Abstract
Programmed cell death (PCD) associated with immunity is triggered when a plant disease resistance (R) protein recognizes a corresponding pathogen virulence protein. In tomato, detection by the host Pto kinase of the Pseudomonas syringae proteins AvrPto or AvrPtoB causes localized PCD. Previously, we reported that both MAPKKKα (mitogen-activated protein kinase kinase kinase) and the tomato 14-3-3 protein 7 (TFT7) positively regulate Pto-mediated PCD in tomato and Nicotiana benthamiana. In addition, in contrast to MAPKKKα, TFT7 is required for PCD mediated by four other R proteins. Here we investigate why TFT7 is required for PCD induced by diverse R proteins in plants. We discovered that a MAPKK, SlMKK2, which acts downstream of SlMAPKKKα, also interacts with TFT7 in plant cells. Gene silencing experiments revealed that the orthologous genes of both SlMKK2 and TFT7 in N. benthamiana are required for PCD mediated by the same set of R proteins. SlMKK2 and its orthologs contain a 14-3-3 binding site in their N terminus, and Thr33 in this site is required for interaction with TFT7 in vivo. Like the structurally similar human 14-3-3ϵ protein, TFT7 forms a homodimer in vivo. Because TFT7 interacts with both SlMAPKKKα and SlMKK2 and also forms a homodimer, we propose that TFT7 may coordinately recruit these client proteins for efficient signal transfer, leading to PCD induction.
Keywords: Cell Death, Gene Silencing, MAP Kinases (MAPKs), Plant, Signal Transduction, 14-3-3 Proteins, Disease Resistance Proteins, Phosphopeptide-binding Proteins
Introduction
Plants have a sophisticated immune system that recognizes virulence proteins, also referred to as effectors, from diverse pathogens, including bacteria, fungi, oomycetes, and viruses. This recognition depends on plant disease resistance (R)2 proteins that either directly or indirectly interact with a cognate pathogen effector, resulting in activation of “effector-triggered immunity” typically associated with a hypersensitive response (1). The hypersensitive response is a rapid, localized, programmed cell death (PCD) at the site of attempted infection that likely plays a role in limiting pathogen spread (2). Pto, a tomato R protein, encodes a serine/threonine protein kinase and exists in a protein complex with the oligomeric Prf protein (3–5). Prf has a nucleotide-binding site and a region of leucine-rich repeats and belongs to a large class of plant R proteins. The Pto-Prf complex confers resistance to bacterial speck disease in both tomato and Nicotiana benthamiana plants by recognizing the presence of the effectors AvrPto or AvrPtoB in Pseudomonas syringae pv. tomato (4).
When Pto is transiently co-expressed in N. benthamiana leaves with either AvrPto or a truncated form of AvrPtoB (AvrPtoB1–387), strong PCD is induced (6, 7). Several other R protein/effector pairs are also known to induce PCD in N. benthamiana. For example, RPS2 and RPP13, Arabidopsis nucleotide-binding and leucine-rich repeat R proteins, recognize the P. syringae effector AvrRpt2 and the oomycete Hyaloperonospora arabidopsidis effector ATR13Emco5, respectively, and induce PCD in N. benthamiana (8, 9). Rx2 and Gpa2, potato nucleotide-binding and leucine-rich repeat R proteins, recognize the potato virus X coat protein and the potato cyst nematode Globodera pallida effector RBP-1, respectively, and also induce PCD in N. benthamiana (10, 11). The induction of PCD by heterologous resistance proteins originating from other plant species implies that N. benthamiana carries well conserved downstream effector-triggered immunity signaling pathways. In addition to this characteristic, N. benthamiana is very amenable to virus-induced gene silencing (VIGS), which is a widely used reverse genetics approach for determining the function of plant genes (12, 13).
Many proteins that act in Pto-mediated signaling pathways have been identified by screening for compromised resistance in tomato (Solanum lycopersicum) or N. benthamiana by loss-of-function approaches, including VIGS (14–16). For example, HSP90, SGT1, and RAR1, which exist as a protein complex and function as chaperones of many plant resistance proteins, are known to be involved in Pto signaling in N. benthamiana (15, 17, 18). Another important class of genes that positively regulates Pto signaling are components of MAPK cascades. A MAPK cascade typically contains a MAPK kinase kinase (MAPKKK) that is phosphorylated by upstream protein kinases for activation (19). Once the MAPKKK is activated, it phosphorylates a downstream MAPKK, which phosphorylates a MAPK to activate its target proteins, such as transcription factors (20). In Pto signal transduction, two MAPKKKs (MAPKKKα and MAPKKKϵ) have been shown to function as positive regulators (16, 21). Based on VIGS assays and immunocomplex kinase assays, two MAPKKs (MEK1 and MEK2) and two MAPKs (salicylic acid-induced protein kinase and wound-induced protein kinase) have been proposed to act downstream of MAPKKKα (16, 22). In addition, on the basis of VIGS assays, MEK2 and wound-induced protein kinase are proposed to act downstream of MAPKKKϵ (21). Interestingly, expression of MAPKKKα, MAPKKKϵ, or a constitutively active form of MEK2 (MEK2DD) in N. benthamiana each induces PCD, further reinforcing their role in Pto-mediated PCD (16, 21, 23).
We recently reported a new component, a 14-3-3 protein TFT7, in Pto signaling that functions as a positive regulator (24). 14-3-3 proteins typically bind to phosphorylated “client” proteins to regulate activities of the client protein. They are known to play a role in regulating many important biological processes, including development and growth, protein trafficking, and PCD in both animals and plants (25–27). Four major modes of action of 14-3-3 proteins have been described, and one of them is to regulate protein stability of client proteins (27, 28).
