Abstract
Insulin is the key regulator of glucose homeostasis in mammals, and glucose-stimulated insulin biosynthesis is essential for maintaining glucose levels in a narrow range in mammals. Glucose specifically promotes the translation of insulin in pancreatic β-islet, and the untranslated regions of insulin mRNA play a role in such regulation. Specific factors in the β-islets bind to the insulin 5′ UTR and regulate its translation. In the present study we identify protein-disulfide isomerase (PDI) as a key regulator of glucose-stimulated insulin biosynthesis. We show that both in vitro and in vivo PDI can specifically associate with the 5′ UTR of insulin mRNA. Immunodepletion of PDI from the islet extract results in loss of glucose-stimulated translation indicating a critical role for PDI in insulin biosynthesis. Similarly, transient overexpression of PDI resulted in specific translation activation by glucose. We show that the RNA binding activity of PDI is mediated through PABP. PDI catalyzes the reduction of the PABP disulfide bond resulting in specific binding of PABP to the insulin 5′ UTR. We also show that glucose stimulation of the islets results in activation of a specific kinase that can phosphorylate PDI. These findings identify PDI and PABP as important players in glucose homeostasis.
Keywords: Gene Regulation, Glucose, Insulin Synthesis, RNA-binding Protein, Translation Control
Introduction
Nutrients regulate insulin biosynthesis, and physiologically the most relevant is glucose. Induction of insulin biosynthesis by glucose involves the regulation at transcriptional translational and post-translational levels (1–5). In the early phase of glucose stimulation, insulin biosynthesis is predominantly regulated at the translational and secretion levels (6, 7). Insulin is stored in pancreatic β cells in large dense secretory granules and is secreted in response to glucose or other nutrient secretagogues. A complementary increase in proinsulin biosynthesis follows this secretion to replenish intracellular stores. Increase in glucose levels results in a greater than 10-fold increase in insulin translation, whereas general protein synthesis increases by a moderate 2-fold (8). Insulin mRNA is selectively recruited to the endoplasmic reticulum (ER)5 with an accompanying increase in the rate of translation (9).
The untranslated regions of the insulin mRNA have evolutionarily conserved features. Structural analysis of the 5′ UTR of insulin mRNA of rat, mouse, and human suggest the presence of a conserved stem-loop structure. A role for the untranslated regions (UTR) of the insulin mRNA has been previously shown in its translation (10–12). The 5′ and 3′ UTRs of insulin mRNA interact synergistically in enhancing its translation (13). A conserved element in the 5′ UTR of rat insulin gene 2 RNA (ppIGE-preproinsulin glucose element), which is necessary and sufficient for translation regulation of insulin mRNA has been reported (14). We have previously described a conserved minimal element in the 5′ UTR of rat insulin gene 1, which is necessary and sufficient to activate translation of insulin mRNA upon glucose stimulation (15). In the present study we have identified protein-disulfide isomerase (PDI) as the trans-acting factor that is necessary for glucose-stimulated insulin biosynthesis.
PDI was first identified as an ER-resident catalyst of native disulfide bond formation (16). Although the N-terminal of PDI contains the ER signal sequence, and the C-terminal contains the -KDEL, an ER retrieval sequence, other cellular localizations have been reported (17). Apart from its catalytic role in reduction, oxidation, and isomerization of protein disulfides during the folding processes of polypeptides, PDI also assists in folding of even those proteins that do not have disulfide bonds (18). PDI is also an essential subunit in the enzyme complexes of collagen prolyl-4-hydroxylase (19, 20) and microsomal triglyceride-transfer protein (21).
The presence of trans-acting factors in islet cytosolic extracts that bind to the insulin mRNA in a glucose-dependent manner and regulate translation have been reported (14, 15). In this study we have identified PDI and PABP as trans-acting factors that bind to the 5′ UTR of rat insulin gene 1 in pancreatic islets and regulate its translation. We show that PDI associates with the insulin 5′ UTR in vitro and in vivo. Functional involvement of PDI in the glucose-stimulated translation of insulin mRNA is demonstrated by the abrogation of translation activation in the presence of anti-PDI antibody, and further substantiated by the translation activation observed in insulin producing β cells by transient overexpression of PDI.
EXPERIMENTAL PROCEDURES
Isolation and Culture of Pancreatic Islets and βTC6, Preparation of Cytosolic Extracts
All animal work involving mouse and rats were performed as per the “Institutional Experimental Animal User's Committee” guidelines and approval. Adult Wistar rats were sacrificed by cervical dislocation after anesthetization by ether. Pancreatic islets were then isolated by pancreatic duct injection of 500 units/ml of collagenase solution followed by digestion at 37 °C for 40 min with mild shaking. Islets were washed several times with Hanks' balanced salt solution and separated from acinar cells on a discontinuous Ficoll 400 gradient. The islets were cultured in RPMI 1640 HGB, supplemented with 10% fetal calf serum (Sigma), 100 units/ml of benzyl penicillin, and 0.1 mg/ml of streptomycin. The islets were cultured free floating for 12 h in 5% CO2 atmosphere at 37 °C.
Min6 and βTC6 cells were cultured in DMEM with 15% FCS in 5% CO2 atmosphere. Rat pancreatic islets and Min6/βTC6 cells were exposed to substimulatory serum-free conditions of low glucose (2.7 mm) for 1.5 h followed by incubation in high glucose (16.6 mm) or low glucose medium for 1 h. After treatment the islets were harvested by centrifugation and lysed in buffer containing 50 mm Tris (pH 7.6), 50 mm NaCl, 1 mm PMSF, 1 mm DTT, 1× protease inhibitor mixture (Sigma). The islet suspension was homogenized using a Dounce homogenizer with 7–10 strokes. The homogenized suspension was passed 2–3 times through a 26-gauge needle. The homogenized suspension was then centrifuged at 100,000 × g for 30 min at 4 °C. The supernatant was collected and the protein concentration of the supernatant was adjusted to 1 mg/ml.
Phosphorylation and Dephosphorylation of the Extracts
100 ng of HG extract was incubated with AP buffer (50 mm Tris (pH 7.6), 50 mm NaCl, 1 mm PMSF, 1 mm DTT, 0.1 mm ZnCl2, 1 mm sodium vanadate) at 25 °C for 20 min with or without 1 unit of calf intestinal alkaline phosphatase (Promega) in a 10-μl reaction. For phosphorylation, 100 ng of LG extract was incubated with 5 mm ATP with or without 0.068 units of protein kinase C (Sigma) in a 10-μl reaction. The extracts were incubated at 4 °C for a further 5–10 min and were used for RNA-EMSA or in vitro transcription reaction.
Phosphorylation of Recombinant PDI by Islet Extracts
LG- or HG-treated islet extracts were incubated with 75 ng of either His-PDI or His-GFP with 3 pm [γ-32P]ATP at 25 °C for 20 min, followed by purification with Ni-NTA-agarose (Invitrogen) beads. In control experiments His-PDI was incubated with islet lysis buffer (as previously described) with radiolabeled ATP followed by Ni-NTA purification. The bound proteins were eluted by SDS loading dye and heated at 95 °C for 5 min and analyzed by 10% SDS-PAGE followed by autoradiography.
