Abstract
T-box transcription factors figure prominently in embryonic cardiac cell lineage specifications. Mesenchymal precursor cells expressing Tbx18 give rise to the heart's pacemaker, the sinoatrial node (SAN). We sought to identify targets of TBX18 transcriptional regulation in the heart by forced adenoviral overexpression in postnatal cardiomyocytes. Neonatal rat cardiomyocytes (NRCMs) transduced with GFP showed sarcolemmal, punctate Cx43 expression. In contrast, TBX18-transduced NRCMs exhibited sparse Cx43 expression. Both the transcript and protein levels of Cx43 were greatly down-regulated within 2 days of TBX18 transduction. Direct injection of TBX18 in the guinea pig heart in vivo inhibited Cx43 expression. The repressor activity of TBX18 on Cx43 was highly specific; protein levels of Cx45 and Cx40, which comprise the main gap junctions in the SAN and conduction system, were unchanged by TBX18. A reporter-based promoter assay demonstrated that TBX18 directly represses the Cx43 promoter. Phenotypically, TBX18-NRCMs exhibited slowed intercellular calcein dye transfer kinetics (421 ± 54 versus control 127 ± 43 ms). Intracellular Ca2+ oscillations in control NRCM monolayers were highly synchronized. In contrast, TBX18 overexpression led to asynchronous Ca2+ oscillations, demonstrating reduced cell-cell coupling. Decreased coupling led to slow electrical propagation; conduction velocity in TBX18 NRCMs slowed by more than 50% relative to control (2.9 ± 0.5 versus 14.3 ± 0.9 cm/s). Taken together, TBX18 specifically and directly represses Cx43 transcript and protein levels. Cx43 suppression leads to significant electrical uncoupling, but the preservation of other gap junction proteins supports slow action potential propagation, recapitulating a key phenotypic hallmark of the SAN.
Keywords: Gap Junctions, Gene Regulation, Gene Therapy, Transcription Factors, Transcription Target Genes, Cardiac Pacemaker, Sinoatrial Node, Tbx Transcription Factors
Introduction
T-box transcription factors play a critical role in heart development, particularly in the differentiation, determination, and regulation of the SA4 nodal gene program (1–6). Tbx18-expressing mesenchymal precursor cells give rise to the SA node (1, 3–5), especially the head region, which represents ∼75% of the SAN volume and originates pacemaker activity in the early embryonic heart (1). In contrast, the surrounding right atrium is negative for Tbx18 and originates from Isl1-expressing second heart field mesodermal progenitors (7). Tbx18 homozygous knock-out mice show a marked shrinkage of the SA nodal head region (1). Tbx18 is thus not only a marker for the SAN but also a functional determinant of its development. However, the transcriptional targets of Tbx18 in heart are largely unknown.
Many traits distinguish the SAN from the atrial and ventricular myocardium. Among those, expression of a unique set of gap junctional proteins in pacemaker tissue is a salient feature that shields the SAN from the hyperpolarizing environment of atrial tissue (8, 9). Electrotonic coupling in human SAN is mediated mainly by Cx45 and, to a lesser degree, by Cx31.9 (10). Gap junctions composed of these proteins display substantially lower single-channel conductances as compared with Cx40 or Cx43, the atrial connexins (11–14). The high intercellular electrical resistance at the core of the SAN is thought to protect the spontaneously oscillating action potentials of pacemaker tissue from the adjacent, much larger, and more hyperpolarized atrial tissue, overcoming the “source-to-sink mismatch” (15). However, at the periphery of SAN, Cx43 as well as Cx45 are expressed, allowing the SAN to drive the atrial muscle (16). The detailed molecular program specifying these differences remains unclear, but Tbx18 is known to lead SA nodal development, and its expression is followed by Tbx3, which in turn is essential for development of the downstream conduction system (17).
We set out to study TBX18 and its regulatory targets in postnatal heart by somatic gene transfer. Tbx18 was chosen given its prominent role in SA nodal development. Postnatal cardiomyocytes were chosen to facilitate the dissection of definitive transcriptional targets of TBX18, which is difficult to assess in an embryonic model due to potential interaction of multiple transcription factors during development (6). Our data indicate that TBX18 overexpression specifically and significantly down-regulates Cx43 in vitro and in vivo, while not affecting other gap junction proteins. This molecular effect, which is at least partially attributable to transcriptional repression, results in reductions of cell-cell coupling and conduction velocity in cardiomyocyte monolayers.
EXPERIMENTAL PROCEDURES
Molecular Cloning
The human TBX18 gene with a C-terminal Myc/FLAG tag was excised from pCMV6-TBX18 (OriGene, Rockville, MD) by digestion with FseI and SalI and then subcloned into a NotI- and XhoI-digested lentiviral expression vector with the desired reporter gene, pLVX-IRES-ZsGreen1 (Clontech), to create pLV-TBX18-IRES-ZsGreen (∼10.1 kb). We utilized ZsGreen1 as the reporter protein for TBX18-transduced cardiomyocytes. We refer to ZsGreen1 as GFP throughout this study because ZsGreen1 has indistinguishable spectral characteristics as the commonly used GFP. The recombinant target gene was then introduced to an adenovirus vector backbone by Gateway recombination cloning using Gateway-adapted vectors (Invitrogen). attL and attR recombination reaction was performed between the entry clone and the destination vector, pAd/CMV/V5-DEST (∼36.7 kb), to generate the desired adenoviral expression construct, pAd-CMV-TBX18-IRES-GFP (∼39.8 kb). Positive constructs were verified to have the correct target gene by DNA sequencing (Laragen, Los Angeles, CA).
