Abstract
Sld3 is essential for the initiation of DNA replication, but Sld3 does not travel with a replication fork. GINS binds to Cdc45 and Mcm2-7 to form the replication fork helicase in eukaryotes. We purified Sld3, Cdc45, GINS, and Mcm2-7 and studied their interaction and assembly into complexes. Sld3 binds tightly to Cdc45 in the presence or absence of cyclin-dependent kinase activity. Furthermore, Sld3 binds tightly to the Mcm2-7 complex, and a ternary complex forms among Cdc45, Mcm2-7, and Sld3, with a 1:1:1 stoichiometry (CMS complex). GINS binds directly to Mcm2-7, and GINS competes with Sld3 for Mcm2-7 binding. GINS also binds directly to Cdc45, and GINS competes with Sld3 for Cdc45 binding. Cdc45, Mcm2-7, and GINS form a ternary complex with a stoichiometry of 1:1:1 (CMG complex). Size exclusion data reveal that when Sld3, Cdc45, Mcm2-7, and GINS are added together, the result is a mixture of CMS and CMG complexes. The data suggest that GINS and Sld3 compete with one another for Mcm2-7 and Cdc45 binding. Our results are consistent with a model wherein GINS trades places with Sld3 at a replication origin, contributing to the activation of the replication fork helicase.
Keywords: Cell Cycle, DNA Replication, Protein Assembly, Protein Kinases, Yeast
Introduction
The Mcm2-7 complex functions with Cdc45 and the four-subunit GINS complex (composed of Psf1, Psf2, Psf3, and Sld5) to unwind DNA at a replication fork in eukaryotic organisms (1–3). The Mcm2-7 complex exhibits weak helicase activity on its own, but it is stimulated by the presence of Cdc45 and GINS, suggesting that Cdc45 and GINS function as helicase-stimulating factors (2, 4). The Mcm2-7, Cdc45, and GINS proteins are all essential for cell growth, suggesting that their function in unwinding DNA at a replication fork is required for DNA replication.
Sld3 is also essential for cell growth, but the role of Sld3 in DNA replication may be limited to the initiation step. Whereas the Mcm2-7, Cdc45, and GINS proteins travel with the replication fork, Sld3 does not travel with the fork (5). Sld3 forms a complex with Cdc45 in vivo, and Sld3 is required for the association of Cdc45 with origins or replication (6). The mechanism of how Sld3 triggers the initiation of DNA replication is not well understood.
The loading of DNA replication proteins as a function of cell cycle is beginning to be elucidated. In late M phase and early G1, the ring-shaped Mcm2-7 complex is loaded onto replication origins (7, 8). ORC, Cdc6, and Cdt1 are responsible for loading two sister Mcm2-7 complexes onto replication origins (8). During M and G1, the Mcm2-7 complex is inactive as a helicase. Sld3 and Cdc45 form a complex that associates with early replication origins in G1 (6). However, it is currently not known how the Sld3-Cdc45 complex is recruited to replication origins.
In S phase, the cyclin-dependent kinase (CDK)2 phosphorylates Sld2, enabling this protein to bind to Dpb11 (9). Sld2, Dpb11, GINS, and Pol ϵ form a transient “preloading complex” that is dependent upon CDK activity (10). CDK also phosphorylates Sld3, and phosphorylation of Sld3 by CDK triggers the formation of an Sld3-Dpb11 complex (11, 12). CDK-dependent formation of the Sld3-Dpb11 complex may be a mechanism to recruit components of the preloading complex to origins of replication in S phase (10).
In S phase, the Dbf4-dependent kinase (DDK) phosphorylates multiple subunits of the Mcm2-7 complex (13–17). DDK phosphorylation of Mcm4 alleviates an inhibitory activity of the N-terminal region of Mcm4, and this event is important for the initiation of DNA replication (18). A rearrangement of subunits may take place upon the initiation of DNA replication to allow the Mcm2-7-Cdc45-GINS complex to migrate away from origins, whereas Sld3 and Dpb11 remain associated with origins until they are eventually cleared from the DNA (2, 5, 13, 19). Recent studies suggest that upon replication initiation, the two sister replisomes travel away from one another and function autonomously (20). It is currently not understood how Sld3 is displaced from components of the replisome as the replication origin is activated.
We purified Sld3, Cdc45, GINS, and Mcm2-7 and studied their ability to form complexes in vitro. We found that Sld3 and Cdc45 form a stable complex that is independent of CDK activity. We also found that Sld3 binds directly to the Mcm2-7 complex, and size exclusion chromatography revealed a stable ternary complex among Cdc45, Mcm2-7, and Sld3, with a stoichiometry of 1:1:1 (CMS complex). We also investigated how Sld3 is released from Cdc45 and Mcm2-7. GINS binds directly to Mcm2-7, and GINS competes with Sld3 for Mcm2-7 binding. Furthermore, GINS binds directly to Cdc45, and GINS competes with Sld3 for Cdc45 binding. A stable ternary complex can form among Cdc45, Mcm2-7, and GINS with a stoichiometry of 1:1:1 (CMG complex). Size exclusion data reveal that when Sld3, Cdc45, Mcm2-7, and GINS are added together, the result is a mixture of CMS and CMG complexes. The results are consistent with a model wherein GINS trades places with Sld3 at a replication origin, contributing to the activation of the replication fork helicase.
EXPERIMENTAL PROCEDURES
Plasmids
CDK protein expression plasmids and purification methods were a generous gift from Dr. H. Araki (9). Full-length Cdc45 or Sld3 PCR product was cloned into the BamHI/XhoI site of either pET 41a or pET 33b to contain a GST tag or PKA recognition sequence at the N terminus of the protein, respectively. The stop codon was removed at the C terminus to allow for a read-through into the His6 tag. Plasmids for the expression of GINS were a generous gift from Mike O'Donnell. These GINS plasmids (pET-Psf3 and Psf5, pLANT-Psf2, pCDF-His-Psf1) were used to co-express all four subunits of GINS simultaneously (21). PKA-GINS has a PKA tag at the N terminus of Psf1.
Protein Expression and Purification
PKA was a generous gift from Dr. Susan Taylor. Mcm2-7 complex and PKA-Mcm2-7 complex (PKA-Mcm3) were purified as described (22, 23). CDK was purified as described (9).
