Abstract
Alzheimer disease is characterized by accumulation of the β-amyloid peptide (Aβ) generated by β- and γ-secretase processing of the amyloid precursor protein (APP). The intake of the polyunsaturated fatty acid docosahexaenoic acid (DHA) has been associated with decreased amyloid deposition and a reduced risk in Alzheimer disease in several epidemiological trials; however, the exact underlying molecular mechanism remains to be elucidated. Here, we systematically investigate the effect of DHA on amyloidogenic and nonamyloidogenic APP processing and the potential cross-links to cholesterol metabolism in vivo and in vitro. DHA reduces amyloidogenic processing by decreasing β- and γ-secretase activity, whereas the expression and protein levels of BACE1 and presenilin1 remain unchanged. In addition, DHA increases protein stability of α-secretase resulting in increased nonamyloidogenic processing. Besides the known effect of DHA to decrease cholesterol de novo synthesis, we found cholesterol distribution in plasma membrane to be altered. In the presence of DHA, cholesterol shifts from raft to non-raft domains, and this is accompanied by a shift in γ-secretase activity and presenilin1 protein levels. Taken together, DHA directs amyloidogenic processing of APP toward nonamyloidogenic processing, effectively reducing Aβ release. DHA has a typical pleiotropic effect; DHA-mediated Aβ reduction is not the consequence of a single major mechanism but is the result of combined multiple effects.
Keywords: Alzheimer Disease, Amyloid, Fatty Acid, Lipid, Neurodegeneration
Introduction
Alzheimer disease (AD)4 is the most common cause of dementia among neurodegenerative diseases in the industrialized nations. Currently, about 35.6 million people worldwide are estimated to suffer from this dementia, a number expected to double in about 20 years (1). Amyloid β peptide (Aβ), a major hallmark of AD that accumulates as senile plaques in the brain, is generated by amyloidogenic processing of the amyloid precursor protein (APP) (2–4). APP is first cleaved by the aspartyl protease BACE1 (β-site APP-cleaving enzyme, β-secretase) releasing the soluble β-cleaved ectodomain (sAPPβ) (5, 6). The remaining C-terminal fragment of 99 amino acids (C99) is processed within the transmembrane domain by γ-secretase, a protein complex consisting of at least four proteins in which presenilin includes the active center, generating Aβ peptides (7–10). APP is also processed in a nonamyloidogenic pathway through α-secretase cleavage within the Aβ domain thus precluding Aβ production and releasing the ectodomain α-secreted APP (sAPPα) (11–13). Subsequent processing of the C-terminal fragment C83 by γ-secretase results in the formation of the peptide p3 (14, 15).
Evidence is mounting that alterations in dietary intake could be beneficial for preventing or treating AD (16, 17). In recent years docosahexaenoic acid (DHA), an ω-3 polyunsaturated fatty acid predominantly found in marine fish and algae, has become of major interest. DHA is an essential fatty acid, although it can be biosynthesized at a very low conversion rate from α-linolenic acid through elongation and desaturation (18). Approximately 60% of the fatty acids in neuronal cell membranes consist of DHA, which is particularly concentrated in synaptic membranes and myelin sheaths (19–21). Intriguingly, it was found that DHA is decreased in AD post-mortem brains in certain brain regions like hippocampus, white matter, frontal gray matter, and pons (22). Moreover, AD patients have reduced brain and serum DHA levels compared with age-matched nondemented controls (23), suggesting that a deficiency in this polyunsaturated fatty acid could play an important role in the development of AD. Several epidemiological studies revealed an inverted relationship between ω-3 uptake and AD incidence or cognitive decline (24–28), whereas clinical studies found no effect or only minor effects in improvement in the very early stage of the disease (29–32). These findings indicate that DHA might be more effective for prevention rather than for treatment of this disease.
Importantly, DHA has shown the efficacy to reduce Aβ peptide production in vitro and in animal models of AD (33–37, 39). To date, the molecular mechanism how DHA interferes with APP processing is not completely understood. In this study, we systematically tested the direct and indirect effects of DHA on the secretases involved in APP processing and cholesterol metabolism.
EXPERIMENTAL PROCEDURES
Animals
Twelve male C57Bl/6J mice (Charles River, Sulzfeld, Germany), aged 9 weeks at arrival, were housed in groups of six animals per cage. The animals were kept in a controlled environment, with temperature at 20–22 °C, humidity at 50–60%, and lights on between 07:00 and 19:00 h. Food and water were freely available throughout the study. All procedures concerning the mice were carried out in accordance with Dutch laws governing the use of laboratory animals.
Diets
Mice were fed for 4 weeks with one of two different diets (Research Diet Services, Wijk bij Duurstede, The Netherlands) that differed in their fatty acid composition. The control diet contained 1.9% soy oil, 0.9% coconut oil, and 2.2% corn oil. The fish oil diet contained 0.1% coconut oil, 1.9% corn oil, and 3.0% fish oil. Both diets were AIN-93 M based (40), isocaloric, and identical with respect to their protein, carbohydrate, fiber, and mineral content.
Tissue Preparation
After the 4-week supplementation period, animals were sacrificed by CO2 gas inhalation and decapitation by guillotine. Brains were rapidly removed from the skull, snap-frozen in liquid nitrogen, and stored at −80 °C until further use. Brains were cut into small pieces and homogenized with 2 ml of water with a Teflon homogenizer.
Cell Culture
SH-SY5Y cells were cultured in DMEM (Sigma) with 10% FBS (PAN Biotech, Aidenbach, Germany) and 0.1 mm (1%) minimal essential medium (Sigma); APP-transfected cells were additionally cultured in 400 μg/ml hygromycin B (PAN Biotech, Aidenbach, Germany). All cells were routinely cultivated at 37 °C in a humidified atmosphere of 5% CO2. Experiments with DHA (Cayman Chemicals, Michigan) were carried out in DMEM with 0.1% FBS and 0.1% fatty acid-free BSA (Sigma). Ethanol was adjusted to the same concentration in controls and did not exceed 0.05%. Cells were treated 24 h with DHA or ethanol-containing medium with a medium exchange after 12 h.
PS1, BACE-1, ADAM17, and HMGCR Protein Quantification
Cell extracts from SH-SY5Y cells were prepared in lysis buffer (1% Nonidet P-40, 1% Triton X-100, 10 mm Tris, 2 mm EDTA) supplemented with protease inhibitor (Roche Diagnostics). Equal amounts of protein (40–70 μg) were separated on 10–20% Tris-Tricine gels, transferred to nitrocellulose membrane, and blocked overnight in 5% nonfat milk in PBS, pH 7.5, or TBST, pH 7.5. Antibodies and dilutions used in this study include ADAM17 ab39162 (1:5000; Abcam, Cambridge, UK), presenilin1 sc-7860 (1:500; Santa Cruz Biotechnology), BACE1 B0806 (1:1000; Sigma), and HMGCR (1:500; Upstate). Quantitative densitometric analyses were performed on digitized images of blots using ImageGauge software.
Immunoprecipitation
For detection of total Aβ and sAPPα levels, equal volumes of conditioned media, adjusted to the same protein amount of the corresponding cell lysates, were immunoprecipitated with 20 μl of protein G-Sepharose (Sigma) and the antibody W02 (41). The immunoprecipitated proteins were separated on 10–20% Tris-Tricine gels (Invitrogen). Western blot (WB) analysis was performed with antibody W02 (1 μg/ml).