RNA-Affinity Purification of Insulin 5′ UTR-binding Proteins
Pancreatic islet lysate (5 mg of protein for each pull down) was incubated with either biotinylated wild type or the 14-16,41-43 deleted fragment of the insulin 5′ UTR along with tRNA (500 μg), nonspecific competitor Min RNA (20 μg), and RNAsin (2000 units/ml) in 1× gel shift buffer for 30 min at 4 °C. 100 μl of μMACs streptavidin microbeads (Milltenyi) were added to the above mixture and further incubated for 10 min. The mixture was applied to a magnetic column and the column was washed with 1× gel shift buffer. Bound proteins were eluted with 1 m NaCl. The eluates were desalted and resolved on 10% SDS-PAGE and proteins were visualized by Coomassie Blue staining.
In Vitro Transcription of the Insulin 5′ UTR Fragments
5′ UTR fragments of rat insulin gene 1 were generated by oligonucleotide-directed transcription. Oligonucleotides corresponding to the complementary sequence of the desired UTR fragment and containing the complementary T7 promoter sequence at the 3′ end were custom synthesized. These oligonucleotides were annealed to the T7 promoter oligonucleotide and partially double-stranded oligonucleotides were used as templates in transcription reactions.
A radiolabeled synthetic transcript of the rat insulin 5′ UTR was prepared by in vitro transcription. 0.15 pmol of annealed double stranded oligonucleotide was transcribed at 37 °C for 1 h using T7 RNA polymerase (New England Biolabs) in the presence of 50 μCi of [α-32P]UTP (BRIT, Hyderabad). Unincorporated nucleotides were removed by passing the transcription reaction through a G-10 Sephadex column.
RNA EMSA and Depletion of PDI from Extracts
[α-32P]UTP-labeled insulin 5′ UTR (38 fmol) was incubated for 30 min at 4 °C with cell extract (4 μg of protein) in 20 μl of reaction mixture containing 1× gel shift buffer (5 mm Tris, pH 7.5, 15 mm KCl, 0.25 mm DTT, 5 mm MgCl2, 4 μg of yeast tRNA, 40 units of RNasin, and 10% glycerol). For supershift experiments, control IgG antibody (1 μg) or PDI monoclonal antibody (Abcam) (1 μg) was then added and the mixture was incubated on ice for a further 15 min. The RNA-protein complex was resolved on a 6% native PAGE in 0.5× TBE. The gel was dried, and the complex formation was assessed by autoradiography. Immunodepletion of PDI from extracts was carried out by addition of 10 μg of PDI antibody or control antibody to 50 μg of protein extracts. Following incubation for 15 min on ice, protein A beads were added to sequester the antibody-antigen complex. Beads were then centrifuged and the supernatant was used as the PDI-depleted extract. To elute PDI from the protein A beads, 0.2 m glycine (pH 2.5) was used and the eluate was neutralized immediately with 1 m Tris (pH 9.0). Each experiment was repeated at least three times with different extract preparations with similar results and a representative example is shown for each experiment.
PDI-RNA Immunoprecipitation
βTC6 cells in a 100-mm Petri dish were washed twice in PBS buffer and formaldehyde (37%, w/w) was added to a final concentration of 1% (w/v) and incubated at room temperature for 10 min with slow rocking followed by the addition of glycine to a final concentration of 0.25 m to neutralize excess formaldehyde. The cells were incubated for 5 min at room temperature and then washed twice with ice-cold PBS and resuspended in 4 ml of RIPA buffer (50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% SDS, 1 mm EDTA, 150 mm NaCl) containing protease inhibitors and 50 units/ml of RNase inhibitor. Cell suspensions were sonicated to lyse the cells and centrifuged at 13,000 × g at 4 °C for 10 min to remove insoluble material. 25% of the supernatant was incubated with 5 μl of anti-PDI or anti-mouse IgG antibodies for 1 h followed by addition of 5 μl of protein A/G-agarose beads at 4 °C for 4 h. The beads were collected by centrifugation at 1,000 × g for 2 min and washed six times with 1 ml of RIPA buffer, at high-salt (1 m NaCl) conditions. The beads after the wash were suspended in 200 μl of 50 mm Tris-HCl (pH 7.0), 5 mm EDTA, 10 mm DTT, 1% SDS followed by incubation at 70 °C for 45 min to reverse the cross-links. The immunoprecipitated RNAs were isolated with TRI Reagent and analyzed by RT-PCR.
UV Cross-linking and IP
After performing the RNA EMSA as described above, the samples were irradiated for 7 min at 4 °C in a UV cross-linker at 1200 microjoules. The reaction mixture was diluted in RIPA buffer and aliquots were incubated with 5 μl of anti-PDI or anti-mouse IgG antibodies followed by addition of 5 μl of protein A/G-agarose beads at 4 °C for 4 h. The beads were collected by centrifugation at 1,000 × g for 2 min and washed six times with 1 ml of RIPA buffer, at high-salt (1 m NaCl) conditions. SDS dye was added to the beads and denatured at 100 °C. The samples were analyzed by 10% SDS-PAGE followed by autoradiography.
Preparation of ins5′UTR-Luc-Ins3′UTR pcDNA3, PDIpcDNA3 Constructs for in Vivo Translation Studies
We had previously cloned the 5′ and 3′ UTR of insulin mRNA in pSP64-LucCp3′UTR-poly(A) plasmid (from Dr. Paul Fox, Lerner Research Institute, Cleveland Clinic Foundation). From the above plasmid the ins5′UTR-Luc-ins3′UTR was removed by HindIII and Ecl136II. The digested product was cloned into pcDNA3 (Promega) at HindIII and EcoRV sites. Two other constructs containing the 5′ UTR deletion mutant and the 5′ UTR minimal sequence along with the full-length 3′ UTR, were also generated similarly from their corresponding pSP64 constructs.
PDI ORF was amplified from pancreatic islet cDNA using primers containing restriction sites BamHI and XhoI sites. The amplified product was then cloned into pcDNA3 to generate the PDI pcDNA3 construct for overexpression of PDI in cells.
The pSP64 Ins5′Luc-3′UTR-poly(A) full-length clones and the clones corresponding to the other mutants were digested with PvuII and the linear constructs were subjected to in vitro transcription with SP6 polymerase (mMessage machine, Ambion) to produce capped transcripts. Control Renilla was made by in vitro transcription of pRL (Promega) by T7 polymerase (Megascript kit). After transcription, the reaction mixture was treated with DNase I followed by phenol-chloroform extraction and ethanol precipitation. Synthesis of full-length cRNA transcripts was verified by electrophoresis on a 1.2% formaldehyde-agarose gel.