TFT7 physically interacts with a C-terminal region of the tomato MAPKKKα (SlMAPKKKα) that contains one of two previously described 14-3-3 binding motifs (R/K-X-X-X-pS/T-X-P), where pS/T indicates a phosphorylated serine or threonine and X indicates a random amino acids (24). An alanine substitution at Ser535 in the 14-3-3 binding motif of SlMAPKKKα reduces protein stability of the kinase and reduces its PCD-eliciting activity. This mechanism is similar to how the human MAPKKK, MEKK3, which is responsible for apoptosis induced by several environmental stimuli in human cells, is regulated by the human 14-3-3ϵ protein (29). Interestingly, of the seven human 14-3-3 proteins, 14-3-3ϵ is the most similar to TFT7 at both the amino acid level and the structural level (24). In addition to TFT7, there is one other example where a 14-3-3 protein has been shown to contribute to plant immunity. Arabidopsis 14-3-3λ interacts with and regulates the function of RPW8, a protein that confers resistance to powdery mildew in Arabidopsis (30).
In addition to its role in Pto signaling, we reported that TFT7 is also involved in defense signaling mediated by other R proteins, including RPS2, RPP13, Rx2, and Gpa2 (24). On the basis of this activity of TFT7 downstream of several PCD elicitors, we hypothesized that TFT7 may associate with other host proteins that are necessary for PCD elicitation. In this study, we report that a tomato MAPKK (SlMKK2), an ortholog of tobacco MEK2 (NtMEK2), is necessary for PCD mediated by the same set of resistance proteins as is TFT7. In addition, we show that TFT7 physically interacts with SlMKK2 and can form a homodimer in vivo.
EXPERIMENTAL PROCEDURES
Bacterial, Yeast, and Plant Species
Agrobacterium tumefaciens strain GV2260 was grown at 30 °C in Luria-Bertani medium with appropriate antibiotics. Yeast strain EGY48(pSH18–34) was grown at 30 °C in synthetic drop-out medium with glucose as the carbon source. N. benthamiana plants were grown in the greenhouse with light for 16 h and at a temperature of 24–26 °C (gene-silenced plants were maintained in the growth chamber, see below).
Site-directed Mutagenesis
For substitution of threonine 33 to alanine in both SlMKK2 and SlMKK2DD, PCR-based site-directed mutagenesis was performed as described previously (31).
VIGS
Gene silencing constructs for TFT7 or NtMEK2 (24) in a tobacco rattle virus (TRV) vector were used for VIGS in N. benthamiana, and an empty TRV vector was used as a control. Two- to three-week-old seedlings were infected with the TRV constructs, and the plants were maintained in a growth chamber with light for 16 h at a temperature of 24 °C (32). After 4–5 weeks in the growth chamber, the gene-silenced plants were used for cell death assays.
Cell Death and Electrolyte Leakage Assays
Seven different cell death elicitors were transiently expressed by Agrobacterium infiltration in leaves of gene-silenced plants as described previously (24). The elicitors used were Pto/AvrPto, SlMAPKKKα full-length, SlMAPKKKα kinase domain only, RPS2/AvrRpt2, Rx2/CP, RPP13/ATR13Emco5Δ40, and Gpa2/RBP-1. Cell death induced by SlMKK2DD or its mutant form, SlMKK2DD(T33A) was compared in wild-type N. benthamiana. In the case of SlMAPKKKα and SlMKK2DD, 2 μm of estradiol was applied to the agro-infiltrated area of leaves 2 days after infiltration. To quantify the degree of cell death, electrolyte leakage was measured as described previously (24). Briefly, two leaf discs (9 mm in diameter) were collected from each agro-infiltrated area and kept in water for 2 h at 25 °C. Ion conductivity was measured by using an Acorn CON 5 ion conductivity meter (Oakton, Vernon Hills, IL). For each cell death elicitor, data were collected from six different agro-infiltrated areas (one per plant) and were analyzed statistically by a t test (p < 0.05).
Yeast Two-hybrid Assay
The full-length genes of SlMKK2 and SlMKK3 were cloned into the bait vector, pEG202, for a yeast two-hybrid assay with the full-length TFT7 gene in the prey vector, pJG4–5 (24). The yeast two-hybrid assay was performed with strain EGY48(pSH18–34), as described in the Yeast Protocols Handbook (Clontech). To check protein expression of SlMKK2 and SlMKK3 in yeast, strains were grown overnight in synthetic drop-out media lacking uracil, histidine, and tryptophan, and total proteins were released by addition of 1× SDS loading buffer.
Co-immunoprecipitation
For co-IP, proteins were transiently co-expressed by Agrobacterium infiltration in leaves of N. benthamiana. Expression of HA-tagged SlMKK2 and its derivatives was controlled by the estradiol-inducible system using the vector pER8. Expression of TFT7 or GFP proteins was controlled by the CaMV 35S promoter in the vector pBTEX. Co-IP assays were performed as described previously (24). Briefly, 24 h after estradiol treatment, 18 leaf discs (9 mm in diameter) for each protein combination were collected for protein extraction with the extraction buffer (GTEN) (10% glycerol, 25 mm Tris (pH 7.5), 1 mm EDTA, 150 mm NaCl), 10 mm DTT, 0.1% Triton X-100, 1× plant protease inhibitor (Sigma), 1× phosphatase inhibitor (Sigma), and 2% w/v polyvinypolylpyrrolidone). For samples involving Pto/AvrPto-mediated PCD, 22 leaf discs (9 mm in diameter) were used for protein extraction. During desalting of the protein extracts, the buffer was exchanged to IP buffer (GTEN, 0.15% Nonidet P-40, 2 mm DTT). Desalted protein extracts were incubated with goat IgG-agarose beads (Rockland, Gilbertville, PA) to remove proteins that bind to agarose beads. Protein extracts were incubated with HA affinity matrix (Roche) or FLAG tag antibody-agarose beads (Sigma) for IP overnight. Finally, beads were collected and washed four to six times with the IP buffer.