Adenovirus Construction and Purification
The expression constructs were digested with PacI to expose inverted terminal repeats before transfecting into 293A cells to produce adenoviral stocks for use in subsequent expression of the transgene. Adenoviruses were plaque-purified, amplified, and affinity column-purified using Adenopure kit (Puresyn, Inc.) and stored at −80 °C.
Myocyte Isolation and Transduction
NRCMs were isolated from 1- to 2-day-old pups and cultured as monolayers as described previously (18). A monolayer of NRCMs was transduced with either Ad-TBX18-IRES-GFP or Ad-GFP (control vector; multiplicity of infection 1–10) 1 day post cell isolation and cultured for 3–5 days.
Intracellular Calcium Recordings and Analyses
For measurements of intracellular Ca2+ oscillations, 2 × 106 NRCMs were plated in 35-mm glass-bottom dishes (MatTek Cultureware) or 22-mm fibronectin-coated glass coverslips, transduced, and analyzed 4 days post-transduction. Cells were loaded with Rhod2-AM (2 μmol/liter) (Molecular Probes) for 18 min, then washed once, and subsequently placed in normal Tyrode solution with 2 mmol/liter calcium. Calcium transients were recorded at 37 °C from Ad-TBX18-IRES-GFP- and Ad-GFP-transduced NRCMs. Images were acquired on an inverted confocal laser-scanning microscope (PerkinElmer Life Sciences/Nikon) or Leica SP5 confocal microscope. Off-line analysis was performed using Ultraview (PerkinElmer Life Sciences) and ImageJ. Whole-cell Ca2+ transients were obtained from confocal line scan images through single NRCMs by averaging the signal of an individual cell. Ca2+ transients are presented as background-subtracted, normalized fluorescence (F/F0). For two-dimensional confocal Ca2+ imaging, calcium transients were obtained by averaging the signal through the entire cell.
Multielectrode Array Measurements
Extracellular field potential signals were recorded from a monolayer of NRCMs cultured on the multielectrode array. The conduction velocity was obtained by comparing the activation time delays between beats of each electrode to a single reference electrode.
Immunostaining
NRCMs are fixed 5 days post adenoviral transduction with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Cells were then incubated with the appropriate primary antibody as follows: Cx43 (Sigma, C6219), Tbx18 (Santa Cruz Biotechnology, Sc17869), FLAG (AbCam, ab1162), and Alexa Fluor-conjugated secondary antibodies (Invitrogen). Western blots were performed using specific antibodies against Cx43 (Sigma, C6219), Cx40 (Invitrogen, 36-4900), and Cx45 (a generous gift provided by Thomas H. Steinberg, Washington University, St. Louis). Briefly, Ad-TBX18- and Ad-GFP-transduced NRCMS were homogenized in RIPA buffer containing a protease inhibitor mixture (Sigma). Protein content was quantified by BCA assay, and cell lysates (15 μg per lane) were run on a 12% SDS-polyacrylamide gel and transferred onto a PVDF membrane. Then the transferred membrane was incubated with a primary antibody overnight at 4 °C, followed by a 1-h incubation with a peroxidase-conjugated secondary antibody. Immunoreactivity was detected by chemiluminescence (ECL Western blotting analysis system, Amersham Biosciences). Equal protein loading of the gels was assessed by re-probing the membrane with monoclonal anti-GAPDH antibody (Chemicon).
Cx43 Promoter Assay
NRCMs were plated in 96-well white tissue culture plates at 60,000 cells per well in 100 μl to obtain 95% confluency. Subsequently, they were transduced with Ad-TBX18 or Ad-GFP (multiplicity of infection, 2–20). After 24 h, cells were co-transfected with 50 ng of connexin43 promoter or control luciferase plasmids and thymidine kinase (TK) promoter-Renilla-luciferase plasmid (SwitchGear Genomics) to control for transfection efficiency using FuGENE 6 (Roche Applied Science) in triplicate. Forty eight hours later, cells were incubated with 100 μl of ONE-Glo luciferase substrate (Promega), and luciferase and Renilla activities were assayed in a 96-well plate luminometer according to the protocol in the Dual-Luciferase kit (Promega). We determined the promoter strength of each DNA fragment by calculating the ratio of luciferase signal to Renilla signal from each transfection to control for well-to-well variation in transfection efficiency.
In Vivo Gene Transfer and Immunohistochemistry
Adenoviruses were injected into the left ventricular apex of guinea pigs. Adult female guinea pigs (weight, 250–300 g; Charles River Laboratories) were anesthetized with 4% isoflurane, intubated, and placed on a ventilator with a vaporizer supplying 1.5–2% isoflurane. After lateral thoracotomy, a 30-gauge needle was inserted at the free wall apex of the left ventricle. 100 μl of adenovirus containing 1 × 109 plaque-forming units of Ad-TBX18-IRES-GFP or Ad-GFP (control group) was injected into the left ventricle apex. Guinea pig hearts were excised 1–2 weeks post-injection, frozen-sectioned, fixed, and immunostained with appropriate primary and secondary antibodies.
RT-PCR for MicroRNAs
TBX18- and GFP-transduced NRCMs were harvested (mirVanaTM microRNA isolation kit, Ambion, Austin, TX) at 4 days post-transduction. The samples were probed for miR-1, miR-133a, and miR-133b and evaluated by TaqMan quantitative PCR assay. Real time PCR was performed on a 7900HT fast real time PCR system (Applied Biosystems/Invitrogen).
