Abstract
The cancer/testis antigens (CTAs) are an important group of heterogeneous proteins that are predominantly expressed in the testis in the normal human adult but are aberrantly expressed in several types of cancers. Prostate-associated gene 4 (PAGE4) is a member of the CT-X family of CTAs that in addition to testis, is highly expressed in the fetal prostate, and may also play an important role both in benign and malignant prostate diseases. However, the function of this gene remains poorly understood. Here, we show that PAGE4 is a highly (100%) intrinsically disordered protein (IDP). The primary protein sequence conforms to the features of a typical IDP sequence and the secondary structure prediction algorithm metaPrDOS strongly supported this prediction. Furthermore, SDS-gel electrophoresis and analytical size exclusion chromatography of the recombinant protein revealed an anomalous behavior characteristic of IDPs. UV circular dichroism (CD) and NMR spectroscopy confirmed that PAGE4 is indeed a highly disordered protein. In further bioinformatic analysis, the PredictNLS algorithm uncovered a potential nuclear localization signal, whereas the algorithm DBS-Pred returned a 99.1% probability that PAGE4 is a DNA-binding protein. Consistent with this prediction, biochemical experiments showed that PAGE4 preferentially binds a GC-rich sequence. Silencing PAGE4 expression induced cell death via apoptosis and in mice carrying PCa xenografts, siRNA-mediated knockdown of the PAGE4 mRNA attenuated tumor growth in vivo. Furthermore, overexpressing PAGE4 protected cells from stress-induced death. To our knowledge, PAGE4 is the first example of a CTA that is an IDP with an anti-apoptotic function.
Keywords: Circular Dichroism (CD), DNA-binding Protein, NMR, Protein Structure, Protein-Protein Interactions, Cancer/Testis Antigen, Intrinsically Disordered Protein, Prostate Cancer, Prostate-associated Gene 4, Stress-response Protein
Introduction
Contrary to the prevailing notion that most proteins are globular with rigid three-dimensional conformations, many biologically active proteins lack stable three-dimensional structures under physiological conditions in vitro. Such proteins, referred to as intrinsically disordered proteins (IDPs),3 are highly abundant in nature and possess a number of unique structural properties such as high stability when exposed to low pH and high temperature, and structural indifference toward unfolding by strong denaturants (1).
Physiologically, IDPs are involved in signaling, and transcriptional regulation, where binding to multiple partners and high specificity/low affinity interactions play a crucial role. They often function by molecular recognition, in which the disordered segments are used for partner recognition (2). The binding segment undergoes induced folding to attain an ordered structure upon binding to a biological target (coupled folding and binding), and in addition, IDPs may structurally adapt to different partners with different functional outcomes, resulting in promiscuity in function termed “moonlighting” (3). Indeed, the overexpression of IDPs is often associated with human diseases, including cancer, cardiovascular disease, amyloidoses, neurodegenerative diseases, and diabetes and thus, the IDPs represent attractive targets for drugs modulating protein-protein interactions (4).
The cancer/testis antigens (CTAs) are an important group of proteins that are predominantly expressed in the testis but aberrantly expressed in several types of cancers (5). Because of their restricted expression in an immune-privileged organ, the CTAs represent attractive immunotherapy targets. Indeed, autoantibodies against several CTAs have been detected in cancer patients and often such patients present with better prognosis (6).
Broadly speaking, the CTAs can be divided into two groups: the CT-X antigens located on the X chromosome and non-X CT antigens located on various autosomes. The non-X CTAs are fairly well conserved with known functions including, transformation (7–9), chromatin remodeling (10–12), transcriptional regulation (13–16), and signaling (17, 18) to name a few. In contrast, the CT-X antigens appear to have evolved fairly recently and most lack orthologues in lower mammals except primates (19). Furthermore, members of the CT-X antigens in particular are typically associated with advanced cancer with poorer outcomes (20–25).
PAGE4 is a member of the PAGE family of CT-X antigens that consists of several members that map to Xp11.21 on the X chromosome. However, unlike a typical CT-X antigen that is predominantly expressed only in the testis, PAGE4 appears to be a developmental gene that is highly up-regulated in the fetal prostate but is present at basal levels in the adult gland, both at the protein4 and mRNA levels. However, PAGE4 is “reactivated” in both malignant (26) and symptomatic but not asymptomatic benign prostatic diseases (27). Despite these dynamic expression patterns that parallel the proliferative growth of the prostate gland during development and disease, the function of PAGE4 remains poorly understood.
EXPERIMENTAL PROCEDURES
Bioinformatics Predictions
Disorder in the PAGE4 structure was predicted using FoldIndex (28), RONN (29), and metaPrDOS (30). Nuclear localization signal (NLS) was predicted using the PredictNLS algorithm (31) and DNA binding was predicted using the algorithm DBS-Pred (32).
Recombinant Protein Production
PAGE4 cDNA was obtained by PCR using a cDNA clone (Origene, Rockville, MD) as template and two gene-specific forward and reverse primers containing NdeI and EcoRI restriction sites, respectively. Following restriction digestion, the cDNA was cloned into the pET28a vector at the NdeI/EcoRI cloning sites resulting in its in-frame fusion with an N-terminal His tag, and transfected into the TOP10 Escherichia coli cells (Invitrogen). Expression of the recombinant protein was induced with isopropyl 1-thio-β-d-galactopyranoside and protein was purified over a nickel column from the supernatant obtained after bacterial lysis. The eluted protein was digested with thrombin (EMD Biosciences, San Diego, CA) to remove the His tag and again passed through a nickel column to capture the His tag.