Western Blotting
Proteins were separated in 10–12% SDS-PAGE gel and transferred onto Immobilon-P membrane (Millipore, Billerica, MA) by an electroblotter (Bio-Rad). Western blotting was carried out with the ECL PLUS Western detection kit (GE Healthcare). A FLAG antibody (Sigma) or a HA antibody (Roche) was used for detecting HA-tagged or FLAG-tagged proteins from N. benthamiana. A LexA antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used for SlMKK2 and SlMKK3 from yeast.
Protein Sequence Alignment
Protein sequences of MAPKKs were obtained from the GenBanktm database: SlMKK2, AAU04434; SlMKK3, AAU04435; AtMKK4, O80397; AtMKK5, NP_188759; AtMKK6, NP_200469; NtMEK1, CAC24705; NtMEK2, AAG53979; OsMKK4, BAH00050; and MtSIMKK, CAC69137. Protein sequence alignment was performed by using the ClustalW method in the MegAlign software (Lasergene, DNAStar, UK).
RESULTS
The N. benthamiana Orthologs of TFT7 and SlMKK2 Are Required for PCD Mediated by Diverse Resistance Proteins
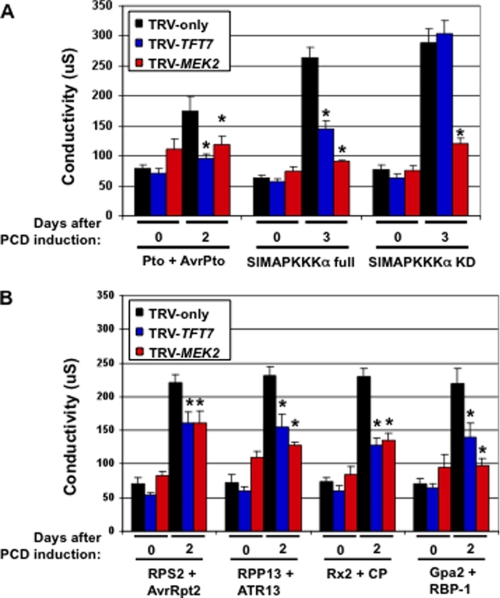
We previously found that silencing of TFT7 compromised PCD caused by diverse R proteins. This observation suggested that TFT7 may regulate a host component that plays a conserved role in PCD signaling. MEK2 is a good candidate for such a component, as it is known to be necessary for PCD mediated by at least two R proteins, Pto and N (33), and expression of its constitutively active form, MEK2DD, in leaves causes strong cell death (23). However, it has not been investigated previously whether MEK2 is necessary for PCD induced by R proteins other than N and Pto. We therefore examined the effect of silencing the MEK2 gene in N. benthamiana on PCD mediated by several diverse R proteins (RPS2, RPP13, Rx2, and Gpa2) that were shown previously to require TFT7 (24). The TFT7 or MEK2 genes were silenced by TRV-based VIGS in N. benthamiana, and the degree of PCD induced by seven different elicitors, as shown in Fig. 1, was compared in both TFT7- or MEK2-silenced plants by measuring electrolyte leakage. TRV-only infected plants were used as the control.
FIGURE 1.
Silencing of either TFT7 or MEK2 (a MAPK kinase) compromises PCD. A, PCD mediated by Pto and SlMAPKKKα. B, PCD mediated by other resistance proteins. TRV was used to knock down expression of the TFT7 or MEK2 genes in N. benthamiana. TRV-only was used as a negative control. To determine the degree of PCD, electrolyte leakage induced by each elicitor was measured. Two leaf discs (9 mm in diameter) were collected from the agro-infiltrated area at the indicated time points and incubated in 2 ml of water for 2 h at 25 °C. Ion conductivity was measured using an Acorn CON 5 ion conductivity meter. The asterisk indicates a significant reduction (t test; p < 0.05) in electrolyte leakage by silencing of TFT7 or MEK2 compared with TRV-only plants. Data presented are means of six infiltrated areas (one per plant). Bars indicate standard errors.
We observed that all seven elicitors caused high electrolyte leakage in the TRV-only plants at day 2 or 3 after PCD was induced (Fig. 1). In contrast, electrolyte leakage associated with Pto/AvrPto, RPS2/AvrRpt2, RPP13/ATR13Emco5Δ40, Rx2/CP, Gpa2/RBP-1, and SlMAPKKKα full-length was significantly reduced in both TFT7- and MEK2-silenced plants. In response to expression of the SlMAPKKKα kinase domain only, which causes very strong PCD, we observed a reduction in electrolyte leakage only in MEK2-silenced plants. These results indicate that TFT7 and MEK2 are required not only for Pto-mediated PCD, but also for PCD mediated by RPS2, RPP13, Rx2, and Gpa2.