Cdc45, Sld3 (GST, PKA, and Native), GINS Expression
The plasmids were transformed into Escherichia coli Rosetta 2(BL21) (EMD Biosciences), and a colony from transformation was used to inoculate 6 liters of ZYM-5052 autoinducing medium (24) containing 100 μg/ml kanamycin and 34 μg/ml chloramphenicol. For GINS, the medium also contained 20 μg/ml streptomycin sulfate and 100 μg/ml ampicillin. The cells were grown at 37 °C until an A600 of 1.0 was reached, then the temperature was then lowered to 14 °C, and the cells were allowed to express protein for 16 h.
GST Protein Purification
Harvested cells were lysed with French press in 200 ml of 50 mm Tris-HCl, pH 8.0, 500 mm NaCl, and 50 mm imidazole. The lysate was then applied to a 20-ml NiSO4-charged chelating Sepharose fast flow resin (GE Healthcare). The column was washed with the same buffer. The protein was then eluted in 50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 10% glycerol, and 250 mm imidazole. Peak fractions were pooled, and 4 ml of glutathione-Sepharose 4B (GE Healthcare) resin was added. The mixture was rotated gently on ice for 1 h and then poured into a column, washed with 50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 10% glycerol and eluted with the same buffer containing 100 mm glutathione. Peak fractions were then pooled, dialyzed against 50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 10% glycerol and applied to an 8-ml Source Q column (GE Healthcare). The protein was then eluted with a 20-ml linear gradient from 50 to 500 mm NaCl. Peak fractions were pooled and frozen. His-tagged proteins were purified using the same procedure as GST proteins, except that the glutathione-Sepharose step was replaced by a size exclusion chromatography purification step (Superose 6).
We analyzed the purity of every purified protein sample with SDS-PAGE followed by Coomassie Blue staining and found that every protein was 99% pure. We also analyzed the purity of every protein sample with agarose gel and ethidium bromide staining. We found no contaminating nucleic acid in any of the purified protein samples. We also determined the A280/260 ratio for every purified protein sample and found that every value was consistent with pure protein with no contaminating nucleic acid.
Kinase Labeling
PKA labeling was performed as described (22). For CDK labeling, a 50-μl reaction contained 20 nm Sld3, 20 ng of CDK, 5 mm cold ATP, 5 μCi of [γ-32P]ATP (6000 Ci/nmol; PerkinElmer Life Sciences) in 5 mm Tris-HCl, pH 8.5, 10 mm MgCl2, 1 mm DTT. Reactions were incubated at 30 °C for 1 h.
Analytical Size Exclusion Chromatography
100 pmol of radiolabeled protein was mixed with equimolar unlabeled partner(s) protein in 200 μl and incubated for 1 h at 30 °C in column buffer (50 mm Tris, pH 7.5, 100 mm NaCl, 1 mm EDTA, 5% glycerol). The protein mixture was then subjected to 24-ml Superose 6 (GE Life Sciences) size exclusion chromatography, in the same column buffer. Each 250-μl fraction was then subjected to SDS-PAGE analysis/separation and quantified using PhosphorImager software.
Pulldown Assays
The 100 μl GST pulldown reaction contained 2 pmol of GST-tagged protein in GST-binding buffer (40 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.1 mm EDTA, 10% glycerol, 0.1% Triton X-100, 1 mm DTT, 0.7 μg/ml pepstatin, 0.1 mm PMSF, 0.1 mg/ml BSA) and varying amounts of radiolabeled protein as described in each figure. Reactions were incubated at room temperature for 1 h. Following incubation, reactions were added to 40 μl of prepared glutathione-Sepharose and gently mixed. Binding of GST-tagged protein to the beads was performed for 20 min, with gentle mixing every few minutes. Once the binding was complete, the reaction mixture was aspirated, and the beads were washed two times with 0.5 ml of GST-binding buffer. After the last wash, 30 μl of 5 × SDS sample buffer was added to each reaction, and the samples were boiled for 10 min. Samples (20 μl) were then analyzed by SDS-PAGE. All GST pulldown-bound material was electrophoresed on the same gel as the experimental input material, as required for proper quantification of bound product. All PKA- and CDK-labeled proteins, as analyzed by SDS/PAGE, appeared as a single band on the phosphorimage. A thin slice is presented in the manuscript figures so as to conserve blank space in the manuscript figure.
RESULTS
Sld3 and Cdc45 Form a Binary Complex
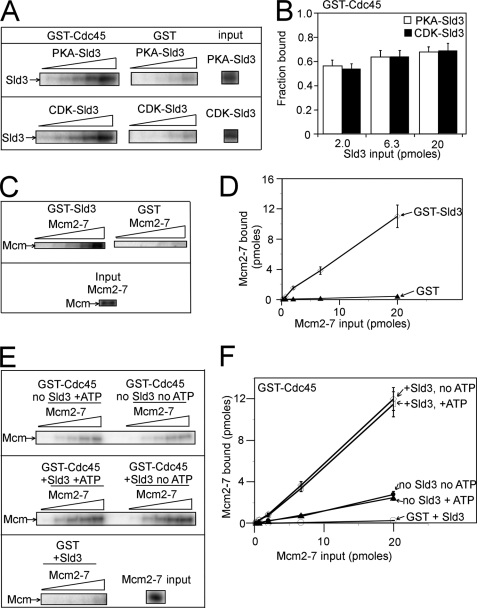
Cdc45 and Sld3 are associated with one another in vivo (6). We first determined whether there is a direct interaction between Sld3 and Cdc45 in vitro. For this experiment, we purified Sld3-containing an N-terminal protein kinase A (PKA) tag from bacterial cells. The PKA tag is used as a tool to radiolabel the protein. We then radiolabeled Sld3 with either PKA (PKA-Sld3) or CDK (CDK-Sld3) and matched PKA-Sld3 with CDK-Sld3 for total radioactive counts and final concentration. We then added varying concentrations of PKA-Sld3 or CDK-Sld3 to 2 pmol of GST-Cdc45 or GST. Bound radiolabeled Sld3 was isolated and analyzed by SDS-PAGE followed by phosphorimaging. At 0.2, 0.63, and 2.0 pmol of input PKA-Sld3 or CDK-Sld3, more than half of the Sld3 was bound by GST-Cdc45 (Fig. 1, A and B). These data suggest that Sld3 and Cdc45 bind tightly and directly to one another and that this interaction is not modified by CDK phosphorylation of Sld3.