SDS-PAGE and Immunoblotting
The eluate was dialyzed against PBS. Purified recombinant PAGE4 protein was analyzed on a NuPAGE 4–12% BisTris gel (Invitrogen) under reducing and denaturing conditions and stained with Coomassie Blue. Immunoblotting was done as described previously (33). Total cellular lysate (50 μg of protein) from LNCaP cells was used to detect endogenous PAGE4. The polyclonal antibody was the same as described previously (25) and was used at a 1:2,500 dilution overnight followed by incubation with secondary antibody at 1:50,000 dilution for 1 h.
Analytical Size Exclusion Chromatography
Samples were injected into a Superdex 200 10/30 AKTA FPLC column (GE Healthcare) using PBS as running buffer. Prior to PAGE4 chromatography, 200 μg each of ovalbumin and ribonuclease A were injected to serve as standards.
Biophysical Measurements
CD spectra were recorded at the following temperatures: 5, 10, 15, 20, and 25 °C. A protein concentration of 0.33 mg/ml was used in 10 mm sodium phosphate buffer containing 150 mm sodium chloride, pH 7.4. CD measurements were made on a JASCO spectropolarimeter using a 0.1-cm path length. One-dimensional 1H NMR spectra of PAGE4 as a function of temperature were recorded on a Bruker DMX-600 spectrometer using a protein concentration of 1 mg/ml and the same buffer conditions as above. A water flip-back pulse sequence was used for solvent suppression.
DNA Binding Studies
To determine whether PAGE4 binds DNA and to identify the sequence to which it may preferentially bind, we employed a technique we developed previously (34) with some modifications. A “library” of 10-mer dsDNA binding sites was constructed using a synthetic oligonucleotide 5′-CGAGGTCGACGGTATCGNNNNNNNNNNGGATCCACTAGTTCTAGAGC-3′ that was converted to dsDNA by PCR. PCR primers had the following sequences: P1, 5′-CGAGGTCGACGGTATCG-3′; and P2, 5′-GCTCTAGAACTAGTGGATC-3′. PCR was performed with GoTaq® Green Master Mix (Promega, Madison, WI). PCR cycling conditions were 95 °C for 30 s, then 95 °C for 10 s, 62 °C for 30 s, and 72 °C for 1 s for 35 cycles. The PAGE4 protein was cloned into a FLAG-tagged-pCMV6 (Origene, Rockville, MD), transiently expressed in HEK293T cells (35), and purified by immunoprecipitation (36) with FLAG-agarose beads (Sigma), using Frackelton buffer (10 mm Tris-HCl, 30 mm Na4P2O7, pH 7.1, 50 mm NaCl, 50 mm NaF, HaltTM Protease Inhibitor Mixture (Thermo Scientific, Rockford, IL)), containing 10 pmol of the dsDNA oligonucleotide library. The agarose beads were washed 5 times with Tris-buffered saline (TBS) and the DNA that co-immunoprecipitated was extracted with a QIAEX II kit (Qiagen, Gaithersburg, MD) and amplified with P1/P2 primers as described above. The resulting PCR product was purified on a 2% agarose gel and used in a new round of selection with PAGE4 protein. After the third round of selection, the PCR product was cloned into pCR NT/T7 TOPO TA vector (Invitrogen), plated on ampicillin-containing agar plates. Twenty-five clones were randomly selected to determine the DNA sequence of the binding sites.
Cell Culture
Prostate cancer cell lines were obtained from the American Type Culture Collection (Rockville, MD). Cells were cultured at 37 °C in routine RPMI (Invitrogen) medium supplemented with 10% fetal bovine serum (FBS) with humidified air and 5% carbon dioxide. The LNCaP-96 cells used in this study were those established at Johns Hopkins University by Pflug et al. (37). They were routinely cultured in phenol-red free RPMI supplemented with 10% charcoal-stripped FBS (HyClone, Logan, UT).
In Vitro Studies
PAGE4 ON-TARGETplus SMARTpool siRNA were obtained from Dharmacon (Lafayette, CO). Approximately 5 × 103 cells were transfected with siRNAs at 50 nm total concentration using 0.2 μl of DharmaFECT-3 of the transfection reagent and plated in a 96-well plate. Drug treatment was applied 24–48 h after transfection.
RNA Isolation and Q-RT PCR
Total RNA was isolated using the RNeasy mini kit (Qiagen, Valencia, CA) following the suppliers protocol. RNA samples were treated with DNase I (Invitrogen), and cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was done using an iCycler iQ Multicolor Real-time PCR Detection system (Bio-Rad). PCR was done with RNA samples from a minimum of three independent experiments and each PCR was set up in triplicate. Target gene expression was compared with the housekeeping gene TATA box-binding protein (TBP) for normalization. The sequences of the PCR primers used were: TBP forward, GAATATAATCCCAAGCGGTTTG; TBP reverse, ACTTCACATCACAGCTCCCC; and PAGE4 forward, CGTAAAGTAGAAGGTGATTG, PAGE4 reverse, ATGCTTAGGATTAGGTGGAG. PCR cycling conditions were 95 °C for 5 min, then 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s for 40 cycles.
Cell Viability and Inhibition Assay
Approximately 1 × 104 cells per well were seeded in 24-well plates. After growing for 1–7 days, the cell proliferation reagent WST-1 (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate) was added to each well, as specified by the supplier (Roche Applied Science). After 4 h of incubation, WST-1 absorbance was measured at 450 nm. The cells were also stained with 0.05% crystal violet plus 20% methanol. In some experiments, 5 × 103 cells per well were seeded in 96-well plates. Seventy-two h after drug treatment, cell viability was analyzed by the WST-1 assay. Alternately, cell viability was evaluated by cell counting using a hemocytometer after excluding dead cells by trypan blue staining. All cell viability and inhibition experiments were biological replicates repeated at least three times.