TFT7 Interacts with SlMKK2 in Vivo Regardless of PCD Induction
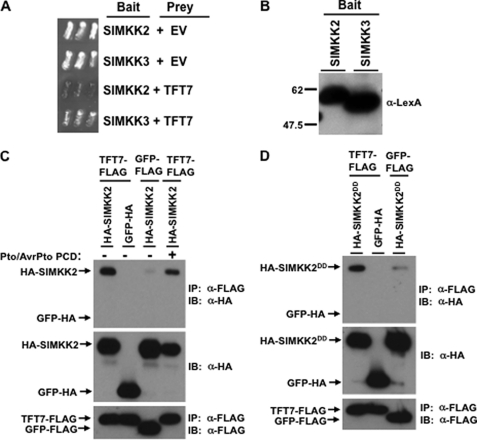
The observation that both TFT7 and MEK2 in N. benthamiana are required for PCD mediated by the same set of R proteins supported the hypothesis that MEK2 is a client protein of TFT7 in tomato. To test this hypothesis further, we examined the possible protein-protein interaction between TFT7 and SlMKK2 by a yeast two-hybrid assay. Previously, SlMKK3, a NtMEK1 ortholog, was also reported to be located downstream of SlMAPKKKα and to be involved in PCD mediated by SlMAPKKKα (16, 22). We therefore included SlMKK3 in our yeast two-hybrid assay. We observed that SlMKK2, but not SlMKK3, interacted with TFT7 in yeast (Fig. 2A), although both proteins were expressed equally in yeast (Fig. 2B).
FIGURE 2.
TFT7 interacts with SlMKK2 in yeast and in vivo. A, the TFT7 gene in the prey vector was analyzed in a yeast two-hybrid system with either SlMKK2 or SlMKK3 genes in the bait vector. Yeast transformants were streaked onto plates with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and photographs were taken 2 days later. Blue patches indicate positive interaction. EV, empty prey vector. B, expression of the SlMKK2 and SlMKK3 proteins in yeast was verified using immunoblotting with an anti-LexA antibody. C and D, for co-IP, the TFT7-FLAG or GFP-FLAG and HA-SlMKK2 or HA-SlMKK2DD proteins were co-expressed in N. benthamiana. Estradiol was applied to the leaves 36 h after agro-infiltration to induce expression of HA-SlMKK2 or HA-SlMKK2DD. Leaf discs were collected for protein extraction 18 h after estradiol treatment, and α-FLAG-agarose beads were used for IP. GFP-HA was used as a negative control. Immunoblotting (IB) was performed with α-HA for SlMKK2 or GFP and with α-FLAG for TFT7. To induce Pto-mediated PCD, AvrPto was expressed by estradiol-inducible system, and leaf discs were sampled when yellowing phenotype appeared but before PCD appeared.
To test the interaction between TFT7 and SlMKK2 in plants, co-IP experiments were performed by co-expressing FLAG-tagged TFT7 and HA-tagged SlMKK2 with or without Pto/AvrPto-mediated PCD in N. benthamiana, followed by immunoprecipitation with FLAG-tag antibody-agarose beads. As shown in Fig. 2C, SlMKK2 co-immunoprecipitated with TFT7 in vivo, regardless of Pto/AvrPto-mediated PCD. The constitutive-active form of SlMKK2, SlMKK2DD, also co-immunoprecipitated with TFT7 in vivo (Fig. 2D). A negative control, GFP-HA, did not co-immunoprecipitate with any of the tomato proteins in this experiment. These results demonstrate that SlMKK2 interacts with TFT7 and is therefore a putative client of this 14-3-3 protein.
SlMKK2 and Its Orthologs Contain a Putative 14-3-3 Binding Site in Their N Terminus
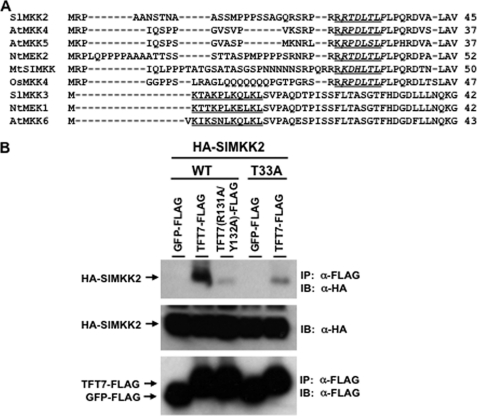
Two conserved 14-3-3 binding motifs have been described in client proteins of 14-3-3 proteins: R/K-X-X-pS/T-X-P and R/K-X-X-X-pS/T-X-P, in which pS/T indicates a phosphorylated serine or threonine and X indicates any amino acid (26). To investigate whether SlMKK2 has a 14-3-3 binding motif, we analyzed the primary amino acid sequence of SlMKK2 and discovered one putative 14-3-3 binding site (R-X-X-X-T-X-P) in the N terminus of SlMKK2. Based on an amino acid alignment, five SlMKK2 orthologs (tobacco NtMEK2, Arabidopsis AtMKK4 and AtMKK5, alfalfa MtSIMKK, and rice OsMKK4) have the same motif. Interestingly, all of these MAPKKs belong to Group C of plant MAPKKs (34). In contrast, SlMKK3 and its two orthologs (tobacco NtMEK1 and Arabidopsis AtMKK6) do not have a 14-3-3 binding motif, consistent with our observation that SlMKK3 and TFT7 do not interact in yeast (Fig. 2A).
MAPKKs phosphorylate downstream MAPKs for signal transduction. For this to occur, MAPKKs physically bind to MAPKs at a MAPK docking site (D site) that is generally present in the MAPKK N terminus. D sites have been experimentally verified only in mammalian MAPKKs, and they have a consensus signature, which is K/R-K/R-X(1–5)-L/I-X-L/I (35). Interestingly, we discovered that a consensus D site is present in SlMKK2 and its orthologs, and, remarkably, it overlaps directly with the putative 14-3-3 binding site (Fig. 3A).