Statistical Analysis
Data were analyzed for mean, standard deviation, and standard error of the mean. The quantitative figures in this work represent the mean ± S.E. of the measurement. p < 0.05 was considered significant unless indicated otherwise.
RESULTS
TBX18 Overexpression Leads to Reduction of Cx43
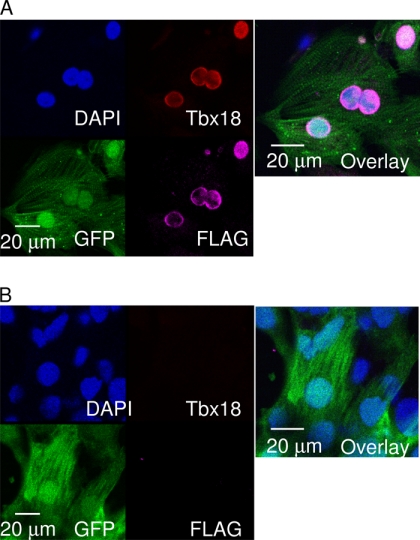
Monolayers of NRCMs were transduced with a bicistronic adenoviral vector constitutively expressing TBX18 with a green fluorescent reporter protein (Ad-TBX18-IRES-GFP). Three days post-transduction, the transduced (GFP-positive) NRCMs displayed robust nuclear expression of TBX18 (Fig. 1A). In contrast, cardiomyocytes transduced with a reporter gene alone (Ad-GFP, control) showed no detectable TBX18 (Fig. 1B). The initial data validated our strategy to study transcriptional targets of exogenous TBX18, which is not expressed endogenously in control myocytes.
FIGURE 1.
Heterologous expression of TBX18 in NRCMs. A short synthetic epitope (FLAG) was fused to the C-terminal end of TBX18 to distinguish the exogenous TBX18 from unknown levels of endogenous TBX18. Immunocytochemistry was performed with antibodies against TBX18 and FLAG in NRCMs 5 days post-adenoviral transduction. A, Ad-TBX18/FLAG-IRES-GFP-transduced NRCMs display nuclear expression of exogenous TBX18, which overlaps with the immunostaining against FLAG. B, Ad-GFP-transduced NRCMs (control) did not show an evidence of endogenous TBX18. Nuclei were co-stained with DAPI. The overlay images reveal nuclear co-localization of TBX18 and FLAG in TBX18-transduced NRCMs.
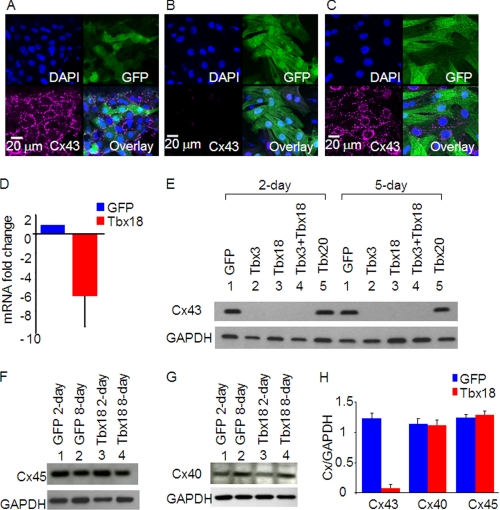
One of the key phenotypic hallmarks of SA nodal tissue is the characteristic high intercellular electrical resistance, which is essential to overcome impedance mismatch with surrounding atrial myocardium (19). Connexin43 is a negative marker of the SAN but is expressed robustly in the surrounding atria (9). Monolayers of NRCMs transduced with GFP only displayed typical punctate expression of Cx43 in the sarcolemma (Fig. 2A). In contrast, NRCMs transduced with TBX18 showed little Cx43 expression either at the sarcolemma or in the cytosol at 5 days post-transduction (Fig. 2B). Furthermore, Cx43 protein expression was substantially down-regulated in NRCMs expressing TBX18, TBX3, or TBX18 + TBX3 at 2- or 5-day post-transduction (Fig. 2E). Parallel to the down-regulation of Cx43 protein, the mRNA level of Cx43 decreased by 6.5-fold (Fig. 2D), indicating that direct transcriptional repression of Cx43 accounts for much, if not all, of the reduction in Cx43 protein. The effects of TBX18 were specific; TBX18 overexpression did not affect the protein levels of Cx45 (Fig. 2, F and H), the gap junction protein normally expressed in pacemaker cells (9, 19), or Cx40, the gap junction protein expressed in the bundle branches as well as in atria (Fig. 2, G and H) (20, 21). In addition, overexpression of TBX20, which underlies the differentiation of ventricular myocardium (22), had no effect on the expression of Cx43 (Fig. 2, C and E). Taken together, the data demonstrate that TBX18 represses Cx43 transcription and thus reduces Cx43 protein level, without affecting other connexins.
FIGURE 2.
TBX18 overexpression leads to marked down-regulation of Cx43. Adenoviral somatic gene transfer of control (A), TBX18 (B), and TBX20 (C) in NRCMs led to specific down-regulation of Cx43 in TBX18-transduced cells but not in GFP control or TBX20-transduced NRCMs at 5 days post-adenoviral transduction. Immunocytochemistry exhibits punctate, sarcolemmal expression of Cx43 in control NRCMs (A) and TBX20-expressing NRCMs (C). In contrast, marked down-regulation of Cx43 is observed in TBX18-overexpressing NRCMS (B). D, real time reverse transcriptase-PCR indicated an average of 6.5-fold down-regulation of Cx43 mRNA level compared with control NRCMs. E, Western blot experiments demonstrated that heterologous expression of TBX18, TBX3, and TBX18 + TBX3 nearly eliminated Cx43 protein expression compared with control and TBX20 NRCMs 2 days post-transduction (lanes 1–5) and 5 days post-transduction (lanes 6–10). No discernable changes are observed in the protein levels of Cx45 and Cx40 among control and TBX18-transduced NRCMs (F, G, and H).
