Abstract
Pax5/B cell lineage specific activator protein (BSAP) is a B lineage-specific regulator that controls the B lineage-specific gene expression program and immunoglobulin gene VH to DJH recombination. Despite extensive studies on its multiple functions, little is known about how the activity of Pax5 is regulated. Here, we show that co-expression of histone acetyltransferase E1A binding protein p300 dramatically enhances Pax5-mediated transcriptional activation. The p300-mediated enhancement is dependent on its intrinsic histone acetyltransferase activity. Moreover, p300 interacts with the C terminus of Pax5 and acetylates multiple lysine residues within the paired box DNA binding domain of Pax5. Mutations of lysine residues 67 and 87/89 to alanine within Pax5 abolish p300-mediated enhancement of Pax5-induced Luc-CD19 reporter expression in HEK293 cells and prevent Pax5 to activate endogenous Cd19 and Blnk expression in Pax5−/− murine pro B cells. These results uncover a novel level of regulation of Pax5 function by p300-mediated acetylation.
Keywords: Coactivator Transcription, Immunology, Lymphocyte, p300, Transcription, B Cell, Pax5, Acetylation
Introduction
Pax5/BSAP is an important regulator for B lineage cell development (1, 2). It belongs to the PAX family of transcription factors with a highly conserved paired box DNA binding domain (PRD)2 (3, 4). The expression of Pax5 is initiated in the common lymphoid progenitor cells, continued through all stages of B lineage cells, and turned off in terminally differentiated plasma cells (1, 2). Pax5 controls the B lineage cell developmental progress at multiple steps. First, Pax5 activates the transcription of many B lineage-specific genes, such as Cd19, Mb1, Blnk, Ebf1, and Aid (1, 2, 5). In Pax5−/− mice, B cell development is blocked at the pro B cell stage (6, 7). Recent studies showed that Pax5 is responsible for the induction of 170 target genes that encode proteins responsible for multiple functions in B lineage cells (8–10). Second, Pax5 specifies the B lineage developmental program through repressing the expression of lineage-inappropriate genes, such as Notch1, M-CSFR/c-fms, and Ccl3 (1, 11, 12). Without these restrictions, Pax5−/− pro B cells are able to differentiate into Natural killer (NK) cells, dendritic cells, macrophages, osteoclasts, granulocytes, or T lineage cells under appropriate conditions (11, 13). Conditionally deleting Pax5 in mature B cells can lead to de-differentiation to uncommitted progenitors back into the bone marrow and then redevelop into T lineage cells (12, 14). Third, Pax5 controls the B lineage differentiation progression through repressing the expression of differentiation stage-inappropriate genes, such as Flt3, Sca-1, Igj, Blimp1, and Cd28 (15, 16). Loss of Pax5 in avian or murine mature B cells also leads to early expression of plasma cell-specific genes (15, 16). Last, Pax5 plays an important role in regulation of the B lineage-specific immunoglobulin heavy chain gene (IgH) VH to DJH recombination. In Pax5−/− pro B cells, although the DH to JH recombination occurs normally, rearrangement of most of the upstream VH7183 and VHJ558 genes is severely compromised, except for a few DH-proximal VH7183 genes (7, 17). Conversely, forced expression of Pax5 transgenes in early T lineage cells can induce the recombination of a few DH-proximal VH7183 genes to the DJH locus (18–20). It has also been shown that Pax5 is required for histone hypomethylation across the IgH locus (21) and for contraction of the IgH locus prior to V(D)J recombination (18, 20), a mechanism that might facilitate the recombination of upstream VHJ558 genes by looping them closer to the DJH locus. Moreover, Pax5 has been found to associate with multiple VH gene coding regions and to interact with the recombination activating gene product (RAG)1/RAG2 protein complexes (22). Through these interactions, Pax5 transactivates RAG-mediated VH to DJH recombination (22).
Pax5 interacts with many cellular factors to exert its multiple biological functions. For example, Pax5 forms a ternary complex with Ets-1 and binds to a complex Pax5/Ets binding site on the Mb-1 promoter to fully activate the Mb-1 gene transcription (23–26). On the other hand, Pax5 interacts with Groucho protein family members of transcriptional co-repressors to repress transcription of target genes (27, 28). The interaction between DAXX and Pax5 also modulates Pax5-mediated functions in different cell types (29).
It has been well documented that acetylation of lysine residues within histone tails provides an important regulatory mechanism for transcription along DNA templates packed into the chromatin structure (30, 31). A group of histone acetyltransferases (HATs), including general control of amino acid synthesis 5 (GCN5/KAT2A), E1A binding protein p300 or Ep300 (p300), CREB-binding protein (CBP), p300/CBP-associating factor, and TATA Box Binding protein associated factor (TAFII250), can transfer acetyl groups to the ϵ-NH2 of lysine residues at the N terminus of histones (30, 31). Besides histones, HATs can acetylate many other cellular proteins, including transcription factors, to modulate their functions (31). For example, acetylation of p53, GATA-1, NFκB p50, STAT6, and Runx1 enhances their binding to cognate DNA binding elements and augments their transcriptional activities (32–36). Conversely, acetylation of Bcl-6 diminishes its DNA binding activity and attenuates Bcl-6-mediated transcriptional repression (37). Acetylation of E2A enhances its transcriptional activation through increasing its nuclear retention (38). Acetylation of p53 excludes polyubiquitination on the same lysine residue and thus stabilizes p53 proteins by preventing them from proteasome-dependent degradation (39).
In searching for potential regulators for Pax5-mediated function, several lines of evidence led us to explore the functional interaction between Pax5 and p300. It has been shown that CBP and p300 are involved in the regulation of numerous transcription factors in B lineage cells, including Pu.1, E47, EBF, and NF-κB (40). In particular, Pax5 interacts with GcN5 and CBP through an adaptor protein, Ada2, and thus recruits GcN5 and CBP as transcriptional co-activators (41). The requirement of p300 during early B lineage cell commitment and development had been demonstrated in conditional p300 knockout mice (p300flox/flox;Mx-Cre mice) (42) as well as in mice carrying KIX domain-deleted p300 proteins (p300KIX/KIX mice) (43). In this study, we focused on the functional interaction between Pax5 and p300. We found that histone acetyltransferase p300 interacts with and acetylates Pax5 and thus acts as a potent co-activator for Pax5.