Quantitative Real Time Experiments
Total RNA was extracted from cells with RNeasyPlus mini kit (Qiagen, Hilden, Germany) or TRIzol reagent (Invitrogen), according to manufacturers' protocols. Reverse transcription of 2 μg of RNA was performed under the usage of high capacity cDNA reverse transcription kits. Quantitative real time PCR analysis was carried out using Fast SYBR Green Master Mix on 7500 fast real time PCR system (7500 Fast System SDS software 1.3.1; Applied Biosystems, Darmstadt, Germany). Results were normalized to β-actin gene expression. Primers were purchased from Eurofins MWG Operon (Ebersberg, Germany). Changes in gene expression were calculated using 2−(ΔΔCt) method (42). The following primer sequences were used: ADAM17, 5′-CTG TGT GCC CTA TGT CGA TG-3′ and 5′-CAG CTG GTC AAT GAA ATC CC-3′; BACE1, 5′-GCA GGG CTA CTA CGT GGA GA-3′ and 5′-TAG TAG CGA TGC AGG AAG GG-3′; PSEN1, 5′-CTC AAT TCT GAA TGC TGC CA-3′ and 5′-GGC ATG GAT GAC CTT ATA GCA-3′; HMGCR, 5′-TCT TCC ACG TGC TTG TGA CT-3′ and 5′-CGT GCA AAT CTG CTA GTG CT-3′; and β-actin, 5′-CTT CCT GGG CAT GGA GTC-3′ and 5′-AGC ACT GTG TTG GCG TAC AG-3′. To verify the results obtained by quantitative real time experiments, samples were separated on 1.5% agarose gels in TBE buffer (90 mm Tris, 90 mm boric acid, 2 mm EDTA, pH 8.0).
Preparation of Purified Membranes
Cells were washed three times using ice-cold phosphate-buffered saline (PBS) and scraped in sucrose buffer (10 mm Tris/HCl, pH 7.4, including 1 mm EDTA and 200 mm sucrose). Mouse brains were transferred into sucrose buffer. Cells and brains were both homogenized using a PotterS (Braun, Melsungen, Germany) at maximum speed (25 strokes) on ice. Protein amount was determined and adjusted according to Smith et al. (43). Samples were centrifuged at 900 relative centrifugal force for 10 min at 4 °C, and the resulting supernatant, containing postnuclear fractions, was transferred to a new tube. The supernatants were centrifuged (Optima MAX Ultracentrifuge, Beckman Coulter, Krefeld, Germany) for 75 min at 55,000 rpm and 4 °C. The resulting supernatant was discarded, and pellets were resuspended using cannulas (10 strokes each cannula) with decreasing diameter (0.6, 0.4, and 0.33 mm) in sucrose buffer.
γ- and β-Secretase Activity Assays
Detection of β- and γ-secretase activity was performed as described before (44). After addition of the γ- or β-secretase-specific substrate (Calbiochem) to the resuspended membrane pellet, the fluorescence was measured continuously at an excitation wavelength of 355 nm (bandwidth 10 nm) and emission wavelength of 440 nm (bandwidth 10 nm) for γ-secretase and at an excitation wavelength of 345 nm (bandwidth 5 nm) and emission wavelength of 500 nm (bandwidth 5 nm) for β-secretase with a fluorometer (Tecan Safire2). The slope of the linear range correlates with γ- and β-secretase activity, respectively.
HMGCR Activity Assay
The HMGCR activity was measured with a commercially available activity kit in accordance to the manufacturer's protocol (Sigma). Briefly, the decrease in absorbance at 340 nm, which represents the oxidation of NADPH by the catalytic subunit of HMGCR in the presence of the substrate HMG-CoA, was detected.
Lipid Raft Preparation
Lipid raft preparation was performed according to Cordy et al. (45) with slight modifications. Briefly, cells were homogenized in MES-buffered saline (MBS: 25 mm MES, pH 6.5, 150 mm NaCl) containing 0.1% Triton X-100 and adjusted to a protein amount of 3 mg/ml. 1.2 ml of MBS, 90% sucrose, 0.1% Triton X-100 was added to 1.2 ml of homogenate followed by a second sucrose layer with 4 ml of 35% sucrose in MBS, 0.1% Triton X-100. Finally, 4.5 ml of 5% sucrose in MBS, 0.1% Triton X-100 was added on top, and buoyant density centrifugation was performed at 35,000 rpm (SW40 rotor, Beckman ultracentrifuge) for 18 h at 4 °C. Fractions of 650 μl were collected and tested by WB analysis for the presence of the lipid raft marker flotillin (610821, 1:250, BD Biosciences) and cadherin (ab6528, 1:1000, Abcam, Cambridge, UK) as well as for presenilin1 (sc-7860, Santa Cruz Biotechnology) and BACE1 (B0806, Sigma) protein levels. Cholesterol levels and γ-and β-secretase activity were measured directly from the fractions.
Iodixanol Gradient Centrifugation
To detect BACE1 in EEA1-positive fractions, cell homogenate was fractionated using an OptiPrep gradient according to Woods et al. (46) with minor modifications. Briefly, cells were washed three times with PBS at 4 °C and scraped off in homogenization buffer (0.25 m sucrose, 140 mm NaCl, 1 mm EDTA, 20 mm Tris/HCl, pH 8.0). Cells were homogenized using a PotterS (B. Braun, Melsungen, Germany) for 20 strokes at 1000 rpm and 4 °C. Protein amount was quantified according to Smith et al. (43). OptiPrep (Progen Biotechnik GmbH, Heidelberg, Germany) gradient was made using 10, 20, 30, and 40% of OptiPrep (OptiPrep stock 60%; diluent buffer: 0.25 m sucrose, 140 mm NaCl, 3 mm EDTA, 60 mm Tris/HCl, pH 8.0). The resulting discontinuous gradient was sealed and carefully rotated to a horizontal position to allow the single layers to diffuse and even out the density discontinuities. 5 mg/ml protein was layered on top of each gradient and centrifuged at 48,000 × g for 18 h at 4 °C (SW41 rotor, Beckman ultracentrifuge). Fractions of 600 μl were collected from top of the gradient and subjected to WB analysis. 20 μl of each fraction were separated using 10–20% Tricine gels (Invitrogen). To detect EEA1-positive fractions, blots were blocked overnight using 10% milk/PBS at 4 °C. EEA1 antibody ab2900 (Abcam, Cambridge, UK; 1:1000 in 5% milk/PBS) was used for 1.5 h at room temperature and detected with anti-rabbit antibody (Promega, Mannheim, Germany; 1:10,000 in 5% milk/PBS, 1 h at room temperature). EEA1-positive fractions (see text) were pooled and again separated on 10–20% Tricine gels (Invitrogen). BACE1 and cadherin detections were performed as described above.
Cholesterol de Novo Synthesis
SH-SY5Y-wt cells were treated with DHA for 24 h following a 6-h incubation with 0.4 μCi/ml [14C]acetate (PerkinElmer Life Sciences) and DHA. Lipids were extracted by a modified method according to Bligh and Dyer (47). Briefly, the cells were homogenized in a buffer containing 8.1 mm Na2HPO4, 1.5 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl and adjusted to a protein amount of 1 mg/ml. 3.75 ml of chloroform/methanol/HCl (1:2:0,06) was added to the sample and vortexed for 1 h. Then 1.25 ml of chloroform was added, and the sample was vortexed for another hour. After addition of 1.25 ml of water and 10 min of mixing, the sample was centrifuged for 15 min at 4700 rpm. The lower organic phase was collected, and 2 ml of chloroform was added to the aqueous phase. The sample was vortexed again for 1 h and centrifuged for 15 min at 4700 rpm. The lower phase was mixed to the former lower phase and evaporated in a N2 atmosphere at 30 °C. The resulting lipid phase was evaporated under a gentle stream of nitrogen and resolved in 200 μl of CHCl3 and 2.5 ml of scintillation solution Ultima Gold (Packard Instruments, Meriden, CT). Incorporation of the radioactive precursor into cholesterol was determined using a scintillation counter (PerkinElmer Life Sciences).
Cholesterol Determination
Cholesterol levels were determined according to the manufacturer's protocol of the Amplex Red® cholesterol assay (Invitrogen). As a negative control, lysis buffer at different concentrations was used, and for positive control, cholesterol (Sigma) at different concentrations was used.
Cytotoxicity Assay
Cell viability and membrane integrity were analyzed using a commercially available lactate dehydrogenase assay (Cayman Chemicals) in accordance to the manufacturer's protocol.