TBX18 Overexpression Down-regulates Cx43 Expression in Adult Ventricular Myocardium in Vivo
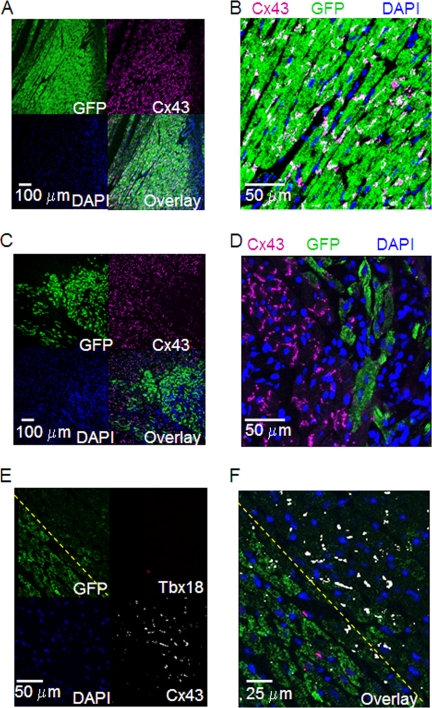
To investigate if the findings in cultured monolayers of NRCMs could be extended to adult myocardium in vivo, we expressed either TBX18 or control adenoviral vector in guinea pig hearts by direct myocardial injection. Immunochemistry on histological sections from the injection site revealed normal Cx43 expression in cardiomyocytes expressing GFP (Fig. 3, A and B). In contrast, sections from animals injected with TBX18 adenovirus showed striking down-regulation of Cx43 in regions where GFP was detected (Fig. 3, C and D). We then asked if Cx43 down-regulation persisted even after repression of TBX18 had faded. We took advantage of previous reports of slow turnover rate for GFP (at least 2 weeks (23)), and our observations that adenoviral vector-mediated expression of TBX18 in guinea pig ventricular myocardium subsides after about 1 week post-injection. At day 10 post-injection of Ad-TBX18-IRES-GFP, sections of the injected myocardium exhibited cardiomyocytes that were positive for GFP and TBX18 as well as cardiomyocytes that were positive for GFP alone. Although TBX18-positive cardiomyocytes were almost always negative for Cx43, some GFP+/TBX18− cardiomyocytes were also negative for Cx43 (Fig. 3, E and F). Taken together, the data demonstrate that suppression of Cx43 by TBX18 occurs in adult ventricular myocardium as well as in neonatal cardiomyocytes, and the down-regulation persists even after TBX18 expression fades.
FIGURE 3.
Heterologous expression of TBX18 in vivo significantly and specifically down-regulates Cx43 expression in guinea pig myocardium. Immunohistochemical staining of GFP-expressing left ventricular apex (control; A and B) and TBX18-expressing left ventricular apex (C and D) 10 days post-adenoviral transduction. Typical punctate Cx43 staining at the sarcolemma is observed in the control myocardium. In contrast, TBX18 expression in ventricular myocardium led to significant down-regulation of Cx43. Double immunostaining for Cx43 and TBX18 (E and F) revealed that the majority of the GFP-positive cardiomyocytes demarcated by the yellow dashed line were Cx43-negative and did not express TBX18.
TBX18 Directly Represses Cx43 Promoter
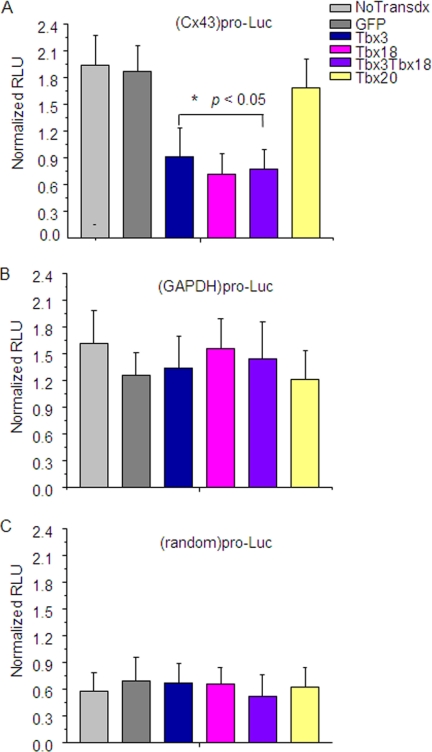
To investigate whether down-regulation of Cx43 in TBX18-overexpressing cells results from transcriptional repression, we transfected adenovirally transduced NRCMs with a reporter construct expressing luciferase gene under the Cx43 promoter. Fig. 4A shows that the relative luciferase activity was repressed in TBX18 (0.48 ± 0.15 RLUFirefly/RLURenilla)-, TBX3 (0.61 ± 0.28 RLUFirefly/RLURenilla)-, and TBX18 + TBX3 (0.41 ± 0.13 RLUFirefly/RLURenilla)-overexpressing NRCMS in comparison with freshly isolated nontransduced (2.0 ± 0.35 RLUFirefly/RLURenilla)- and GFP (1.8 ± 0.36 RLUFirefly/RLURenilla)-overexpressing NRCMs. In contrast, no change in reporter activity was observed in NRCMs overexpressing TBX20 (1.9 ± 0.42), which has been suggested to specify embryonic development of ventricular chambers (Fig. 4) (22). The promoter assay demonstrates that the repression of Cx43 by TBX18 and TBX3 is highly specific and occurs at the transcriptional level.