EXPERIMENTAL PROCEDURES
Cell Cultures
HEK293 or ΦNX cells were maintained in DMEM (Cellgro, Herndon, VA) supplemented with 10% heat-inactive fetal bovine serum, 25 mm HEPES, 1 mm sodium pyruvate, 100 units/ml penicillin, and 100 μg/ml streptomycin. Human B lineage EU12 cells, Abelson retroviral transformed mouse pre B 2A cells, and Pax5−/− G5 pro B cells were maintained in RPMI medium (Cellgro) supplemented with 10% heat-inactive fetal bovine serum, 12.5 μm β-mercaptoethanol, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Plasmids
The Luc-CD19 reporter construct and expression vectors for human Pax5, ΔHD, and ΔPRD constructs were kindly provided by the laboratory of Dr. Meinrad Busslinger. The wild-type p300 expression vector was obtained from the laboratory of Dr. Gary Nabel. The p300ΔHAT expression vector was obtained from the laboratory of Dr. Tso-Peng Yao. The Pax5 lysine-to-alanine mutant constructs were generated from the FLAG-Pax5 expression vector by a PCR-based mutagenesis approach using specific primers listed in the supplemental list of oligonucleotide sequences. All the mutant sites were confirmed by DNA sequencing. The coding regions of wild-type Pax5 and different Pax5 lysine-to-alanine mutants were subcloned into the pMI retroviral vectors in front of the Internal Ribosome Entry Site-GFP expression cassette to generate retroviral vectors for recombinant retrovirus production.
Luciferase Reporter Assays
The Luc-CD19 reporter constructs (0.5 μg) were transfected into HEK293 cells together with different combinations of expression vectors using the Fugene 6 (Roche) reagents following the manufacturer's instructions. A pCMV-Renilla luciferase expression vector (0.01 μg) was always included in each transient transfection to monitor the transfection efficiency. One day after transfection, cells were lysed, and luciferase activities were measured using the dual luciferase assay kit (Promega) on a luminometer (Turner Design or BMG).
Immunoprecipitation and Western Blotting
Nuclear extracts were prepared from EU12, 2A, or G5 cells, or HEK293 cells transiently transfected with different Pax5 constructs as described elsewhere (22). After preclearance with non-relevant goat or rabbit IgG and protein A/G-agarose beads, immunoprecipitation was performed in HEGN150 buffer (10 mm Hepes, 0.25 mm EDTA, 10% glycerol, 150 mm NaCl, 0.1 mm DTT, 0.1 mm PMSF) using antibodies specific for Pax5 (N-19 and C-20, Santa Cruz Biotechnology) or p300 (C-20, Santa Cruz biotechnology) at 4 °C overnight with slow rotation. EtBr (10 μg/ml) was always included in the immunoprecipitation reaction to avoid nonspecific DNA binding. Immunoprecipitates were collected by additional incubation with protein G-agarose (50 μl) for 30 min. After washing four times with HEGN150 buffer, samples were eluted by boiling with 1× SDS gel loading buffer and analyzed by Western blotting. For detection of the full-length Pax5, monoclonal anti-Pax5 (A11, Santa Cruz Biotechnology) was used. For detection of different Pax5 truncation products, goat polyclonal anti-Pax5 antibodies (Santa Cruz Biotechnology), either the C-20 antibody recognizing the C terminus of Pax5 or the N-19 antibody recognizing the N terminus of Pax5, were used. Anti-GST antibody (Sigma) was used to detect the GST fusion Pax5. For Western blotting, the primary antibodies were diluted in Tris-Buffered Saline Tween-20 (TBST) with 5% nonfat dry milk, except for the anti-acetyl lysine antibodies (4G12, Upstate, Lake Placid, NY), which were diluted in TBST (1:200). Bound antibodies were detected using HRP-conjugated secondary antibodies, followed by standard ECL detection procedures. For co-immunoprecipitation assays, Trueblot secondary antibodies (eBioscience, San Diego, CA) were used in co-immunoprecipitation experiments to avoid detection of the immunoprecipitation antibodies.
GST Fusion Protein Pull-down Assay
GST fusion full-length Pax5 protein or different Pax5 truncation constructs were expressed and purified from HEK293 cells using glutathione-agarose beads. After extensive washing with HEGN600 buffer (10 mm HEPES, 0.25 mm EDTA, 10% glycerol, 600 mm NaCl, 0.1 mm DTT, 0.1 mm PMSF), different GST-Pax5 constructs or GST protein retained on the beads was incubated with purified recombinant p300 (200 ng) (Active Motif, Carlsbad, CA) in 30 μl of HEGN150 buffer for 2 h at 4 °C. DNase (0.1 units/ml) and ethidium bromide (10 μg/ml) were included in the binding reaction to reduce nonspecific DNA binding. After washing four times with 1 ml HEGN150 buffer, bound proteins were eluted by boiling in 1× SDS gel loading buffer and analyzed by Western blotting.
Tandem Mass Spectroscopy
Tandem mass spectral analyses were performed at the University of Alabama at Birmingham Mass Spectroscopy Core and at the University of Nebraska Medical Center Mass Spectrometry Core Facility. Briefly, GST-Pax5 protein was purified from about 100 × 106 HEK293 cells co-transfected with GST-Pax5 and p300 expression vectors. Purified GST-Pax5 proteins (1–5 μg) were separated on premade SDS-PAGE gels (Bio-Rad) and stained with Coomassie Blue. A single band that correlates with the size of GST-Pax5 protein was excised and subjected to tryptic digestion at 37 °C for 16 h. The resulting peptides were purified and analyzed with the Q-TOF2 mass spectrometer (Micromass, Manchester, UK) at the University of Alabama at Birmingham or the LTQ Orbitrap XL ETD (Thermo Fisher Scientific) at the University of Nebraska Medical Center. The obtained results at the University of Alabama at Birmingham were analyzed manually to identify the acetylated peptides derived from the Pax5 protein. The results obtained at the University of Nebraska Medical Center were analyzed by Mascot via automated database searching of Mascot generic format files against the Human IPI protein database version 3.52.
In Vitro Acetylation Assays
For in vitro acetylation assays, purified GST fusion Pax5 (1 μg) from HEK293 cells was incubated with full-length recombinant p300 (Active Motif) in the presence of 5 mm Tris HCl (pH8.0), 1% glycerol, 0.1 mm DTT, 0.1 mm PMSF, 1 mm sodium butyrate, and 12 mm acetyl CoA at room temperature for 1 h with gentle shaking. Reaction samples were separated by SDS-PAGE, and acetylated proteins were detected by monoclonal anti-acetyl-lysine antibodies (4G12, Upstate).
Recombinant Retrovirus-mediated Transduction
Recombinant retroviruses were produced by co-transfection of the retroviral expression vectors together with the pGP and pEco booster vectors into semi-confluent ΦNX packaging cells using the Fugene 6 reagent (Roche). Culture supernatant containing recombinant retroviruses was collected 48 h after transfection and filtered through a 0.45-μm filter. The resulting live viruses were used to transduce Pax5−/− mouse pro B cells with the help of Polybrene (4 μg/ml). The culture medium was changed to Polybrene-free RPMI growth medium the following day. Transduction efficiency was monitored by FACS analysis evaluating the frequency of GFP-positive cells. All Pax5 constructs are expressed together with GFP from a bicistronic RNA transcript. The expression of Pax5 constructs can be tracked by analyzing GFP+ cells.
