Abstract
Hyperpolarization-activated cyclic nucleotide-gated (HCN) channels are expressed in the brain and heart and are essential for physiological functions in cardiac and nervous systems. We identified two Hcn4 mRNA variants with different transcription start sites and differential expression patterns in mouse brain and heart. Only one mRNA variant was detected in the brain, whereas both variants were found in the heart. Patch clamp recordings of these two variants in HEK293H cells revealed different electrophysiological properties in channel activation. Mutagenesis studies showed that three positively charged amino acids (Arg-9, Lys-10, and Lys-22) contribute to the functional difference. Our results demonstrate that HCN4 channels are expressed in different patterns in mouse brain and heart and that the N terminus is important for HCN4 channel activation.
Keywords: Brain, Gene Expression, Heart, Ion Channels, Kinetics, Membrane Biophysics, Membrane Proteins, Neurobiology, HCN Channels, Pacemaker Channels
Introduction
Hyperpolarization-activated cyclic nucleotide-gated (HCN)2 channels play essential roles in the pacemaking activities in cardiac and nervous systems (1–4). Four subtypes of HCN channels, named HCN1–HCN4, form either homomeric or heteromeric tetramers. Each HCN channel subunit is composed of three major regions: a transmembrane domain and cytoplasmic N and C termini. The transmembrane domain exhibits several structural characteristics conserved in other voltage-gated ion channels, including a six-transmembrane segment structure, an S5-S6 pore formation, and an S4 voltage sensor. The C terminus contains a six-helix linker motif and a cyclic nucleotide-binding domain (5). A region at the end of the N-terminal domain is conserved among HCN channel subtypes and has been found to be important for channel trafficking (6).
Two major functional characteristics of HCN channels are activation by membrane hyperpolarization and modulation by cAMP to positively shift the voltage dependence of activation (7). Among the four HCN channels, the HCN4 channel is unique in several aspects. Structurally, it has the longest N-terminal sequence (262 amino acids). Functionally, it has the slowest activation and deactivation kinetics and is most sensitive to cAMP among the four HCN channel subtypes (8). HCN4 is the predominant HCN channel expressed in the sinoatrial node to set the heart rhythm (9, 10). In the brain, it is expressed predominantly in the thalamus and olfactory bulb (10, 11).
We have studied the expression of HCN4 channels in mouse brain and heart. We identified two Hcn4 mRNA variants with different transcription start sites that can result in different translation start sites in the proteins. Only one mRNA variant was detected in the brain, whereas both variants were found in the heart. Patch clamp recordings of the two HCN4 variants in HEK293H cells showed they have distinct properties in voltage dependence and kinetics of channel activation. Furthermore, using mutagenesis, we examined the amino acids in the beginning of the N terminus where the two HCN4 variants differ and found that three positively charged residues (Arg-9, Lys-10, and Lys-22) contribute to the functional difference between these two channels. Our data demonstrate HCN4 channel mRNA variants with distinct transcription start sites in mouse brain and heart and that the N terminus of the protein is important for HCN4 channel activation.
EXPERIMENTAL PROCEDURES
Molecular Biology
Three-week-old C57 mice were obtained from Charles River, and their brains and hearts were taken and pooled for RNA isolation. Three mice were used for total RNA isolation for rapid amplification of cDNA ends (5′-RACE), and another two were used for RT-PCR. All procedures were approved by the University of Texas Animal Use Committee.
