREK-1 (Leu174) to line the inner pore (14, 29). However, in our computer models for the open state, Phe125 does not line the pore. An interaction of this residue with A1899 is only suggested in the closed-state model (supplemental Fig. S3). We therefore propose that the M2 segment is slightly rotated in the open state as compared with the closed-state, leading to a displacement of the aromatic residue of Phe125. The importance of the exact orientation of the corresponding phenylalanine in human ERG has also been discussed by Chen et al. (28).
Based on their TREK-1 model, Treptow and Klein (29) suggest that two leucines, one in the M2 and one in the M4 segment of TREK-1 (Leu174 and Leu289), delimit the narrowest part of the inner pore and form a hydrophobic gate. These leucine residues align to Phe125 and Leu239 of TASK-1. Our data indicate that TASK-1 channels do not have the same or conserved hydrophobic gating ring. The hydrophobic gating ring might be the molecular correlate to the difference between the gating mechanisms of TREK-1 and TASK channels.
The selective TASK-1 blocker A1899 described here is a powerful tool for studying the physiological role of TASK-1 channels in native tissues. We took advantage of the blocker to probe the direct interaction with the amino acids of the inner pore and created the first model for a mammalian K2P channel with experimentally validated orientation of the inner helices. Thus, using A1899 as a chemical probe, we validated a model for the mammalian K2P channel TASK-1 in the open state and identified the first complete drug binding site in a K2P channel. The docking of A1899 into this open-pore homology model and the subsequent molecular dynamics simulations confirmed the amino acids of the drug binding site identified by our alanine scan. Our data provide an excellent starting point for future studies on the molecular pharmacology of other K2P channels. In addition, our data should influence the rational drug design of selective blockers of other K2P channels. As K2P channels have emerged as novel drug targets, the identification of a TASK-1-specific blocker together with the identification of a high-affinity drug binding site in a K2P channel is a promising finding for future drug design and for basic research in physiology.
Acknowledgments
We thank K. Ramlow for excellent technical support and members of the M. S. P. Sansom laboratory for helpful discussions. The compound A1899 was obtained from Sanofi Aventis GmbH Germany.
This work was supported by Kempkes Stiftung Grant 12/08 (to N. D.) and by Deutsche Forschungsgemeinschaft Grants BA-1793/5-1 (to T. B.), DE-1482/3-1 (to N. D. and J. D.), and DE-1482/2-1 (to N. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S4.
- K2P
- two-pore domain potassium channel
- TASK
- TWIK-related acid-sensitive K+
- MD
- molecular dynamics
- HRE
- halothane response element
- TWIK
- tandem of P domains in a weak inwardly rectifying K+ channel
- ERG
- ether-à-go-go-related gene
- dORK
- Drosophila open rectifier K+ channel.
REFERENCES
- 1. Duprat F., Lesage F., Fink M., Reyes R., Heurteaux C., Lazdunski M. (1997) EMBO J. 16, 5464–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rajan S., Wischmeyer E., Xin Liu G., Preisig-Müller R., Daut J., Karschin A., Derst C. (2000) J. Biol. Chem. 275, 16650–16657 [DOI] [PubMed] [Google Scholar]
- 3. Ashmole I., Goodwin P. A., Stanfield P. R. (2001) Pflügers Arch. 442, 828–833 [DOI] [PubMed] [Google Scholar]
- 4. Decher N., Maier M., Dittrich W., Gassenhuber J., Brüggemann A., Busch A. E., Steinmeyer K. (2001) FEBS Lett. 492, 84–89 [DOI] [PubMed] [Google Scholar]