FACS Analysis and Sorting
After washing with FACS buffer (PBS supplied with 2% fetal bovine serum), cells (106) were stained with phycoerythin (PE) conjugated anti-mouse CD19 antibodies (BD Biosciences) on ice for 30 min. Cells were washed twice with 1 ml FACS buffer and resuspended in 500 μl FACS buffer containing propidium iodide (1 μg/ml). Samples were analyzed on a FACScalibur (BD Biosciences), and FACS data were analyzed using the Winmid 2.8 software.
RNA Extraction and Real-time RT-PCR
Pax5−/− pro B cells (G5) were reconstituted with retroviruses expressing wild-type or different mutant Pax5 constructs. The reconstituted (GFP+) cells were purified on a MoFlow FACS sorter (BD Biosciences) and directly collected in TRIzol reagent (Invitrogen). Total RNA was purified following the manufacturer's protocol. First-strand cDNA was synthesized using the Superscript III kit (Invitrogen). The cDNA samples were subjected to real-time PCR analysis on an ABI Prism 7900 sequence detection system (Applied Biosystems) using the SYBRGreen PCR master mix (SuperArray). Primers used are listed in the supplemental list of oligonucleotide sequences.
EMSA
The BSAP binding probe, derived from the Pax5 binding site within the Cd19 promoter, was labeled with Alexa Fluor 488 at the 5′ end. Nuclear extracts were prepared from HEK293 cells transiently transfected with expression vectors for the wild-type Pax5 or different lysine-to-alanine mutants without or with co-transfection of p300 expression vectors. Binding reactions were carried out in 20 μl of buffer containing 1 μg/μl poly-dI/dC, 10 mm HEPES (pH 7.9), 50 mm KCl, 10% glycerol, 0.2 mg/ml BSA, 0.1 mm DTT, and 0.1 mm PMSF at room temperature for 20 min. Samples were separated on 5% native polyacrylamide gel at 4 °C with Tris borate-EDTA buffer (8.9 mm Tris, 8.9 mm boric acid, 0.2 mm EDTA). Wet gels were directly analyzed with a FluorChemQ Multi Imager III (Alpha Innotech).
RESULTS
Overexpression of Histone Acetyltransferase p300 Dramatically Enhances Pax5-mediated Transcriptional Activity
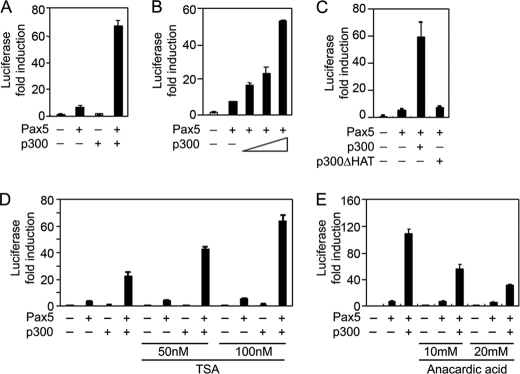
Pax5/BSAP is a B lineage-specific transcription factor that activates many B lineage-specific genes (40). On the other hand, it has been shown that histone acetyltransferase p300 plays an important role during early B lineage cell commitment and development (42, 43). To explore the potential functional interaction between p300 and Pax5, we performed luciferase reporter assays in HEK293 cells using the Pax5-responsive Luc-CD19 reporter construct with different combinations of the Pax5 and p300 expression vectors. Co-expression of Pax5 with the Luc-CD19 reporter construct in HEK293 cells resulted in a 7–10-fold induction of luciferase activities, which was dramatically enhanced by co-expression of p300 together with Pax5 (60-fold higher than that of the basal level) but not by expression of p300 alone (Fig. 1A). The p300-mediated enhancement is dependent on the level of p300 expression, as shown in a series of transient transfection assays using increasing amounts of p300 expression vectors (Fig. 1B and supplemental Fig. 1A). Overexpression of p300 did not affect Pax5 protein expression levels in these transient transfection assays (supplemental Fig. 1A).
FIGURE 1.
Co-expression of p300 dramatically enhances Pax5-mediated induction of the Luc-CD19 reporter expression. Luciferase reporter assays were performed in HEK293 cells by transient transfection with different combinations of vectors as follows: A, Luc-CD19 reporter (0.5 μg), Pax5 (0.5 μg), and p300 (0.5 μg). B, Luc-CD19 reporter (0.5 μg) and Pax5 (0.5 μg) with increasing amounts (0.1, 0.25, or 0.5 μg) p300 expression vectors. C, Luc-CD19 reporter (0.5 μg) and Pax5 (0.5 μg) with the wild-type p300 (0.5 μg) or the p300ΔHAT (0.5 μg) constructs. D, Luc-CD19 reporter (0.3 μg), Pax5 (0.3 μg), and p300 (0.3 μg). After transfection, cells were treated with different concentrations of trichostatin A for 24 h. E, Luc-CD19 reporter (0.3 μg), Pax5 (0.3 μg), and p300 (0.3 μg). After transfection, cells were treated with different concentrations of anacardic acid for 24 h. All inhibitors were added 6 h after transfection. Results presented are the mean values of relative fold induction above the basal level. Error bars indicate mean ± S.D. for triplicate experiments.
It has been well characterized that p300 possesses intrinsic HAT activity, which is required for its co-activator function (30). Therefore, we compared the effects of the wild-type p300 and p300ΔHAT constructs on Pax5-mediated function. In contrast to the wild-type p300, the p300ΔHAT construct failed to enhance Pax5-mediated induction of the Luc-CD19 reporter expression (Fig. 1C), indicating that the p300 HAT activity is required for enhancing Pax5 function.
In addition, the p300-mediated enhancement of Pax5 transcriptional activity was further augmented by blocking histone deacetylase activity with trichostatin A treatment (Fig. 1D). Conversely, treatment of transfected cells with anacardic acid to inhibit HAT activity attenuated p300-mediated enhancement (Fig. 1E). The p300-mediated enhancement is dependent on the DNA binding domain and transcriptional activation domain of Pax5 (supplemental Fig. 1B). Taken together, these results show that histone acetyltransferase p300 acts as a transcriptional co-activator for Pax5 and that the intrinsic HAT activity of p300 is essential for enhancing the transcriptional activity of Pax5.
Pax5 Interacts with p300
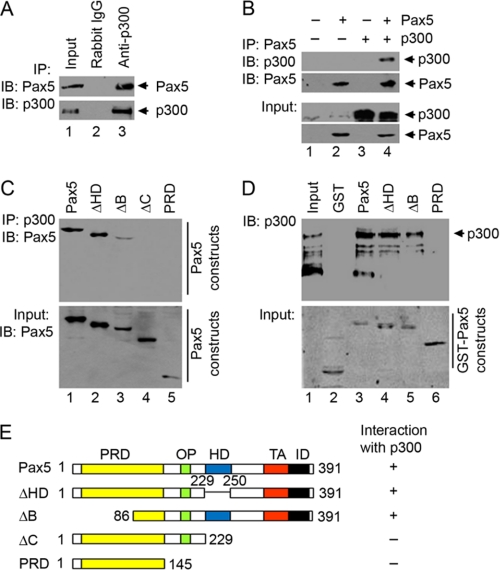
It has been shown that Pax5 can recruit histone acetyltransferase CBP and Gcn5 as co-activators for transactional regulation (41). Next, we performed co-immunoprecipitation assays to determine whether Pax5 interacts with p300. Using anti-p300 antibodies but not the control rabbit IgG, we were able to immunoprecipitate Pax5 from crude nuclear extracts prepared from human B lineage EU12 cells containing endogenous Pax5 and p300 proteins (Fig. 2A). Using anti-Pax5 antibodies, we were also able to immunoprecipitate p300 from HEK293 cells transiently transfected with the Pax5 and p300 expression vectors (Fig. 2B).