Total RNA from mouse tissue was isolated with RNA STAT-60 reagent (Tel-Test) and used in 5′-RACE and RT-PCR. 5′-RACE was performed as described previously (12, 13). The two gene-specific primers used in 5′-RACE were mHCN4+550R and mHCN4+377R. The products were gel-extracted and cloned into a pCR2.1 vector (Invitrogen). RT-PCRs were performed with total RNA isolated from regions dissected from hearts and brains. Reverse transcription was carried out with RT-PCR primer mHCN4+992R and reverse transcriptase (Invitrogen). PCRs were then performed with mHCN4–33F/mHCN4+815R targeted for mHCN4L only or mHCN4+325F/mHCN4+815R for both mHcn4L and mHcn4S. The sequences of the primers used for 5′RACE are as follows: mHCN4+550R, CCGAGGGCTGCTCGCAGGAGGCAGA; and mHCN4+377R, GCGGAGTCATGCAGGTGCCCGAGAC. Those used for RT-PCR are as follows: mHCN4+992R, TGCGGGTCAAGGATGATTTCTGTGT; mHCN4–33F, CCGCACCGCTGCCCCCGGCCCAATC; mHCN4+325F, GGCAGTGGTGGAGCAGGGGGCGGCA; and mHCN4+815R, AGCAACAGCATCGTCAGGTCCCAGT.
Electrophysiology
The full-length mouse Hcn4 (mHcn4) clone was obtained from Open Biosystems (catalog no. 9088182) and subcloned into pcDNA6 (Invitrogen) for expression in HEK293H cells. The mHcn4S plasmid was constructed by deletion of the first 74 nucleotides of mHcn4L sequence. Either one of the two mHcn4 expression plasmids was cotransfected with a GFP expression plasmid into HEK293H cells with Lipofectamine 2000 reagent (Invitrogen) and incubated at 37 °C with 5% CO2 for 24 h before recording. The HCN4 channels were recorded with a patch clamp in the inside-out configuration.
Currents conducted by HCN4 channels were elicited by 2.5-s voltage steps from −6 to −190 mV in 10-mV increments at 22 °C. A 10-s interval at 0 mV between steps was used for complete channel deactivation. Currents were recorded with an Axopatch 200A amplifier with a 1-kHz filter. The pipette solution contained 107 mm KCl, 5 mm NaCl, 1 mm MgCl2, and 10 mm HEPES (pH 7.3 with KOH). The internal solution was either the same as the pipette solution or with 100 μm cAMP (Sigma) added. Recorded current traces were zero-subtracted and leak-subtracted, and the steady-state currents (average of the last 250 μs of the activation current) were used to calculate the conductance, assuming a liner single-channel current-voltage relationship and a reversal potential of 0 mV. Conductance was normalized to the maximal value recorded, plotted against voltage, and then fitted with a Boltzmann equation to obtain V1/2. Activation time constants were obtained by monoexponential fits of current traces. Deactivation kinetics were measured and compared by fitting a single exponential equation to the tail currents at different voltages (−90 to −30 mV in 10-mV increments) following the activation of HCN current at −160 mV for 3 s. A two-tailed t test was used for statistical comparisons.
RESULTS
5′-RACE Reveals Two mRNA Variants of Hcn4 with Different Transcription Start Sites
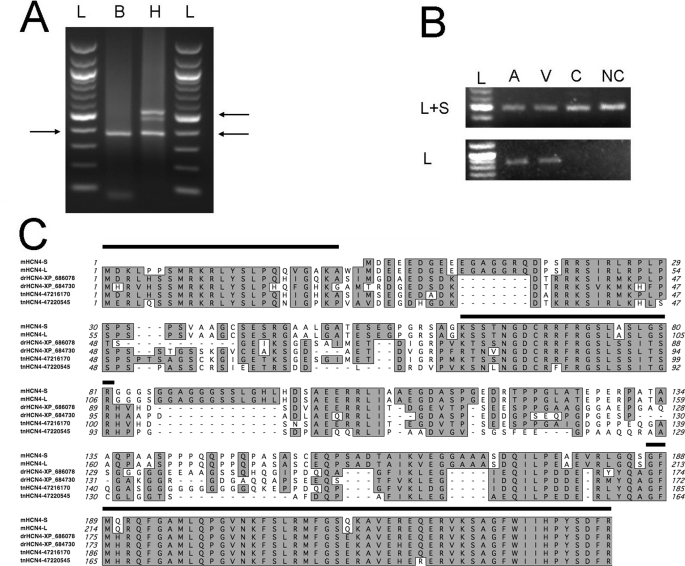