FIGURE 1.
Sld3 stimulates the ability of GST-Cdc45 to pull down Mcm2-7. A, Sld3 containing an N-terminal PKA tag was radiolabeled with either PKA or CDK. PKA-Sld3 and CDK-Sld3 were then matched for radioactive counts and total Sld3 concentration. Varying amounts of Sld3 (0.02, 0.06, 0.2, 0.6, or 2.0 pmol) were then incubated with 2.0 pmol of GST-Cdc45 or GST alone, and the bound radioactive Sld3 was analyzed by SDS-PAGE followed by phosphorimaging. B, results from experiments like A were quantified, averaged, and plotted. C, Mcm2-7 complex was assembled from recombinant subunits as described (23). The Mcm3 subunit contains an N-terminal PKA site, and the Mcm2-7 complex was radiolabeled with PKA as described (22). 2.0 pmol of GST-Sld3 or GST was incubated with radiolabeled Mcm2-7 complex (0.02, 0.06, 0.2, 0.6, or 2.0 pmol), and the bound radioactive Mcm was analyzed by SDS-PAGE followed by phosphorimaging. D, results from experiments like C were quantified, averaged, and plotted. E, radiolabeled Mcm2-7 (0.02, 0.06, 0.2, 0.6, or 2.0 pmol) was incubated with 2.0 pmol of GST-Cdc45 with no Sld3, 2.0 pmol of GST-Cdc45, and 2.0 pmol of Sld3 or 2.0 pmol of GST with 2.0 pmol of Sld3. These experiments were performed in the presence or absence of 5 mm ATP. F, results from experiments like E were quantified, averaged, and plotted.
Sld3 Binds Directly to Mcm2-7
The Sld3-Cdc45 complex associates with early origins in G1, after the loading of Mcm2-7, but prior to the arrival of the preloading complex (Dp11, Sld2, GINS, and Pol ϵ) (6, 10). Thus, we hypothesized that Sld3 and/or Cdc45 may bind directly to Mcm2-7. We purified these proteins and studied their interaction in vitro. The Mcm2-7 complex was reconstituted from individually purified proteins and radiolabeled as described (22, 23). We first studied the interaction between Sld3 and Mcm2-7. Varying concentrations of radiolabeled Mcm2-7 were incubated with GST-Sld3 or GST, and the bound Mcm2-7 complex was analyzed by SDS-PAGE followed by phosphorimaging. For inputs of 0.2, 0.63, and 2.0 pmol of Mcm2-7, at least half of the input Mcm2-7 was bound by Sld3 (Fig. 1, C and D). These data suggest that Sld3 forms a direct interaction with Mcm2-7.
Sld3 Stimulates the Ability of GST-Cdc45 to Pull Down Mcm2-7
We next investigated the interaction between Cdc45 and Mcm2-7. Varying amounts of radiolabeled Mcm2-7 were incubated with 2.0 pmol of GST-Cdc45, and bound Mcm2-7 was isolated and analyzed by SDS/PAGE followed by phosphorimaging (Fig. 1, E and F). In the absence of Sld3, a weak interaction was detected between Cdc45 and Mcm2-7, with ∼10% of input Mcm2-7 bound by Cdc45. However, in the presence of Sld3, more than half of the input Mcm2-7 was bound by Cdc45 at 2.0, 0.63, and 0.2 pmol input Mcm2-7 (Fig. 1, E and F). These data suggest that Sld3 stimulates the ability of GST-Cdc45 to pull down Mcm2-7.
The Mcm2-7 complex uses the energy derived from ATP binding and hydrolysis to power its motion (4, 23, 25). We therefore studied the interaction of Sld3 and Cdc45 with Mcm2-7 in the presence and absence of 5 mm ATP. In the absence or presence of ATP, Sld3 stimulated the ability of GST-Cdc45 to pull down radiolabeled Mcm2-7 (Fig. 1, E and F).
Size Exclusion Chromatography Reveals a Ternary Complex among Cdc45, Mcm2-7, and Sld3 (CMS Complex)
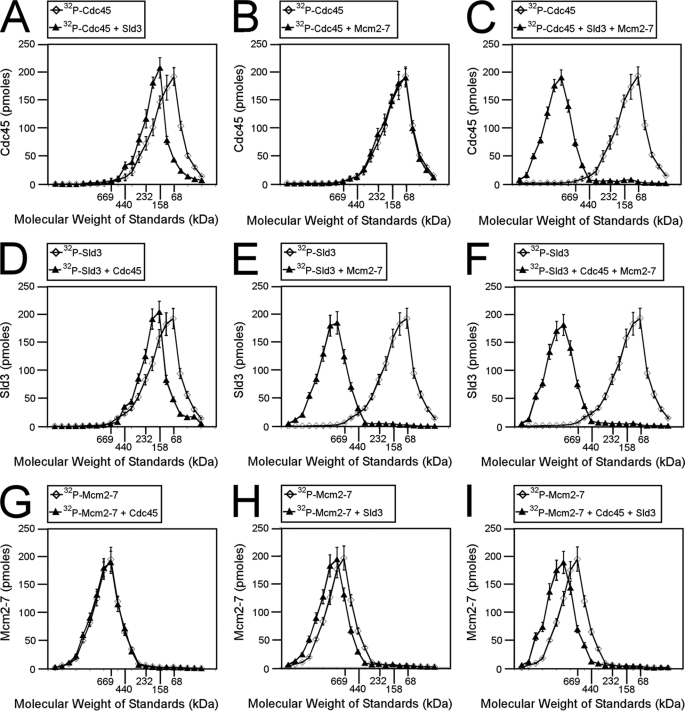
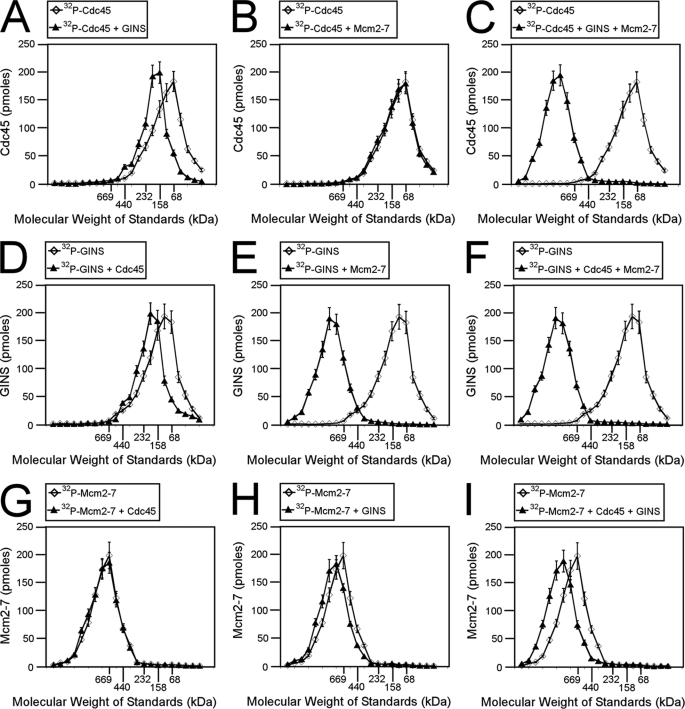
We also used size exclusion chromatography to investigate the interaction among Cdc45, Sld3, and Mcm2-7 proteins (Fig. 2). The proteins in these studies had no GST tags. We radiolabeled Cdc45 and subjected the protein to size exclusion chromatography in the presence or absence of Sld3 (Fig. 2A). The Cdc45 was radiolabeled to quantify the elution profile accurately. The elution peak of Cdc45 was in a fraction that corresponded to the elution profile of a 68-kDa standard. In the presence of Sld3, the Cdc45 peak shifted to that of a 158-kDa protein, suggesting that a complex formed between Sld3 and Cdc45. The elution profile of Cdc45 was not affected by Mcm2-7 alone (Fig. 1B); however, in the presence of Sld3 and Mcm2-7, Cdc45 elutes at a peak of ∼800 kDa (Fig. 1C). These data suggest that Sld3, Cdc45, and Mcm2-7 form a ternary complex.