In Vitro Translation of cRNA Constructs by Reticulocyte Lysate
For in vitro translation, 30 fmol of cRNA containing the insulin 5′ UTR-Luciferase was added to 17.5 μl of rabbit reticulocyte lysate (Promega), 20 μm amino acid mixture, 40 units of RNAsin, and 2 μg of high glucose or low glucose-treated pancreatic islet extract and 1 μl of α-PDI or α-mouse IgG and 1 μg of PDI-GST in a total volume of 25 μl. The translation reaction was carried out for 30 min at 30 °C. cRNA encoding Renilla luciferase was added to each reaction as a translation efficiency control. After the translation reaction, a dual luciferase assay was performed to check for translated products.
PDI Overexpression
Plasmids ins5′UTR-Luc-Ins3′UTRpcDNA3, ins5′UTRdel-Luc-Ins3′UTRpcDNA3, ins5′UTRcon1-Luc-Ins3′UTRpcDNA3, and either PDIpcDNA3 or empty pcDNA3, were transfected (total DNA, 250 ng) using Lipofectamine (Invitrogen). Renilla luciferase was added (0.25 ng) to each transfection as control. 2 h after transfection, complete medium was added for recovery of cells. Following recovery in complete medium for 24 h, cells were incubated with low glucose medium (1.67 mm) for 4 h followed by a further incubation in either low or high glucose (16.6 mm) medium for 2 h and luciferase activity was measured using the Dual Luciferase assay (Promega) following the manufacturer's instructions.
Western Blot
10 μg of protein extract was resolved on 10% SDS-PAGE, transferred to PVDF membrane, and the blots were probed with α-PDI (Abcam) and then stripped and reprobed with α-actin polyclonal (Abcam) for loading control.
Statistics
The experiments were performed at least three times, and wherever appropriate the gels were scanned and the band intensities were quantified by Adobe photoshop after inversion of the image. The results were analyzed by Sigmaplot and the p values were calculated by pairwise Student's t test.
RESULTS
Isolation of Insulin mRNA 5′ UTR-binding Proteins
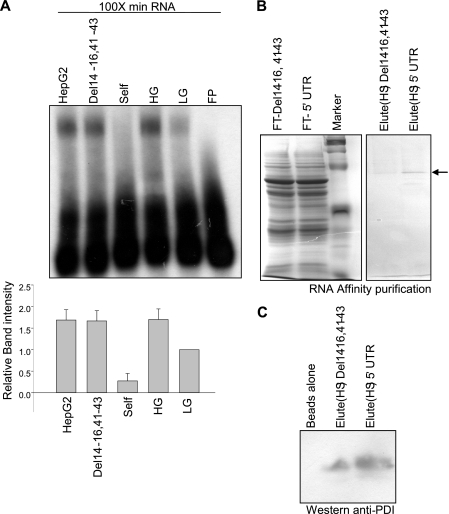
We have previously shown that specific factors in the cytoplasmic extracts of the rat pancreatic islets bind to the 5′ UTR of insulin mRNA and this binding correlates with translation activation upon glucose stimulation. To isolate factors binding to the insulin 5′ UTR we prepared cytoplasmic extracts prepared from rat pancreatic islets, treated with 2.7 mm (LG, low glucose) or 16.7 mm (HG, high glucose) of glucose for 1 h. The cytosolic extracts were then incubated with the radiolabeled 5′ UTR RNA of rat insulin gene 1 in the presence of a 100-fold excess of nonspecific Min RNA (Min is derived by deleting bases 1–7, 23–36, and 54–57 in the 5′ UTR sequence of rat insulin gene 1) to assess the presence of specific RNA binding factors by RNA-EMSA. A specific RNA-protein complex was formed with β cell extracts and the 5′ UTR of rat insulin gene 1 RNA. A 70-fold molar excess of an unlabeled full-length 5′ UTR RNA was able to compete out the labeled complex, but a similar fold excess of Del14-23,41-43 RNA or HepG2 total RNA did not compete out the labeled complex (Fig. 1A). The extracts that showed specific binding to the rat insulin 5′ UTR were then used for purification and identification of the RNA-binding protein(s).
FIGURE 1.
Specific binding of islet proteins to the rat insulin 5′ UTR. A, competitive RNA-EMSA was performed as described under “Experimental Procedures.” The sample in each lane is as indicated: the radiolabeled insulin 5′ UTR (FP), extract from low glucose (2.7 mm)-treated islets with no competitor (LG), extract from high glucose (16.7 mm)-treated islets with no competitor (HG), HG with the unlabeled 5′ UTR as a competitor (Self), HG with unlabeled in vitro synthesized nonspecific RNA as competitor (Del14-16,41-43), and HG with total RNA from HepG2 cells as competitor (HepG2). All lanes contain 100-fold molar excess of Min RNA. The gel-shifted bands were quantitated densitometrically, and expressed with respect to LG extracts set to 1. The graph represents the average of four independent experiments and the error bar indicates the mean ± S.E. calculated by σ plot (lower panel). B, RNA affinity pulldown was carried out as described under “Experimental Procedures.” The protein(s) bound to RNA was eluted using 1 m NaCl. The flow-through (FT) and the high salt (HS)-eluted fractions were resolved on a 10% SDS-PAGE and detected by Coomassie staining and also by immunoblotting with PDI antibody (C).
Insulin 5′ UTR-binding proteins were isolated by RNA-affinity purification using biotinylated insulin 5′ UTR. We also took the biotinylated deletion fragment of the insulin 5′ UTR (del14-16,41-43) that is inactive with respect to mRNP formation and translational activation as the nonspecific control (15). The bound proteins were eluted with high salt buffer, and the purified proteins were resolved by SDS-PAGE and stained with Coomassie Blue. A major specific protein band at about 60 kDa was preferentially eluted from the insulin 5′ UTR column (Fig. 1B) as compared with the column with nonspecific del14-16,41-43 RNA. Protein present in this band was identified by mass spectrometry and PDI was identified as one of the proteins. The presence of PDI in this 60-kDa band was further confirmed by Western blotting of the eluates with anti-PDI antibody (Fig. 1C).