Using 5′-RACE, we identified two mRNA variants of Hcn4 in mouse heart that can code for two mHCN4 channels with different lengths of the N-terminal domain (Fig. 1A). One mRNA variant starts from −182 bp of the mHcn4 cDNA sequence in the GenBankTM Data Bank (accession number NM_001081192), and its protein product is named mHCN4L. The other skips the first start codon of mHcn4L and starts from +35 bp of the GenBankTM mHcn4 sequence, therefore most likely coding for a shorter protein product, named mHCN4S. Interestingly, only one mRNA variant (Hcn4S) was identified in the brain, whereas both mHcn4L and mHcn4S were found in the heart (Fig. 1A). We then performed RT-PCR to confirm the differential expression pattern in the brain and heart. A pair of primers aimed at only mHcn4L (−33 to +815 bp) but not mHcn4S generated RT-PCR products in both atria and ventricles from the heart, but not in the brain (Fig. 1B). However, another primer pair aimed at a common sequence in mHcn4l and mHcn4S (+325 to +815 bp of mHcn4L) generated RT-PCR products in all the samples. In both reactions, the primers were designed in two exons to avoid false-positive products from DNA contamination.
FIGURE 1.
Identification of two Hcn4 mRNA variants in mouse brain and heart. A, 5′-RACE revealed differential expression of the Hcn4 channel in the brain (B) and heart (H). Only mHcn4S was detected in the brain, but both variants were found in the heart. Arrows point to the PCR products of the two variants, 380 and 550 bp, respectively, as indicated by the DNA ladder (L). B, RT-PCR confirmed the differential expression pattern of the two Hcn4 isoforms with primers targeted to a common area in the two variants (upper panel) or to the specific 5′-end of mHcn4L (lower panel). mHcn4L was detected in both the atria (A) and ventricles (V), but not in the cortex (C) or non-cortex (NC) regions in the brain. C, sequence alignment of mHCN4S and mHCN4L with four predicted fish HCN4 channels shows that the beginning of the N terminus is conserved across species, suggesting its importance to channel functions. The N-terminal sequences before the transmembrane domain are aligned. Three long stretches of conserved regions are marked above the sequence.
Compared with the mHCN4S protein, mHCN4L has an extended 25-amino acid segment in the N terminus. Protein sequence analyses suggested that this segment is hydrophilic; thus, it is likely located inside the cell. We aligned the mHCN4L protein sequence with two predicted HCN4 channel sequences derived from the zebrafish genome (GenBankTM accession numbers XP_686078 and XP_684730) and two predicted HCN4 channel sequences from the pufferfish genome (GenBankTM accession numbers 47216170 and 47220545). Three conserved regions (Fig. 1C) were seen in the N termini. The longest stretch of conserved sequence, located at the end of the N terminus and immediately preceding the transmembrane domain, has been found to be important for channel trafficking (6). Another long stretch is at the beginning of the N terminus where mHCN4L and mHCN4S differ. The sequence similarity between mouse HCN4 and evolutionarily distant fish homologs suggests that this extended N-terminal segment is under positive selection during evolution and may therefore be important for the channel function.