FIGURE 2.
Cdc45, Mcm2-7, and Sld3 form a ternary complex (CMS complex). A–C, 100 pmol of 32P-labeled Cdc45 was incubated with equimolar Sld3 (A), Mcm2-7 (B), or Sld3 and Mcm2-7 (C) and subjected to Superose 6 size exclusion chromatography as described under “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of Cdc45 in each fraction. Cdc45 (pmol) was then plotted versus the elution of molecular mass standards. D–F, 100 pmol of 32P-labeled Sld3 was incubated with equimolar Cdc45 (D), Mcm2-7 (E), or Cdc45 and Mcm2-7 (F) and subjected to size exclusion chromatography as described under “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of Sld3 in each fraction. Sld3 (pmol) was then plotted versus the elution of molecular mass standards. G–I, 100 pmol of 32P-labeled Mcm2-7 was incubated with equimolar Cdc45 (G), Sld3 (H), or Cdc45 and Sld3 (I) and subjected to size exclusion chromatography as described under “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of Mcm2-7 in each fraction. Mcm2-7 (pmol) was then plotted versus the elution of molecular mass standards.
We then radiolabeled Sld3 and incubated the protein with Cdc45 (Fig. 2D), Mcm2-7 (Fig. 2E), or Cdc45 and Mcm2-7 (Fig. 2F). Sld3 elution was slightly shifted in the presence of Cdc45, as expected (Fig. 2D). The elution profile of Sld3 was shifted substantially by the presence of Mcm2-7, suggesting that Sld3 binds directly to Mcm2-7 (Fig. 2E). Sld3 elution was also shifted substantially in the presence of Cdc45 and Mcm2-7 (Fig. 2F), consistent with the formation of a ternary complex among Sld3, Cdc45, and Mcm2-7.
Mcm2-7 was then radiolabeled and incubated with Cdc45 (Fig. 2G), Sld3 (Fig. 2H), or Cdc45 and Sld3 (Fig. 2I). The elution of Mcm2-7 was not affected by Cdc45 (Fig. 2G), but the elution of Mcm2-7 was slightly shifted by either Sld3 or Sld3 and Cdc45 (Fig. 2, H and I). Taken together, the data from Fig. 2 suggest that a stable complex can form among Cdc45, Mcm2-7, and Sld3, and the stoichiometry of the ternary complex is 1:1:1 (Cdc45:Mcm2-7:Sld3, CMS complex). Furthermore, the data suggest that Sld3 binds directly to Cdc45 and Sld3 binds directly to Mcm2-7.
GINS Binds to Mcm2-7
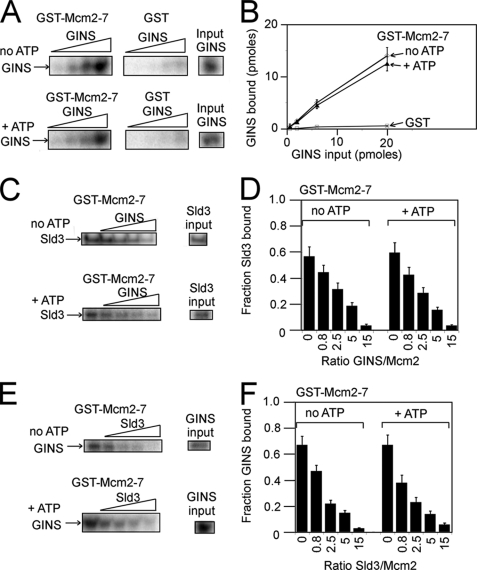
GINS is one of the last replication initiation complexes to arrive at a replication fork (10, 26), and it forms a complex with Mcm2-7 and Cdc45 in the cell (1–3). The Cdc45-Mcm2-7-GINS complex is the replication fork helicase in eukaryotic cells (1–3). We first determined whether GINS can bind directly to the Mcm2-7 complex. At input amounts of 0.2, 0.6, or 2.0 pmol of GINS and in the absence or presence of 5 mm ATP, more than half of the input GINS complex was bound by GST-Mcm2-7 (Fig. 3, A and B). These data suggest that in the absence or presence of ATP, GINS binds tightly and directly to Mcm2-7.
FIGURE 3.