Flow Cytometry
Apoptosis and cell cycle were detected on a Guva system (Guava Technologies, Hayward, CA) using the Terminal Transferase dUTP Nick End Labeling (TUNEL) kit (Guava Technologies, Billerica, MA) and Guava Cell Cycle Reagent (Guava Technologies), respectively.
Immunofluorescence
Twenty-five thousand cells were seeded in chamber slides. Seventy-two h after siRNA treatment, cells were washed with PBS and fixed by 4% paraformalin, and then incubated with phospho-histone H2A.X (γ-H2A.X) antibody (Millipore, Billerica, MA) at room temperature for 1 h followed by FITC-conjugated secondary antibodies. Slides were observed with a Nikon Eclipse TE2000E using the GFP-BP filter (Ex 460–500, DM 5005, DA:510–560) and analyzed with NIS Elements AR 3.00 Software.
PAGE4 Overexpression
The entry vector containing the PAGE4 open reading frame with C-terminal fusion of the DDK tag, pCMV6-Entry, and destination vector pCMV6-AC-GFP were purchased from Origene (Rockville, MD). The expression vector pCMV6-PAGE4-GFP was generated by first digesting pCMV6-Entry with SgfI and MluI digestion to extract the PAGE4 cDNA followed by in-frame ligation with destination vector pCMV6-AC-GFP. Sequences of all constructs were verified by SeqWright DNA Technology Services (Houston, TX). These constructs were expressed in CWR22rv1 or HEK-293T cells by transfection with FuGENE HD (Roche Applied Science).
In Vivo Treatments
For creating xenografts, ∼2 × 106 LNCaP cells suspended in 75 μl of PBS were mixed with an equal volume of Matrigel and inoculated in the flank of 6-week-old SCID mice (Harlan Sprague-Dawley). When tumor volumes reached 200 mm3, mice were divided into 3 groups with 7 animals per group and administered PAGE4 siRNA, scrambled siRNA, or PBS treatment. siRNA was delivered using the AteloGeneTM in vivo siRNA transfection kit (KOKEN, Japan), and administrated intratumorally following the suppliers protocol every 5 days for a total of 3 times. Tumor growth was measured every 3 days and the relative tumor volumes were calculated as described (38). Two days after the last treatment of siRNA, 2 mice in each group were sacrificed under ether anesthesia, and tumors were dissected and frozen at −80 °C in RNAlater until further use. All treatment protocols were approved by the animal care and use committee of the Johns Hopkins University.
Statistical Analysis
Statistical comparisons were made using the Student's t test. Two-sided p < 0.05 was considered significant. All experiments were biological replicates repeated a minimum of three times except in the case of the in vivo experiments where they were done twice.
RESULTS AND DISCUSSION
Predicting Disorder in PAGE4
Based on the relationships among intrinsic disorder, sequence complexity as measured by Shannon's entropy, and amino acid composition, IDPs are typically thought to display a bias toward polar amino acids and a bias away from non-polar amino acids (39). Indeed, as shown in Fig. 1A, whereas the PAGE4 protein sequence was enriched in polar and charged amino acids such as Gln, Ser, Pro, Glu, and Lys, there was a distinct lack of bulky hydrophobic residues such as Leu, Met, Phe, Trp, and Tyr. Thus, whereas the charged residues constitute ∼53% of the total protein, the hydrophobic residues make up <10% suggesting that PAGE4 may represent an IDP (supplemental Table S1).
FIGURE 1.
Bioinformatic predictions of disorder in the PAGE4 protein structure. A, amino acid sequence of PAGE4. Polar residues are indicated in green and non-polar residues are in red. The putative NLS and DNA-binding region are indicated by asterisks and underlines, respectively. Disorder in the PAGE4 protein was predicted by FoldIndex (B), RONN (C), and metaPrDOS (D) as described under “Experimental Procedures.”
To further investigate this possibility, we applied two different protein secondary structure prediction algorithms namely, FoldIndex and Regional Order Neural Network (RONN) to discern disorder. FoldIndex developed by Prilusky et al. (28) implements the algorithm described by Uversky et al. (40) to make a calculation based on average net charge and average hydrophobicity of the sequence to predict whether a given sequence is ordered or disordered. In contrast, RONN uses a neural network technique to predict whether any given residue is likely to be ordered or disordered in the context of the surrounding amino acid sequence. Although the physical properties of amino acids are the fundamental basis in determining disorder, the neural network used in RONN avoids explicit parameterization of amino acids in this way. Instead it uses non-gapped sequence alignment to measure “distances” between windows of sequence for the unknown protein and windowed sequences for known folded proteins derived from the Protein Data base (29). However, both FoldIndex and RONN predicted 100% disorder in the PAGE4 protein structure (Fig. 1, B and C, respectively).
To rule out any bias in the algorithms, we also used an additional prediction method namely, metaPrDOS. MetaPrDOS uses a meta approach that does not predict disordered regions from amino acid sequence directly but predicts them by integrating the results of eight different prediction methods (30). Consistent with the previous two algorithms, metaPrDOS also predicted 100% disorder in the PAGE4 protein structure (Fig. 1D). Taken together, these data provided good evidence that PAGE4 is an IDP.