Two HCN4 Channels Differ in Activation Voltage Dependence and Activation Kinetics
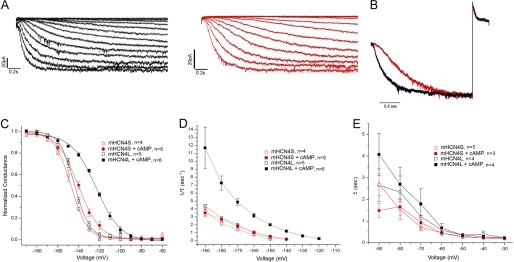
To examine whether the extended N-terminal segment causes any functional difference, we expressed the cDNA constructs corresponding to these two mRNA variants in HEK293H cells and recorded their currents with an inside-out patch clamp. We compared the voltage dependence of channel activation and the kinetics of activation and deactivation of the two variants with or without saturating cAMP (100 μm) in the intracellular solution (Fig. 2 and Table 1). Without cAMP, the two mHCN4 variants behaved similarly in activation voltage dependence, but mHCN4L activated slightly faster than mHCN4S (τ = −180 mV, p < 0.05). However, when cAMP was present, mHCN4L activated at voltages significantly less negative than mHCN4S (p < 0.001) (Fig. 2C) and ∼3-fold faster (−160 and −180 mV, p < 0.001) (Figs. 2, B and D, and 3I). In terms of cAMP modulation, mHCN4S channel activation was only slightly modulated by 100 μm cAMP in the voltage dependence (p < 0.05) (Fig. 2C) and in kinetics (−180 mV, p < 0.05) (Figs. 2D and 3I). On the other hand, mHCN4L was greatly modulated by saturating cAMP. The V1/2 of activation was right-shifted by 21 mV (p < 0.001) (Fig. 2C), which covers almost half of the range of channel activation. It also greatly sped the rate of activation (−180 mV, p < 0.001) (Figs. 2D and 3I). Regardless of the presence of cAMP, the two HCN4 variants did not differ in deactivation kinetics (Fig. 2, B and E).
FIGURE 2.
Comparison of activation and inactivation properties of the two mHCN4 variants. A, representative activation currents of mHCN4L (left panel) and mHCN4S (right panel) elicited by 2.5-s voltage steps from −60 to −190 mV in 10-mV increments. Both recordings shown were performed with 100 μm cAMP in the intracellular solution. B, representative normalized mHCN4L (black) and mHCN4S (red) currents activated at −160 mV and then deactivated at +30 mV with cAMP present showing the difference in the speed of activation, but not in deactivation. The differences between time constants of the representative traces and the group averages are <1%. C, conductance-voltage relationships of the mHCN4 channels with or without 100 μm cAMP. D, activation time constants of the two mHCN4 channels with or without cAMP. E, deactivation time constants of the two mHCN4 channels with or without cAMP.
TABLE 1.
Properties of mHCN4 channels and mutants
All recordings were performed with 100 μm cAMP in the intracellular solution, except as indicated in the first two rows. Statistical values from t tests for the mutants were obtained by comparison with mHCN4L (see Footnotes b–d).
| na | V1/2 | τactivation |
||
|---|---|---|---|---|
| at −180 mV | at −50 mV (n) | |||
| mHCN4L (no cAMP) | 5 | −144.02 ± 1.23 | 0.33 ± 0.02 | 0.35 ± 0.03 (4) |
| mHCN4S (no cAMP) | 4 | −147.59 ± 1.26 | 0.48 ± 0.05 | 0.26 ± 0.04 (5) |
| mHCN4L | 6 | −123.17 ± 0.78 | 0.15 ± 0.02 | 0.35 ± 0.07 (4) |
| mHCN4S | 6 | −140.97 ± 1.98 | 0.35 ± 0.01 | 0.30 ± 0.07 (3) |
| Del7 | 7 | −129.88 ± 2.25b | 0.26 ± 0.02 | |
| Del12 | 6 | −141.64 ± 2.77c | 0.49 ± 0.16b | |
| RKR to AAA | 5 | −133.77 ± 2.84d | 0.33 ± 0.03c | |
| K3A | 4 | −120.56 ± 3.85 | 0.15 ± 0.02 | |
| R9A | 5 | −137.82 ± 1.93c | 0.54 ± 0.12d | |
| K10A | 6 | −132.43 ± 3.35b | 0.39 ± 0.11 | |
| R11A | 6 | −119.33 ± 3.11 | 0.21 ± 0.01b | |
| K22A | 5 | −131.59 ± 3.69b | 0.27 ± 0.04b | |
a These numbers are for activation recordings; the numbers for deactivation (τ) are listed in parentheses after the τ value.
b p < 0.05.
c p < 0.001.
d p < 0.01.