PDI Is Necessary for RNA-Protein Complex Formation
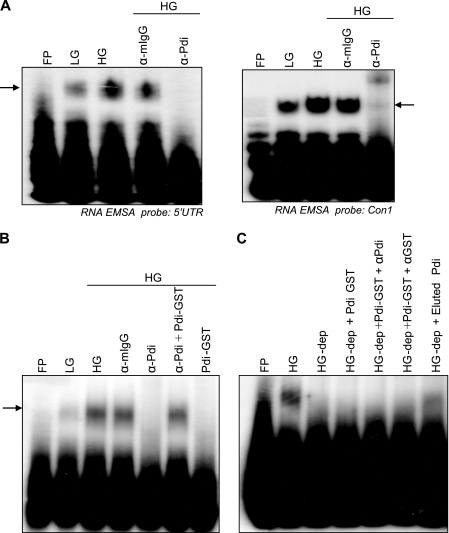
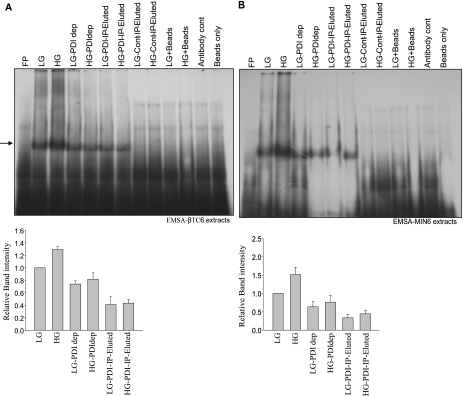
PDI is normally an ER lumen protein that is responsible for the oxidative folding of newly synthesized proteins. A role of PDI in translation regulation was reported in the plant system where PDI associates with the PsbA RNA and regulates translation in response to light (22). We now show that in rat pancreatic islets, PDI is necessary for the formation of the insulin 5′ UTR RNA-protein complex. The 5′ UTR of the rat insulin gene 1 was incubated with glucose-treated extracts in an EMSA reaction followed by addition of anti-PDI antibodies. Formation of specific RNA-protein complexes by islet extracts and the 5′ UTR of the rat insulin gene were inhibited in the presence of anti-PDI antibody, whereas the control antibody had no significant effect on complex formation (Fig. 2A). The complex formed was shifted in mobility in the presence of anti-PDI antibody when a shorter minimal RNA probe was used for the RNA mobility shift experiment. Furthermore, when bacterially expressed recombinant PDI-GST was added along with antibody the complex formation was restored due to sequestering of the antibody by the recombinant protein (Fig. 2B), whereas the recombinant PDI on its own was not able to form a specific complex with the insulin 5′ UTR. Specific immunodepletion of PDI the extracts resulted in loss of the complex forming ability, and the complex forming ability was not restored by addition of bacterially expressed recombinant PDI, but only by the addition of the immunoprecipitated PDI from β islets (Fig. 2C). These results suggest a requirement of specific modifications and/or the necessity of other proteins for complex formation. We also tested whether this PDI-5′UTR RNA complex formation was restricted to rat pancreatic islets or a similar mechanism operating in other systems. We prepared cytoplasmic extracts from two insulin producing mouse cell lines, MIN6 or βTC-6, which show glucose-stimulated insulin synthesis. We observed that immunodepletion of PDI resulted in reduced RNA-protein complex formation and the PDI-immunoprecipitates contain factors that can form a specific complex with the insulin 5′ UTR (Fig. 3, A and B).
FIGURE 2.
PDI is part of the RNA-protein complex. A, supershift RNA EMSA with the full-length insulin 5′ UTR and con1 probe were performed. Extract from LG- or HG-treated islets were incubated in the presence or absence of control IgG antibody (α-mIgG) or PDI antibody (α-Pdi). B, RNA-EMSA with PDI-depleted extracts. Lanes are as indicated with HG-dep referring to the PDI-depleted HG extract. C, presence of PDI in the RNA-protein complex. EMSA was performed with the insulin 5′ UTR probe. The extracts and the other reagents used are described. Extracts from 2.7 mm glucose-treated islets, 16.6 mm glucose-treated islets, HG with control IgG antibody (α-mIgG), HG with PDI antibody (α-Pdi), HG with α-PDI and PDI-GST, and PDI GST alone.
FIGURE 3.
PDI is essential for RNA-protein complex formation. EMSA was performed with immunoprecipitated proteins and the insulin 5′ UTR. PDI was immunoprecipitated from either: LG or HG extracts and the bound proteins were eluted and used for EMSA. Lanes in both A (βTC6 cells) and B (Min cells) are as follows: extract from 2.7 mm glucose-treated cells (LG), extract from 16.6 mm glucose-treated cells (HG), LG or HG extract after depletion with PDI antibody (LG-PDI-dep and HG-PDI-dep), LG or HG immunoprecipitated with PDI antibody (LG-PDI IP-Elute, HG-PDI IP-Elute), LG or HG immunoprecipitated with control IgG (LG-control IP-elute, HG-control IP-Elute), LG or HG with beads only (LG-Bead, HG-Bead), and with only PDI antibody. The gel-shifted bands (indicated by the arrow) were quantitated densitometrically, and expressed with respect to LG extracts set to 1 (lower panel). The graph represents the average of three independent experiments and the error bar indicates the mean ± S.E. calculated by σ plot.
PDI Binds Specifically to Insulin 5′ UTR RNA
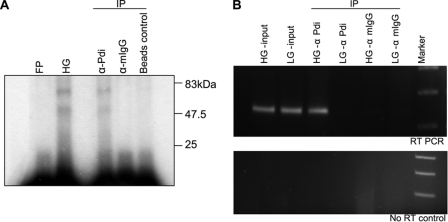
Specific interaction of PDI with the insulin 5′ UTR was further confirmed by UV cross-linking of the labeled insulin 5′ UTR-RNA to the protein followed by immunoprecipitation. Antibodies against PDI specifically pulled down cross-linked RNA-protein complex, whereas the control IgG or beads alone did not (Fig. 4A). The in vivo interaction of PDI with the rat insulin 5′ UTR was shown by RNA immunoprecipitation. The RNA-protein complex was cross-linked by treating the cells with formaldehyde and the PDI-associated RNA was immunoprecipitated and analyzed by RT-PCR. Insulin mRNA was found to be specifically associated with PDI and the amount of insulin mRNA associated with PDI increases when the cells are treated with higher levels of glucose (Fig. 4B) without any significant alteration in the total insulin RNA or PDI (supplemental Fig. S1).
FIGURE 4.
PDI associates with the insulin 5′ UTR. A, immunoprecipitation (IP) of UV cross-linked PDI. Lanes are free probe (FP), high glucose-treated (HG) extracts after UV cross-linking before IP, immunoprecipitation with α-PDI (α-PDI) or mouse IgG (α-mIgG), or beads alone control. B, immunoprecipitation of RNA associated with PDI. Islets were treated with formaldehyde and the cross-linked RNA associated with PDI was immunoprecipitated with PDI antibody and purified. The RNA was assayed for the presence of insulin mRNA by RT-PCR using insulin mRNA-specific primers. The RT-PCR product from the HG- and LG-treated cell input (HG- and LG-input), IP with α-PDI (α-PDI), and control mouse IgG (α-mIgG) of these extracts. Lower panel shows the no RT control.