ADAM17 Stability
For pulse-chase experiments, SH-SY5Y-wt cells were cultured to confluency in 10-cm dishes, then washed three times with DMEM without methionine (Sigma), and radiolabeled for 1.5 h with 48.16 μCi/ml [35S]methionine (PerkinElmer Life Sciences) in 2.5 ml of the same medium. Cells were then washed three times with DMEM containing 0.1% FBS and 0.1% fatty acid-free BSA and incubated with 100 μm DHA for 4 h. Afterward, cells were washed twice with PBS, lysed in 1 ml of lysis buffer with protease inhibitor, and adjusted to the same protein amount. Protein G-Sepharose and 3 μg/ml ADAM17 antibody (ab39162, Abcam, Cambridge, UK) were added to each sample as well as 2 μg/ml actin antibody (sc-8432, Santa Cruz Biotechnology) in a second approach as a control. Following an overnight incubation at 4 °C on an end-over-end shaker, the beads were washed, and [35S]methionine incorporation was measured by scintillation counting. A second set of samples was applied on a Tris-Tricine gel and blotted on a nitrocellulose membrane. The membrane was exposed to Kodak (XAR-5) film at −80 °C for 70 h (48). Densitometric scanning was used to quantitate ADAM17 protein amount. Samples were normalized to actin protein half-lives.
FACS Analysis
For detection of intra- and extracellular BACE1 levels, confluent cells were washed with PBS and decollated by 50 mm EDTA in PBS. For detection of intracellular BACE1, extracellular proteins were split off by a short exposure to trypsin. Cells were repeatedly washed in FACS buffer (2% FCS in PBS) and stained with antibody BACE1 B0806 (1:200; Sigma) and ab6717 (1:200; Abcam, Cambridge, UK) in FACS buffer. The cells were fixed with 2% paraformaldehyde. Extracellular levels of BACE1 were calculated by deviation of intracellular and total BACE1 levels. FACS analysis was performed in FACSCanto II (BD Biosciences) and FACSDiva 6.1 software. Shown histograms represent an average of four independent experiments.
Data Analysis
All quantified data represent an average of at least three independent experiments. In figures, the error bars represent standard deviation of the mean. Statistical significance was determined by two-tailed Student's t test; significance was set at *, p ≤ 0.05; **, p ≤ 0.01, and ***, p ≤ 0.001; n.s. indicates not significant.
RESULTS
DHA Decreases Aβ Production in SH-SY5Y Cells
We evaluated the effect of DHA on Aβ production in the human neuroblastoma cell line SH-SY5Y stably expressing human APP695, the major APP isoform expressed in neuronal cells (49, 50). Aβ levels were decreased in a dose-dependent manner for the tested DHA concentrations at 25, 50, and 100 μm (Fig. 1A), which corresponds to physiological DHA concentrations in the brain (51). The applied experimental conditions did not induce cytotoxicity or impair membrane integrity (supplemental Fig. S1). Lactate dehydrogenase assay for the highest DHA concentration revealed no increased mortality in treated cells. Gas chromatographic analysis showed that 98.53 μg of DHA per mg of protein was taken up into SH-SY5Y cells reflecting an uptake of ∼77% of total DHA supplied (supplemental Fig. S2).
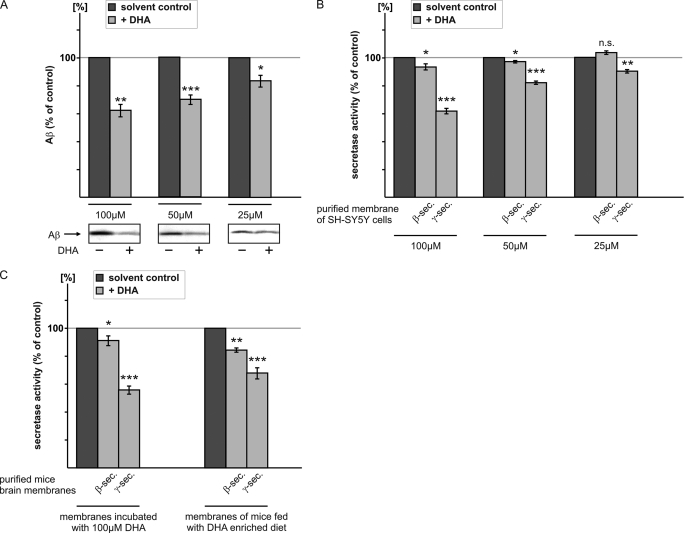
FIGURE 1.
Influence of DHA on β- and γ-secretase activity. A, SH-SY5Y cells stably expressing APP695 were incubated with 25, 50, and 100 μm DHA or solvent control. Equal volumes of conditioned media were immunoprecipitated with antibody W02, recognizing an epitope between amino acids 1 and 10 of Aβ. Immunoprecipitated Aβ peptides were detected by WB analysis with W02. DHA decreases dose-dependent Aβ generation. B, purified membranes of SH-SY5Y-wt cells were incubated in vitro with 25, 50, and 100 μm DHA or solvent control, and β- and γ-secretase activities were determined by a fluorometric assay. DHA directly decreases β- and γ-secretase activity. C, left, purified membranes of mouse brain were incubated with 100 μm DHA or solvent control, and β- and γ-secretase activities were determined. DHA also decreases β- and γ-secretase activities in membranes of mouse brain. Right, membranes of mice fed a DHA-enriched diet were prepared, and β- and γ-secretase activities were measured. Membranes of mouse brain fed the DHA-enriched diet show reduced β- and γ-secretase activities compared with membranes of mouse brain fed a calorie-matched control diet. Purified membranes were prepared as described under “Experimental Procedures.”
DHA Reduces Amyloidogenic Processing by Decreasing γ- and β-Secretase Activity
In addition to the indirect analysis of γ- and β-secretase activities by detection of Aβ levels, we examined the direct effect of DHA on secretases, by measuring γ- and β-secretase activities of SH-SY5Y-wt membrane extracts in the presence of DHA and a solvent control. In agreement with decreased Aβ levels, the γ-secretase activity was significantly reduced in a dose-dependent manner for all tested DHA concentrations (Fig. 1B). Furthermore, β-secretase activity measurement of DHA-treated cell membrane extracts resulted in a significant reduction for 100 and 50 μm DHA, but the effect magnitude was smaller than for γ-secretase (Fig. 1B). Subsequently, we purified membranes from mouse brain to examine the effect of DHA on γ- and β-secretase activities in brain. Both γ- and β-secretase activity were significantly decreased following DHA treatment (Fig. 1C). Again, the reduction in γ-secretase activity was more pronounced than in β-secretase activity. To address a possible dietary effect of DHA on secretase activities, mice were fed for 4 weeks with a DHA-enriched diet or a control diet and assayed for their brain γ- and β-secretase activities. In agreement with the in vitro and ex vivo experiments, the in vivo γ- and β-secretase activities were significantly decreased (Fig. 1C). These results suggest that DHA directly affects γ- and β-secretase activity. Whereas in vitro and ex vivo the direct influence of DHA on β- and γ-secretase activity was monitored, the secretase activity in vivo might be influenced by additional indirect mechanisms such as gene expression and protein stability.
DHA Does Not Alter PS1 and BACE1 Gene and Protein Expression in SH-SY5Y Cells
To address the question of whether DHA-induced Aβ reduction was provoked by additional indirect effects, we examined the protein and mRNA levels of PS1 and BACE1 in SH-SY5Y-wt cells treated with DHA. Both PS1 and BACE1 protein levels as well as mRNA levels were not altered in the presence of DHA (Fig. 2A). In line with the in vitro secretase assays, these results confirm that the influence of DHA on γ- and β-secretase activity can be explained by a direct effect.
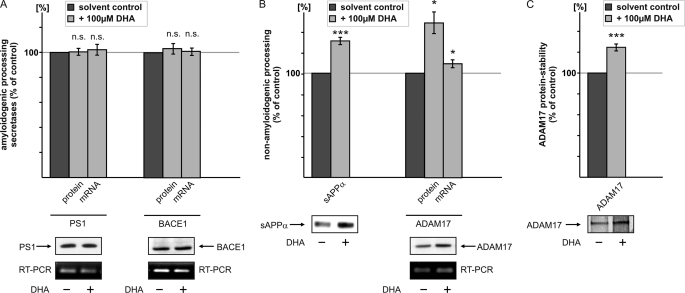
FIGURE 2.