FIGURE 3.
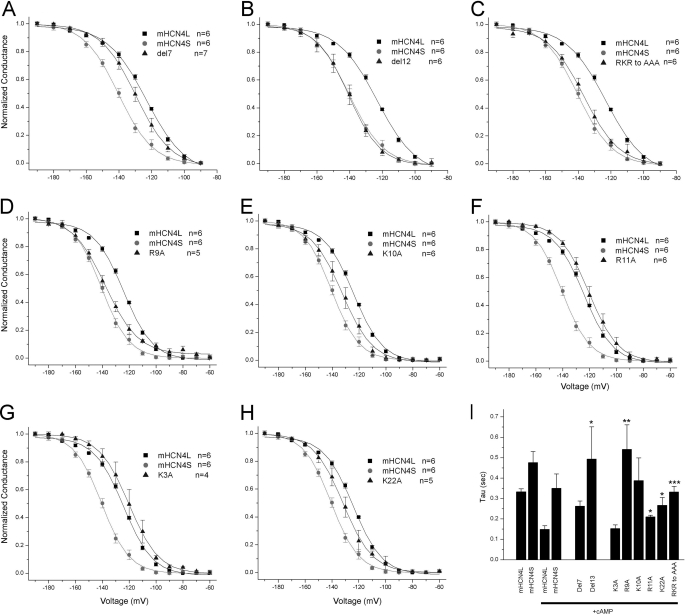
Positively charged residues affect HCN4 channel activation. The functional difference between mHCN4L and mHCN4S was partially changed by deletion of the first seven amino acids (A) but fully abolished by deletion of the first 12 amino acids (B). Mutation of three charged residues (Arg-9, Lys-10, and Arg-11) to alanines also strongly shifted the activation voltage dependence (C). Single mutations (D–H) show that Arg-9, Lys-10, and Lys-22 contribute to the difference, whereas Arg-11 and Lys-3 do not. The activation time constants at −180 mV of the mutants show similar effects (I). *, p < 0.05; **, p < 0.01; ***, p < 0.001 (p values in t tests compared with mHCN4L + cAMP).
Positive Charges in the Extended N Terminus Contribute to the Functional Difference
Because the only difference between two mHCN4 variants is the 25-amino acid segment, the structural basis of the functional difference must be located within these residues. We performed a series of mutagenesis studies and compared the activation properties of the mutants with those of the wild-type channels in the presence of cAMP. First, we made two mutants with shortened N-terminal segments. Deletion of the first seven residues moved the activation of mHCN4L half-way toward mHCN4S (p < 0.05 compared with mHCN4L) (Fig. 3A), whereas deletion of the first 12 residues (with the addition of an extra methionine encoded by the start codon) almost fully abolished the effect of the extended N terminus (p < 0.001 compared with mHCN4L) (Fig. 3B), suggesting that some key residues are located among the residues 8–12. Interestingly, three out of these four amino acids (Arg-9, Lys-10, and Arg-11) are positively charged. The predicted fish HCN4 channels also show identical amino acids (Fig. 1C). These findings led us to focus on the effect of the positively charged residues in the sequence. First, mutation of all three of these positively charged residues (Arg-9, Lys-10, and Arg-11) to alanines together shifted the voltage dependence toward mHCN4S (p < 0.01 compared with mHCN4L) (Fig. 3C). Mutations of each single residue suggest that Arg-9 and Lys-10 both contribute to the effect (p < 0.001 and 0.05 compared with mHCN4L) (Fig. 3, D and E) but that Arg-11 does not (Fig. 3F). In addition, we also examined the properties of two other positively charged residues in this region (Lys-3 and Lys-22). The K3A mutation did not shift the voltage dependence of activation (Fig. 3G). However, K22A shifted the voltage dependence of activation (p < 0.05 compared with mHCN4L) (Fig. 3H), suggesting that this residue is also necessary for the functional difference. We also compared the activation kinetics of the mutants with those of the wild-type channels; the results are consistent with the comparison of steady-state activation (Fig. 3I).