PDI Activates Glucose-stimulated Insulin Translation
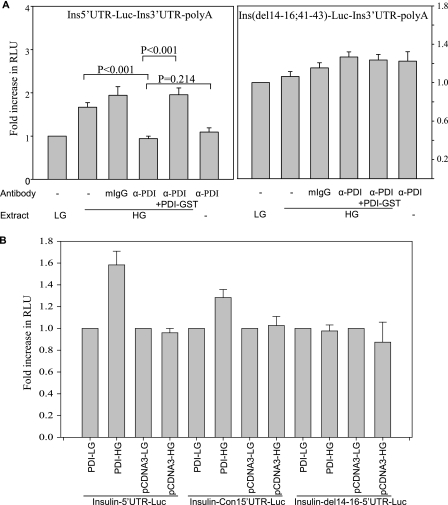
The role of PDI on the insulin 5′ UTR-mediated translation regulation was assessed using luciferase reporter containing the insulin 5′ UTR. In vitro translation was performed in the presence of extracts from β islets treated with either low glucose or high glucose. We have previously shown that the extracts from high glucose-treated islets show 50% increases in the insulin 5′ UTR-dependent stimulation in translation activity (15). Here also we observed a specific increase in relative luciferase activity in the presence of the high glucose extracts when compared with translation in the presence of the low glucose extract. However, this increase in translation was abolished by the addition of antibodies against PDI, whereas the addition of nonspecific IgG had no significant effect (Fig. 5A). The glucose-dependent translation activation was restored when the PDI antibody was neutralized using recombinant PDI-GST. These results show that PDI is essential for translation activation of the insulin mRNA by glucose.
FIGURE 5.
Insulin 5′ UTR-dependent translation regulation by PDI. A, in vitro translation of the chimeric ins-5′UTR-Luc-ins3′UTR in the presence of low (LG, 2.7 mm) or high glucose (HG, 16.7 mm) extract and PDI antibody or control mouse IgG (mIgG) or PDI-GST. Renilla luciferase mRNA was added to each reaction for normalization of translation activity. Translation of luciferase was normalized with Renilla levels and expressed as a fraction of translation of the insulin 5′ UTR containing RNA in the presence of the low glucose extract. The experiments were done in triplicate and the graph represents the average of three experiments. The error bars indicate mean ± S.E. B, translation regulation of the insulin 5′ UTR by PDI in vivo. Transfections of luciferase constructs containing the full-length 5′ UTR or con1 or deletion mutant before luciferase ORF along with PDI-pcDNA3 were done in βTC6. LG (2.7 mm) and HG (16.7 mm) indicate the glucose treatments, and the transfected plasmids are indicated. Renilla luciferase plasmid was cotransfected for normalization of transfection. Translation of luciferase was normalized with Renilla levels and expressed as the fraction of translation of the insulin 5′ UTR-containing plasmid in low glucose conditions. The experiments were done in triplicate and the graph represents the average of four independent experiments. The p values were estimated by paired t test using σ plot.
To determine whether PDI has a role in translational regulation in vivo, transfections were done in βTC6 and HeLa cell lines. βTC6 is a glucose responsive mouse insulinoma cell line (23) and βTC6 extracts show a similar glucose induced binding with the 5′ UTR of the rat insulin gene 1 (15). PDI was overexpressed along with the luciferase construct having the insulin 5′ UTR. There was about 60% increase in relative luciferase activity upon glucose stimulation when PDI was co-expressed with the full-length insulin 5′ UTR or the minimal con1 5′ UTR luciferase constructs, whereas no change was observed with the mutant nonbinding 14-16,41-43 deletion construct (Fig. 5B). The translation regulation was specific to insulin producing cells, as no changes in relative luciferase expression were observed in insulin nonproducing HeLa cells (supplemental Fig. S2). The levels of overexpressed PDI in the transfected cells were similar and remain unaltered with low or high glucose treatments (supplemental Fig. S3).
A role of PDI in translation regulation was reported in the plant system where PDI associates with the PsbA RNA and regulates translation in response to light (22). The nuclear-encoded PDI is localized to the chloroplast and associates with PABP and modulates its binding to the 5′ UTR of PsbA mRNA (22). PDI alters the redox status of cytoplasmic PABP thus modulating its ability to associate with the 5′ UTR of PsbA RNA (24), and the activity of PDI itself is regulated by ADP-dependent phosphorylation (25). A similar role for PDI in regulating the translation of insulin mRNA could be envisaged, like the light-regulated PsbA expression, translation of insulin mRNA is also increased very rapidly by severalfold in response to glucose.
PABP Binds Specifically to Insulin 5′ UTR
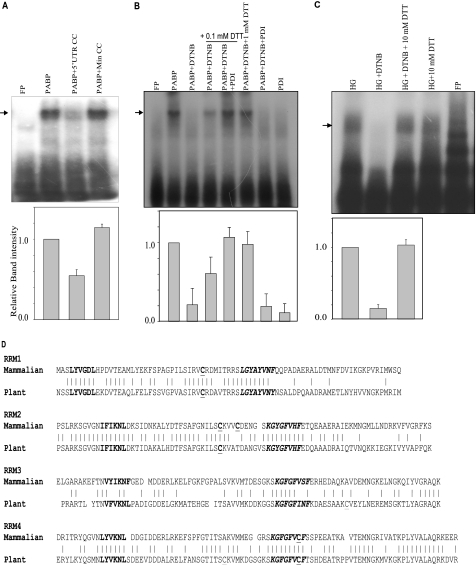
We analyzed the insulin 5′ UTR binding activity of PABP by RNA-EMSA using bacterially expressed recombinant PABP. Recombinant PABP was able to bind specifically to the insulin 5′ UTR (Fig. 6A). The complex was competed out by unlabeled insulin 5′ UTR but not by the mutant non-binding insulin 5′ UTR. We also assessed the regulation of the insulin 5′ UTR RNA binding activity of PABP by oxidation/reduction of disulfide bonds. Recombinant PABP was oxidized with 3 mm DTNB and binding to the insulin 5′ UTR was assessed (Fig. 6B). Oxidation of cysteines in PABP by DTNB causes loss of binding to the insulin 5′ UTR, which can be restored by addition of excess DTT. Similar results were obtained with the islet extract (Fig. 6C). The binding activity can be enhanced by the addition of limiting amounts of DTT and recombinant PDI. These results suggest that as in the plant chloroplast system the RNA binding activity that is responsible for translation regulation is predominantly through PABP and PDI rearranges/reduces the disulfide linkages to regulate the RNA binding activity. We also assessed the modifications to PABP by DTNB by resolving the oxidized products in acid urea gels. These results indicate that PABP is modified by DTNB and can be reduced by DTT (supplemental Fig. S4).
FIGURE 6.
PABP bind to the insulin 5′ UTR. A, competitive RNA-EMSA was performed as described under “Experimental Procedures.” The sample in each lane is as indicated. The radiolabeled insulin 5′ UTR (FP) was incubated with bacterially expressed recombinant PABP (100 ng) in the presence of the unlabeled insulin 5′ UTR or mutant 5′ UTR RNA as competitor. The gel-shifted bands were quantitated densitometrically, and expressed with respect to only PABP without competitor set to 1 (lower panel). The graph represents the average of three independent experiments and the error bar indicates the mean ± S.E. calculated by σ plot. B, RNA-EMSA was performed to assess the role of free cysteines in the RNA binding activity. The radiolabeled insulin con1 probe was incubated with PABP, PABP oxidized with DTNB, and the oxidized PABP reduced with varying amounts of DTT or DTT and PDI (200 ng). The gel-shifted bands were quantitated densitometrically, and expressed with respect to only PABP without competitor set to 1 (lower panel). The graph represents the average of three independent experiments and the error bar indicates the mean ± S.E. calculated by σ plot. C, the radiolabeled con1 insulin 5′ UTR were incubated with islet extracts, extracts oxidized with DTNB, and oxidized extracts reduced with DTT. The gel-shifted bands of HG, HG-DTNB, and HG + DTNB + DTT were quantitated densitometrically, and expressed with respect to the HG extract set to 1 (lower panel). The graph represents the average of four independent experiments and the error bar indicates the mean ± S.E. calculated by σ plot. D, amino acid sequence comparison of the RRMs of PABP from mammals and plants. The conserved hexamer (bold) and octamer (bold italics) in RRMs, as well as the cysteine residues (bold underlines) are indicated.