FIGURE 4.
TBX18 suppresses Cx43 promoter activity. A, relative Cx43 promoter activity in TBX3-, TBX18-, TBX3 + TBX18-, and TBX20-overexpressing NRCMs compared with the controls (freshly isolated NRCMs and GFP-NRCMs). *, p < 0.05. No change between the groups in GAPDH promoter activity; B, random promoter activity (negative control; C) was observed. Luc, luciferase.
Cx43 Down-regulation and Gap Junctional Uncoupling in TBX18-overexpressing Myocytes Lead to Lower Gap Junctional Permeability and Slow Conduction
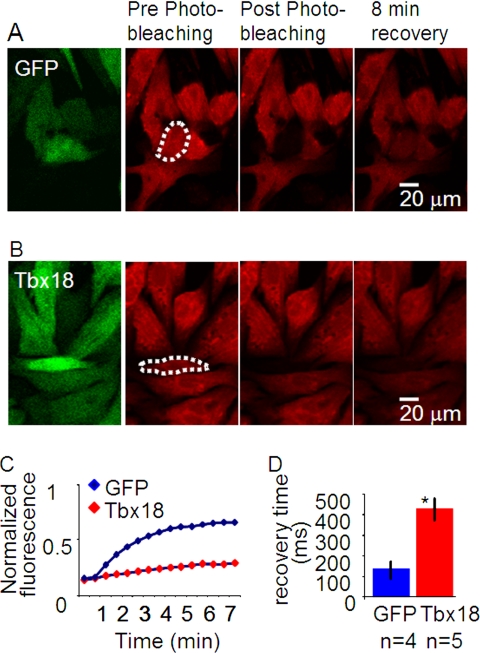
We next examined electrophysiological consequences of the Cx43 down-regulation. NRCM monolayers transduced with TBX18 were loaded with calcein orange, a membrane-impermeant fluorescent dye that has been shown to passively cross gap junctions (24). Focused laser illumination of single fluorescent myocytes completely photobleached the target cells, which were surrounded by nonphotobleached, fluorescent myocytes. Fluorescence recovery after photobleaching provides a quantitative index of gap junctional permeability (25). TBX18-transduced myocytes exhibited a time constant of fluorescence recovery after photobleaching, which was prolonged by more than 3-fold relative to control (420.9 ± 54.3 ms, n = 5, versus 127.3 ± 42.5 ms, n = 4; p < 0.05; Fig. 5).
FIGURE 5.
TBX18 expression decreases dye transfer kinetics between cells on a monolayer. Cell-cell gap junctional coupling was assayed by calcein orange dye transfer kinetics using fluorescence recovery after photobleaching. A, control (Ad-GFP-transduced NRCMs); B, TBX18-transduced NRCMs. Dotted area outlines the photobleached cell. C, time course of calcein orange fluorescence recovery after photobleaching in GFP- and TBX18-transduced NRCMs. D, summary of the recovery kinetics (tau) after photobleaching indicates the recovery time in TBX18-transduced NRCMs (420.9 ± 54.3 ms; n = 5) is significantly larger than the control (127.3 ± 42.5 ms; n = 4; p < 0.05).
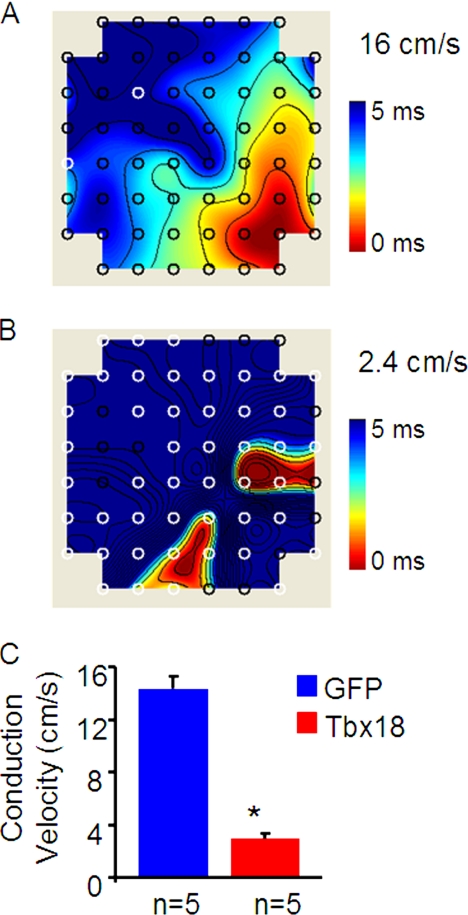
Reduced cell-cell coupling would be expected to delay propagation of action potentials in NRCM monolayers. To gauge electrophysiological effects of the Cx43 down-regulation, extracellular field potentials were recorded in NRCM monolayers with multielectrode array. Analyses of isochrone maps by plotting field potential activation times at each of the 64 electrodes demonstrate that TBX18-transduced NRCM monolayers exhibit marked slowing of conduction velocity (2.9 ± 0.5 cm/s, n = 5) compared with controls (14.3 ± 0.9 cm/s, n = 5; p < 0.005; Fig. 6), recapitulating the slow action potential propagation documented within the SAN (26).
FIGURE 6.