GINS and Sld3 compete with one another for binding to Mcm2-7. A, GINS containing an N-terminal PKA tag on Psf1 was radiolabeled with PKA. Varying amounts of radiolabeled GINS (0.06, 0.2, 0.6, or 2.0 pmol) were then incubated with 2.0 pmol of GST-Mcm2-7 or GST alone, and the bound radioactive GINS was analyzed by SDS-PAGE followed by phosphorimaging. The experiments were performed in the absence or presence of 5 mm ATP. B, results from experiments like A were quantified, averaged, and plotted. C, 2.0 pmol of PKA-radiolabeled Sld3 was incubated with 2.0 pmol of GST-Mcm2-7 and GINS (0, 1.6, 5.0, 10, or 30 pmol) in the absence or presence of 5 mm ATP. The bound radioactive Sld3 was analyzed by SDS-PAGE followed by phosphorimaging. D, results from experiments like C were quantified, averaged, and plotted. E, 2.0 pmol of PKA-radiolabeled GINS was incubated with 2.0 pmol of GST-Mcm2-7 and Sld3 (0, 1.6, 5.0, 10, or 30 pmol) in the absence or presence of 5 mm ATP. The bound radioactive GINS was analyzed by SDS-PAGE followed by phosphorimaging. F, results from experiments like E were quantified, averaged, and plotted.
GINS Competes with Sld3 for Mcm2-7 Binding
Sld3 binds tightly to the Mcm2-7 (Figs. 1 and 2), but Sld3 does not travel with the Mcm2-7 complex after the helicase departs from the origin (5). Thus, there may be a mechanism to dislodge Sld3 from Mcm2-7 as the helicase prepares to leave the origin. GINS is one of the last complexes to arrive at the origin (10, 26), and GINS binds to the Mcm2-7 complex (Fig. 3, A and B). These data prompted us to investigate whether GINS can compete with Sld3 for Mcm2-7 binding. GST-Mcm2-7 was used to pull down radiolabeled Sld3 in the absence or presence of GINS (Fig. 3, C and D). As the concentration of GINS increased in the reaction, the fraction of Sld3 bound by GST-Mcm2-7 decreased (Fig. 3, C and D). This result occurred in the absence or presence of 5 mm ATP (Fig. 3, C and D). These data suggest that GINS competes with Sld3 for Mcm2-7 binding. The arrival of GINS at a replication origin during S phase may help release Sld3 from Mcm2-7.
Sld3 Competes with GINS for Mcm2-7 Binding
The observation that GINS competes with Sld3 for Mcm2-7 binding prompted us to investigate whether Sld3 competes with GINS for Mcm2-7 binding. GST-Mcm2-7 pulls down more than 60% of input GINS in the presence or absence of ATP (Fig. 3, E and F). As Sld3 is added in increasing amounts to the reaction, the fraction of GINS pulled down by GST-Mcm2-7 progressively decreases (Fig. 3, E and F). These data suggest that Sld3 competes with GINS for Mcm2-7 binding. BecauseSld3 competes with GINS for Mcm2-7 binding, and GINS competes with Sld3 for Mcm2-7 binding, Sld3 and GINS may be competing for the same binding site on Mcm2-7. Sld3 may help block the activation of the replication fork helicase by blocking the binding site of GINS for Mcm2-7.
GINS Competes with Sld3 for Cdc45 Binding
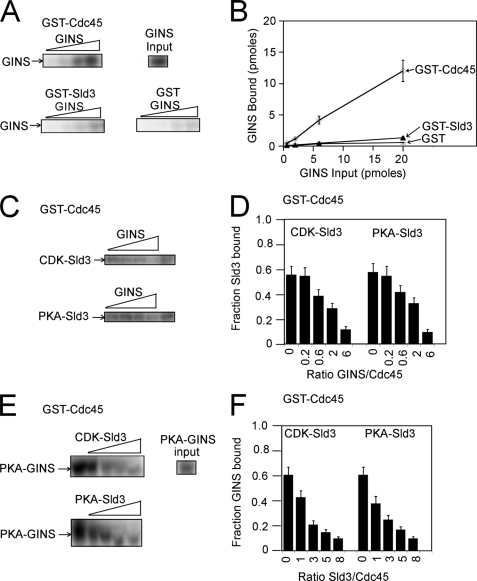
We next examined whether GINS can bind directly to Sld3 or Cdc45 (Fig. 4, A and B). GST-Cdc45 pulled down more than half of the GINS at input amounts of 0.2, 0.6, and 2.0 pmol GINS, suggesting a direct and tight interaction between Cdc45 and GINS (Fig. 4, A and B). However, GST-Sld3 pulled down <5% of input GINS at similar input amounts of GINS, suggesting that GINS does not bind tightly to Sld3 (Fig. 4, A and B).
FIGURE 4.
GINS and Sld3 compete with one another for binding to Cdc45. A, GINS containing an N-terminal PKA tag on Psf1 was radiolabeled with PKA. Varying amounts of radiolabeled GINS (0.06, 0.2, 0.6, or 2.0 pmol) were then incubated with 2.0 pmol of GST-Cdc45, GST-Sld3, or GST alone, and the bound radioactive GINS was analyzed by SDS-PAGE followed by phosphorimaging. B, results from experiments like A were quantified, averaged, and plotted. C, 2.0 pmol of PKA-radiolabeled Sld3 or CDK-radiolabeled Sld3 was incubated with 2.0 pmol of GST-Cdc45 and GINS (0, 0.4, 1.2, 4.0, or 12 pmol). The bound radioactive Sld3 was analyzed by SDS-PAGE followed by phosphorimaging. D, results from experiments like C were quantified, averaged, and plotted. E, 2.0 pmol of PKA-radiolabeled GINS was incubated with 2.0 pmol of GST-Cdc45 and Sld3 (0, 2.0, 6.0, 10, or 16 pmol) in the absence or presence of 5 mm ATP. The bound radioactive GINS was analyzed by SDS-PAGE followed by phosphorimaging. F, results from experiments like E were quantified, averaged, and plotted.
Cdc45 travels with the replisome, whereas Sld3 does not (5). Thus, the tight interaction observed between Sld3 and Cdc45 is likely to be broken during replication initiation. We therefore investigated whether GINS competes with Sld3 for Cdc45 binding because GINS is one of the last replication initiation proteins to arrive at a replication fork and because GINS binds to Cdc45. Using the GST pulldown assay, we observed that GINS competes with Sld3 for Cdc45 binding in a concentration-dependent manner (Fig. 4, C and D). Furthermore, GINS competed with Sld3 for Cdc45 binding regardless of whether Sld3 is phosphorylated by CDK (Fig. 4, C and D). These data suggest that GINS may allow for the separation of Cdc45 and Sld3 that must arise during the initiation of DNA replication.