PABP contains four well characterized RNA recognition motifs (RRMs) in the N terminus of the protein (Fig. 6D) (26). In the plant system the disulfide bond formation between RRM2 and RRM3 is responsible for regulating its RNA binding activity (27). In mammals the cysteine residue in the RRM3 is not conserved, whereas RRM2 contains an additional cysteine residue that is conserved in PABP from mammals. We believe that the RNA binding activity and the translation of insulin mRNA is regulated by oxidoreduction of these two cysteines.
Role of Phosphorylation in Glucose-stimulated Insulin Translation
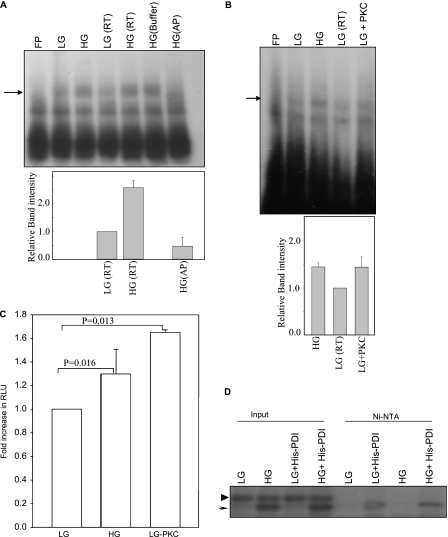
We assessed the role of phosphorylation on the insulin 5′ UTR RNA binding activity. We performed RNA-EMSA experiments with alkaline phosphatase-treated extracts. Phosphatase treatment of the extract resulted in complete loss of the insulin 5′ UTR-specific complex formation (Fig. 7A). Because phosphatase treatment resulted in loss of RNA binding activity we tested whether phosphorylation of extracts would result in increased binding activity. Phosphorylation of the extracts with purified casein kinase did not alter the RNA binding activity of the extract. When extracts from low glucose-treated cells were phosphorylated with protein kinase C (PKC), it resulted in increased RNA-protein complex formation (Fig. 7B). Furthermore, when these phosphorylated extracts were used in the in vitro translation assay it resulted in increased translation (Fig. 7C). The increase in translation observed with extracts from low glucose-treated cells phosphorylated with PKC was similar to the one obtained with extracts from high glucose-treated extracts (Fig. 7C). We then addressed whether glucose activates specific kinase in pancreatic islets. Kinase activity of the extracts treated with either 2.7 (LG) or 16.7 mm (HG) glucose was determined using recombinant PDI. The extracts were incubated with bacterially expressed recombinant His-PDI and radiolabeled ATP. HG-treated extracts showed a specific labeled product at about 60 kDa (Fig. 7D, indicated by an arrow), whereas a nonspecific labeled product of about 70 kDa (Fig. 7D, indicated by an arrowhead) was observed in both LG- and HG-treated samples (Fig. 7D, lanes 1–4). Purification of the recombinant His-PDI from this reaction using the Ni-NTA column resulted in isolation of a specifically labeled product corresponding to the 60-kDa band with an increased labeled product in the HG-treated extract when compared with LG-treated extracts (Fig. 7D, lanes 5–8). These results indicate that upon glucose stimulation, a specific kinase is activated that can phosphorylate PDI. Phosphorylated PDI then associates with insulin mRNA and activates its translation.
FIGURE 7.
Role of phosphorylation in insulin translation. A, RNA-EMSA was performed with extracts treated with phosphatase. Indicated lanes are: extract from LG-treated islets (LG), extract from HG-treated islets (HG), LG extract kept at RT (LG-RT), HG extract kept at RT (HG-RT), HG extract incubated with only phosphatase buffer (HG buffer), and HG extract treated with alkaline phosphatase (HG-AP). The gel-shifted bands in LG(RT), HG(RT), and HG(AP) lanes were quantitated densitometrically, and expressed with respect to LG extract set to 1 (lower panel). The graph represents the average of three independent experiments and the error bar indicates the mean ± S.E. calculated by σ plot. B, RNA-EMSA was performed with extracts treated with protein kinase C (PKC). Lanes are extract from low glucose-treated islets (LG), high glucose-treated islets (HG), LG extract incubated at RT (LG-RT), LG extract incubated with PKC (LG-PKC), and probe incubated with only PKC (FP-PKC). The gel-shifted bands in LG(RT), HG(RT), and LG + PKC-treated lanes were quantitated densitometrically, and expressed with respect to LG extract set to 1 (lower panel). The graph represents the average of three independent experiments and the error bar indicates mean ± S.E. calculated by σ plot. C, in vitro translation of chimeric ins-5′UTR-Luc-ins3′UTR in the presence of low (LG, 2.7 mm) or high glucose (HG, 16.7 mm) extract or low (LG, 2.7 mm) extract treated with PKC. Renilla luciferase mRNA was added to each reaction for normalization of translation activity. Translation of luciferase was normalized with Renilla levels and expressed as a fraction of translation of the insulin 5′ UTR containing RNA in the presence of low glucose extract. The experiments were done in triplicate and the graph represents the average of three experiments. Error bars indicates the mean ± S.E. The p values were estimated by paired t test using σ plot. D, activation of PDI-specific kinase by glucose stimulation. Extracts from 2.7 (LG) or 16.6 mm (HG) glucose-treated islets were incubated with recombinant His-PDI in the presence of [γ-32P]ATP (3 pmol). Phosphorylated proteins were resolved on SDS-PAGE before or after purification by Ni-NTA resin. Lanes are as indicated. LG extracts with His-PDI (LG + His-PDI), HG extracts with His-PDI (HG + His-PDI) are indicated.