DISCUSSION
HCN4 channels are expressed in the brain and play important roles in synaptic transmission and dendritic integration (4). HCN4 is the predominant channel responsible for If in normal human heart and setting cardiac rhythm (4). Its mRNA and protein levels are up-regulated in failing hearts (14). Therefore, it is important to identify its molecular expression profiles and regulation mechanisms in the brain and heart. We identified two Hcn4 mRNA variants that can be translated into proteins with different lengths in N termini. The two mHcn4 variants show differential expression profiles in the brain and heart. When the two channels were expressed in HEK293H cells, patch clamp recordings showed their distinct channel activation properties in voltage dependence and kinetics. Furthermore, using mutagenesis, we found that three positively charged residues (Arg-9, Lys-10, and Lys-22) contribute to the functional difference between these two channels.
Both mHcn4L and mHcn4S are expressed in heart, but mHcn4S is the only form found in the brain. The two mRNA variants differ at the 5′-end and are thus most likely the results of differential transcription start sites, which are driven by differential tissue-specific transcription regulation mechanisms involving distinct sets of transcription factors. In an early report, an Hcn4 channel homologous to mHcn4S was cloned from rabbit but was regarded as a partial transcript (15). Based on our findings, it was likely a cloning product from the rabbit Hcn4S homolog. Different start sites of mRNA were also seen when we compared the sequences of Hcn4 screened from human cDNA libraries (GenBankTM accession numbers AJ132429 and AJ238850) and a recently published 5′-RACE product with human heart RNA (GenBankTM accession number DQ854815) (16). These results suggest that differential transcription regulations of the Hcn4 gene may be conserved in mammals. Our preliminary results from functional promoter analysis show that the upstream region contains both promoters and repressor elements, behaving in tissue-specific ways in cardiac and neuron representative cell lines. Further experiments are needed to identify the tissue-specific transcription mechanisms.
The properties of mHCN4L we recorded from HEK293H cells are consistent with previous reports (8, 17). However, mHCN4S showed distinctively different properties, indicating that the N terminus of the HCN4 channel affects channel activation. Previously, replacement of both the N and C termini of mHCN4 with the termini of mHCN2 shifted the activation and deactivation kinetics recorded with an inside-out patch clamp on Xenopus oocytes, suggesting that the intracellular domains contribute to the channel functions (17). Deletion of the first 214 amino acid residues and the last 422 amino acid residues of rabbit HCN4 shifted the voltage dependence of activation by −10 mV and significantly slowed the kinetics of activation (18), but not of deactivation (19), in whole-cell recordings of COS-7 cells. This is consistent with our findings. However, the chimera replacement of the mHCN4 N terminus with the mHCN1 N terminus showed no functional change in whole-cell recordings (20). One explanation is that functional motifs in the mHCN4 N terminus can be replaced by their counterparts in mHCN1.
Analysis of alanine mutants showed that three positively charged residues contribute to the functional difference. These three charged residues, along with other residues in the middle part of the extended N-terminal segment, are identical to the two predicted HCN4 channel sequences in zebrafish and the two predicted HCN4 channel sequences in pufferfish. The high level of sequence similarity suggests positive selection and therefore the importance of these residues for proper channel function. During evolution, a genome duplication event in teleost fish caused two genes corresponding to every gene in mammals (21, 22). Two predicted Hcn4 genes are found in the genomes of zebrafish and pufferfish, respectively. Whether or not during evolution these two genes diverged in function and expression pattern, like what was seen in Na+ channels (13, 23), remains an interesting question.