Anomalous Behavior of PAGE4 Protein
To experimentally confirm the extent of disorder, a recombinant form of PAGE4 was produced in bacteria with an estimated molecular weight of 11,153 calculated from the conceptual ORF. However, its mobility in SDS-PAGE suggested a size of 25 kDa (Fig. 2A). Size exclusion chromatography under non-denaturing conditions indicated an apparent molecular mass of 26 kDa and the presence of another species of ∼44 kDa suggesting potential dimerization (Fig. 2, C and D). Such anomalous mobility of IDPs is not uncommon (2). In SDS-PAGE, their mobility appears to be retarded because of abnormally low SDS binding to hydrophilic residues in the IDPs and, in size exclusion columns, in contrast to the behavior of globular proteins, they present extended conformations resulting in a larger than expected Stokes radius. Anomalous behavior of the endogenous PAGE4 protein as well as the purified recombinant protein was also confirmed by Western blotting with an anti-PAGE4-specific antibody. As shown in Fig. 2B, the endogenous protein exhibited a molecular mass of ∼30 kDa, whereas that of the recombinant protein was ∼25 kDa. The increase in the weight of the endogenous protein presumably reflects some kind of post-translational modification(s).
FIGURE 2.
Anomalous behavior of PAGE4 protein in gel electrophoresis and analytical gel filtration chromatography. A, SDS-PAGE of recombinant PAGE4 synthesized in bacteria. B, immunoblot showing anomalous migration of the endogenous PAGE4 protein obtained from LNCaP cells (lane 1, 50 μg) and 5 μg of recombinant PAGE4 (lane 2) using a anti-PAGE4-specific antibody. C and D, analytical size exclusion chromatography. Absorptions at 280 and 220 nm were monitored as a function of retention volume and plotted with units shown on the left and right axis, respectively.
Biophysical Studies Confirm That PAGE4 Is an IDP
The structural properties of PAGE4 in aqueous solution were characterized qualitatively using circular dichroism (CD) and NMR spectroscopy. CD spectra were recorded as a function of temperature from 5 to 25 °C (Fig. 3A). These spectra showed that the PAGE4 polypeptide chain contains no significant α-helical or β-strand secondary structural elements over this temperature range as evidenced by low ellipticity values in the 215–230 nm region. One-dimensional 1H NMR spectra displayed a narrow chemical shift dispersion of amide proton signals, with most resonances in the 7.0–8.5 ppm range (Fig. 3B). Narrow dispersion of peaks was also present in the aliphatic methyl region of the spectrum (data not shown). Resonances in the amide and methyl regions of folded proteins are more spread out due to ordered packing effects. Thus, the lack of signal dispersion is strongly indicative of a polypeptide chain with little or no defined tertiary structure. Taken together, the CD and NMR data show that PAGE4 does not have detectable secondary and tertiary structure, consistent with an IDP.
FIGURE 3.
Biophysical characterization of PAGE4 disorder. A, circular dichroism spectra of PAGE4 as a function of temperature. CD spectra were recorded at the following temperatures: 5 (cyan), 10 (orange), 15 (green), 20 (red), and 25 °C (blue). A protein concentration of 0.33 mg/ml was used in 10 mm sodium phosphate buffer containing 150 mm sodium chloride, pH 7.4. CD measurements were made using a 0.1-cm path length. B, one-dimensional 1H NMR spectra of PAGE4 as a function of temperature. The amide proton region is shown. Spectra were recorded on a Bruker DMX-600 using a protein concentration of 1 mg/ml and the same buffer conditions as above. A water flip-back pulse sequence was used for solvent suppression.
PAGE4 Contains a NLS and Is a Putative DNA-binding Protein
The PredictNLS algorithm (31) identified a NLS in the N terminus of PAGE4 (Fig. 1A, underlined) suggesting that PAGE4 can potentially translocate into the nucleus, and the algorithm DBS-Pred (32) returned a 99.1% probability that PAGE4 is a DNA-binding protein. Furthermore, the DNA binding region precisely overlapped with the NLS (Fig. 1A, Arg residues are indicated by asterisks above the underline).
To confirm the DBS-Pred prediction that PAGE4 indeed binds DNA, we challenged immunopurified PAGE4 protein with a library of 10-mer DNA binding sites as described under “Experimental Procedures.” Indeed, as shown in Fig. 4A, whereas there was no “selection” of a dsDNA binding motif with the GFP protein (lane 1), we observed enrichment of PAGE4 preferential binding sites when the PAGE4-FLAG protein was present (lane 3) as well as FLAG-p53, which was used as a positive control. DNA sequence analysis revealed that PAGE4 preferentially binds to a GC-rich sequence, 5′-GCCGCGGGG-3′ (Fig. 4). Thus, it is possible that PAGE4 is a transcription factor or a sequence-specific DNA-binding protein that is involved in aborting stress-induced apoptosis. Interestingly, in a microarray analysis, Iavarone et al. (26) observed that the expression of lipoprotein lipase, a gene frequently deleted in prostate cancer, is down-regulated in a cell line that overexpresses PAGE4. It is thus plausible that PAGE4 could transcriptionally regulate lipoprotein lipase expression in these cells. However, additional experiments are required to uncover its precise function.
FIGURE 4.
PAGE4 binds to a GC-rich DNA sequence. The DNA-binding assay using purified FLAG-tagged PAGE4 protein was done as described. A, dsDNA obtained by PCR. Lane 1, vector control; lane 2, PAGE4; and lane 3, p53 positive control. B, the GC-rich consensus sequences using the WebLogo software obtained from sequencing 33 independent clones.
Decreasing PAGE4 Attenuates Cell Survival
Up-regulation of several CTAs including PAGE4 is associated with drug resistance in cancer cells (41–44). Given the intrinsic nature of IDPs to engage in promiscuous interactions and the presence of a putative DNA-binding region in PAGE4, we hypothesized that PAGE4 may play a critical role in cancer cells by mitigating the genotoxic stress caused by cytotoxic drugs either by interacting with key proteins or by directly binding DNA (or both). To test this hypothesis, we determined the effect of down-regulating PAGE4 expression on cell proliferation. First, we determined the levels of PAGE4 mRNA in several normal and transformed prostate cells by quantitative PCR and (supplemental Fig. S1) and selected the following prostate cancer cell lines, LNCaP, LN96, CWR22rv1, an embryonic cell line, HEK293T, and the BPH cell line, BRF55, that express significant levels of PAGE4. Transfecting these cells with a PAGE4-specific siRNA significantly down-regulated (85–97.5%) PAGE4 mRNA compared with the scrambled (control) siRNA (supplemental Fig. S2A). Furthermore, by immunoblotting experiments using a FLAG-tagged construct of PAGE4, we confirmed that the knockdown resulted in decreased PAGE4 protein levels as well (supplemental Fig. S2B).