TBX18 expression slows conduction velocity in cardiomyocyte monolayers. Multielectrode array (MEA) recordings of conduction velocities demonstrate uniform and repetitive propagation of action potentials with a conduction velocity of 16 cm/s (A). In contrast, TBX18-transduced NRCM monolayers exhibited multiple foci of action potential propagation origin with a significantly lower conduction velocity of 2.4 cm/s (B). C, averaged data show decrease in the conduction velocity in TBX18-transduced NRCM monolayers (2.9 ± 0.5 cm/s; n = 5) in comparison with GFP-transduced NRCMs (14.3 ± 0.9 cm/s; n = 5; p < 0.005).
Unsynchronized Ca2+ Transients
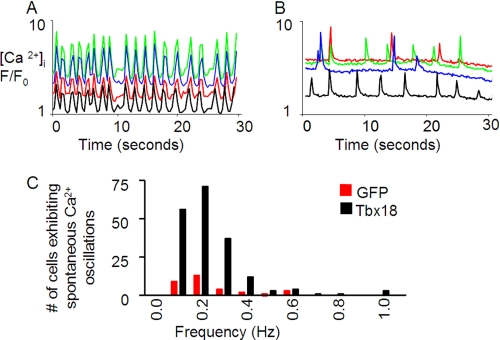
Intracellular Ca2+ oscillations were examined by confocal microscopy to further assay the extent and physiological consequences of electrical uncoupling. Monolayers of control NRCMs loaded with a Ca2+-sensitive dye, Rhod-2AM, exhibited rhythmic contractions and synchronous Ca2+ transients (Fig. 7A). In contrast, multiple asynchronous and intracellular Ca2+ oscillations were observed in densely packed neighboring myocytes within TBX18-transduced NRCM monolayers (Fig. 7B). These findings complement the electrical recordings that demonstrated propagation, albeit limited in velocity, of field potentials in TBX18-transduced NRCM monolayers.
FIGURE 7.
TBX18 expression leads to cell-cell uncoupling in NRCM monolayers. Two-dimensional confocal microscopy with Rhod-2AM was employed to acquire spatially averaged F/F0 plots of spontaneous oscillations in intracellular Ca2+ concentrations. In control, synchronous calcium transients were observed capturing the entire monolayer (A). In contrast, TBX18-transduced NRCMs revealed frequent episodes of asynchronous and independent (nonsyncitial) Ca2+ transients in individual cells (B). C, number of cells that exhibit spontaneous Ca2+ oscillations were significantly higher in the TBX18-transduced NRCM monolayers in comparison with GFP-transduced NRCMS upon the treatment with a gap-junction uncoupler, palmitoleic acid (40 μm).
The asynchronous intracellular Ca2+ oscillations observed in TBX18-transduced NRCM monolayers could be mimicked in control monolayers by exposing the myocytes to a specific gap junction uncoupler, palmitoleic acid (27). Addition of palmitoleic acid (40 μm) led to multiple spontaneous and asynchronous intracellular Ca2+ oscillations in control monolayers, reminiscent of TBX18-transduced NRCM monolayers. Interestingly, the number of TBX18-transduced myocytes displaying spontaneous Ca2+ oscillations was about 6-fold greater than that of control myocytes in normal Tyrode solution with palmitoleic acid (40 μm) (Fig. 7C).
DISCUSSION
We have demonstrated that TBX18 specifically down-regulates Cx43 at both the mRNA and protein levels, while having no effect on Cx40 or Cx45. Additionally, Cx43 promoter activity is directly repressed by TBX18 and TBX3, but not by TBX20, which plays a role during ventricular chamber formation (28). Reduced Cx43 is predicted to weaken cell-cell electrotonic coupling. Indeed, we observed slowed dye transfer kinetics through gap junctions and substantially decreased conduction velocity in TBX18-expressing cardiomyocyte monolayers.
Our in vitro promoter assay data indicate that the down-regulation of Cx43 upon TBX18 expression is mediated by direct transcriptional repression (Fig. 4). TBX18 caused complete repression of Cx43 promoter activity because the level of the Cx43 promoter-driven luciferase activity was similar to that of the negative control group. Co-expression of TBX18 with TBX3 did not cause further suppression of the Cx43 promoter activity, reflecting that expression of TBX18 alone already caused maximum suppression. In addition, a consensus T-box-binding motif (TCACAC) is present in the promoter region of Cx43 in zebrafish, mouse, rat, and human (29). Based on these data, we conclude that direct transcriptional repression accounts for the specific down-regulation of Cx43 observed in our culture system. Nevertheless, these in vitro data do not exclude the possibility that TBX18-mediated modulation of gap junctional coupling could be sustained through indirect and/or long range pathways even after the heterologous expression has expired. Because cultures of NRCM are viable up to about 1 week, we injected TBX18 into guinea pig ventricular myocardium in vivo. Similar to the in vitro data, we routinely observed stark down-regulation of Cx43 in cardiomyocytes that expressed TBX18 shortly after the gene transfer (2–4 days post-injection). In some sections, Cx43 down-regulation persisted even after transient expression of TBX18 had faded away (10 days after gene injection, Fig. 3, E and F). Given the transient nature of adenoviral gene transfer and the very short half-lives of Cx43 proteins (1.5–2 h for NRCMs and 1.3 h for adult rat ventricular myocardium) (30, 31), the long term down-regulation of Cx43 seems likely to be due to the cell autonomous effects after the cessation of heterologous TBX18 expression.