The intracellular domains are important in the modulation of many ion channels. Some are through electrostatic interaction (24–26), and some are through cysteine disulfide bonds (27) especially in the cyclic nucleotide-gated ion channels, closely related to HCN channels (28, 29). In mHCN4, the first 25-amino acid segment does not contain cysteine, and three of five positively charged residues affect channel function; thus, the N terminus probably modulates the channel function through electrostatic interactions. The interaction partner may locate within the C terminus, as seen in Kv2.1 channel (30). Alternatively, the partner may be one of the transmembrane domains. The C-linker region of HCN channels interacts with the core transmembrane domain (31, 32). The N terminus may be involved in this interaction or may interact with other parts of the transmembrane domain. Stretches of charged residues are involved in recruiting associate proteins, such as the five positively charged residues in the SK2 channel (33). The 25-amino acid segment in mHCN4 channels may affect the channel function in a similar manner.
Footnotes
- HCN
- hyperpolarization-activated cyclic nucleotide-gated
- 5′-RACE
- rapid amplification of cDNA ends
- mHCN4
- mouse HCN4.
REFERENCES
- 1. DiFrancesco D. (1991) J. Physiol. 434, 23–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Accili E. A., Redaelli G., DiFrancesco D. (1997) J. Physiol. 500, 643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DiFrancesco D., Ferroni A., Mazzanti M., Tromba C. (1986) J. Physiol. 377, 61–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robinson R. B., Siegelbaum S. A. (2003) Annu. Rev. Physiol. 65, 453–480 [DOI] [PubMed] [Google Scholar]
- 5. Zagotta W. N., Olivier N. B., Black K. D., Young E. C., Olson R., Gouaux E. (2003) Nature 425, 200–205 [DOI] [PubMed] [Google Scholar]
- 6. Tran N., Proenza C., Macri V., Petigara F., Sloan E., Samler S., Accili E. A. (2002) J. Biol. Chem. 277, 43588–43592 [DOI] [PubMed] [Google Scholar]
- 7. Ludwig A., Zong X., Jeglitsch M., Hofmann F., Biel M. (1998) Nature 393, 587–591 [DOI] [PubMed] [Google Scholar]
- 8. Ludwig A., Zong X., Stieber J., Hullin R., Hofmann F., Biel M. (1999) EMBO J. 18, 2323–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi W., Wymore R., Yu H., Wu J., Wymore R. T., Pan Z., Robinson R. B., Dixon J. E., McKinnon D., Cohen I. S. (1999) Circ. Res. 85, e1–e6 [DOI] [PubMed] [Google Scholar]
- 10. Moosmang S., Biel M., Hofmann F., Ludwig A. (1999) Biol. Chem. 380, 975–980 [DOI] [PubMed] [Google Scholar]
- 11. Monteggia L. M., Eisch A. J., Tang M. D., Kaczmarek L. K., Nestler E. J. (2000) Brain Res. Mol. Brain Res. 81, 129–139 [DOI] [PubMed] [Google Scholar]
- 12. Liu H., Wu M. M., Zakon H. H. (2007) Dev. Neurobiol. 67, 1289–1304 [DOI] [PubMed] [Google Scholar]
- 13. Liu H., Wu M. M., Zakon H. H. (2008) J. Neurosci. 28, 9173–9182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stillitano F., Lonardo G., Zicha S., Varro A., Cerbai E., Mugelli A., Nattel S. (2008) J. Mol. Cell. Cardiol. 45, 289–299 [DOI] [PubMed] [Google Scholar]