Influence of DHA on mRNA and protein levels of PS1, BACE1, and ADAM17. A, SH-SY5Y-wt cells were incubated with 100 μm DHA or solvent control. Protein levels of PS1 and BACE1 were analyzed by WB analysis. Equal amounts of protein were loaded on Tris-Tricine gels, and PS1 was detected with the antibody sc-7860 and BACE1 with the antibody B0806. mRNA levels of DHA-treated SH-SY5Y cells were analyzed by RT-PCR analysis. No significant changes could be detected in protein and mRNA levels of PS1 and BACE1 in DHA-treated cells compared with cells treated with solvent control. B, left, conditioned media of SH-SY5Y-wt cells, which were incubated with 100 μm DHA, were immunoprecipitated with antibody W02. sAPPα levels were detected with the antibody W02. sAPPα levels of DHA-treated cells were significantly increased compared with cells incubated with solvent control. Right, SH-SY5Y cells were incubated with 100 μm DHA, and mRNA and protein levels of ADAM17 were determined. Protein levels of ADAM17 were analyzed by the use of an ADAM17-specific antibody by WB analysis. mRNA analysis of ADAM17 was performed by RT-PCR analysis. ADAM17 protein levels are significantly increased, whereas mRNA levels are only slightly but significantly increased. C, pulse-chase experiment of SH-SY5Y-wt cells in the presence of DHA or solvent control. [35S]Methionine-labeled ADAM17 protein was immunoprecipitated with an ADAM17-specific antibody.
DHA Increases Nonamyloidogenic Processing and Elevates sAPPα and ADAM17 Protein Level
Aside from influencing γ- and β-secretase, DHA also might act on the nonamyloidogenic processing of APP. To evaluate the effect of DHA on α-secretase activity, we first analyzed sAPPα secretion in SH-SY5Y-wt cells. After DHA treatment, sAPPα protein levels were significantly increased (Fig. 2B). The zinc metalloproteinases ADAM10 and ADAM17 represent the main α-secretases and cleave APP to release sAPPα and C83 (11–13). Inhibition or knock-out of ADAM17 was shown to decrease the regulated α-secretase cleavage of APP (11). Thus, we analyzed ADAM17 protein and mRNA levels in SH-SY5Y cells. Protein levels were significantly increased to 148.5% (Fig. 2B), whereas mRNA levels showed a small but significant increase to 109.4% (Fig. 2B) in the presence of DHA. This suggests that increased gene expression alone cannot account for increased ADAM17 protein expression, resulting in increased α-secretase cleavage.
DHA Increases ADAM17 Stability in SH-SY5Y Cells
The discrepancy between ADAM17 protein and mRNA levels led us to investigate the effect of DHA on ADAM17 protein stability in SH-SY5Y cells. A significant elevation in ADAM17 protein stability was observed in pulse-chase experiments with DHA-treated cells (Fig. 2C). These findings indicate that the nonamyloidogenic processing of APP, in contrast to the amyloidogenic processing, is influenced indirectly by DHA. The increased α-secretase activity derives from increased ADAM17 protein levels, which in turn results from a slight increase in gene expression and decreased ADAM17 protein degradation.
DHA Influences Cholesterol de Novo Synthesis and HMGCR Activity
The effect of cholesterol on degenerative processes in AD is well known. Elevated cholesterol levels are linked to increased amyloid plaque formation in animal models and Aβ aggregation in vitro (52–60). On the cellular level, cholesterol increases Aβ production by increasing β- and γ-secretase cleavage (44). The interplay between cholesterol and DHA in membrane stabilization and fluidity appears to be a critical factor influencing the development of AD. To investigate the impact of DHA on cholesterol metabolism, we first analyzed cholesterol de novo synthesis in DHA-treated SH-SY5Y cells. DHA reduced the incorporation of [14C]acetate into cholesterol by 33% (Fig. 3A).
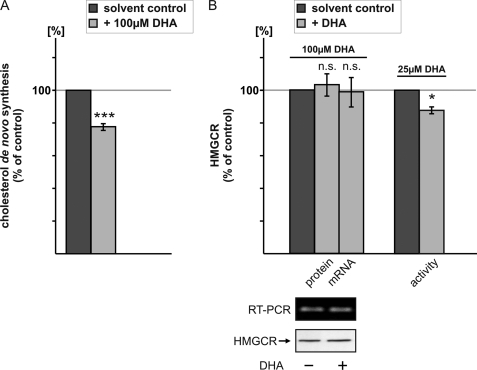
FIGURE 3.
Influence of DHA on cholesterol homeostasis. A, SH-SY5Y-wt cells were treated with 100 μm DHA for 24 h following a 6-h incubation with 0.4 μCi/ml [14C]acetate (PerkinElmer Life Sciences) and DHA. Incorporation of the radioactive precursor into cholesterol was determined as described under “Experimental Procedures.” DHA significantly reduced cholesterol de novo synthesis compared with solvent control. B, left, SH-SY5Y-wt cells were incubated with 100 μm DHA or solvent control. For the detection of HMGCR, an equal protein amount was loaded on a Tris-Tricine gel, and HMGCR was detected by WB analysis with a HMGCR-specific antibody. mRNA levels of HMGCR were determined by RT-PCR. DHA incubated cells show no differences in HMGCR protein level and gene expression. Right, in vitro the catalytic subunit of HMGCR was incubated with 25 μm DHA, and HMGCR activity was measured by a fluorometric assay. DHA directly decreases HMGCR activity.
The committed step reaction in cholesterol biosynthesis is catalyzed by HMGCR. Protein and mRNA levels of HMGCR were unchanged (Fig. 3B), indicating that decreased cholesterol de novo synthesis in the presence of DHA is not caused by reduced HMGCR expression. In line with this observation, an activity assay with the catalytic subunit of HMGCR with and without DHA revealed decreased HMGCR activity, indicating that the effect of DHA on HMGCR is direct (Fig. 3B). As a consequence, DHA affects Aβ production by altering cholesterol homeostasis.
DHA Influences Cholesterol Distribution in Plasma Membrane and Hence γ-Secretase Activity
Cholesterol is strongly enriched in detergent-resistant cholesterol- and sphingolipid-rich microdomains of the plasma membrane, also called rafts (61). Interestingly, β- and γ-secretase are discussed to be present in these plasma membrane microdomains (62–67), and cholesterol increases Aβ generation (44, 58, 60, 68, 69). We analyzed whether DHA also affects cholesterol distribution with respect to the detergent-resistant microdomains. Raft preparation of DHA-treated cells showed that in the presence of DHA cholesterol was shifted toward non-raft microdomains of the plasma membrane, leading to an increased relative distribution of cholesterol to non-raft fractions in the presence of DHA (Fig. 4A). Notably, the shift toward non-raft fractions was observed in the same way for γ-secretase activity (Fig. 4B) in DHA-treated cells, indicating that DHA decreases γ-secretase activity by inducing a redistribution of cholesterol out of rafts, where the amyloidogenic processing of APP preferentially occurs.
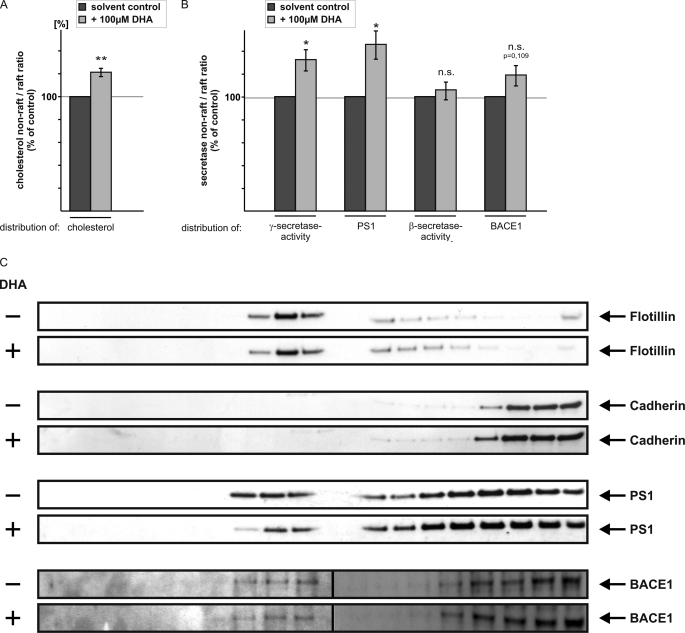
FIGURE 4.