Next, the effect of silencing PAGE4 expression on proliferation of the siRNA-transfected cells was determined. As shown in Fig. 5A and supplemental Fig. S3A, a WST-1 assay demonstrated that siRNA-mediated down-regulation of PAGE4 significantly impaired the proliferation of these cells in comparison to the scrambled siRNA. The decrease in proliferation was also confirmed by staining cells with crystal violet. As shown in Fig. 5B, whereas there was no discernable decrease in crystal violet staining in cells transfected with the scrambled siRNA, there was a significant decrease in cells that were transfected with the PAGE4-specific siRNA when compared with non-transfected control cells. We also determined the effects of using a decreased dose of pooled PAGE4-specific siRNA (supplemental Fig. S3B) or the individual siRNAs from the siRNA pool (supplemental Fig. S3C). Indeed, both low concentrations of the PAGE4-specific siRNA pool and individual siRNAs that target different PAGE4 mRNA sequences showed a similar effect in attenuating cell growth suggesting that they specifically target PAGE4. Furthermore, we also found that PAGE4 siRNA treatment showed little effect on the growth of PC3 and LAPC4 cells both of which have extremely low levels of PAGE4 mRNA (supplemental Fig. S3D). Taken together, the data strongly suggest that siRNAs used in the present study specifically target PAGE4 with no discernable “off-target” effects.
FIGURE 5.
Silencing PAGE4 expression inhibits cell survival and enhances chemo-cytotoxicity in prostate cancer cells. A, LNCaP, LNCaP-96, and CWR22rv1 cells were transfected with 50 nm PAGE4 SMARTpool siRNA. Cell viability was evaluated by WST-1 assay 3–7 days after transfection. B, crystal violet staining of CWR22rv1 cells 5 days after siRNA transfection. C, LNCaP cells was stained with propidium iodide 72 h after siRNA transfection and subjected to cell cycle analysis. D, apoptosis of LNCaP cells was evaluated by TUNEL assay 72 h after siRNA transfection. E, immunofluorescence staining of γ-H2A.X in LNCaP cells 72 h after siRNA transfection. Magnification is ×200 in all panels. F, the whole lysate of LNCaP cells were subjected to immunoblotting for p21, phospho-p53 (Ser15) (pS-p53), and γ-H2A.X 72 h after siRNA transfection. Actin was used as a control for protein loading. G, LNCaP, LNCaP-96, and CWR22rv1 cells were transfected with 50 nm PAGE4 SMARTpool siRNA. Forty-eight hours after transfection, cells were treated with ADM at the indicated dose for 4 h, and then moved back to normal medium for another 16 h. Cell viability was evaluated by WST-1 assay. All experiments were biological replicates repeated a minimum of three times. Data are represented as mean ± S.D. *, p < 0.05 in comparison to scrambled-siRNA; #, p < 0.05 in comparison to normal controls without any treatment.
Silencing PAGE4 Results in Cell Death via Apoptosis
To characterize the inhibition of cell growth by PAGE4 depletion, cell cycle progression was then analyzed. Although the scrambled siRNA did not affect the cell cycle profile 3 days post-transfection, PAGE4-specific siRNA increased the percentage of cells in the G1/G0 phase by ∼20% but decreased the percentage of cells in the S phase by ∼30% (Fig. 5C), suggesting that PAGE4-depleted cells do not progress beyond this restriction checkpoint and undergo apoptosis if they failed to reenter the cell cycle. To examine whether apoptosis occurred following PAGE4 knockdown, the number of TUNEL-positive cells were determined. Indeed, TUNEL-positive cells were 5-fold higher following PAGE4 knockdown than that seen with the scrambled siRNA (Fig. 5D) confirming apoptosis. Furthermore, a significant accumulation of the variant histone γH2A.X, a hallmark of double strand breakage in cells depleted of PAGE4 with the specific siRNA (Fig. 5E) but not in scrambled siRNA-transfected cells provided good evidence that in the absence of PAGE4, the arrested cells undergo apoptosis.
To gain further insight into the mechanism of apoptosis following suppression of PAGE4 expression, immunoblotting analyses were performed for p21, phosphorylated p53 as well as γH2A.X. Phosphorylation of γH2A.X at serine 139 correlates well with DNA damage and protein p21 is a potent cyclin-dependent kinase inhibitor that binds to and inhibits the activity of cyclin-CDK2 or -CDK4 complexes acting as a regulator of cell cycle progression at G1. The expression of this gene is tightly controlled by the tumor suppressor protein p53, through which it mediates the p53-dependent cell cycle G1 phase arrest in response to a variety of stress stimuli. As shown in Fig. 5F, there was a significant increase in the levels of p21 and phospho-p53 upon PAGE4 suppression with a concomitant increase in the accumulation of γH2A.X suggesting that PAGE4 plays roles in protecting against stress and anti-apoptosis.
Silencing PAGE4 Expression Enhances Sensitivity of Prostate Cancer Cells to Adriamycin
Because silencing PAGE4 expression alone resulted in significant cell death due to DNA damage, a combined effect of siRNA-mediated knockdown and cytotoxic drug treatment was evaluated. PCa cell lines that were depleted of PAGE4 expression were treated with various concentrations of adriamycin, a commonly used cytotoxic drug used to treat cancer. As shown in Fig. 5G, we observed a significant effect in cell survival. This effect was most pronounced in LNCaP cells wherein, >90% of the cells were killed at the highest drug dose compared with control siRNA transfected or mock-transfected controls.