DISCUSSION
Glucose-stimulated insulin synthesis/secretion is important for glucose homeostasis in mammals. Although specific translation activation of insulin in response to glucose was reported several decades ago, the precise molecular mechanism of such a regulation has not yet been elucidated. The 5′ UTR of insulin mRNA was shown to play an important role in this regulation and cytoplasmic factors in the islets that bind to the 5′ UTR of the insulin RNA have been reported earlier. Identification of these trans-acting factors involved in the regulation of insulin mRNA by glucose will help in understanding the mechanistic details of nutrient-mediated regulation of insulin biosynthesis. In addition it may give new insights into some of the causes for Type II diabetes, as there is indeed a deregulation of insulin production in this metabolic disorder. In our earlier studies we characterized the elements in the RNA involved in this regulation and identified a conserved stem-loop structure with key residues that are necessary and sufficient for the activation. In this study we characterize the trans-acting factor interacting with the minimal element. We have isolated the proteins that bind to the insulin 5′ UTR, and identified it to be PDI by mass spectroscopy. The results of mass spectrometry were confirmed by Western analysis of PDI. Furthermore, α-PDI abrogates complex formation with the 5′ UTR probe and also can immunoprecipitate the labeled insulin 5′ UTR RNA after UV cross-linking, suggesting direct binding with the RNA probe. The full-length recombinant PDI was unable to bind to the RNA and the reasons could be that stable binding of PDI to RNA requires other interacting partners or possibly specific modifications, for binding to the insulin 5′ UTR. We demonstrate that association of PDI with insulin RNA is regulated by glucose levels and increases upon glucose treatment in βTC6 cells, suggesting a role for RNA-protein association in the translation activation. Results from the in vitro translation experiments support the idea that PDI is an activator of translation and is further affirmed by the in vivo experiments in βTC6 cells, where overexpression of PDI results in glucose-mediated stimulation. Although βTC6 is a glucose responsive cell line (23), we did not see any appreciable level of induction without overexpression of PDI. This could be due to low levels of PDI in βTC6 cells, which is not sufficient to show an appreciable change in translated levels of luciferase RNA, which are synthesized in large amounts.
PDI is predominantly an ER resident protein but has been known to be secreted from hepatocytes (28), pancreatic exocrine cells (29), endothelial cells (30), and platelets (31). PDI has also been localized to the cytosol of human and monkey liver cells (32). Nuclear localization of PDI with specific association with the nuclear matrix and DNA has also been reported (33–35). The physiological function of PDI in these non-ER locations is not very clear. In insulin producing cells, PDI is predominantly localized to cytosolic fractions as assessed by biochemical fractionation (supplemental Fig. S5). We also find that the insulin 5′ UTR binding activity is also predominantly in the cytoplasmic fraction indicating that activation of PDI does not cause any major redistribution of the protein.
Multiple activities for PDI have been reported previously, in plants, nuclear encoded PDI is localized to the chloroplast and associates with chloroplast PABP and modulates its binding to the 5′ UTR of PsbA mRNA (22). PDI alters the redox status of cPABP thus modulating its ability to associate with the 5′ UTR of PsbA RNA (24), and the activity of PDI itself is regulated by ADP-dependent phosphorylation (25, 36) and a similar role for PDI in regulating the translation of insulin mRNA could be envisaged. Similar to light-regulated PsbA expression, translation of insulin mRNA is also increased very rapidly by severalfold in response to glucose.
PDI is also the β subunit of prolyl-4-hydroxylase, an enzyme that catalyzes the formation of 4-hydroxyproline on proteins (20). Thus PDI has alternate enzymatic functions depending on its associated protein partners. Interestingly, Argonaute2 (Ago2), an important player in micro-RNA-mediated translation repression has been shown to be hydroxylated by prolyl-4-hydroxylase. PDI knockdown results in loss of prolyl-4-hydroxylase activity resulting in the reduced stability of Ago2 and thus regulating the micro-RNA functions in the cells (37). Thus the mechanism of PDI-mediated insulin mRNA translation regulation could also involve Ago2 function and needs to be explored. Recently it was reported that overexpression of PDI increased the steady-state intracellular proinsulin levels in the insulin producing cell line (38). Our results suggest that the increase in steady-state proinsulin levels is likely to occur by increased translation of insulin as a result of PDI overexpression.
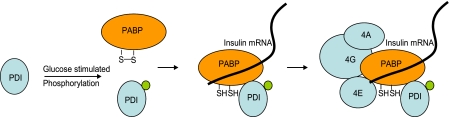
PDI consists of four structural domains (a-b-b′-a′) with homology to the thioredoxin family of proteins and an acidic C-terminal domain containing the ER retention signal (39). The a and a′ are the catalytic domains and have the CXXC motif in the active site, whereas the b and b′ domains do not contain this motif and presumably lack the catalytic function and are thought to be important for peptide binding and substrate recognition. The annular organization of the four domains was revealed by atomic force microscopy (40). The crystal structure of the yeast PDI confirmed the modular arrangement of the domains and also a possible cooperative interaction between the catalytically active a′ and the C-terminal domains was indicated (41). Although, bacterially expressed recombinant full-length PDI was unable to bind to the insulin 5′ UTR RNA, the a′ domain was able to bind specifically to the insulin 5′ UTR RNA with weak affinity, suggesting that PDI on its own can bind to insulin mRNA with very low affinity and PDI can catalyze the oxidoreduction of the sulfhydryl groups of PABP thereby regulating its association with insulin mRNA. Interaction with specific factors may make the a′ domain more accessible, which then associates with the insulin 5′ UTR. Apart from its role in translation, PDI can also function as a chaperone for the nascent insulin peptide that emerges from the translating ribosome thereby increasing the effective insulin levels. Although the mechanism of how the binding of PABP to the insulin 5′ UTR activates translation is yet to be elucidated, but it is reasonable to expect that PABP can recruit the eIF4F complex to the insulin mRNA 5′ end resulting in increased translation. The mechanistic details of glucose-mediated insulin translation regulation have yet to be worked out. Our experimental results indicate that PDI functions as an activator of translation and glucose stimulation of the cells might activate the enzymatic action of PDI by stimulating its phosphorylation. Activated PDI interact with PABP and rearranges/reduces the disulfide bonds of the PABP resulting in specific association with insulin mRNA (Fig. 8). PDI-PABP may also increase the rate of ribosome recycling by promoting the interaction between the 5′ and 3′ UTRs through its interaction with other translation factors resulting in the observed cooperativity between the 5′ and 3′ UTRs of insulin mRNA during glucose stimulation.
FIGURE 8.
Mechanism of the PABP- and PDI-mediated glucose-induced translation of insulin.
Acknowledgments
We thank Dr. Ramanamurthy of the experimental animal facility, NCCS, for help with animal work. We thank Arti V. Bhosle for technical assistance. We also thank Dr. Joseph and his group members for discussions and suggestions.
This work was supported in part by Department of Biotechnology, India, Grant BT/PR5813/BRB/10/411/2005 and a intramural grant from the National Centre for Cell Science, India (to V. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- ER
- endoplasmic reticulum
- Ni-NTA
- nickel-nitrilotriacetic acid
- PDI
- protein-disulfide isomerase
- DTNB
- 5,5′-dithiobis(nitrobenzoic acid)
- RRM
- RNA recognition motifs
- HG
- high glucose
- LG
- low glucose
- PABP
- poly(A) binding protein.