Isolation of raft and non-raft microdomains from SH-SY5Y-wt cells. A, SH-SY5Y-wt cells were incubated with 100 μm DHA, solubilized in Triton X-100, and subjected to sucrose gradient fractionation for the isolation of raft and non-raft microdomains. The cholesterol levels of the collected fractions of the sucrose gradient were analyzed by a H2O2 Amplex Red-based assay. DHA-incubated cells show a significant increase in the ratio of non-raft/raft cholesterol, indicating that DHA displaces cholesterol out of the raft microdomains. B, same fractions were subjected to the γ-secretase activity assay. A similar shift in the ratio of non-raft/raft γ-secretase activity and PS1 protein levels were observed in the presence of DHA compared with cells treated with solvent control. Gradient fractions were also analyzed for β-secretase activity and BACE1 protein levels. C, representative WB analysis of the collected gradient fractions for the proteins flotillin, cadherin, PS1, and BACE1. Each fraction was analyzed by SDS-PAGE and immunoblotted with the respective antibodies. Flotillin-positive fractions represent detergent-insoluble raft fractions. Cadherin-positive fractions represent detergent-soluble non-raft microdomains. In the presence of DHA, PS1 protein level was reduced in flotillin-positive fractions.
DHA Reduces PS1 Protein Level in Lipid Rafts
Our results indicate that DHA influences amyloidogenic processing of APP in raft membrane microdomains. We therefore analyzed whether DHA also affects the distribution of PS1 and BACE1 in raft microdomains. To determine raft localization of PS1 and BACE1, SH-SY5Y-wt cells were solubilized in Triton X-100 and separated by buoyant density centrifugation. To identify raft and non-raft membrane microdomains, gradient fractions were immunoblotted with an antibody to flotillin, a raft marker protein, and an antibody to cadherin to determine gradient fractions representing non-raft microdomains. In the presence of DHA, PS1 protein levels were reduced in flotillin-positive fractions, whereas the distribution of the marker proteins flotillin and cadherin is not affected in the presence of DHA (Fig. 4C). The observed reduced PS1 protein level in raft fractions is accompanied by a shift of PS1 to non-raft microdomains, indicating that amyloidogenic processing of APP in lipid rafts is reduced in the presence of DHA, resulting in a shift of γ-secretase activity to non-raft microdomains (Fig. 4, B and C). In contrast to PS1, the shift of β-secretase BACE1 protein and activity to non-raft microdomains is not significantly affected in the presence of DHA (Fig. 4, B and C).
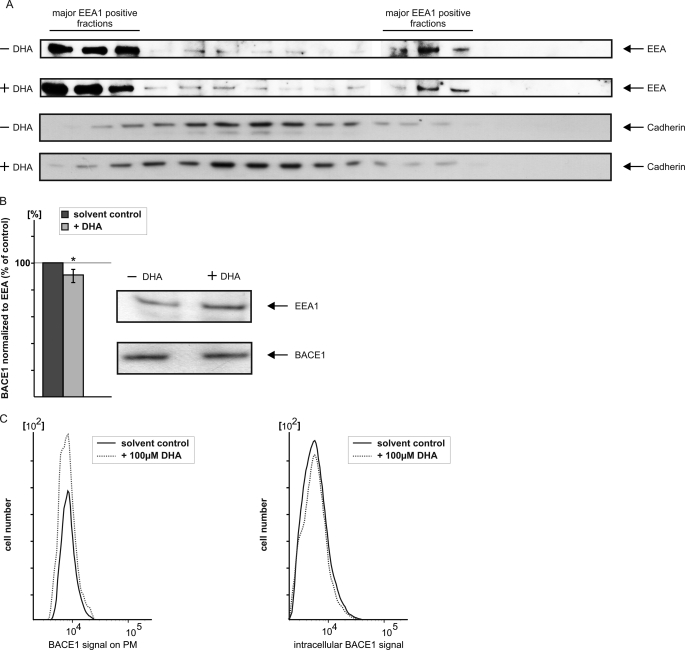
Effect of DHA on Internalization of BACE1 to Endosomes
BACE1 transits the secretory pathway before initially targeting the plasma membrane. Subsequently, BACE1 is internalized to the endosomal system (70, 71). As an aspartyl protease, BACE1 has a mildly acidic pH optimum, making endosomes the most likely location for the amyloidogenic activity of the protein. To address the question whether DHA reduces β-secretase activity by affecting internalization of BACE1 to the endosomal system, SH-SY5Y-wt cells were homogenized and subjected to discontinuous density centrifugation using OptiPrep gradient (46). Collected gradient fractions were immunoblotted with the early endosomal marker protein EEA1 to identify fractions containing early endosomal membranes (Fig. 5A). The purification of endosomal membranes was further validated by the distribution of cadherin as a marker for plasma membrane fractions. The BACE1 level of the EEA1-positive fractions was determined by WB analysis and normalized to the EEA1 protein level (Fig. 5B). In the presence of DHA, the BACE1/EEA1 protein level ratio was moderately decreased, indicating that DHA impairs internalization of BACE1 to the endosomal system. To validate this altered BACE1 distribution, we performed FACS analysis of DHA-treated cells and control cells. In agreement with the previous result, BACE1 protein at the cell surface is increased, whereas the signal for intracellular BACE1 is decreased upon DHA treatment (Fig. 5C).
FIGURE 5.
Analysis of BACE1 internalization in early endosomal membranes. A, SH-SY5Y-wt cells were homogenized and subjected to density centrifugation using OptiPrep gradient. Collected fractions were analyzed by SDS-PAGE and immunoblotted with early endosomal antigen1 (EEA1) antibody and cadherin for detection of fractions containing plasma membranes. B, determination of BACE1 in early endosomal membranes. EEA1-positive fractions were pooled, and WB analysis was performed with BACE1 antibody. BACE1 protein level was normalized to the EEA1 protein level. C, FACS analysis of BACE1 present at plasma membrane and intracellularly. In the presence of DHA, more BACE1 is present at plasma membrane, whereas BACE1 is decreased intracellularly.
DISCUSSION
DHA, an essential ω-3 fatty acid, is discussed to decrease Aβ production and has been found to be associated with reduced risk of AD (25). In this study, we have systematically elucidated the cellular effects of DHA, which lead to decreased generation of Aβ peptides. The experiments performed suggest that DHA reduces Aβ generation not by a single predominant mechanism but rather by moderately modulating multiple cellular functions at once. All these multiple changes decreased amyloidogenic processing of APP and increased the nonamyloidogenic pathway, suggesting a directed change in cellular properties. Therefore, DHA acts via pleiotropic mechanisms on the release of Aβ, involving both direct and indirect effects on the processing of APP.
Direct measurement of β- and γ-secretase activity of SH-SY5Y-wt membrane extracts and purified membranes of mouse brain showed decreased β- and γ-secretase activities, which is in line with reduced Aβ levels in DHA-treated SH-SY5Y cells, stably expressing APP695. Whereas the reduction in β-secretase activity in vitro, although clearly present, is rather small, this was not the case when mice were fed a DHA-containing diet. Here, the reduction in β-secretase activity is considerably stronger. Because RNA and protein levels of BACE1 and PS1 are unaltered in the presence of DHA, the observed effect of DHA on β- and γ-secretase seems to be direct. In contrast, reduced steady-state levels of PS1 have been described for a 3×Tg-AD mouse model (72) fed with a diet supplemented with DHA. However, taking into consideration that gene or protein expression cannot account for the observed effect by direct measuring β- and γ-secretase activity in membrane extracts of cells or mouse brain, one can conclude that DHA directly reduces β- and γ-secretase activity. Diminished Aβ release cannot only be provoked by reducing β- and γ-secretase activity but also by increasing the nonamyloidogenic pathway of APP processing. Indeed, SH-SY5Y-wt cells incubated with DHA showed significantly increased sAPPα levels. The ADAM17 protein level, one leading candidate for α-secretase processing of APP (11), was also significantly increased in the presence of DHA, whereas the RNA level was only marginally elevated. This putative contradictory result can be explained by impaired ADAM17 protein degradation. Pulse-chase experiments of SH-SY5Y-wt cells showed increased ADAM17 protein stability in the presence of DHA. In contrast to the direct effect of DHA on β- and γ-secretase, DHA increases the nonamyloidogenic processing of APP indirectly by increasing ADAM17 gene transcription and by impairing ADAM17 protein degradation.