Knocking Down PAGE4 Attenuates Tumor Growth in Vivo
To determine the in vivo effect of silencing PAGE4, we determined the effect of PAGE4 down-regulation on the growth of PCa xenografts in SCID mice. After the tumors were allowed to develop for 4 weeks, siRNA was administered intratumorally as described under “Experimental Procedures.” Tumors that received either PBS without any siRNA or those that included the scrambled siRNA continued to grow, with tumor volume reaching 8 × 102 mm3 (Fig. 6A). In contrast, there was significant impairment in the growth of tumors that received PAGE4-specific siRNA. To confirm that the decrease in tumor volume was indeed due to PAGE4 silencing, PAGE4 mRNA levels in the tumors recovered from the animals 2 days after the last administration of siRNA were determined by quantitative PCR. As shown in Fig. 6B, there was a dramatic decrease in PAGE4 mRNA in the tumor samples from animals treated with the specific siRNA compared with the scrambled control. Taken together, these in vitro and in vivo data suggest that PAGE4 may play an essential role in prostate cell survival.
FIGURE 6.
PAGE4 siRNA administration inhibits cell growth of LNCaP xenografts in SCID mice. A, SCID mice (seven in each group) were subcutaneously injected with 2 × 106 LNCaP cells suspended in PBS and Matrigel, and treated with PAGE4 siRNA by local injection every 5 days for 3 times. Mean relative tumor volume of each group was plotted over time. B, two mice in each group were sacrificed 2 days after the last administration of siRNA. Tumor tissues were removed and subjected to RT-PCR to evaluate PAGE4 mRNA expression. Data are represented as mean ± S.D. *, p < 0.05 in comparison to scrambled siRNA treatment; #, p < 0.05 in comparison to PBS treatment.
Overexpressing PAGE4 Protects Cell Death
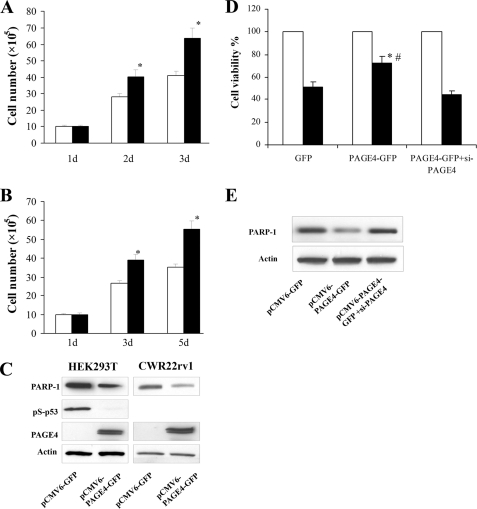
To elucidate the effect of PAGE4 up-regulation under stress, we determined the viability of CWR22rv1 and HEK-293T cells that were transiently transfected with either a PAGE4-green fluorescent protein (PAGE4-GFP) construct or the empty vector carrying only the GFP cDNA. In agreement with previous knockdown experiments in that down-regulating PAGE4 levels decreased cell survival, overexpression of PAGE4 significantly increased cell viability in CWR22rv1 and HEK-293T cells following transient transfection (Fig. 7A). Furthermore, overexpression of PAGE4 blocked the increase of poly(ADP-ribose) polymerase (PARP-1) protein in the transfected cells, as well as activation of p53 in HEK-293T cells, both of which indicate a stress-response suppression (18, 20) (Fig. 7B). To further evaluate the stress protection of PAGE4 overexpression, we treated cells with adriamycin. As expected, treating the empty vector-transfected cells with the drug decreased their survival by 50%, whereas in those transfected with the PAGE4 construct, cell viability was decreased by 25% indicating a 50% increase in survival. In contrast, when PAGE4 expression was silenced using an siRNA, survival dropped to 50% of the empty vector controls (Fig. 7C). At the same time, the decrease of PARP-1 following drug treatment in PAGE4 overexpressed cells was also reversed by siRNA treatment suggesting that the stress protective status evidenced by the reduced PARP-1 level in these cells is likely due to the enhanced expression of PAGE4 (Fig. 7D). Taken together, the result provides good evidence that PAGE4 favors cell survival and protects cytotoxic stress in cells that typically overexpress it.
FIGURE 7.
Overexpression of PAGE4 protected cells from stress-induced death. HEK293T (A) and CWR22rv1 (B) cells were seeded in 6-well plates and transfected with 4 μg of pCMV6-PAGE4-GFP vector (■) or empty vector pCMV6-GFP (□). The number of living cells was counted using a hemocytometer after excluding dead cells by trypan blue staining 1–5 days after transfection. C, Western blot for PARP-1, pS-p53, and PAGE4 protein in the cell lysate of the two cell lines 72 h after transfection. Actin was blotted as loading control. D, CWR22rv1 cells were seeded in 12-well plates and transfected with pCMV6-GFP, pCMV6-PAGE4-GFP, or pCMV6-PAGE4-GFP followed by 50 nm PAGE4 siRNA. Forty-eight hours after transfection, cells were treated with 1 μg/ml of ADM for 4 h, and then moved back to normal medium for another 19 h. Cell viability was evaluated by WST-1 assay. ■, ADM treatment; □, normal medium control. *, p < 0.05 as compared with GFP-transfected cells; #, p < 0.05 as compared with PAGE4-GFP plus PAGE4 siRNA-transfected cells. E, cells were lysed 24 h after ADM treatment and subjected to Western blot for PARP-1. Data are represented as mean ± S.D. All experiments were biological replicates repeated a minimum of three times.