- 15. Ishii T. M., Takano M., Xie L. H., Noma A., Ohmori H. (1999) J. Biol. Chem. 274, 12835–12839 [DOI] [PubMed] [Google Scholar]
- 16. Lin H., Xiao J., Luo X., Chen G., Wang Z. (2009) Cell Physiol. Biochem. 23, 317–326 [DOI] [PubMed] [Google Scholar]
- 17. Wicks N. L., Chan K. S., Madden Z., Santoro B., Young E. C. (2009) Pflugers Arch. 458, 877–889 [DOI] [PubMed] [Google Scholar]
- 18. Ishii T. M., Takano M., Ohmori H. (2001) J. Physiol. 537, 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishii T. M., Nakashima N., Takatsuka K., Ohmori H. (2007) Biochem. Biophys. Res. Commun. 359, 592–598 [DOI] [PubMed] [Google Scholar]
- 20. Viscomi C., Altomare C., Bucchi A., Camatini E., Baruscotti M., Moroni A., DiFrancesco D. (2001) J. Biol. Chem. 276, 29930–29934 [DOI] [PubMed] [Google Scholar]
- 21. Amores A., Force A., Yan Y. L., Joly L., Amemiya C., Fritz A., Ho R. K., Langeland J., Prince V., Wang Y. L., Westerfield M., Ekker M., Postlethwait J. H. (1998) Science 282, 1711–1714 [DOI] [PubMed] [Google Scholar]
- 22. Jaillon O., Aury J. M., Brunet F., Petit J. L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Nicaud S., Jaffe D., Fisher S., Lutfalla G., Dossat C., Segurens B., Dasilva C., Salanoubat M., Levy M., Boudet N., Castellano S., Anthouard V., Jubin C., Castelli V., Katinka M., Vacherie B., Biémont C., Skalli Z., Cattolico L., Poulain J., De Berardinis V., Cruaud C., Duprat S., Brottier P., Coutanceau J. P., Gouzy J., Parra G., Lardier G., Chapple C., McKernan K. J., McEwan P., Bosak S., Kellis M., Volff J. N., Guigó R., Zody M. C., Mesirov J., Lindblad-Toh K., Birren B., Nusbaum C., Kahn D., Robinson-Rechavi M., Laudet V., Schachter V., Quétier F., Saurin W., Scarpelli C., Wincker P., Lander E. S., Weissenbach J., Roest Crollius H. (2004) Nature 431, 946–957 [DOI] [PubMed] [Google Scholar]
- 23. Zakon H. H., Zwickl D. J., Lu Y., Hillis D. M. (2008) J. Exp. Biol. 211, 1814–1818 [DOI] [PubMed] [Google Scholar]
- 24. Sala F., Mulet J., Sala S., Gerber S., Criado M. (2005) J. Biol. Chem. 280, 6642–6647 [DOI] [PubMed] [Google Scholar]
- 25. Zeng W., Yuan J. P., Kim M. S., Choi Y. J., Huang G. N., Worley P. F., Muallem S. (2008) Mol. Cell 32, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Todorovic J., Welsh B. T., Bertaccini E. J., Trudell J. R., Mihic S. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schulteis C. T., Nagaya N., Papazian D. M. (1996) Biochemistry 35, 12133–12140 [DOI] [PubMed] [Google Scholar]
- 28. Gordon S. E., Varnum M. D., Zagotta W. N. (1997) Neuron 19, 431–441 [DOI] [PubMed] [Google Scholar]
- 29. Zheng J., Varnum M. D., Zagotta W. N. (2003) J. Neurosci. 23, 8167–8175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mohapatra D. P., Misonou H., Pan S. J., Held J. E., Surmeier D. J., Trimmer J. S. (2009) Channels 3, 46–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang J., Chen S., Siegelbaum S. A. (2001) J. Gen. Physiol. 118, 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Decher N., Chen J., Sanguinetti M. C. (2004) J. Biol. Chem. 279, 13859–13865 [DOI] [PubMed] [Google Scholar]
- 33. Allen D., Fakler B., Maylie J., Adelman J. P. (2007) J. Neurosci. 27, 2369–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]