Because GINS can compete with Sld3 for Cdc45 binding, we next investigated whether Sld3 can compete with GINS for Cdc45 binding. GST-Cdc45 pulldown of radiolabeled GINS was challenged with increasing concentrations of Sld3 (Fig. 4, E and F). As the concentration of Sld3 was increased in this reaction, there was a progressive decrease in the interaction observed between GST-Cdc45 and radiolabeled GINS (Fig. 4, E and F). Similar results were observed if Sld3 was phosphorylated by CDK or PKA (Fig. 4, E and F). These data suggest that Sld3 and GINS compete with one another for binding to Cdc45. In Fig. 4E the PKA-GINS band is migrating nonuniformly and lower with increasing PKA-Sld3. However, this migration change is not reproducible, and we attribute the migration change to an electrophoretic anomaly.
A Ternary Complex Can Form among Cdc45, Mcm2-7, and GINS (CMG Complex)
The CMG complex is the replication fork helicase in eukaryotic organisms, and we next determined whether the CMG complex can be reconstituted from our purified proteins using size exclusion chromatography (Fig. 5). The proteins did not have GST tags in these experiments. Size exclusion chromatography detected a binary complex between GINS and Mcm2-7 and a binary complex between Cdc45 and GINS (Fig. 5). Furthermore, a ternary complex can form among Cdc45, Mcm2-7, and GINS (CMG complex), with a stoichiometry of 1:1:1 (Fig. 5).
FIGURE 5.
Cdc45, Mcm2-7, and GINS form a ternary complex (CMG complex). A–C, 100 pmol of 32P-labeled Cdc45 was incubated with equimolar GINS (A), Mcm2-7 (B), or GINS and Mcm2-7 (C) and subjected to size exclusion chromatography as described under “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of Cdc45 in each fraction. Cdc45 (pmol) was then plotted versus the elution of molecular mass standards. D–F, 100 pmol of 32P-labeled GINS was incubated with equimolar Cdc45 (D), Mcm2-7 (E), or Cdc45 and Mcm2-7 (F) and subjected to size exclusion chromatography as described under “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of GINS in each fraction. GINS (pmol) was then plotted versus the elution of molecular mass standards. G–I, 100 pmol of 32P-labeled Mcm2-7 was incubated with equimolar Cdc45 (G), GINS (H), or Cdc45 and GINS (I) and subjected to size exclusion chromatography as described under “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of Mcm2-7 in each fraction. Mcm2-7 (pmol) was then plotted versus the elution of molecular mass standards.
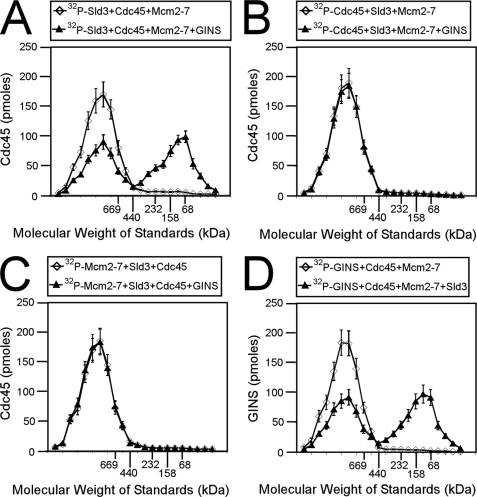
When Sld3, Cdc45, GINS, and Mcm2-7 Are Added Together, the Result Is a Mixture of CMG and CMS Complexes
We found that the addition of Cdc45, Mcm2-7, and Sld3 results in a CMS complex with 1:1:1 stoichiometry (Fig. 2), and the addition of Cdc45, Mcm2-7, and GINS results in a CMG complex with a 1:1:1 stoichiometry (Fig. 5). We also found that GINS competes with Sld3 for Mcm2-7 binding (Fig. 3), and GINS competes with Sld3 for Cdc45 binding (Fig. 4). We next mixed equimolar Sld3, Cdc45, GINS, and Mcm2-7 and analyzed the mixture by size exclusion chromatography (Fig. 6). We first radiolabeled Sld3 and mixed it with equimolar Cdc45, Mcm2-7, and GINS (Fig. 6A). The result was a mixture of high and low molecular mass forms of Sld3 (Fig. 6A). We then radiolabeled Cdc45 and mixed it with equimolar Sld3, Mcm2-7, and GINS (Fig. 6B). The result was only high molecular mass forms of Cdc45 (Fig. 6B). We then radiolabeled Mcm2-7 and mixed it with equimolar Sld3, Cdc45, and GINS (Fig. 6C), and the result was only high molecular mass forms of Mcm2-7 (Fig. 6C). Finally, we radiolabeled GINS, and added equimolar Sld3, Cdc45, and Mcm2-7 (Fig. 6D). The resulted was a mixture of low and high molecular weight forms of GINS (Fig. 6D). Taken together, the data suggest that when Sld3, Cdc45, Mcm2-7, and GINS are added together in equimolar amounts, the result is a roughly equimolar mixture of CMS and CMG complexes. These data also suggest that Sld3 and GINS compete with one another for Mcm2-7 and Cdc45 binding.
FIGURE 6.
When Sld3, Cdc45, GINS, and Mcm2-7 are added together, the result is a mixture of CMG and CMS complexes. A, 100 pmol of 32P-labeled Sld3 was incubated with equimolar Cdc45 and Mcm2-7 or equimolar Cdc45, Mcm2-7, and GINS and subjected to size exclusion chromatography as described under “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of Sld3 in each fraction. Sld3 (pmol) was then plotted versus the elution of molecular mass standards. B, 100 pmol of 32P-labeled Cdc45 was incubated with equimolar Sld3 and Mcm2-7 or equimolar Sld3, Mcm2-7, and GINS and subjected to size exclusion chromatography as described under “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of Cdc45 in each fraction. Cdc45 (pmol) was then plotted versus the elution of molecular mass standards. C, 100 pmol of 32P-labeled Mcm2-7 was incubated with equimolar Sld3 and Cdc45 or equimolar Sld3, Cdc45, and GINS and subjected to size exclusion chromatography as described under “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of Mcm2-7 in each fraction. Mcm2-7 (pmol) was then plotted versus the elution of molecular mass standards. D, 100 pmol of 32P-labeled GINS was incubated with equimolar Cdc45 and Mcm2-7 or equimolar Cdc45, Mcm2-7, and Sld3 and subjected to size exclusion chromatography as described under “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of GINS in each fraction. GINS (pmol) was then plotted versus the elution of molecular mass standards.