CONCLUSION
To our knowledge, the present report is the first example of a CTA that is an IDP. The inherent nature of IDPs to engage in promiscuous interactions when present in increased concentrations is thought to be the mechanism underlying their toxic/pathologic effect (45). Furthermore, increasing disorder can also tune the binding affinity of an IDP to maximize the specificity of promiscuous interactions (46).
Recently, Liu et al. (46) applying thermodynamic models of protein interactions in which IDPs are characterized by positive folding free energies presented a quantitative theory predicting the role of intrinsic disorder in protein structure and function. The authors predicted that both catalytic and low-affinity binding (Kd ≥ 10−7 m) proteins prefer ordered structures, whereas only high-affinity binding proteins (found mostly in eukaryotes) can tolerate disorder. Given the remarkable degree of disorder in PAGE4, it is possible that by initiating promiscuous interactions, PAGE4 may play a critical role in the disease process. Future experiments that identify the interacting proteins should permit further biophysical studies of the physiologically relevant conformations accessed by PAGE4.
Acknowledgments
We thank Prof. Pamela Bjorkman, Max Delbruck Professor of Biology, California Institute of Technology, for support and interest in this project and Prof. Gaetano Montelione, Department of Molecular Biology and Biochemistry, Rutgers University, for pointing out that PAGE4 may be an IDP.
This work was supported, in whole or in part, by National Institutes of Health Grant SPORE 2P50CA058236-16 from the NCI, O'Brien Grant P50DK082998 from the NIDDK, and the Patrick C. Walsh Prostate Cancer Research Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
R. H. Getzenberg, unpublished data.
- IDP
- intrinsically disordered protein
- CTA
- cancer/testis antigen
- PAGE4
- prostate-associated gene 4
- NLS
- nuclear localization signal
- RONN
- regional order neural network
- WST-1
- 4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate
- PARP-1
- poly(ADP-ribose) polymerase 1
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Uversky V. N. (2010) J. Biomed. Biotechnol. 568–068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hazy E., Tompa P. (2009) ChemPhysChem 10, 1415–1419 [DOI] [PubMed] [Google Scholar]
- 3. Tompa P., Szász C., Buday L. (2005) Trends Biochem. Sci. 30, 484–489 [DOI] [PubMed] [Google Scholar]
- 4. Uversky V. N., Oldfield C. J., Dunker A. K. (2008) Annu. Rev. Biophys. 37, 215–246 [DOI] [PubMed] [Google Scholar]
- 5. Scanlan M. J., Simpson A. J., Old L. J. (2004) Cancer Immun. 4, 1. [PubMed] [Google Scholar]
- 6. Scanlan M. J., Gure A. O., Jungbluth A. A., Old L. J., Chen Y. T. (2002) Immunol. Rev. 188, 22–32 [DOI] [PubMed] [Google Scholar]
- 7. Lee J. H., Schütte D., Wulf G., Füzesi L., Radzun H. J., Schweyer S., Engel W., Nayernia K. (2006) Hum. Mol. Genet. 15, 201–211 [DOI] [PubMed] [Google Scholar]
- 8. Kanehira M., Katagiri T., Shimo A., Takata R., Shuin T., Miki T., Fujioka T., Nakamura Y. (2007) Cancer Res. 67, 3276–3285 [DOI] [PubMed] [Google Scholar]
- 9. Por E., Byun H. J., Lee E. J., Lim J. H., Jung S. Y., Park I., Kim Y. M., Jeoung D. I., Lee H. (2010) J. Biol. Chem. 285, 14475–14485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y. T., Venditti C. A., Theiler G., Stevenson B. J., Iseli C., Gure A. O., Jongeneel C. V., Old L. J., Simpson A. J. (2005) Cancer Immun. 5, 9. [PubMed] [Google Scholar]
- 11. Barrett A., Santangelo S., Tan K., Catchpole S., Roberts K., Spencer-Dene B., Hall D., Scibetta A., Burchell J., Verdin E., Freemont P., Taylor-Papadimitriou J. (2007) Int. J. Cancer 121, 265–275 [DOI] [PubMed] [Google Scholar]
- 12. Morinière J., Rousseaux S., Steuerwald U., Soler-López M., Curtet S., Vitte A. L., Govin J., Gaucher J., Sadoul K., Hart D. J., Krijgsveld J., Khochbin S., Müller C. W., Petosa C. (2009) Nature 461, 664–668 [DOI] [PubMed] [Google Scholar]
- 13. Simpson A. J., Caballero O. L., Jungbluth A., Chen Y. T., Old L. J. (2005) Nat. Rev. Cancer 5, 615–625 [DOI] [PubMed] [Google Scholar]
- 14. Bai S., He B., Wilson E. M. (2005) Mol. Cell. Biol. 25, 1238–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sargeant J. K., Adey T., McGregor F., Pearce P., Quinn D., Milev R., Renaud S., Skakum K., Dada N. (2010) Can. J. Psychiatry 55, 1–20 [PubMed] [Google Scholar]
- 16. McKenzie L., King S., Marcar L., Nicol S., Dias S. S., Schumm K., Robertson P., Bourdon J. C., Perkins N., Fuller-Pace F., Meek D. W. (2010) Cell Cycle 9, 4200–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bai S., Wilson E. M. (2008) Mol. Cell. Biol. 28, 1947–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ajiro M., Katagiri T., Ueda K., Nakagawa H., Fukukawa C., Lin M. L., Park J. H., Nishidate T., Daigo Y., Nakamura Y. (2009) Int. J. Oncol. 35, 673–681 [PubMed] [Google Scholar]