Tbx18 functions as a transcriptional repressor by forming homo- or heterodimers with other T-box proteins or with other families of transcription factors (32). For example, Tbx18 can bind to the zinc finger domain of Gata4 or the homeobox domain of NKx2.5 and repress the cardiac expression of Nppa (natriuretic peptide precursor type a, also known as atrial natriuretic factor) (32). Thus, heterodimerization of Tbx18 with other transcription factors could provide multilayer mechanisms of regulation of cardiac pacemaker gene expression and explain the indirect and long range effects of TBX18 we observed in adult ventricular myocardium. Our observations of long term, gap junctional modeling effects of TBX18 are corroborated by the recent and significant reports of cellular reprogramming by transient expression of embryonic transcription factors as follows: 1) induced pluripotent stem cells derived from reprogramming of somatic cells (33), and 2) conversion of pancreatic nonislet cells into insulin-producing islet cells in situ (34). Although a deeper understanding of our in vivo observations is beyond the scope of this study, the data warrant further investigations to study other significant targets of Tbx18 and the possibility of permanent reprogramming.
Our findings also hint at the possibility that TBX18 may help to shape other features of pacemaker cells. For example, TBX18 increased the number of cardiomyocytes exhibiting spontaneous intracellular Ca2+ oscillations (Fig. 7C). These data motivate future investigation of the effects of TBX18 expression on “Ca2+ and membrane clock” processes in the generation of SA nodal pacemaker activity (35–37).
Up-regulation of a muscle-specific microRNA, miR-1, has been shown to down-regulate Cx43 and Kir2.1 (38). We examined TBX18-transduced NRCMs and their possible interaction with miR-1. NRCMs transduced with TBX18 or control vector produced similar amounts of miR-1 by RT-PCR (supplemental Tables 1 and 2), suggesting that the mechanisms of Cx43 down-regulation by TBX18 and miR-1 are likely independent of each other.
Aside from the essential role of Tbx18 in cardiac pacemaker specification, Tbx18 has also been suggested to be expressed in pools of proepicardial progenitor cells and contribute to cardiac ventricular lineage (39), although the findings have been debated (40). We looked for but did not identify any potential cardiac progenitors in TBX18-transduced NRCMs by immunostaining for stem cell antigens c-Kit and Isl-1 (data not shown).
In summary, postnatal re-expression of TBX18 targets Cx43 via transcriptional repression and thereby weakens gap junctional coupling, recapitulating a key phenotypic feature of pacemaker tissue. This effect was specific; Cx45 and Cx40 were not modified by TBX18 (Fig. 2, F and G). Thus, TBX18 expression recapitulates at least some cardinal features of the SA node. Combining TBX18 with other transcription factors may facilitate definitive trans-differentiation of ventricular cardiomyocytes to pacemaker cells.
Acknowledgments
We thank Prabhav Anand and Baiming Sun for technical assistance and Dr. Thomas H. Steinberg (Washington University, St. Louis) for the generous gift of Cx45 primary antibody.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1, Tables 1 and 2, and an additional reference.
- SA
- sinoatrial
- SAN
- sinoatrial node
- NRCM
- neonatal rat cardiomyocyte
- RLU
- relative luciferase unit.
REFERENCES
- 1. Wiese C., Grieskamp T., Airik R., Mommersteeg M. T., Gardiwal A., de Gier-de Vries C., Schuster-Gossler K., Moorman A. F., Kispert A., Christoffels V. M. (2009) Circ. Res. 104, 388–397 [DOI] [PubMed] [Google Scholar]
- 2. McNally E. M., Svensson E. C. (2009) Circ. Res. 104, 285–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hoogaars W. M., Engel A., Brons J. F., Verkerk A. O., de Lange F. J., Wong L. Y., Bakker M. L., Clout D. E., Wakker V., Barnett P., Ravesloot J. H., Moorman A. F., Verheijck E. E., Christoffels V. M. (2007) Genes Dev. 21, 1098–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hoogaars W. M., Barnett P., Moorman A. F., Christoffels V. M. (2007) Cell. Mol. Life Sci. 64, 646–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mommersteeg M. T., Dominguez J. N., Wiese C., Norden J., de Gier-de Vries C., Burch J. B., Kispert A., Brown N. A., Moorman A. F., Christoffels V. M. (2010) Cardiovasc. Res., [DOI] [PubMed] [Google Scholar]
- 6. Christoffels V. M., Smits G. J., Kispert A., Moorman A. F. (2010) Circ. Res. 106, 240–254 [DOI] [PubMed] [Google Scholar]
- 7. Snarr B. S., O'Neal J. L., Chintalapudi M. R., Wirrig E. E., Phelps A. L., Kubalak S. W., Wessels A. (2007) Circ. Res. 101, 971–974 [DOI] [PubMed] [Google Scholar]
- 8. Chandler N. J., Greener I. D., Tellez J. O., Inada S., Musa H., Molenaar P., Difrancesco D., Baruscotti M., Longhi R., Anderson R. H., Billeter R., Sharma V., Sigg D. C., Boyett M. R., Dobrzynski H. (2009) Circulation 119, 1562–1575 [DOI] [PubMed] [Google Scholar]