Besides DHA, cholesterol has been shown to affect Aβ generation, and cholesterol is discussed to be a risk factor for the development of AD (52, 73, 74). Cholesterol is concentrated in detergent-resistant membranes, which are also rich in sphingolipids (61). DHA-containing phospholipids also incorporate in cellular membranes (75) and have been shown to change the organization of sphingolipid/cholesterol lipid-raft membrane domains (76). The analysis of cholesterol distribution in raft and non-raft microdomains, which were isolated by sucrose density centrifugation, showed that DHA displaces cholesterol out of rafts. Interestingly, raft microdomains have been shown to be involved in the amyloidogenic processing of APP. As we observed changes in γ-secretase activity in raft and non-raft microdomains after DHA incubation, one can conclude that DHA reduces Aβ generation by displacing cholesterol out of rafts, thus reducing the enzyme activities responsible for amyloidogenic processing of APP. In addition, DHA reduces the amount of PS1 protein localized in lipid raft microdomains, causing a reduction in raft-associated γ-secretase processing of APP, indicating that membrane microdomain switching of PS1 is one cause for reduced γ-secretase processing in the presence of DHA. Although DHA-containing phospholipids have been shown to form unique non-raft membrane domains in model membranes, in the cellular model DHA directly incorporates into lipid rafts (77, 78) and modulates protein activity (78) consistent with our findings in SH-SY5Y cells. In contrast to γ-secretase activity, the effect of DHA on β-secretase activity was less pronounced, making the identification of the underlying mechanism more difficult. The raft to non-raft shift of BACE1 and β-secretase activity did not reach a significant level. However, in EEA1-positive membrane fractions, BACE1 protein level normalized to the EEA1 signal is decreased in the presence of DHA, and BACE1 levels found on the cell surface are increased, and the intracellular BACE1 signal is slightly decreased. These findings would support a model in which DHA shifts the raft/non-raft ratio of BACE1 and impairs the internalization of BACE1, finally resulting in reduced β-secretase processing of APP in endosomes in the presence of DHA. Accordingly, DHA apparently alters amyloidogenic APP processing at least by altering the subcellular distribution of the secretases. Moreover, cholesterol might be involved in this. Recently, it has been reported that cholesterol increases β-secretase processing of APP by triggering APP-BACE1 clustering into lipid rafts, preceded by rapid endocytosis (79). Indeed, we found that DHA changes the raft/non-raft distribution of cholesterol, and thus one might speculate that DHA reduces secretase activities by affecting cholesterol levels in lipid raft microdomains, resulting in reduced internalization to the endosomal system, which is discussed to be important for amyloidogenic processing of APP (80). Additionally, DHA alters cholesterol level by directly influencing HMGCR activity, the rate-limiting enzyme in cholesterol de novo synthesis. This enzyme can be inhibited by statins, which decrease the production of Aβ and are discussed to have protective effects on AD (54, 55). However, clinical trials have yielded mixed results regarding the benefits of statin treatment in AD (81–83). Interestingly, we found that DHA directly influences HMGCR enzyme activity without affecting HMGCR gene expression and total protein level. HMGCR enzyme activity was reduced in the presence of DHA, thus leading to reduced cholesterol de novo synthesis and hence reduced Aβ levels as observed in our cell culture experiments and in studies with DHA-enriched diets.
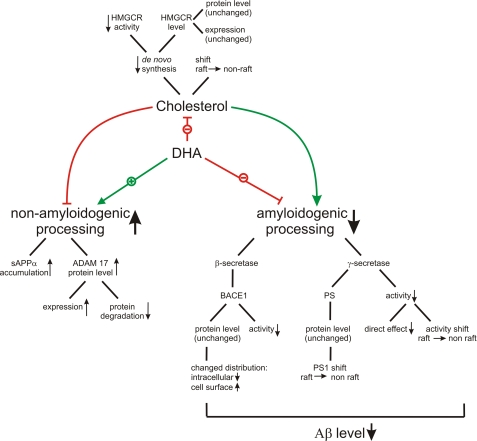
Taken together, the ω-3 polyunsaturated fatty acid DHA, which is highly enriched in fish oil, effectively reduces Aβ generation by a concerted and pleiotropic manner. Multiple steps directly and indirectly involved in APP processing act toward increased nonamyloidogenic processing in parallel to reduced amyloidogenic processing. Aβ is generated by a cascade of several steps that are affected by DHA (Fig. 6); the impact of DHA on Aβ generation is remarkable, although the effects on the single steps are rather small. DHA might therefore represent an internal regulator of the balance between amyloidogenic versus nonamyloidogenic processing of APP. Although DHA is not as effective in reducing Aβ production as γ-secretase inhibitors, for example, the combination of several small effects can be assumed to present a safe and tolerable approach for long term treatment in AD prevention. This is in line with the established safety of DHA in preclinical and clinical trials (38). Effectiveness in AD prevention might also be further supported by other neuroprotective activities of DHA, but specifically designed trials will be needed to answer this question in the future.
FIGURE 6.
Schematic overview, DHA decreases Aβ production by multiple ways. In the presence of DHA, amyloidogenic processing is decreased by a reduced β- and γ-secretase activity, whereas the protein and expression levels of BACE1 and PS1 are unchanged. Nonamyloidogenic processing is increased by an elevated ADAM17 protein level, caused by a decreased protein degradation and an increased expression level. Besides the direct effect on APP processing, DHA affects Aβ production by a cross-link to cholesterol homeostasis. DHA decreases cholesterol de novo synthesis by decreasing HMGCR activity, whereas HMGCR protein level and expression are unchanged. Besides the reduced cholesterol production, DHA causes a shift of cholesterol from the raft to the non-raft fraction, which is accompanied by a shift in γ-secretase activity and PS1 protein.
This work was supported by European Union FP7 Project LipiDiDiet Grant 211696 (to T. H.), Deutsche Forschungsgemeinschaft, Bundesministerium für Bildung, Forschung, Wissenschaft, und Technologie via NGFNplus (to T. H.), and Kompetenznetz Degenerative Demenzen (KNDD) (to T. H.), HOMFOR 2008 and HOMFOR 2009 Saarland University Research Grants (to M. O. W. G. and T. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and an additional reference.
- AD
- Alzheimer disease
- APP
- amyloid precursor protein
- sAPP
- soluble APP
- Aβ
- β-amyloid peptide
- DHA
- docosahexanoic acid
- HMGCR
- 3-hydroxy-3-methylglutaryl-coenzyme-A reductase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- WB
- Western blot.