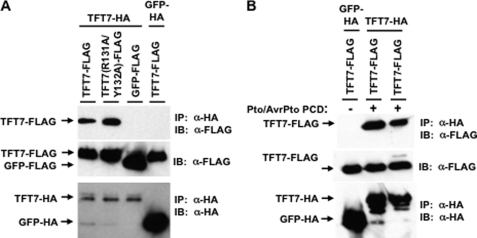
FIGURE 3.
The Thr33 residue of SlMKK2 in its putative 14-3-3 binding site is critical for interaction with TFT7. A, alignment of amino acid sequences by Clustal W method. SlMKK2 and its orthologs contain a putative 14-3-3 binding site in their N termini. A putative 14-3-3 binding site (R-X-X-X-S/T-X-P) was found (italicized) at the same region as a proposed MAPK docking site (K/R-K/R-X(1–5)-L/I-X-L/I) (underlined). B, co-IP) assay. The Thr33 residue of SlMKK2 was substituted with the alanine. For co-IP, the TFT7-FLAG or TFT7(R131A/Y132A)-FLAG and HA-SlMKK2 or HA-SlMKK2(T33A) proteins were co-expressed in N. benthamiana. Estradiol was applied to the leaves 36 h after agro-infiltration to induce expression of HA-SlMKK2. Leaf discs were collected for protein extraction 24 h after estradiol treatment, and α-FLAG-agarose beads were used for IP. GFP-FLAG was used as a negative control. Immunoblotting (IB) was performed with α-HA for SlMKK2 and with α-FLAG for TFT7 or GFP.
The Thr33 Residue in the 14-3-3 Binding Site of SlMKK2 Is Required for Strong Interaction with TFT7 in Vivo
A phosphorylated serine or threonine in the 14-3-3 binding sites of client proteins is typically required for interaction with 14-3-3 proteins (29). We therefore substituted Thr33 of SlMKK2 with alanine to test whether its putative 14-3-3 binding site is critical for interaction with TFT7. Co-IP experiments were carried out by co-expressing both FLAG-tagged TFT7 and HA-tagged SlMKK2 or SlMKK2(T33A) in N. benthamiana, followed by immunoprecipitation with FLAG-tag antibody-agarose beads. Both SlMKK2 and SlMKK2(T33A) were expressed equally well in plant cells. As shown in Fig. 3B, the in vivo interaction of TFT7 with SlMKK2(T33A) was markedly reduced as compared with the interaction of TFT7 with wild-type SlMKK2. This result indicates that the conserved 14-3-3 binding site of SlMKK2 is required for strong interaction with TFT7 in plant cells.
The Amino Acid Residues of TFT7 That Are Required for Binding to SlMAPKKKα Are Also Required for Interaction with SlMKK2 in Vivo
Previously, we reported that two amino acid residues (Arg131 and Tyr132) in the phosphopeptide binding motif of TFT7 are critical for interaction with SlMAPKKKα (24). We therefore tested whether the same TFT7 residues are involved in the interaction with SlMKK2 by co-IP experiments. As shown in Fig. 3B, the interaction in plant cells between the variant TFT7(R131A/Y132A) and SlMKK2 was dramatically reduced compared with interaction between the wild-type TFT7 and SlMKK2. The same phosphopeptide binding pocket in TFT7 is therefore required for interaction with two different client proteins.
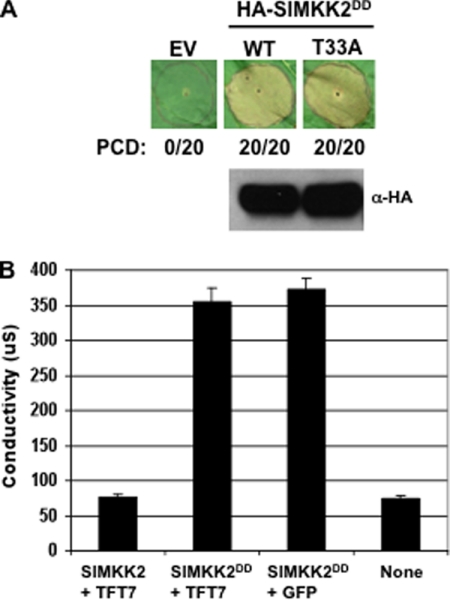
The T33A Substitution in SlMKK2DD Does Not Affect the PCD-eliciting Activity or Protein Stability of the Kinase
Previously, we found that Ser535 in SlMAPKKKα is important for its interaction with TFT7 in vivo and also that the interaction with TFT7 affected both stability and PCD-eliciting activity of SlMAPKKKα in N. benthamiana (24). Because the interaction between TFT7 and SlMKK2 shares a similarity to that between TFT7 and SlMAPKKKα, we tested if Thr33 of SlMKK2 contributes to protein stability and PCD-eliciting activity of SlMKK2. For this, the T33A substitution was introduced into SlMKK2DD, and the degree of SlMKK2DD(T33A)-mediated cell death was compared with that of SlMKK2DD-mediated cell death by expressing the protein in leaves of N. benthamiana. We observed no difference in the degree of PCD induced by the wild-type or the T33A variant of SlMKK2DD (Fig. 4A). Moreover, the abundance of these two proteins in the leaf tissue was the same at 48 h after agro-infiltration (Fig. 4). These results indicate that, in contrast to Ser535 in SlMAPKKKα, Thr33 in SlMKK2 does not contribute to the PCD-eliciting activity or protein stability of this MAPKK.
FIGURE 4.