- 19. Stevenson B. J., Iseli C., Panji S., Zahn-Zabal M., Hide W., Old L. J., Simpson A. J., Jongeneel C. V. (2007) BMC Genomics 8, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gure A. O., Chua R., Williamson B., Gonen M., Ferrera C. A., Gnjatic S., Ritter G., Simpson A. J., Chen Y. T., Old L. J., Altorki N. K. (2005) Clin. Cancer Res. 11, 8055–8062 [DOI] [PubMed] [Google Scholar]
- 21. Velazquez E. F., Jungbluth A. A., Yancovitz M., Gnjatic S., Adams S., O'Neill D., Zavilevich K., Albukh T., Christos P., Mazumdar M., Pavlick A., Polsky D., Shapiro R., Berman R., Spira J., Busam K., Osman I., Bhardwaj N. (2007) Cancer Immun. 7, 11. [PMC free article] [PubMed] [Google Scholar]
- 22. Andrade V. C., Vettore A. L., Felix R. S., Almeida M. S., Carvalho F., Oliveira J. S., Chauffaille M. L., Andriolo A., Caballero O. L., Zago M. A., Colleoni G. W. (2008) Cancer Immun. 8, 2. [PMC free article] [PubMed] [Google Scholar]
- 23. Napoletano C., Bellati F., Tarquini E., Tomao F., Taurino F., Spagnoli G., Rughetti A., Muzii L., Nuti M., Benedetti Panici P. (2008) Am. J. Obstet. Gynecol. 198, 99 e1–7 [DOI] [PubMed] [Google Scholar]
- 24. Suyama T., Shiraishi T., Zeng Y., Yu W., Parekh N., Vessella R. L., Luo J., Getzenberg R. H., Kulkarni P. (2010) Prostate 70, 1778–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grigoriadis A., Caballero O. L., Hoek K. S., da Silva L., Chen Y. T., Shin S. J., Jungbluth A. A., Miller L. D., Clouston D., Cebon J., Old L. J., Lakhani S. R., Simpson A. J., Neville A. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13493–13498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iavarone C., Wolfgang C., Kumar V., Duray P., Willingham M., Pastan I., Bera T. K. (2002) Mol. Cancer Ther. 1, 329–335 [PubMed] [Google Scholar]
- 27. Prakash K., Pirozzi G., Elashoff M., Munger W., Waga I., Dhir R., Kakehi Y., Getzenberg R. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7598–7603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prilusky J., Felder C. E., Zeev-Ben-Mordehai T., Rydberg E. H., Man O., Beckmann J. S., Silman I., Sussman J. L. (2005) Bioinformatics 21, 3435–3438 [DOI] [PubMed] [Google Scholar]
- 29. Yang Z. R., Thomson R., McNeil P., Esnouf R. M. (2005) Bioinformatics 21, 3369–3376 [DOI] [PubMed] [Google Scholar]
- 30. Ishida T., Kinoshita K. (2008) Bioinformatics 24, 1344–1348 [DOI] [PubMed] [Google Scholar]
- 31. Cokol M., Ozbay F., Rodriguez-Esteban R. (2008) EMBO Rep. 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ahmad S., Gromiha M. M., Sarai A. (2004) Bioinformatics 20, 477–486 [DOI] [PubMed] [Google Scholar]
- 33. Zeng Y., Kulkarni P., Inoue T., Getzenberg R. H. (2009) J. Cell. Biochem. 107, 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nallur G. N., Prakash K., Weissman S. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1184–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mooney S. M., Grande J. P., Salisbury J. L., Janknecht R. (2010) Biochemistry 49, 1–10 [DOI] [PubMed] [Google Scholar]
- 36. Mooney S. M., Goel A., D'Assoro A. B., Salisbury J. L., Janknecht R. (2010) J. Biol. Chem. 285, 30443–30452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pflug B. R., Reiter R. E., Nelson J. B. (1999) Prostate 40, 269–273 [DOI] [PubMed] [Google Scholar]
- 38. Zeng Y., Wu X. X., Fiscella M., Shimada O., Humphreys R., Albert V., Kakehi Y. (2006) Int. J. Oncol. 28, 421–430 [DOI] [PubMed] [Google Scholar]
- 39. Romero P., Obradovic Z., Li X., Garner E. C., Brown C. J., Dunker A. K. (2001) Proteins 42, 38–48 [DOI] [PubMed] [Google Scholar]
- 40. Uversky V. N., Gillespie J. R., Fink A. L. (2000) Proteins 41, 415–427 [DOI] [PubMed] [Google Scholar]
- 41. Cilensek Z. M., Yehiely F., Kular R. K., Deiss L. P. (2002) Cancer Biol. Ther. 1, 380–387 [PubMed] [Google Scholar]
- 42. Duan Z., Duan Y., Lamendola D. E., Yusuf R. Z., Naeem R., Penson R. T., Seiden M. V. (2003) Clin. Cancer Res. 9, 2778–2785 [PubMed] [Google Scholar]
- 43. Kasuga C., Nakahara Y., Ueda S., Hawkins C., Taylor M. D., Smith C. A., Rutka J. T. (2008) J. Neurosurg. Pediatr. 1, 305–313 [DOI] [PubMed] [Google Scholar]
- 44. Kim Y., Park H., Park D., Lee Y. S., Choe J., Hahn J. H., Lee H., Kim Y. M., Jeoung D. (2010) J. Biol. Chem. 285, 25957–25968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vavouri T., Semple J. I., Garcia-Verdugo R., Lehner B. (2009) Cell 138, 198–208 [DOI] [PubMed] [Google Scholar]
- 46. Liu J., Faeder J. R., Camacho C. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19819–19823 [DOI] [PMC free article] [PubMed] [Google Scholar]