FIGURE 2.
Pax5 interacts with p300. A, immunoprecipitation (IP) was performed using nuclear proteins extracted from human B lineage EU12 cells with non-relevant control rabbit IgG or anti-p300 antibodies. Input (10% of sample) or immunoprecipitated samples were analyzed by Western blotting (IB) using mouse monoclonal anti-Pax5 antibodies or rabbit polyclonal anti-p300 antibodies. B, immunoprecipitation was performed using nuclear proteins extracted from HEK293 cells transiently transfected with different combinations of Pax5 and p300 expression vectors. Immunoprecipitated samples were analyzed by Western blotting using monoclonal anti-Pax5 antibodies or rabbit polyclonal anti-p300 antibodies. The expression levels of Pax5 and p300 in the nuclear extracts were also analyzed by Western blotting and shown as Input. C, interaction of p300 with different Pax5 constructs. Nuclear extracts were prepared from HEK293 cells co-expressing p300 with different Pax5 constructs and subjected to immunoprecipitation using anti-p300 antibodies. Immunoprecipitated samples were analyzed by Western blotting using goat polyclonal anti-Pax5 antibodies N-19 and C-20 or rabbit polyclonal anti-p300 antibodies. The expression of different Pax5 constructs was also analyzed by Western blot and shown as input. D, different GST fusion Pax5 constructs were expressed and purified from HEK293 cells with or without co-expression of p300 and retained on the glutathione-agarose beads for interaction with purified recombinant p300 protein. Bound p300 proteins were separated by SDS-PAGE and detected by rabbit polyclonal anti-p300 antibodies. Different purified GST-Pax5 truncations were detected by Coomassie Blue staining (shown as Input). E, diagrams of different Pax5 constructs with functional domains. Their abilities to interact with p300 are summarized. PRD, paired box DNA binding domain; OP, Oct-peptide; HD, homeolike domain; TA, transactivation domain; ID, inhibition domain.
To further dissect the domains of Pax5 responsible for interaction with p300, we performed co-immunoprecipitation assays using a series of Pax5 truncation constructs transiently expressed in HEK293 cells. Using anti-p300 antibodies, we were able to immunoprecipitate Pax5 and the ΔHD proteins and, to a lesser extent, the ΔB construct. Under the same experimental conditions, we could not detect the ΔC and PRD constructs in immunoprecipitated samples (Fig. 2C).
To confirm the physical interaction between Pax5 and p300, we performed GST fusion protein pull-down assays. Different GST fusion Pax5 constructs were expressed and purified from HEK293 cells and used to interact with purified full-length p300 proteins. The p300 proteins can be efficiently pulled down by GST-Pax5, ΔHD, and ΔB constructs but not by the GST-PRD construct (Fig. 2D). These results are consistent with our co-immunoprecipitation results and demonstrate that the C terminus of Pax5 is required for interaction with p300 (Fig. 2E).
p300 Acetylates Pax5 on Multiple Lysine Residues within the PRD DNA Binding Domain
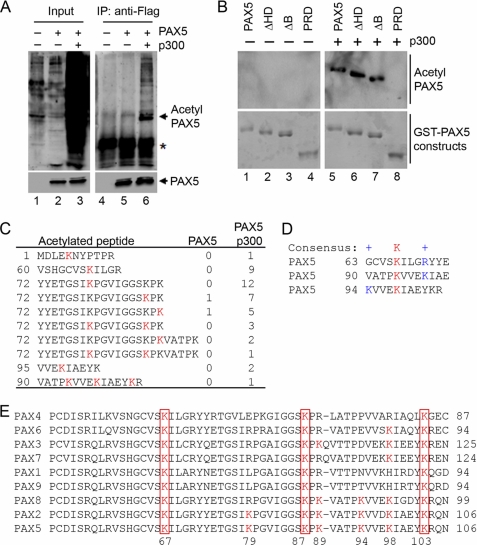
Originally identified as a histone acetyltransferase, p300 can also acetylate many non-histone proteins, such as transcription factors, to modulate their functions. Next, we analyzed if overexpression of p300 enhances Pax5 acetylation. In our transient transfection, overexpression of p300 resulted in global acetylation of many proteins (Fig. 3A, lane 3). Purified Pax5 proteins from cells overexpressing p300 were acetylated, as detected by a monoclonal anti-acetyl lysine antibody (Fig. 3A, lane 6). When different Pax5 truncation constructs were analyzed, overexpression of p300 resulted in acetylation of GST fusion Pax5, ΔHD, and ΔB proteins, but not the PRD construct in HEK293 cells (Fig. 3B).
FIGURE 3.
p300 acetylates Pax5 on multiple lysine residues within the PRD domain. A, HEK293 cells were transfected with different combinations of expression vectors of FLAG-tagged Pax5 and p300. Crude nuclear extracts were used as Input. FLAG-tagged Pax5 was purified using anti-FLAG antibodies. Acetylated proteins were analyzed by Western blotting using monoclonal anti-acetyl lysine antibodies. The asterisk indicates the position of the light chain in the immunoprecipitated samples. B, different GST-Pax5 constructs are listed in Fig. 2E. HEK293 cells were transfected with different GST-Pax5 expression constructs with or without p300 expression vectors. GST-Pax5 was purified using glutathione-agarose beads, and acetylated Pax5 was detected by Western blotting using monoclonal anti-acetyl lysine antibodies. The same batch of purified GST fusion proteins was separated by SDS-PAGE and stained with Coomassie Blue to determine the amounts of purified Pax5 truncations. C, purified GST-Pax5 proteins from HEK293 cells overexpressing p300 were completely digested with trypsin and analyzed by electrospray ionization tandem mass spectrometry. The sequences of the recovered peptides containing acetylated lysine residues are listed. The acetylated lysine (K) residues were highlighted and underlined in red. Numbers indicated the recovered acetylated peptides in samples prepared from cells expressing Pax5 only or Pax5 and p300. D, sequence alignments of acetylated lysine residues and their flanking regions with the consensus sequence for lysine acetylation. The acetylated lysine residues are highlighted in red. The positions of positively charged residues are indicated by + and highlighted in blue. E, partial sequence alignment of all human PAX proteins. The conserved lysine residues among all the PAX proteins correlating with the identified acetylated lysine residues within PAX5 are highlighted with red boxes.