TFT7 binding does not affect the PCD-eliciting activity or protein stability of SlMKK2DD. A, the effect of the T33A substitution in SlMKK2. The Agrobacterium strains carrying genes encoding SlMKK2DD (WT) or SlMKK2DD(T33A) were infiltrated into N. benthamiana leaves, and estradiol was applied to the leaves 48 h after agro-infiltration to induce expression of SlMKK2DD. The SlMKK2DD-mediated PCD was visually scored 60 h after estradiol treatment. The numbers indicate the number of spots showing full PCD/the number of total infiltrated spots. Protein abundance in leaves of SlMKK2DD or SlMKK2DD(T33A) was determined by immunoblotting with α-HA antibody. The protein was extracted from the agro-infiltrated area 24 h after estradiol treatment. EV, empty vector. B, the effect of TFT7 over-expression. SlMKK2 or SlMKK2DD was co-expressed with TFT7 or GFP by Agrobacterium-mediated transient assay in N. benthamiana leaves, as described above. The electrolyte leakage was measured 36 h after estradiol treatment, as described in Fig. 1.
Previously, we reported that co-expression of TFT7 with SlMAPKKKα enhances SlMAPKKKα-mediated PCD in N. benthamiana (24). We determined whether TFT7 can enhance SlMKK2DD-mediated PCD by co-expressing TFT7 with SlMKK2DD. However, the degree of SlMKK2DD-mediated PCD was not changed regardless of co-expression of TFT7 (Fig. 4B). This is consistent with the observation that binding of TFT7 does not affect the PCD-eliciting activity of SlMKK2.
TFT7 Can Form a Homodimer in Vivo
In previous work (24) and here, we have observed that TFT7 interacts with SlMAPKKKα and SlMKK2 in vivo. Furthermore, SlMKK2 acts downstream of SlMAPKKKα (5, 6), and 14-3-3 proteins are known to form homodimers or heterodimers (36–39). Based on these considerations, we hypothesized that TFT7 may form a homodimer that associates coordinately with SlMAPKKKα and SlMKK2. To test this possibility, co-IP experiments were performed by co-expressing FLAG-tagged TFT7 and HA-tagged TFT7 in leaves of N. benthamiana, followed by immunoprecipitation with an anti-HA affinity matrix. As shown in Fig. 5A, TFT7 could form a homodimer in vivo. We also examined whether PCD induction affects this dimerization. PCD was induced by Pto/AvrPto, and co-IP experiments were performed 48 h after PCD induction. Pto-mediated PCD did not affect dimerization of TFT7 in vivo (Fig. 5B). Thus TFT7 can form a homodimer in both naïve and PCD-induced leaf tissues.
FIGURE 5.
TFT7 can form a homodimer in vivo. A, dimerization in uninoculated leaves. B, dimerization during Pto-mediated PCD. To check dimerization, a co-IP assay was used. For co-IP, FLAG-tagged TFT7 and HA-tagged TFT7 proteins were co-expressed in N. benthamiana. Leaf discs were collected for protein extraction 48 h after agro-infiltration, and an α-HA affinity matrix was used for IP. Both GFP-FLAG and GFP-HA were used as negative controls. Immunoblotting was performed with α-HA for HA-tagged TFT7 or GFP and with α-FLAG for FLAG-tagged TFT7 or GFP. In the case of treatment with Pto/AvrPto-mediated PCD, leaf discs were sampled when yellowing phenotype appeared but before PCD appeared.
Based on the tertiary structures of human 14-3-3 proteins, different amino acid residues are known to be responsible for binding of a 14-3-3 protein to its phosphorylated client proteins or for its dimerization (39). We investigated whether TFT7(R131A/Y132A), a variant with substitutions in its phosphopeptide binding site, is able to dimerize by using co-IP experiments involving FLAG-tagged TFT7(R131A/Y132A) and HA-tagged TFT7 in N. benthamiana. The variant form of TFT7 was still able to form a homodimer with wild-type TFT7 (Fig. 5A). These results indicate that different amino acid residues of TFT7 is responsible for binding to target proteins and for dimerization.
DISCUSSION
We have found that SlMKK2, a MAPKK, is a second client protein of a tomato 14-3-3 protein 7, TFT7. Like SlMAPKKKα, SlMKK2 contains a 14-3-3 binding site, which was shown to be necessary for binding to TFT7. However, unlike SlMAPKKKα, a mutation in the 14-3-3 binding site of SlMKK2 did not affect either the protein stability or PCD-eliciting activity of the kinase. These observations suggest that the underlying mechanism of TFT7 function in regulating SlMKK2 is different from that involved with SlMAPKKKα.
Because TFT7 binds to both SlMAPKKKα and SlMKK2 and also forms a homodimer, we hypothesize that a TFT7 homodimer functions as a scaffold to promote the interaction of activated SlMAPKKKα with SlMKK2 (supplemental Fig. 1). In one possible model, when a SlMAPKKK is activated (SlMAPKKKα for Pto and RPP13 and an uncharacterized SlMAPKKK for RPS2, Rx2, and Gpa2), one TFT7 in the homodimer binds to the activated SlMAPKKK to increase its protein stability. At the same time, the other TFT7 in the homodimer quickly recruits SlMKK2 for efficient signal transfer from SlMAPKKK to SlMKK2, and the signal is then transferred to SlMAPKs, resulting in PCD induction.