REFERENCES
- 1. AD International, World Alzheimer Report (2009) in Alzheimer's Disease International Consortium (Prince M., Jackson J. eds) pp. 1–20, Alzheimer's Disease International, London [Google Scholar]
- 2. Masters C. L., Simms G., Weinman N. A., Multhaup G., McDonald B. L., Beyreuther K. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4245–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Selkoe D. J. (2001) Physiol. Rev. 81, 741–766 [DOI] [PubMed] [Google Scholar]
- 4. Sisodia S. S., St George-Hyslop P. H. (2002) Nat. Rev. Neurosci. 3, 281–290 [DOI] [PubMed] [Google Scholar]
- 5. Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. (1999) Science 286, 735–741 [DOI] [PubMed] [Google Scholar]
- 6. Sinha S., Anderson J. P., Barbour R., Basi G. S., Caccavello R., Davis D., Doan M., Dovey H. F., Frigon N., Hong J., Jacobson-Croak K., Jewett N., Keim P., Knops J., Lieberburg I., Power M., Tan H., Tatsuno G., Tung J., Schenk D., Seubert P., Suomensaari S. M., Wang S., Walker D., Zhao J., McConlogue L., John V. (1999) Nature 402, 537–540 [DOI] [PubMed] [Google Scholar]
- 7. Takasugi N., Tomita T., Hayashi I., Tsuruoka M., Niimura M., Takahashi Y., Thinakaran G., Iwatsubo T. (2003) Nature 422, 438–441 [DOI] [PubMed] [Google Scholar]
- 8. Kimberly W. T., LaVoie M. J., Ostaszewski B. L., Ye W., Wolfe M. S., Selkoe D. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6382–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim S. H., Ikeuchi T., Yu C., Sisodia S. S. (2003) J. Biol. Chem. 278, 33992–34002 [DOI] [PubMed] [Google Scholar]
- 10. Wakabayashi T., De Strooper B. (2008) Physiology 23, 194–204 [DOI] [PubMed] [Google Scholar]
- 11. Buxbaum J. D., Liu K. N., Luo Y., Slack J. L., Stocking K. L., Peschon J. J., Johnson R. S., Castner B. J., Cerretti D. P., Black R. A. (1998) J. Biol. Chem. 273, 27765–27767 [DOI] [PubMed] [Google Scholar]
- 12. Lammich S., Kojro E., Postina R., Gilbert S., Pfeiffer R., Jasionowski M., Haass C., Fahrenholz F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3922–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Allinson T. M., Parkin E. T., Condon T. P., Schwager S. L., Sturrock E. D., Turner A. J., Hooper N. M. (2004) Eur. J. Biochem. 271, 2539–2547 [DOI] [PubMed] [Google Scholar]
- 14. Haass C., Schlossmacher M. G., Hung A. Y., Vigo-Pelfrey C., Mellon A., Ostaszewski B. L., Lieberburg I., Koo E. H., Schenk D., Teplow D. B., et al. (1992) Nature 359, 322–325 [DOI] [PubMed] [Google Scholar]
- 15. Haass C., Hung A. Y., Schlossmacher M. G., Teplow D. B., Selkoe D. J. (1993) J. Biol. Chem. 268, 3021–3024 [PubMed] [Google Scholar]
- 16. Kamphuis P. J., Scheltens P. (2010) J. Alzheimers Dis. 20, 765–775 [DOI] [PubMed] [Google Scholar]
- 17. Scheltens P., Kamphuis P. J., Verhey F. R., Olde Rikkert M. G., Wurtman R. J., Wilkinson D., Twisk J. W., Kurz A. (2010) Alzheimers Dement. 6, 1–10.e1 [DOI] [PubMed] [Google Scholar]
- 18. Pawlosky R. J., Hibbeln J. R., Novotny J. A., Salem N., Jr. (2001) J. Lipid Res. 42, 1257–1265 [PubMed] [Google Scholar]
- 19. Bazan N. G., Scott B. L. (1990) Ups J. Med. Sci. Suppl. 48, 97–107 [PubMed] [Google Scholar]
- 20. Ansari K. A., Shoeman D. W. (1990) Neurochem. Res. 15, 7–11 [DOI] [PubMed] [Google Scholar]
- 21. Horrocks L. A., Farooqui A. A. (2004) Prostaglandins Leukot. Essent. Fatty Acids 70, 361–372 [DOI] [PubMed] [Google Scholar]
- 22. Söderberg M., Edlund C., Kristensson K., Dallner G. (1991) Lipids 26, 421–425 [DOI] [PubMed] [Google Scholar]
- 23. Tully A. M., Roche H. M., Doyle R., Fallon C., Bruce I., Lawlor B., Coakley D., Gibney M. J. (2003) Br. J. Nutr. 89, 483–489 [DOI] [PubMed] [Google Scholar]
- 24. Barberger-Gateau P., Letenneur L., Deschamps V., Pérès K., Dartigues J. F., Renaud S. (2002) BMJ 325, 932–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morris M. C., Evans D. A., Bienias J. L., Tangney C. C., Bennett D. A., Wilson R. S., Aggarwal N., Schneider J. (2003) Arch. Neurol. 60, 940–946 [DOI] [PubMed] [Google Scholar]
- 26. Schaefer E. J., Bongard V., Beiser A. S., Lamon-Fava S., Robins S. J., Au R., Tucker K. L., Kyle D. J., Wilson P. W., Wolf P. A. (2006) Arch. Neurol. 63, 1545–1550 [DOI] [PubMed] [Google Scholar]
- 27. Albanese E., Dangour A. D., Uauy R., Acosta D., Guerra M., Guerra S. S., Huang Y., Jacob K. S., de Rodriguez J. L., Noriega L. H., Salas A., Sosa A. L., Sousa R. M., Williams J., Ferri C. P., Prince M. J. (2009) Am. J. Clin. Nutr. 90, 392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Gelder B. M., Tijhuis M., Kalmijn S., Kromhout D. (2007) Am. J. Clin. Nutr. 85, 1142–1147 [DOI] [PubMed] [Google Scholar]
- 29. Freund-Levi Y., Eriksdotter-Jönhagen M., Cederholm T., Basun H., Faxén-Irving G., Garlind A., Vedin I., Vessby B., Wahlund L. O., Palmblad J. (2006) Arch. Neurol. 63, 1402–1408 [DOI] [PubMed] [Google Scholar]
- 30. Kotani S., Sakaguchi E., Warashina S., Matsukawa N., Ishikura Y., Kiso Y., Sakakibara M., Yoshimoto T., Guo J., Yamashima T. (2006) Neurosci. Res. 56, 159–164 [DOI] [PubMed] [Google Scholar]
- 31. Chiu C. C., Su K. P., Cheng T. C., Liu H. C., Chang C. J., Dewey M. E., Stewart R., Huang S. Y. (2008) Prog. Neuropsychopharmacol. Biol. Psychiatry 32, 1538–1544 [DOI] [PubMed] [Google Scholar]
- 32. van de Rest O., Geleijnse J. M., Kok F. J., van Staveren W. A., Dullemeijer C., Olderikkert M. G., Beekman A. T., de Groot C. P. (2008) Neurology 71, 430–438 [DOI] [PubMed] [Google Scholar]
- 33. Lukiw W. J., Cui J. G., Marcheselli V. L., Bodker M., Botkjaer A., Gotlinger K., Serhan C. N., Bazan N. G. (2005) J. Clin. Invest. 115, 2774–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perez S. E., Berg B. M., Moore K. A., He B., Counts S. E., Fritz J. J., Hu Y. S., Lazarov O., Lah J. J., Mufson E. J. (2010) J. Neurosci. Res. 88, 1026–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oksman M., Iivonen H., Hogyes E., Amtul Z., Penke B., Leenders I., Broersen L., Lütjohann D., Hartmann T., Tanila H. (2006) Neurobiol. Dis. 23, 563–572 [DOI] [PubMed] [Google Scholar]
- 36. Calon F., Lim G. P., Yang F., Morihara T., Teter B., Ubeda O., Rostaing P., Triller A., Salem N., Jr., Ashe K. H., Frautschy S. A., Cole G. M. (2004) Neuron 43, 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hooijmans C. R., Rutters F., Dederen P. J., Gambarota G., Veltien A., van Groen T., Broersen L. M., Lütjohann D., Heerschap A., Tanila H., Kiliaan A. J. (2007) Neurobiol. Dis. 28, 16–29 [DOI] [PubMed] [Google Scholar]
- 38. Lien E. L. (2009) Prostaglandins Leukot. Essent. Fatty Acids 81, 125–132 [DOI] [PubMed] [Google Scholar]
- 39. Cole G. M., Ma Q. L., Frautschy S. A. (2009) Prostaglandins Leukot. Essent. Fatty Acids 81, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reeves P. G., Nielsen F. H., Fahey G. C., Jr. (1993) J. Nutr. 123, 1939–1951 [DOI] [PubMed] [Google Scholar]