DISCUSSION
We investigated how Sld3, Cdc45, and GINS interact with one another and with the Mcm2-7 complex in budding yeast. We found that Sld3 forms a tight, binary complex with Cdc45 that is independent of CDK activity. Although Cdc45 alone binds weakly to Mcm2-7, Sld3 binds directly and tightly to Mcm2-7. Furthermore, size exclusion chromatography reveals a stable, ternary complex among Cdc45, Mcm2-7, and Sld3 with a stoichiometry of 1:1:1 (CMS complex). Thus, Sld3 may mediate the initial recruitment of Cdc45 to replication origins. We next studied how the Sld3-Mcm2-7 interaction may be disrupted becauseSld3 does not migrate with a replication fork. GINS competes with Sld3 for Mcm2-7 binding, suggesting a mechanism to ensure that Sld3 does not travel with the replication fork. It may be important that Sld3 does not travel with the replication fork because Sld3 may inhibit activation of the replication fork helicase. Furthermore, GINS inhibits the interaction between Sld3 and Cdc45, suggesting a mechanism whereby recruitment of GINS to the preinitiation complex helps dislodge Sld3 from Cdc45. Cdc45, Mcm2-7, and GINS form a stable ternary complex, with a stoichiometry of 1:1:1 (CMG complex). The addition of Sld3, Cdc45, Mcm2-7, and GINS results in a mixture of CMS and CMG complexes.
Role of Sld3 in the Initiation of DNA Replication
Sld3 is important for the recruitment of Dpb11 to replication origins (11, 12). Dpb11 is part of the preloading complex, consisting of Dpb11, Sld2, Pol ϵ, and GINS. Recruitment of the preloading complex to a replication origin is an essential function of Sld3 that is regulated by CDK (10). In addition to this important, essential function of Sld3, we find that Sld3 interaction with Mcm2-7 is important for the formation of a complex composed of Cdc45, Mcm2-7, and Sld3 (CMS complex). Sld3 interaction with Mcm2-7 inhibits the interaction between Mcm2-7 and GINS, suggesting a mechanism to inhibit activation of the replication fork helicase. In the future it will be useful to investigate whether Sld3 modulates the catalytic functions or nucleic acid binding properties of Mcm2-7. The human homolog of Sld3 is not known with certainty, but Treslin and Ticrr have recently been identified as possible Sld3 homologs in higher eukaryotes (27, 28). Future work may reveal whether Treslin or Ticrr bind directly to Mcm2-7 or whether some other protein fulfills this function in humans.
GINS and Sld3 Compete with One Another for Mcm2-7 and Cdc45 Binding
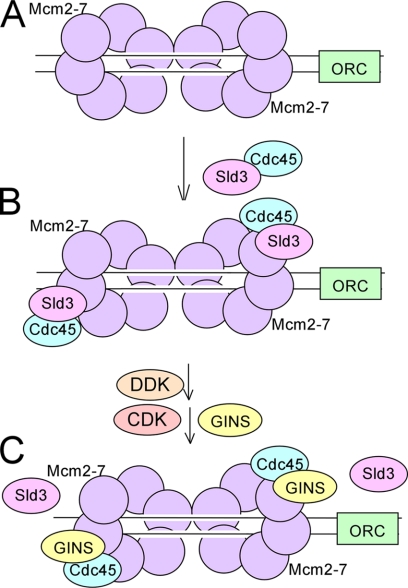
A ternary complex can form among Cdc45, Mcm2-7, and Sld3 (CMS complex). Furthermore, a ternary complex can form among Cdc45, Mcm2-7 and GINS (CMG complex). GINS competes with Sld3 for Mcm2-7 binding, and GINS competes with Sld3 for Cdc45 binding. Furthermore, adding Sld3, Cdc45, Mcm2-7, and GINS results in a mixture of CMS and CMG complexes. Taken together, the data suggest that GINS may promote the dissociation of Sld3 from Mcm2-7 and Cdc45, allowing the formation of the CMG complex. Our results are consistent with a hypothesis that GINS may be trading places with Sld3 at the origin of replication, substituting as the new bridge between Mcm2-7 and Cdc45 (29) (Fig. 7). Our data show that Sld3 and GINS are roughly equally efficient in forming CMS or CMG complexes, respectively. An additional factor in the cell, not identified in this paper, may promote the disassembly of CMS complexes and the formation of CMG complexes during S phase. In the future it will be useful to perform protein interaction site mapping for Sld3, Mcm2-7, Cdc45, and GINS. This work may help elucidate the mechanism underlying the observed binding competition.
FIGURE 7.
GINS may trade places with Sld3 at a replication origin, contributing to the activation of the replication fork helicase. A, sister Mcm2-7 complexes are loaded onto an origin in late M or G1 phase (8). B, for an early replicating origin, Sld3-Cdc45 binds to replication origins in G1 (10). This recruitment may be mediated by direct interaction between Sld3 and Mcm2-7 (Figs. 1 and 2). C, during S phase, DDK alleviates an inhibitory activity of Mcm4 (18). Also during S phase, CDK promotes the recruitment of GINS to a replication origin. GINS competes with Sld3 for binding to Mcm2-7 (Fig. 3), and GINS competes with Sld3 for binding to Cdc45 (Fig. 4). This may result in the conversion of a CMS complex to a CMG complex (Figs. 5 and 6), although an additional cellular factor may be required to accomplish this transition. The sister replication fork helicases separate upon origin activation (20). ORC, origin recognition complex.
GINS is recruited to origins in response to CDK activity because GINS is part of the preloading complex consisting of GINS, Pol ϵ, Dpb11, and Sld2. Thus, when CDK is activated, GINS will be recruited to a replication origin. Data in this paper suggest that GINS recruitment to replication origins remodels the architecture of the preinitiation complex. To be more specific, GINS recruitment competes with the Sld3-Cdc45 interaction, and GINS recruitment competes with the Sld3-Mcm2-7 interaction. Furthermore, GINS can bind to both Mcm2-7 and Cdc45, forming a CMG complex. GINS and Cdc45 stimulate DNA unwinding activity catalyzed by the Mcm2-7 complex (2). Thus, one function of CDK in recruiting GINS to origins of replication is to remodel the architecture of the preinitiation complex, and the inactive CMS complex becomes an active CMG complex.