We attempted to detect an interaction between SlMAPKKKα and SlMKK2 in plant cells with and without co-overexpression of TFT7 (data not shown). No interaction of the kinases was observed under either of these conditions. If our model is correct, these negative results may be due to the transient nature of the SlMAPKKKα/TFT7-TFT7/SlMKK2 complex or to the large mass of the complex (estimated at 180 kDa), making it difficult to immunoprecipitate intact. Further experiments, including use of protein cross-linkers, will be needed to more critically test the possible TFT7-based interaction of SlMAPKKKα and SlMKK2.
We also attempted to detect an interaction between SlMAPKKKα and SlMKK2 in a yeast two-hybrid system but never observed an interaction, although appropriate controls worked as expected (data not shown). Furthermore, introduction of TFT7 into the two-hybrid system did not promote an interaction. These results may indicate that SlMAPKKKα requires other plant proteins (e.g. an adaptor or chaperone) for transient binding to SlMKK2.
We have shown that both TFT7 and MEK2 play a role in PCD mediated by five plant resistance proteins, Pto, RPS2, RPP13, Rx2, and Gpa2 in N. benthamiana. Previously, we reported that SlMAPKKKα is responsible for PCD mediated by only Pto and RPP13 among these five resistance proteins (24). This suggests that there may be one or more other SlMAPKKKs acting upstream of SlMKK2. To date, two additional MAPKKKs, SlMAPKKKϵ and NPK1, in tomato and tobacco, respectively, have been shown to contribute to PCD (21, 40). Silencing of NPK1 in N. benthamiana compromised disease resistance induced by N, Bs2, and Rx resistance proteins, but not by Pto or Cf4 proteins, whereas silencing of SlMAPKKKϵ in N. benthamiana compromised PCD induced by Pto, Cf4, and Cf9.
We have found that NPK1 has two 14-3-3 binding motifs (K-X-X-S-X-P), with the serines located at amino acid positions 141 and 179. SlMAPKKKϵ also has two 14-3-3 binding sites, with the serines located at position 270 (K-X-X-X-S-X-P) and 794 (R-X-X-S-X-P) (data not shown). Interestingly, like NPK1, NtMEK2 is involved in PCD mediated by the N resistance protein (33). Moreover, SlMAPKKKϵ was proposed to act upstream of the MEK2/WIPK module (equivalent to SlMKK2/SlMPK3). These data raise the possibility that both NPK1 and SlMAPKKKϵ are client proteins of TFT7 in N. benthamiana. If that is the case, it would explain how TFT7 and MEK2 regulate PCD mediated by RPS2, Rx2, and Gpa2. In the future, it will be interesting to determine to what extent plant 14-3-3 proteins are involved in regulation of immunity-associated MAPKKKs and MAPKKs.
When a MAPKK is activated by certain upstream MAPKKK(s), it becomes activated and phosphorylates downstream MAPK(s), resulting in their activation. Based on analysis of two MAPKs in vertebrates, ERK and JNK, a conserved MAPK D site has been proposed to occur in mammalian MAPKK proteins: K/R-K/R-X(1–5)-L/I-X-L/I (35, 41). A d-site motif has also been found in plant MAPKKs including AtMKK4 and AtMKK5 (42), although to date there is no experimental evidence that this site is responsible for contact with MAPKs in plant MAPKKs. Interestingly, we observed here that the proposed D site exactly overlaps with the 14-3-3 binding site present in SlMKK2 and all of its orthologs (Fig. 3A). Because co-expression of TFT7 did not affect SlMKK2DD-mediated PCD, TFT7 binding to this MAPKK does not seem to enhance or suppress SlMKK2 function, implying that active SlMKK2 still makes a normal contact with downstream MAPK for activation, even in the presence of over-expressed TFT7. In the future, it will be interesting to determine whether the D site in SlMKK2 and its orthologs is responsible for interaction with downstream MAPKs and, if yes, how both a 14-3-3 protein and a MAPK are able to bind to the same site in these MAPKKs.
Acknowledgment
We thank Dr. Hong Gu Kang for critical comments.
This work was supported by grant IS-4159-08C from the Israel-USA Binational Agriculture Research and Development Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- R
- resistance
- PCD
- programmed cell death
- MAPKKK
- MAPK kinase kinase
- VIGS
- virus-induced gene silencing
- TRV
- tobacco rattle virus
- IP
- immunoprecipitation
- D site
- docking site.