The efficient acetylation of GST-Pax5 in HEK293 cells overexpressing p300 allowed us to purify GST-Pax5 proteins to identify the acetylated lysine residues by mass spectroscopy. From total of four independent experiments, we recovered 43 Pax5-derived peptides containing acetylated lysine residues (10 unique peptides) from cells co-expressing Pax5 and p300 (Fig. 3C). Detailed analysis identified that lysine residues 5, 67, 79, 87, 89, 94, 98, and 103 were modified by acetylation. In contrast, we only identified two acetylated peptides from HEK293 cells expressing Pax5 alone; one contains acetylated lysine 87 and another contains acetylated lysine 89 residue. These results are consistent with the Western blotting results obtained with anti-acetyl lysine antibodies, indicating that overexpression of p300 induced acetylation on Pax5.
It has been shown that acetylation preferentially occurs on lysine residues that have a positively charged residue three or four amino acids up- or downstream (44). Here, the alignment of the acetylated Pax5 peptides show that the lysine residues 67, 94, and 98 fit nicely with such criteria (Fig. 3D). Taken together, these results identified Pax5 as a novel target for p300-mediated acetylation.
Comparison of the amino acid sequences of all the human PAX family members revealed that the Lys-67, Lys-87, and Lys-103 residues are conserved among all eight PAX proteins; the Lys-89, Lys-94, and Lys-98 residues are conserved among the PAX2, 5, and 8 proteins; and the Lys-79 residue is conserved between Pax2 and 5 (Fig. 3E). The conservation of the lysine residues and their surrounding regions within the PRD domain suggest that all the PAX members are potential targets for p300-mediated acetylation.
Acetylation of Pax5 in Vitro and in Vivo
To determine whether p300 acetylates Pax5 directly, we performed in vitro acetylation assays using purified p300 and GST-Pax5. After incubation of purified GST-Pax5 with p300 and acetyl CoA, acetylation of the Pax5 protein can be detected with the monoclonal anti-acetyl lysine antibodies (Fig. 4A). As negative controls, no acetylated Pax5 signal was detected in reaction without p300 or without acetyl CoA (Fig. 4A).
FIGURE 4.
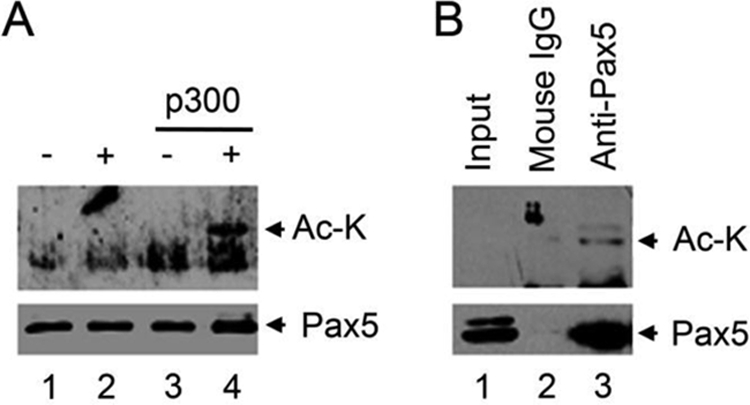
Acetylation of Pax5 in vitro and in vivo. A, in vitro acetylation of purified GST-Pax5 protein using the purified p300 enzyme and acetyl CoA. B, Pax5 is acetylated in Abelson retroviral transformed murine pre B 2A cells. Endogenous Pax5 protein was purified by immunoprecipitation from 2A cells and analyzed by monoclonal anti-acetyl lysine antibodies. The levels of input and purified Pax5 proteins are also analyzed using anti-Pax5 antibodies. Input, nuclear extract (10% of that used for purification of Pax5; Ac-k, protein recognized by the anti-acetyl-lysine antibody.
To determine whether Pax5 is acetylated in murine B lineage cells, we purified Pax5 protein from Abelson retroviral transformed murine 2A cells. The purified Pax5 proteins can be detected by the monoclonal anti-acetyl lysine antibodies (Fig. 4B). These results confirmed that Pax5 is acetylated in murine pre B cells.
The Lysine 67, 87, 89, 103, and 142 Residues within Pax5 Are Required for p300-mediated Enhancement
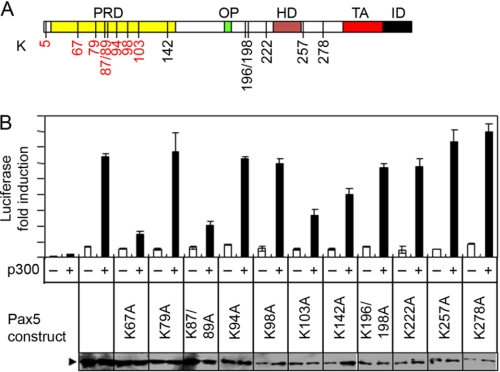
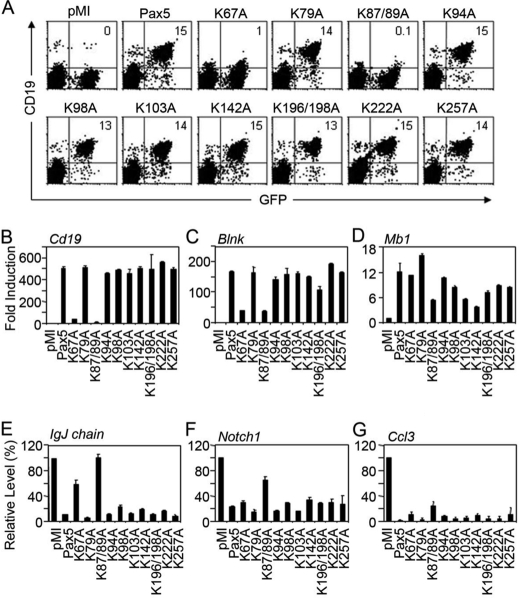
To determine the biological significance of acetylation on different lysine residues within Pax5, we generated a series of lysine-to-alanine mutant Pax5 constructs and tested them in luciferase reporter assays with or without coexpression of p300. Among these lysine-to-alanine mutant constructs, the K67A, K87/89A, and K103A Pax5 mutants showed dramatically reduced p300-mediated enhancement of luciferase activity; the K142A mutant showed marginal reduction of p300-mediated enhancement; and the rest of the Pax5 mutants had no effect (Fig. 5B). All lysine-to-alanine mutants showed similar basal activities on the induction of the Luc-CD19 reporter expression because they were expressed at similar levels (Fig. 5B) and displayed similar DNA binding activities (supplemental Fig. 2). The K67/87/89A triple mutant Pax5 construct expresses normally in the nuclei of transiently transfected HEK293 cells, but it lost its DNA binding activity and was not tested in other functional analyses (supplemental Fig. 3, A and B). Among the five lysine residues that are required for p300-mediated enhancement, the Lys-67, Lys-87, Lys-89, and Lys-103 residues were found to be acetylated in HEK293 cells upon overexpression of p300. These results suggest that acetylation on the Lys-67, Lys-87, Lys-89, and Lys-103 residues within Pax5 are required for p300-mediated enhancement.