It has been shown that Cdc45 binds tightly to the Mcm2-7 complex in budding yeast cells when Mcm4 is hyperphosphorylated by DDK (13). DDK hyperphosphorylation of Mcm4 alleviates an inhibitory mechanism of Mcm4, thereby activating the CMG complex (18). DDK may also phosphorylate Mcm6 as part of a mechanism to activate the CMG complex (17). Thus, upon activation by DDK, the Mcm2-7 complex may bind directly to Cdc45. The functional importance of conversion of the CMS complex to the CMG complex is activation of the replication fork helicase, a key event in the initiation of DNA replication.
Implications for the Initiation of DNA Replication
Sister Mcm2-7 complexes load onto replication origins in late M phase and during G1 (8) (Fig. 7A). Sld3-Cdc45 binds to early replication origins in G1, prior to activation of the replication fork helicase (6, 10). Studies herein suggest that this recruitment is mediated by direct interaction between Sld3 and Mcm2-7 (Fig. 7B). Sld3 blocks the interaction between GINS and Mcm2-7, and Sld3 may therefore help ensure that the replication fork helicase is not activated prematurely. GINS is one of the last replication fork protein complexes to arrive at an origin (10, 26). Although it is widely accepted that the function of GINS arrival at a fork is to activate the replication fork helicase, studies in this paper suggest additional functions for GINS. GINS competes with Sld3 for Mcm2-7 binding, and this may be critical to disrupt an inactivating interaction between Sld3 and Mcm2-7. Furthermore, GINS competes with Sld3 for Cdc45 interaction, which may be important to ensure that Sld3 does not travel with the replication fork. Finally, the architecture of the replication origin is changed upon GINS binding, and this architectural change may help convert the Mcm2-7 complex from an inactive state to an active state (Fig. 7C). GINS and Cdc45 stimulate DNA unwinding activity catalyzed by the Mcm2-7 complex (2). The sister replication fork helicases separate upon origin activation (20).
Acknowledgments
We thank Hiroyuki Araki for the expression construct for S-CDK, Mike O'Donnell for the expression construct for GINS, and Susan Taylor for providing purified PKA.
This work was supported by a Vanderbilt University Discovery grant (to D. L. K.) and American Cancer Society Research Scholar Grant RSG-08-124-01-CCG (to D. L. K.).
- CDK
- cyclin-dependent kinase
- CMG complex
- Cdc45, Mcm2-7, and GINS
- CMS complex
- Cdc45, Mcm2-7, and Sld3
- DDK
- Dbf4-dependent kinase.
REFERENCES
- 1. Pacek M., Tutter A. V., Kubota Y., Takisawa H., Walter J. C. (2006) Mol. Cell 21, 581–587 [DOI] [PubMed] [Google Scholar]
- 2. Ilves I., Petojevic T., Pesavento J. J., Botchan M. R. (2010) Mol. Cell 37, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 4. Bochman M. L., Schwacha A. (2008) Mol. Cell 31, 287–293 [DOI] [PubMed] [Google Scholar]
- 5. Kanemaki M., Labib K. (2006) EMBO J. 25, 1753–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamimura Y., Tak Y. S., Sugino A., Araki H. (2001) EMBO J. 20, 2097–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Randell J. C., Bowers J. L., Rodríguez H. K., Bell S. P. (2006) Mol. Cell 21, 29–39 [DOI] [PubMed] [Google Scholar]
- 8. Remus D., Beuron F., Tolun G., Griffith J. D., Morris E. P., Diffley J. F. (2009) Cell 139, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tak Y. S., Tanaka Y., Endo S., Kamimura Y., Araki H. (2006) EMBO J. 25, 1987–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muramatsu S., Hirai K., Tak Y. S., Kamimura Y., Araki H. (2010) Genes Dev. 24, 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. (2007) Nature 445, 328–332 [DOI] [PubMed] [Google Scholar]
- 12. Zegerman P., Diffley J. F. (2007) Nature 445, 281–285 [DOI] [PubMed] [Google Scholar]
- 13. Sheu Y. J., Stillman B. (2006) Mol. Cell 24, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Francis L. I., Randell J. C., Takara T. J., Uchima L., Bell S. P. (2009) Genes Dev. 23, 643–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lei M., Kawasaki Y., Young M. R., Kihara M., Sugino A., Tye B. K. (1997) Genes Dev. 11, 3365–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Masai H., Taniyama C., Ogino K., Matsui E., Kakusho N., Matsumoto S., Kim J. M., Ishii A., Tanaka T., Kobayashi T., Tamai K., Ohtani K., Arai K. (2006) J. Biol. Chem. 281, 39249–39261 [DOI] [PubMed] [Google Scholar]
- 17. Randell J. C., Fan A., Chan C., Francis L. I., Heller R. C., Galani K., Bell S. P. (2010) Mol. Cell 40, 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sheu Y. J., Stillman B. (2010) Nature 463, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Masumoto H., Sugino A., Araki H. (2000) Mol. Cell. Biol. 20, 2809–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yardimci H., Loveland A. B., Habuchi S., van Oijen A. M., Walter J. C. (2010) Mol. Cell 40, 834–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finkelstein J., Antony E., Hingorani M. M., O'Donnell M. (2003) Anal. Biochem. 319, 78–87 [DOI] [PubMed] [Google Scholar]
- 22. Bruck I., Kaplan D. (2009) J. Biol. Chem. 284, 28823–28831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Davey M. J., Indiani C., O'Donnell M. (2003) J. Biol. Chem. 278, 4491–4499 [DOI] [PubMed] [Google Scholar]
- 24. Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 25. Schwacha A., Bell S. P. (2001) Mol. Cell 8, 1093–1104 [DOI] [PubMed] [Google Scholar]
- 26. Takayama Y., Kamimura Y., Okawa M., Muramatsu S., Sugino A., Araki H. (2003) Genes Dev. 17, 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G. (2010) Cell 140, 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sansam C. L., Cruz N. M., Danielian P. S., Amsterdam A., Lau M. L., Hopkins N., Lees J. A. (2010) Genes Dev. 24, 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. MacNeill S. A. (2010) Biochem. J. 425, 489–500 [DOI] [PubMed] [Google Scholar]