- 9. Vozzi C., Dupont E., Coppen S. R., Yeh H. I., Severs N. J. (1999) J. Mol. Cell. Cardiol. 31, 991–1003 [DOI] [PubMed] [Google Scholar]
- 10. Davis L. M., Rodefeld M. E., Green K., Beyer E. C., Saffitz J. E. (1995) J. Cardiovasc. Electrophysiol. 6, 813–822 [DOI] [PubMed] [Google Scholar]
- 11. Burt J. M., Spray D. C. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 3431–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Valiunas V., Weingart R., Brink P. R. (2000) Circ. Res. 86, E42–E49 [DOI] [PubMed] [Google Scholar]
- 13. Moreno A. P., Laing J. G., Beyer E. C., Spray D. C. (1995) Am. J. Physiol. 268, C356–C365 [DOI] [PubMed] [Google Scholar]
- 14. Valiunas V. (2002) J. Gen. Physiol. 119, 147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Joyner R. W., van Capelle F. J. (1986) Biophys. J. 50, 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coppen S. R., Kodama I., Boyett M. R., Dobrzynski H., Takagishi Y., Honjo H., Yeh H. I., Severs N. J. (1999) J. Histochem. Cytochem. 47, 907–918 [DOI] [PubMed] [Google Scholar]
- 17. Bakker M. L., Boukens B. J., Mommersteeg M. T., Brons J. F., Wakker V., Moorman A. F., Christoffels V. M. (2008) Circ. Res. 102, 1340–1349 [DOI] [PubMed] [Google Scholar]
- 18. Sekar R. B., Kizana E., Cho H. C., Molitoris J. M., Hesketh G. G., Eaton B. P., Marbán E., Tung L. (2009) Circ. Res. 104, 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verheijck E. E., van Kempen M. J., Veereschild M., Lurvink J., Jongsma H. J., Bouman L. N. (2001) Cardiovasc. Res. 52, 40–50 [DOI] [PubMed] [Google Scholar]
- 20. Leaf D. E., Feig J. E., Vasquez C., Riva P. L., Yu C., Lader J. M., Kontogeorgis A., Baron E. L., Peters N. S., Fisher E. A., Gutstein D. E., Morley G. E. (2008) Circ. Res. 103, 1001–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saffitz J. E., Schuessler R. B. (2000) Circ. Res. 87, 835–836 [DOI] [PubMed] [Google Scholar]
- 22. Singh M. K., Christoffels V. M., Dias J. M., Trowe M. O., Petry M., Schuster-Gossler K., Bürger A., Ericson J., Kispert A. (2005) Development 132, 2697–2707 [DOI] [PubMed] [Google Scholar]
- 23. Leutenegger A., D'Angelo C., Matz M. V., Denzel A., Oswald F., Salih A., Nienhaus G. U., Wiedenmann J. (2007) FEBS J. 274, 2496–2505 [DOI] [PubMed] [Google Scholar]
- 24. Elzarrad M. K., Haroon A., Willecke K., Dobrowolski R., Gillespie M. N., Al-Mehdi A. B. (2008) BMC Med. 6, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wade M. H., Trosko J. E., Schindler M. (1986) Science 232, 525–528 [DOI] [PubMed] [Google Scholar]
- 26. Fedorov V. V., Schuessler R. B., Hemphill M., Ambrosi C. M., Chang R., Voloshina A. S., Brown K., Hucker W. J., Efimov I. R. (2009) Circ. Res. 104, 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burt J. M., Massey K. D., Minnich B. N. (1991) Am. J. Physiol. 260, C439–C448 [DOI] [PubMed] [Google Scholar]
- 28. Brown D. D., Martz S. N., Binder O., Goetz S. C., Price B. M., Smith J. C., Conlon F. L. (2005) Development 132, 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chatterjee B., Chin A. J., Valdimarsson G., Finis C., Sonntag J. M., Choi B. Y., Tao L., Balasubramanian K., Bell C., Krufka A., Kozlowski D. J., Johnson R. G., Lo C. W. (2005) Dev. Dyn. 233, 890–906 [DOI] [PubMed] [Google Scholar]
- 30. Beardslee M. A., Laing J. G., Beyer E. C., Saffitz J. E. (1998) Circ. Res. 83, 629–635 [DOI] [PubMed] [Google Scholar]
- 31. Laird D. W., Puranam K. L., Revel J. P. (1991) Biochem. J. 273, 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Farin H. F., Bussen M., Schmidt M. K., Singh M. K., Schuster-Gossler K., Kispert A. (2007) J. Biol. Chem. 282, 25748–25759 [DOI] [PubMed] [Google Scholar]
- 33. Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. (2008) Science, [DOI] [PubMed] [Google Scholar]
- 34. Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D. A. (2008) Nature 455, 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joung B., Ogawa M., Lin S. F., Chen P. S. (2009) Korean Circ. J. 39, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maltsev V. A., Lakatta E. G. (2008) Cardiovasc. Res. 77, 274–284 [DOI] [PubMed] [Google Scholar]
- 37. Vinogradova T. M., Maltsev V. A., Bogdanov K. Y., Lyashkov A. E., Lakatta E. G. (2005) Ann. N. Y. Acad. Sci. 1047, 138–156 [DOI] [PubMed] [Google Scholar]
- 38. Yang B., Lin H., Xiao J., Lu Y., Luo X., Li B., Zhang Y., Xu C., Bai Y., Wang H., Chen G., Wang Z. (2007) Nat. Med. 13, 486–491 [DOI] [PubMed] [Google Scholar]
- 39. Cai C. L., Martin J. C., Sun Y., Cui L., Wang L., Ouyang K., Yang L., Bu L., Liang X., Zhang X., Stallcup W. B., Denton C. P., McCulloch A., Chen J., Evans S. M. (2008) Nature 454, 104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Christoffels V. M., Grieskamp T., Norden J., Mommersteeg M. T., Rudat C., Kispert A. (2009) Nature 458, E8–E9; discussion E9–E10 [DOI] [PubMed] [Google Scholar]