FIGURE 5.
Lysine 67 and 87/89 residues within Pax5 are required for p300-mediated enhancement of transcription activation. A, diagram of the functional domains of Pax5 and the relative positions of the acetylated lysine residues. The relative positions of all the lysine residues within Pax5 protein are indicated by numbers, and the identified acetylated lysine residues are shown in red. PRD, paired box DNA binding domain; OP, Oct-peptide; HD, homeolike domain; TA, transactivation domain; ID, inhibition domain. B, different lysine-to-alanine Pax5 mutants were analyzed in luciferase assays in HEK293 cells using the Luc-CD19 reporter construct with or without co-transfection of the p300 expression vector. Results are presented as the mean values of relative fold inductions ± S.D. from triplicate experiments. The expression levels of the Pax5 mutant constructs in each transfection were determined by Western blotting and are shown at the bottom.
The Lysine 67, 87, and 89 Residues within Pax5 Are Essential for Activation of Endogenous Cd19 and Blnk Expression in Murine Pro B Cells
To further determine the biological significance of Pax5 acetylation in B lineage cells, we reconstituted Abelson retrovirus-transformed Pax5−/− mouse pro B cells (G5) with the wild-type Pax5 or different lysine-to-alanine Pax5 mutants using recombinant retroviruses. All the Pax5 constructs are co-expressed with the GFP reporter from the same bicistronic transcript, and thus the cells expressing different Pax5 constructs can be tracked by their GFP expression (Fig. 6A). Expression of wild-type Pax5 in G5 cells restored endogenous Cd19 expression, as monitored by FACS analyses of cell surface CD19 expression on the GFP+ cells (Fig. 6A) and by real-time RT-PCR analyses of Cd19 mRNA levels in purified GFP+ cells (B). Consistent with our luciferase reporter assay results, mutations of the Lys-67 and Lys-87/89 residues within Pax5 completely abolished its ability to activate endogenous Cd19 gene expression, whereas mutations of other lysine residues had little or no effect (Fig. 6, A and B). G5 cells reconstituted with the K67A and K87/89A Pax5 mutants also failed to activate endogenous Blnk expression (Fig. 6C). However, Pax5-mediated induction of Mb1 gene expression was not affected by mutation of the Lys-67 residue and was only slightly affected by mutation of the Lys-87/89, Lys-103, or Lys-142 residues (Fig. 6D). These results show that the Lys-67, Lys-87/89 residues within Pax5 are essential for activation of endogenous Cd19 and Blnk in murine pro B cells.
FIGURE 6.
Differential requirements of the Lysine 67, 87, and 89 residues for activation or repression of Pax5 target genes. Pax5−/− mouse pro B cells (G5) were reconstituted with recombinant retrovirus expressing the wild-type Pax5 or different lysine-to-alanine Pax5 mutants. A, cell surface CD19 expression was monitored by FACS analysis. Transduced cells expressing different Pax5 constructs can be tracked by gating on GFP+ cells. The percentages of CD19+GFP+ cells are indicated. Results shown are representatives from three independent transduction experiments. B–G, the GFP+ cells were purified by FACS sorting, and the expression levels of Cd19 (B), Blnk (C), Mb1 (D), IgJ chain (E), Notch1 (F), and Ccl3 (G) were analyzed by real-time RT-PCR. For induction of the Cd19, Blnk, and Mb1 genes, the results are presented as relative fold induction over that in cells only expressing GFP. For repression of IgJ chain, Notch1, and Ccl3, the results are presented as relative expression levels compared with that in cells only expressing GFP (set as 100). Results presented are mean ± S.D. from triplicate PCR amplifications. The reconstitution experiments were performed three times.
Besides activation of B lineage-specific gene expression, Pax5 suppresses the expression of many lineage or differentiation stage-inappropriate genes. Restoration of wild-type Pax5 expression in G5 cells efficiently repressed IgJ chain, Notch1, and Ccl3 expression (Fig. 6E). However, cells expressing the K67A or K87/89A mutant Pax5 failed to shut down IgJ chain expression (Fig. 6E). Cells expressing the K87/K89A mutant Pax5 failed to repress Notch1 expression (Fig. 6F). In contrast, cells reconstituted with different Pax5 constructs all have reduced Ccl3 expression, despite the fact that cells reconstituted with the K67A or K87/89A Pax5 mutants have slightly higher levels of Ccl3 expression (Fig. 6G). These results show that different lysine residues within Pax5 are required differentially for activation or repression of specific target genes.
DISCUSSION
The important functions of Pax5 in B lineage cell development have been well studied; however, little is known about how Pax5 activity is regulated. Our initial studies showed that co-expression of p300 dramatically enhanced Pax5-mediated induction of the Luc-CD19 reporter expression. Such enhancement is dependent on the intrinsic HAT domain of p300 and the C-terminal of the Pax5 protein. Indeed, results from the co-immunoprecipitation and GST fusion protein pull-down experiments indicated that the C terminus of Pax5 is required for interaction with the p300 protein. Moreover, our Western blot analyses and the mass spectroscopy analyses showed that p300 acetylates multiple lysine residues within the N-terminal PRD domain of Pax5. Last, mutation of the Lys-67, Lys-87, Lys-89, or Lys-103 residues affected Pax5-mediated induction of endogenous Pax5 target genes in murine pro B cells. Taken together, these results demonstrated that p300 acts as a potent transcriptional co-activator for Pax5-mediated transcriptional activation through interaction with Pax5 and acetylation of Pax5. These findings provide a novel level of regulation for Pax5-mediated function.
Acetylation on Pax5 was first uncovered by Western blotting in HEK293 cells overexpressing p300 and then confirmed in vitro acetylation assays using purified p300 and Pax5 proteins. Mass spectrometry analyses identified eight lysine residues within the Pax5 PRD DNA binding domain that can be acetylated upon p300 overexpression. In HEK293 cells without p300 overexpression, there is a minimal level of acetylation on Pax5. These results are consistent with the luciferase assay results that overexpression of p300 strongly enhances Pax5-mediated activation of the Luc-CD19 reporter expression. In addition, detailed analysis of the interaction between Pax5 and p300 and the subsequent acetylation of Pax5 also suggested a two-step functional interaction between the Pax5 and p300 proteins. p300 first binds to the C terminus of Pax5 and acetylates the N terminus of Pax5. Without interaction with p300, the GST-PRD construct could not be acetylated.
Sequence alignment of the acetylated lysine residues and their flanking regions from different p300 acetylation substrates revealed the preferred presence of positively charged residues three or four residues up- or downstream of the acetylation site (44). Within Pax5, there are eight lysine residues that can be acetylated by p300 overexpression. Among them, the flanking regions of the Lys-67, Lys-94, and Lys-98 residues fit nicely with this consensus for p300 targets. Moreover, the Lys-67, Lys-87, and Lys-103 residues and their surrounding regions are highly conserved among all PAX protein members, identifying that all PAX proteins are potential targets for p300-mediated acetylation.