REFERENCES
- 1. Campbell I. L., Hellquist L. N., Taylor K. W. (1982) Clin. Sci. 62, 449–455 [DOI] [PubMed] [Google Scholar]
- 2. Jahr H., Schröder D., Ziegler B., Ziegler M., Zühlke H. (1980) Eur. J. Biochem. 110, 499–505 [DOI] [PubMed] [Google Scholar]
- 3. Permutt M. A. (1974) J. Biol. Chem. 249, 2738–2742 [PubMed] [Google Scholar]
- 4. Permutt M. A., Kipnis D. M. (1975) Fed. Proc. 34, 1549–1555 [PubMed] [Google Scholar]
- 5. Leibiger B., Wahlander K., Berggren P. O., Leibiger I. B. (2000) J. Biol. Chem. 275, 30153–30156 [DOI] [PubMed] [Google Scholar]
- 6. Itoh N., Sei T., Nose K., Okamoto H. (1978) FEBS Lett. 93, 343–347 [DOI] [PubMed] [Google Scholar]
- 7. Welsh M., Scherberg N., Gilmore R., Steiner D. F. (1986) Biochem. J. 235, 459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Itoh N., Okamoto H. (1980) Nature 283, 100–102 [DOI] [PubMed] [Google Scholar]
- 9. Greenman I. C., Gomez E., Moore C. E., Herbert T. P. (2005) Biochem. J. 391, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hernández-Sánchez C., Mansilla A., de la Rosa E. J., Pollerberg G. E., Martínez-Salas E., de Pablo F. (2003) EMBO J. 22, 5582–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shalev A., Blair P. J., Hoffmann S. C., Hirshberg B., Peculis B. A., Harlan D. M. (2002) Endocrinology 143, 2541–2547 [DOI] [PubMed] [Google Scholar]
- 12. Tillmar L., Carlsson C., Welsh N. (2002) J. Biol. Chem. 277, 1099–1106 [DOI] [PubMed] [Google Scholar]
- 13. Wicksteed B., Herbert T. P., Alarcon C., Lingohr M. K., Moss L. G., Rhodes C. J. (2001) J. Biol. Chem. 276, 22553–22558 [DOI] [PubMed] [Google Scholar]
- 14. Wicksteed B., Uchizono Y., Alarcon C., McCuaig J. F., Shalev A., Rhodes C. J. (2007) Cell Metab. 5, 221–227 [DOI] [PubMed] [Google Scholar]
- 15. Muralidharan B., Bakthavachalu B., Pathak A., Seshadri V. (2007) FEBS Lett. 581, 4103–4108 [DOI] [PubMed] [Google Scholar]
- 16. Anfinsen C. B. (1973) Science 181, 223–230 [DOI] [PubMed] [Google Scholar]
- 17. Turano C., Coppari S., Altieri F., Ferraro A. (2002) J. Cell Physiol. 193, 154–163 [DOI] [PubMed] [Google Scholar]
- 18. Yao Y., Zhou Y., Wang C. (1997) EMBO J. 16, 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koivu J., Myllylä R., Helaakoski T., Pihlajaniemi T., Tasanen K., Kivirikko K. I. (1987) J. Biol. Chem. 262, 6447–6449 [PubMed] [Google Scholar]
- 20. Pihlajaniemi T., Helaakoski T., Tasanen K., Myllylä R., Huhtala M. L., Koivu J., Kivirikko K. I. (1987) EMBO J. 6, 643–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wetterau J. R., Combs K. A., Spinner S. N., Joiner B. J. (1990) J. Biol. Chem. 265, 9800–9807 [PubMed] [Google Scholar]
- 22. Kim J., Mayfield S. P. (1997) Science 278, 1954–1957 [DOI] [PubMed] [Google Scholar]
- 23. Knaack D., Fiore D. M., Surana M., Leiser M., Laurance M., Fusco-DeMane D., Hegre O. D., Fleischer N., Efrat S. (1994) Diabetes 43, 1413–1417 [DOI] [PubMed] [Google Scholar]
- 24. Kim J., Mayfield S. P. (2002) Plant Cell Physiol. 43, 1238–1243 [DOI] [PubMed] [Google Scholar]
- 25. Danon A., Mayfield S. P. (1994) EMBO J. 13, 2227–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deo R. C., Bonanno J. B., Sonenberg N., Burley S. K. (1999) Cell 98, 835–845 [DOI] [PubMed] [Google Scholar]
- 27. Fong C. L., Lentz A., Mayfield S. P. (2000) J. Biol. Chem. 275, 8275–8278 [DOI] [PubMed] [Google Scholar]
- 28. Terada K., Manchikalapudi P., Noiva R., Jauregui H. O., Stockert R. J., Schilsky M. L. (1995) J. Biol. Chem. 270, 20410–20416 [DOI] [PubMed] [Google Scholar]
- 29. Yoshimori T., Semba T., Takemoto H., Akagi S., Yamamoto A., Tashiro Y. (1990) J. Biol. Chem. 265, 15984–15990 [PubMed] [Google Scholar]
- 30. Hotchkiss K. A., Matthias L. J., Hogg P. J. (1998) Biochim. Biophys. Acta 1388, 478–488 [DOI] [PubMed] [Google Scholar]
- 31. Chen K., Lin Y., Detwiler T. C. (1992) Blood 79, 2226–2228 [PubMed] [Google Scholar]
- 32. Wroblewski V. J., Masnyk M., Khambatta S. S., Becker G. W. (1992) Diabetes 41, 539–547 [DOI] [PubMed] [Google Scholar]
- 33. Gerner C., Holzmann K., Meissner M., Gotzmann J., Grimm R., Sauermann G. (1999) J. Cell Biochem. 74, 145–151 [PubMed] [Google Scholar]
- 34. Coppari S., Altieri F., Ferraro A., Chichiarelli S., Eufemi M., Turano C. (2002) J. Cell Biochem. 85, 325–333 [DOI] [PubMed] [Google Scholar]
- 35. VanderWaal R. P., Spitz D. R., Griffith C. L., Higashikubo R., Roti Roti J. L. (2002) J. Cell Biochem. 85, 689–702 [DOI] [PubMed] [Google Scholar]
- 36. Danon A., Mayfield S. P. (1994) Science 266, 1717–1719 [DOI] [PubMed] [Google Scholar]
- 37. Qi H. H., Ongusaha P. P., Myllyharju J., Cheng D., Pakkanen O., Shi Y., Lee S. W., Peng J., Shi Y. (2008) Nature 455, 421–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang L., Lai E., Teodoro T., Volchuk A. (2009) J. Biol. Chem. 284, 5289–5298 [DOI] [PubMed] [Google Scholar]
- 39. Alanen H. I., Salo K. E., Pekkala M., Siekkinen H. M., Pirneskoski A., Ruddock L. W. (2003) Antioxid. Redox Signal. 5, 367–374 [DOI] [PubMed] [Google Scholar]
- 40. Li S. J., Hong X. G., Shi Y. Y., Li H., Wang C. C. (2006) J. Biol. Chem. 281, 6581–6588 [DOI] [PubMed] [Google Scholar]
- 41. Tian G., Xiang S., Noiva R., Lennarz W. J., Schindelin H. (2006) Cell 124, 61–73 [DOI] [PubMed] [Google Scholar]