REFERENCES
- 1. Dangl J. L., Jones J. D. G. (2001) Nature 411, 826–833 [DOI] [PubMed] [Google Scholar]
- 2. van Doorn W. G., Woltering E. J. (2005) Trends Plant Sci. 10, 117–122 [DOI] [PubMed] [Google Scholar]
- 3. Martin G. B., Brommonschenkel S. H., Chunwongse J., Frary A., Ganal M. W., Spivey R., Wu T., Earle E. D., Tanksley S. D. (1993) Science 262, 1432–1436 [DOI] [PubMed] [Google Scholar]
- 4. Mucyn T. S., Clemente A., Andriotis V. M., Balmuth A. L., Oldroyd G. E., Staskawicz B. J., Rathjen J. P. (2006) Plant Cell 18, 2792–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gutierrez J. R., Balmuth A. L., Ntoukakis V., Mucyn T. S., Gimenez-Ibanez S., Jones A. M., Rathjen J. P. (2010) Plant J. 61, 507–518 [DOI] [PubMed] [Google Scholar]
- 6. Tang X., Frederick R. D., Zhou J., Halterman D. A., Jia Y., Martin G. B. (1996) Science 274, 2060–2063 [DOI] [PubMed] [Google Scholar]
- 7. Kang H. G., Oh C. S., Sato M., Katagiri F., Glazebrook J., Takahashi H., Kachroo P., Martin G. B., Klessig D. F. (2010) Plant Cell 22, 918–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Day B., Dahlbeck D., Huang J., Chisholm S. T., Li D., Staskawicz B. J. (2005) Plant Cell 17, 1292–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rentel M. C., Leonelli L., Dahlbeck D., Zhao B., Staskawicz B. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bendahmane A., Querci M., Kanyuka K., Baulcombe D. C. (2000) Plant J. 21, 73–81 [DOI] [PubMed] [Google Scholar]
- 11. Sacco M. A., Koropacka K., Grenier E., Jaubert M. J., Blanchard A., Goverse A., Smant G., Moffett P. (2009) PLoS Pathog. 5, e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Y., Schiff M., Marathe R., Dinesh-Kumar S. P. (2002) Plant J. 30, 415–429 [DOI] [PubMed] [Google Scholar]
- 13. Brigneti G., Martín-Hernández A. M., Jin H., Chen J., Baulcombe D. C., Baker B., Jones J. D. (2004) Plant J. 39, 264–272 [DOI] [PubMed] [Google Scholar]
- 14. Oh C.-S., Martin G. B. (2011) Trends Plant Sci. 16, 132–140 [DOI] [PubMed] [Google Scholar]
- 15. Ekengren S. K., Liu Y., Schiff M., Dinesh-Kumar S. P., Martin G. B. (2003) Plant J. 36, 905–917 [DOI] [PubMed] [Google Scholar]
- 16. del Pozo O., Pedley K. F., Martin G. B. (2004) EMBO. J. 23, 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peart J. R., Lu R., Sadanandom A., Malcuit I., Moffett P., Brice D. C., Schauser L., Jaggard D. A., Xiao S., Coleman M. J., Dow M., Jones J. D., Shirasu K., Baulcombe D. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu R., Malcuit I., Moffett P., Ruiz M. T., Peart J., Wu A. J., Rathjen J. P., Bendahmane A., Day L., Baulcombe D. C. (2003) EMBO J. 22, 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rodriguez M. C., Petersen M., Mundy J. (2010) Annu. Rev. Plant Biol. 61, 621–649 [DOI] [PubMed] [Google Scholar]
- 20. Pedley K. F., Martin G. B. (2005) Curr. Opin. Plant Biol. 8, 541–547 [DOI] [PubMed] [Google Scholar]
- 21. Melech-Bonfil S., Sessa G. (2010) Plant J. 64, 379–391 [DOI] [PubMed] [Google Scholar]
- 22. Pedley K. F., Martin G. B. (2004) J. Biol. Chem. 279, 49229–49235 [DOI] [PubMed] [Google Scholar]
- 23. Yang K. Y., Liu Y., Zhang S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oh C. S., Pedley K. F., Martin G. B. (2010) Plant Cell 22, 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roberts M. R. (2003) Trends Plant Sci. 8, 218–223 [DOI] [PubMed] [Google Scholar]
- 26. Sehnke P. C., DeLille J. M., Ferl R. J. (2002) Plant Cell 14, S339–S354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Darling D. L., Yingling J., Wynshaw-Boris A. (2005) Curr. Top. Dev. Biol. 68, 281–315 [DOI] [PubMed] [Google Scholar]
- 28. Oh C.-S. (2010) Plant Pathol. J. 26, 1–7 [Google Scholar]
- 29. Fritz A., Brayer K. J., McCormick N., Adams D. G., Wadzinski B. E., Vaillancourt R. R. (2006) J. Biol. Chem. 281, 6236–6245 [DOI] [PubMed] [Google Scholar]
- 30. Yang X., Wang W., Coleman M., Orgil U., Feng J., Ma X., Ferl R., Turner J. G., Xiao S. (2009) Plant J. 60, 539–550 [DOI] [PubMed] [Google Scholar]
- 31. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 32. Velasquez A. C., Chakravarthy S., Martin G. B. (2009) J. Vis. Exp. 28, doi 10.3791/1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jin H., Liu Y., Yang K. Y., Kim C. Y., Baker B., Zhang S. (2003) Plant J. 33, 719–731 [DOI] [PubMed] [Google Scholar]
- 34. Ichimura K., et al. (2002) Trends Plant Sci. 7, 301–308 [DOI] [PubMed] [Google Scholar]
- 35. Bardwell A. J., Frankson E., Bardwell L. (2009) J. Biol. Chem. 284, 13165–13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tzivion G., Luo Z., Avruch J. (1998) Nature 394, 88–92 [DOI] [PubMed] [Google Scholar]
- 37. Bridges D., Moorhead G. B. G. (2005) Sci. STKE re10, 2–10 [DOI] [PubMed] [Google Scholar]
- 38. Chaudhri M., Scarabel M., Aitken A. (2003) Biochem. Biophys. Res. Commun. 300, 679–685 [DOI] [PubMed] [Google Scholar]
- 39. Yang X., Lee W. H., Sobott F., Papagrigoriou E., Robinson C. V., Grossmann J. G., Sundström M., Doyle D. A., Elkins J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17237–17242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jin H., Axtell M. J., Dahlbeck D., Ekwenna O., Zhang S., Staskawicz B., Baker B. (2002) Dev. Cell 3, 291–297 [DOI] [PubMed] [Google Scholar]
- 41. Jacobs D., Glossip D., Xing H., Muslin A. J., Kornfeld K. (1999) Genes Dev. 13, 163–175 [PMC free article] [PubMed] [Google Scholar]
- 42. Kiegerl S., Cardinale F., Siligan C., Gross A., Baudouin E., Liwosz A., Eklöf S., Till S., Bögre L., Hirt H., Meskiene I. (2000) Plant Cell. 12, 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]