The biological significance of lysine acetylation on Pax5-mediated biological function was further investigated by reconstitution of Pax5 expression using the wild-type or different lysine-to-alanine mutant Pax5 constructs in Abelson retroviral-transformed Pax5−/− G5 mouse pro B cells. Restoration of wild-type Pax5 expression in G5 cells efficiently activated endogenous Cd19 and Blnk expression. Restoration of Pax5 expression in G5 cells resulted in almost 500-fold induction of the endogenous Cd19 transcripts. Such a level of induction is comparable with our observed luciferase reporter assay results in HEK293 cells co-expressing Pax5 and p300. Moreover, mutations of the Lys-67 and Lys-87/Lys-89 residues within Pax5 dramatically affected p300-mediated enhancement in our luciferase reporter assays and almost completely abolished Pax5-mediated induction of endogenous Cd19 in G5 cells. We speculate that acetylation of Pax5 is required for p300-mediated enhancement of Pax5 function and for Pax5 to fully activate its target genes in murine pro B cells.
One of the interesting findings is that mutation of the Lys-67 residue did not affect Pax5-mediated induction of endogenous Mb1 expression, but mutation of the Lys-87/Lys-89 and Lys-142 residues partially affected Mb1 expression. It has been demonstrated that Pax5 and Ets-1 form a complex to bind to a complex Pax5/Ets-1 binding site within the Mb1 gene promoter region to activate Mb1 expression (23–25). The PRD domain alone is sufficient to activate the Mb1 expression (23–25). Here we show that the PRD construct did not associate with p300 and could not be acetylated by p300. It is plausible that acetylation of Pax5 on Lys-67 is not required for induction of Mb1 expression in murine B cells. We speculate that acetylation on different lysine residues within Pax5 could specify its action on different target genes. Previous studies had illustrated that the Lys-67 residue of Pax5 is contacting the major groove of the Pax5-Ets1 complex binding site within the Mb1 gene promoter region (25). In this study, changing from Lys-67 to Ala affected neither Pax5 binding to the Pax5 binding site derived from the Cd19 gene promoter nor the induction of Mb1 gene expression in murine pro B cells. We could not rule out the possibility that Lys-67 is essential for contacting different Pax5 binding sites for other target genes.
Results from our reconstitution studies also revealed the differential requirements of the Lys-67 and Lys-87/Lys-89 residues in Pax5-mediated suppression of endogenous IgJ chain, Notch1, and Ccl3 expression in murine pro B cells. Mutation of the Lys-67 or Lys-87/89 residues almost diminished Pax5-mediated repression of IgJ chain expression, whereas mutation of the Lys-87/Lys-89 residues but not the Lys-67 residue affected Pax5-mediated repression of Notch1 expression. In contrast, none of the lysine mutants affected Pax5-mediated repression of Ccl3 expression. Again, these result show that different lysine residues are differentially required for different target genes. Given the fact that these three genes are differentially expressed during B cell developmental stages, it would be interesting and important to determine whether individual lysine residues within Pax5 are acetylated differentially at different stages to specifically control downstream target genes.
Collectively, this study demonstrates that p300 acts as a potent transcriptional co-activator for Pax5-mediated transcriptional function through acetylation of Pax5 on multiple lysine residues within the PRD DNA binding domain. Given the important function of Pax5 in early B lineage cell development and later stages of B cell activation and differentiation, modulation of Pax5 transcriptional activity by acetylation on different lysine residues could provide multiple levels of regulation for Pax5-mediated function at different stages. For example, at the early pro B cell commitment stage, acetylation of Pax5 might be essential for Pax5 to activate a broad spectrum of Pax5 target genes and mean time to suppress non-B lineage gene expression to initiate and specify the B lineage development pathway. Recent studies showed that the levels of p300 protein and total histone acetyltransferase activity are elevated in activated B cells, which resulted in the induction of the Btk gene (45). Elevated p300 expression could also enhance Pax5 acetylation and thus induce Pax5 target gene expression during B cell activation, as many of the Pax5 target genes are involved in B cell receptor (BCR) signaling or cell adhesion. Conversely, at the plasma cell terminal differentiation step, the transcriptional activity of Pax5 might be attenuated or even extinguished through deacetylation. Such changes could be controlled by the combinatorial action of HATs and histone deacetylases. Clearly, understanding the dynamic regulation of Pax5 acetylation during B cell development and differentiation is an important and interesting question that should be further investigated.
Acknowledgments
We thank Drs. Meinrad Busslinger (Institute of Molecular Pathology (IMP), Vienna, Austria) for the Pax5 expression vectors and the Luc-CD19 reporter construct; Berry Sleckman (Washington University, St. Louis, MO) for the Abelson retroviral transformed wild-type murine pre B cell line 2A; Mark Schlissel (University of California at Berkeley) for the Abelson retroviral transformed Pax5−/− mouse pro B cell line G5; Gary Nabel (NIAID, National Institutes of Health) for the wild-type p300 expression vectors; Tso-Peng Yao (Duke University) for the p300ΔHAT expression vectors; Larry Gartland and Marion Spells (University of Alabama at Birmingham) for cell sorting; Drs. Mellissa Thal, Scott Swindle, Christopher Klug, Miles D. Lange, and Peter Burrows for help with and discussions of this study.
This work was supported, in whole or in part, by National Institutes of Health Grants AR048592, AI073174, AI074948, and AI076475 (to Z. Z.). This work was also supported by funding from the University of Alabama at Birmingham and the University of Nebraska Medical Center and the Eppley Cancer Institute (to Z. Z.). Purchase of the Q-TOF mass spectrometer in the Shared Facility came from funds provided by the National Center for Research Resources (NCRR) Shared Instrumentation Grants S10 RR13795 and the University of Alabama at Birmingham Health Services Foundation General Endowment Fund (Water/Micromass Q-TOF). The Mass Spectrometry and Proteomics Core Facility at the University of Nebraska Medical Center is supported by the Nebraska Research Initiative.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and a list of oligonucleotide sequences.
- PRD
- paired box DNA binding domain
- Luc
- luciferase
- HAT
- histone acetyltransferase
- CBP
- CREB-binding protein.