- 5. Reyes R., Duprat F., Lesage F., Fink M., Salinas M., Farman N., Lazdunski M. (1998) J. Biol. Chem. 273, 30863–30869 [DOI] [PubMed] [Google Scholar]
- 6. Putzke C., Wemhöner K., Sachse F. B., Rinné S., Schlichthörl G., Li X. T., Jaé L., Eckhardt I., Wischmeyer E., Wulf H., Preisig-Müller R., Daut J., Decher N. (2007) Cardiovasc. Res. 75, 59–68 [DOI] [PubMed] [Google Scholar]
- 7. Budde T., Coulon P., Pawlowski M., Meuth P., Kanyshkova T., Japes A., Meuth S. G., Pape H. C. (2008) Pflügers Arch. 456, 1061–1073 [DOI] [PubMed] [Google Scholar]
- 8. Czirják G., Fischer T., Spät A., Lesage F., Enyedi P. (2000) Mol. Endocrinol. 14, 863–874 [DOI] [PubMed] [Google Scholar]
- 9. Trapp S., Aller M. I., Wisden W., Gourine A. V. (2008) J. Neurosci. 28, 8844–8850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meuth S. G., Bittner S., Meuth P., Simon O. J., Budde T., Wiendl H. (2008) J. Biol. Chem. 283, 14559–14570 [DOI] [PubMed] [Google Scholar]
- 11. Patel A. J., Honoré E., Lesage F., Fink M., Romey G., Lazdunski M. (1999) Nat. Neurosci. 2, 422–426 [DOI] [PubMed] [Google Scholar]
- 12. Meadows H. J., Randall A. D. (2001) Neuropharmacology 40, 551–559 [DOI] [PubMed] [Google Scholar]
- 13. Maingret F., Patel A. J., Lazdunski M., Honoré E. (2001) EMBO J. 20, 47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kollewe A., Lau A. Y., Sullivan A., Roux B., Goldstein S. A. (2009) J. Gen. Physiol. 134, 53–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Armstrong C. M. (1971) J. Gen. Physiol. 58, 413–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barrows B., Cheung K., Bialobrzeski T., Foster J., Schulze J., Miller A. (2009) Channels 3, 239–248 [PubMed] [Google Scholar]
- 17. Decher N., Kumar P., Gonzalez T., Pirard B., Sanguinetti M. C. (2006) Mol. Pharmacol. 70, 1204–1211 [DOI] [PubMed] [Google Scholar]
- 18. Dilmac N., Hilliard N., Hockerman G. H. (2004) Mol. Pharmacol. 66, 1236–1247 [DOI] [PubMed] [Google Scholar]
- 19. Mitcheson J. S., Chen J., Lin M., Culberson C., Sanguinetti M. C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12329–12333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lerche C., Bruhova I., Lerche H., Steinmeyer K., Wei A. D., Strutz-Seebohm N., Lang F., Busch A. E., Zhorov B. S., Seebohm G. (2007) Mol. Pharmacol. 71, 1503–1511 [DOI] [PubMed] [Google Scholar]
- 21. Strutz-Seebohm N., Gutcher I., Decher N., Steinmeyer K., Lang F., Seebohm G. (2007) Cell Physiol. Biochem. 20, 791–800 [DOI] [PubMed] [Google Scholar]
- 22. Faraldo-Gómez J. D., Kutluay E., Jogini V., Zhao Y., Heginbotham L., Roux B. (2007) J. Mol. Biol. 365, 649–662 [DOI] [PubMed] [Google Scholar]
- 23. Bruhova I., Zhorov B. S. (2007) BMC Struct. Biol. 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. (1998) Science 280, 69–77 [DOI] [PubMed] [Google Scholar]
- 25. Long S. B., Tao X., Campbell E. B., MacKinnon R. (2007) Nature 450, 376–382 [DOI] [PubMed] [Google Scholar]
- 26. Kuo A., Gulbis J. M., Antcliff J. F., Rahman T., Lowe E. D., Zimmer J., Cuthbertson J., Ashcroft F. M., Ezaki T., Doyle D. A. (2003) Science 300, 1922–1926 [DOI] [PubMed] [Google Scholar]
- 27. Chatelain F. C., Alagem N., Xu Q., Pancaroglu R., Reuveny E., Minor D. L., Jr. (2005) Neuron 47, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J., Seebohm G., Sanguinetti M. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12461–12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Treptow W., Klein M. L. (2010) J. Am. Chem. Soc. 132, 8145–8151 [DOI] [PubMed] [Google Scholar]