- 41. Ida N., Hartmann T., Pantel J., Schröder J., Zerfass R., Förstl H., Sandbrink R., Masters C. L., Beyreuther K. (1996) J. Biol. Chem. 271, 22908–22914 [DOI] [PubMed] [Google Scholar]
- 42. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 43. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 44. Grimm M. O., Grimm H. S., Tomic I., Beyreuther K., Hartmann T., Bergmann C. (2008) J. Biol. Chem. 283, 11302–11311 [DOI] [PubMed] [Google Scholar]
- 45. Cordy J. M., Hussain I., Dingwall C., Hooper N. M., Turner A. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11735–11740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Woods A. J., White D. P., Caswell P. T., Norman J. C. (2004) EMBO J. 23, 2531–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 48. Cladera J. L., Bryant P. J. (1985) Dev. Genet. 6, 27–37 [Google Scholar]
- 49. Kang J., Lemaire H. G., Unterbeck A., Salbaum J. M., Masters C. L., Grzeschik K. H., Multhaup G., Beyreuther K., Müller-Hill B. (1987) Nature 325, 733–736 [DOI] [PubMed] [Google Scholar]
- 50. Kang J., Müller-Hill B. (1990) Biochem. Biophys. Res. Commun. 166, 1192–1200 [DOI] [PubMed] [Google Scholar]
- 51. Rapoport S. I., Rao J. S., Igarashi M. (2007) Prostaglandins Leukot. Essent. Fatty Acids 77, 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Simons M., Keller P., De Strooper B., Beyreuther K., Dotti C. G., Simons K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6460–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Frears E. R., Stephens D. J., Walters C. E., Davies H., Austen B. M. (1999) Neuroreport 10, 1699–1705 [DOI] [PubMed] [Google Scholar]
- 54. Fassbender K., Simons M., Bergmann C., Stroick M., Lutjohann D., Keller P., Runz H., Kuhl S., Bertsch T., von Bergmann K., Hennerici M., Beyreuther K., Hartmann T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5856–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lu F., Li X., Suo A. Q., Zhang J. W. (2010) Chin. Med. J. 123, 1864–1870 [PubMed] [Google Scholar]
- 56. Solomon A., Sippola R., Soininen H., Wolozin B., Tuomilehto J., Laatikainen T., Kivipelto M. (2010) Neurodegener. Dis. 7, 180–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pappolla M. A., Bryant-Thomas T. K., Herbert D., Pacheco J., Fabra Garcia M., Manjon M., Girones X., Henry T. L., Matsubara E., Zambon D., Wolozin B., Sano M., Cruz-Sanchez F. F., Thal L. J., Petanceska S. S., Refolo L. M. (2003) Neurology 61, 199–205 [DOI] [PubMed] [Google Scholar]
- 58. Guardia-Laguarta C., Coma M., Pera M., Clarimón J., Sereno L., Agulló J. M., Molina-Porcel L., Gallardo E., Deng A., Berezovska O., Hyman B. T., Blesa R., Gómez-Isla T., Lleó A. (2009) J. Neurochem. 110, 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu W. W., Todd S., Coulson D. T., Irvine G. B., Passmore A. P., McGuinness B., McConville M., Craig D., Johnston J. A. (2009) J. Neurochem. 108, 341–349 [DOI] [PubMed] [Google Scholar]
- 60. Schneider A., Schulz-Schaeffer W., Hartmann T., Schulz J. B., Simons M. (2006) Neurobiol. Dis. 23, 573–577 [DOI] [PubMed] [Google Scholar]
- 61. Simons K., Ikonen E. (1997) Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 62. Hur J. Y., Welander H., Behbahani H., Aoki M., Frånberg J., Winblad B., Frykman S., Tjernberg L. O. (2008) FEBS J. 275, 1174–1187 [DOI] [PubMed] [Google Scholar]
- 63. Tun H., Marlow L., Pinnix I., Kinsey R., Sambamurti K. (2002) J. Mol. Neurosci. 19, 31–35 [DOI] [PubMed] [Google Scholar]
- 64. Vetrivel K. S., Meckler X., Chen Y., Nguyen P. D., Seidah N. G., Vassar R., Wong P. C., Fukata M., Kounnas M. Z., Thinakaran G. (2009) J. Biol. Chem. 284, 3793–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ehehalt R., Keller P., Haass C., Thiele C., Simons K. (2003) J. Cell Biol. 160, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sakurai T., Kaneko K., Okuno M., Wada K., Kashiyama T., Shimizu H., Akagi T., Hashikawa T., Nukina N. (2008) J. Cell Biol. 183, 339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cheng H., Vetrivel K. S., Drisdel R. C., Meckler X., Gong P., Leem J. Y., Li T., Carter M., Chen Y., Nguyen P., Iwatsubo T., Tomita T., Wong P. C., Green W. N., Kounnas M. Z., Thinakaran G. (2009) J. Biol. Chem. 284, 1373–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wahrle S., Das P., Nyborg A. C., McLendon C., Shoji M., Kawarabayashi T., Younkin L. H., Younkin S. G., Golde T. E. (2002) Neurobiol. Dis. 9, 11–23 [DOI] [PubMed] [Google Scholar]
- 69. Shie F. S., Jin L. W., Cook D. G., Leverenz J. B., LeBoeuf R. C. (2002) Neuroreport 13, 455–459 [DOI] [PubMed] [Google Scholar]
- 70. Huse J. T., Pijak D. S., Leslie G. J., Lee V. M., Doms R. W. (2000) J. Biol. Chem. 275, 33729–33737 [DOI] [PubMed] [Google Scholar]
- 71. Hunt C. E., Turner A. J. (2009) FEBS J. 276, 1845–1859 [DOI] [PubMed] [Google Scholar]
- 72. Green K. N., Martinez-Coria H., Khashwji H., Hall E. B., Yurko-Mauro K. A., Ellis L., LaFerla F. M. (2007) J. Neurosci. 27, 4385–4395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Abad-Rodriguez J., Ledesma M. D., Craessaerts K., Perga S., Medina M., Delacourte A., Dingwall C., De Strooper B., Dotti C. G. (2004) J. Cell Biol. 167, 953–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Grimm M. O., Grimm H. S., Pätzold A. J., Zinser E. G., Halonen R., Duering M., Tschäpe J. A., De Strooper B., Müller U., Shen J., Hartmann T. (2005) Nat. Cell Biol. 7, 1118–1123 [DOI] [PubMed] [Google Scholar]
- 75. Langelier B., Linard A., Bordat C., Lavialle M., Heberden C. (2010) J. Cell. Biochem. 110, 1356–1364 [DOI] [PubMed] [Google Scholar]
- 76. Shaikh S. R., Locascio D. S., Soni S. P., Wassall S. R., Stillwell W. (2009) Biochim. Biophys. Acta 1788, 2421–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shaikh S. R., Rockett B. D., Salameh M., Carraway K. (2009) J. Nutr. 139, 1632–1639 [DOI] [PubMed] [Google Scholar]
- 78. Raza Shaikh S. (2010) Prostaglandins Leukot. Essent. Fatty Acids 82, 159–164 [DOI] [PubMed] [Google Scholar]
- 79. Marquer C., Devauges V., Cossec J. C., Liot G., Lecart S., Saudou F., Duyckaerts C., Leveque-Fort S., Potier M. C. (2011) FASEB J., 10.1096/fj.10-168633 [DOI] [PubMed] [Google Scholar]
- 80. Tate B. A., Mathews P. M. (2006) Sci. Aging Knowledge Environ. 2006, re2. [DOI] [PubMed] [Google Scholar]
- 81. Simons M., Schwärzler F., Lütjohann D., von Bergmann K., Beyreuther K., Dichgans J., Wormstall H., Hartmann T., Schulz J. B. (2002) Ann. Neurol. 52, 346–350 [DOI] [PubMed] [Google Scholar]
- 82. Sparks D. L., Sabbagh M. N., Connor D. J., Lopez J., Launer L. J., Browne P., Wasser D., Johnson-Traver S., Lochhead J., Ziolwolski C. (2005) Arch. Neurol. 62, 753–757 [DOI] [PubMed] [Google Scholar]
- 83. Feldman H. H., Doody R. S., Kivipelto M., Sparks D. L., Waters D. D., Jones R. W., Schwam E., Schindler R., Hey-Hadavi J., DeMicco D. A., Breazna A. (2010) Neurology 74, 956–964 [DOI] [PubMed] [Google Scholar]