REFERENCES
- 1. Cobaleda C., Schebesta A., Delogu A., Busslinger M. (2007) Nat. Immunol. 8, 463–470 [DOI] [PubMed] [Google Scholar]
- 2. Schebesta M., Heavey B., Busslinger M. (2002) Curr. Opin. Immunol. 14, 216–223 [DOI] [PubMed] [Google Scholar]
- 3. Czerny T., Schaffner G., Busslinger M. (1993) Genes Dev. 7, 2048–2061 [DOI] [PubMed] [Google Scholar]
- 4. Czerny T., Busslinger M. (1995) Mol. Cell. Biol. 15, 2858–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nutt S. L., Morrison A. M., Dörfler P., Rolink A., Busslinger M. (1998) EMBO J. 17, 2319–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Urbánek P., Wang Z. Q., Fetka I., Wagner E. F., Busslinger M. (1994) Cell 79, 901–912 [DOI] [PubMed] [Google Scholar]
- 7. Nutt S. L., Urbánek P., Rolink A., Busslinger M. (1997) Genes Dev. 11, 476–491 [DOI] [PubMed] [Google Scholar]
- 8. Schebesta A., McManus S., Salvagiotto G., Delogu A., Busslinger G. A., Busslinger M. (2007) Immunity. 27, 49–63 [DOI] [PubMed] [Google Scholar]
- 9. Pridans C., Holmes M. L., Polli M., Wettenhall J. M., Dakic A., Corcoran L. M., Smyth G. K., Nutt S. L. (2008) J. Immunol. 180, 1719–1728 [DOI] [PubMed] [Google Scholar]
- 10. Fuxa M., Busslinger M. (2007) J. Immunol. 178, 8222–8228 [DOI] [PubMed] [Google Scholar]
- 11. Nutt S. L., Heavey B., Rolink A. G., Busslinger M. (1999) Nature 401, 556–562 [DOI] [PubMed] [Google Scholar]
- 12. Cobaleda C., Busslinger M. (2008) Curr. Opin. Immunol. 20, 139–148 [DOI] [PubMed] [Google Scholar]
- 13. Rolink A. G., Nutt S. L., Melchers F., Busslinger M. (1999) Nature 401, 603–606 [DOI] [PubMed] [Google Scholar]
- 14. Nutt S. L. (2008) N. Engl. J. Med. 358, 82–83 [DOI] [PubMed] [Google Scholar]
- 15. Delogu A., Schebesta A., Sun Q., Aschenbrenner K., Perlot T., Busslinger M. (2006) Immunity 24, 269–281 [DOI] [PubMed] [Google Scholar]
- 16. Nera K. P., Kohonen P., Narvi E., Peippo A., Mustonen L., Terho P., Koskela K., Buerstedde J. M., Lassila O. (2006) Immunity 24, 283–293 [DOI] [PubMed] [Google Scholar]
- 17. Hesslein D. G., Pflugh D. L., Chowdhury D., Bothwell A. L., Sen R., Schatz D. G. (2003) Genes Dev. 17, 37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fuxa M., Skok J., Souabni A., Salvagiotto G., Roldan E., Busslinger M. (2004) Genes Dev. 18, 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsu L. Y., Liang H. E., Johnson K., Kang C., Schlissel M. S. (2004) J. Exp. Med. 199, 825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roldán E., Fuxa M., Chong W., Martinez D., Novatchkova M., Busslinger M., Skok J. A. (2005) Nat. Immunol. 6, 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson K., Pflugh D. L., Yu D., Hesslein D. G., Lin K. I., Bothwell A. L., Thomas-Tikhonenko A., Schatz D. G., Calame K. (2004) Nat. Immunol. 5, 853–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Z., Espinoza C. R., Yu Z., Stephan R., He T., Williams G. S., Burrows P. D., Hagman J., Feeney A. J., Cooper M. D. (2006) Nat. Immunol. 7, 616–624 [DOI] [PubMed] [Google Scholar]
- 23. Fitzsimmons D., Hodsdon W., Wheat W., Maira S. M., Wasylyk B., Hagman J. (1996) Genes Dev. 10, 2198–2211 [DOI] [PubMed] [Google Scholar]
- 24. Fitzsimmons D., Lutz R., Wheat W., Chamberlin H. M., Hagman J. (2001) Nucleic Acids Res. 29, 4154–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garvie C. W., Hagman J., Wolberger C. (2001) Mol. Cell 8, 1267–1276 [DOI] [PubMed] [Google Scholar]
- 26. Sigvardsson M., Clark D. R., Fitzsimmons D., Doyle M., Akerblad P., Breslin T., Bilke S., Li R., Yeamans C., Zhang G., Hagman J. (2002) Mol. Cell. Biol. 22, 8539–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eberhard D., Busslinger M. (1999) Cancer Res. 59, 1716s–1724s [PubMed] [Google Scholar]
- 28. Linderson Y., Eberhard D., Malin S., Johansson A., Busslinger M., Pettersson S. (2004) EMBO Rep. 5, 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emelyanov A. V., Kovac C. R., Sepulveda M. A., Birshtein B. K. (2002) J. Biol. Chem. 277, 11156–11164 [DOI] [PubMed] [Google Scholar]
- 30. Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y. (1996) Cell 87, 953–959 [DOI] [PubMed] [Google Scholar]
- 31. Yang X. J., Seto E. (2008) Mol. Cell 31, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boyes J., Byfield P., Nakatani Y., Ogryzko V. (1998) Nature 396, 594–598 [DOI] [PubMed] [Google Scholar]
- 33. Gu W., Roeder R. G. (1997) Cell 90, 595–606 [DOI] [PubMed] [Google Scholar]
- 34. Hayakawa F., Abe A., Kitabayashi I., Pandolfi P. P., Naoe T. (2008) J. Biol. Chem. 283, 24420–24425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gingras S., Simard J., Groner B., Pfitzner E. (1999) Nucleic Acids Res. 27, 2722–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sheppard K. A., Rose D. W., Haque Z. K., Kurokawa R., McInerney E., Westin S., Thanos D., Rosenfeld M. G., Glass C. K., Collins T. (1999) Mol. Cell. Biol. 19, 6367–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen L. F., Williams S. A., Mu Y., Nakano H., Duerr J. M., Buckbinder L., Greene W. C. (2005) Mol. Cell. Biol. 25, 7966–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bradney C., Hjelmeland M., Komatsu Y., Yoshida M., Yao T. P., Zhuang Y. (2003) J. Biol. Chem. 278, 2370–2376 [DOI] [PubMed] [Google Scholar]
- 39. Nakamura S., Roth J. A., Mukhopadhyay T. (2000) Mol. Cell. Biol. 20, 9391–9398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Busslinger M. (2004) Annu. Rev. Immunol. 22, 55–79 [DOI] [PubMed] [Google Scholar]
- 41. Barlev N. A., Emelyanov A. V., Castagnino P., Zegerman P., Bannister A. J., Sepulveda M. A., Robert F., Tora L., Kouzarides T., Birshtein B. K., Berger S. L. (2003) Mol. Cell. Biol. 23, 6944–6957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu W., Fukuyama T., Ney P. A., Wang D., Rehg J., Boyd K., van Deursen J. M., Brindle P. K. (2006) Blood 107, 4407–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kasper L. H., Boussouar F., Ney P. A., Jackson C. W., Rehg J., van Deursen J. M., Brindle P. K. (2002) Nature 419, 738–743 [DOI] [PubMed] [Google Scholar]
- 44. Liu X., Wang L., Zhao K., Thompson P. R., Hwang Y., Marmorstein R., Cole P. A. (2008) Nature 451, 846–850 [DOI] [PubMed] [Google Scholar]
- 45. Liu Z., Mai A., Sun J. (2010) J. Immunol. 184, 244–254 [DOI] [PubMed] [Google Scholar]