Abstract
Flaviviruses, such as dengue virus (DENV), depend on the host endoplasmic reticulum for translation, replication, and packaging of their genomes. Here we report that DENV-2 infection modulates the unfolded protein response in a time-dependent manner. We show that early DENV-2 infection triggers and then suppresses PERK-mediated eIF2α phosphorylation and that in mid and late DENV-2 infection, the IRE1-XBP1 and ATF6 pathways are activated, respectively. Activation of IRE1-XBP1 correlated with induction of downstream targets GRP78, CHOP, and GADD34. Furthermore, induction of CHOP did not induce apoptotic markers, such as suppression of anti-apoptotic protein Bcl-2, activation of caspase-9 or caspase-3, and cleavage of poly(ADP-ribose) polymerase. Finally, we show that DENV-2 replication is affected in PERK−/− and IRE1−/− mouse embryo fibroblasts when compared with wild-type mouse embryo fibroblasts. These results demonstrate that time-dependent activation of the unfolded protein response by DENV-2 can override inhibition of translation, prevent apoptosis, and prolong the viral life cycle.
Keywords: ER Stress, Flavi Viruses, Positive-strand RNA Viruses, Protein Synthesis, RNA Viruses, Signal Transduction, Translation Control, Dengue Virus, Integrated Stress Response, Unfolded Protein Response
Introduction
Dengue virus (DENV)3 is a positive-sense, enveloped RNA flavivirus, with a genome of 10.7 kilobases (kb) in length, composed of four serotypes (DENV-1 to -4). Upon entry into the host cell, the genomic RNA functions as a messenger RNA (mRNA) and is translated as a single polyprotein, which is processed by cellular and viral proteases into three structural proteins (C, M, and E) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (1). DENV and other members of the Flaviviridae family are endoplasmic reticulum (ER)-tropic viruses that are dependent on the host ER to translate, replicate, and package their genome (1). The ER is an extensive membranous network that serves various specialized functions such as calcium storage and intracellular signal transduction. Additionally, most secreted and transmembrane proteins enter the lumen of the ER to mature prior to being secreted. To maintain quality control and ensure that only properly folded proteins are secreted, the ER has evolved three mechanistically distinct pathways collectively called the unfolded protein response (UPR). The UPR consists of the integrated stress response (ISR), along with the inositol-requiring protein-1 (IRE1), and activating transcription factor-6 (ATF6) pathways (2–5).
The ISR is the arm of the UPR that functions as a first-response mechanism induced by various stressors and attenuates global protein synthesis (2). This is achieved through phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α) by one of four known kinases (6). The double-stranded RNA (dsRNA) protein kinase R (PKR) is activated by the presence of dsRNA intermediates during viral replication and by interferon (IFN)-α/β. Activation of PKR is induced through binding of dsRNA; the protein then dimerizes and autophosphorylates, leading to PKR-mediated eIF2α phosphorylation. PKR-like endoplasmic reticulum kinase (PERK) also phosphorylates eIF2α (6). PERK is an ER-localized type I transmembrane protein whose lumenal domain senses an excess of unfolded proteins that enter the ER. This excess of misfolded proteins hinders the capacity of the ER to successfully fold nascent proteins. This imbalance (ER stress) is mitigated by transient adaptation achieved by decreasing global protein synthesis via eIF2α phosphorylation (6). In addition, two other kinases, general control non-derepressible-2 kinase (GCN2) and heme-regulated eIF2α kinase, respond to amino acid starvation and heme deficiency, respectively, by phosphorylating eIF2α (6). Thus, phosphorylation of eIF2α is the nexus for an array of cellular responses that are elicited through the ISR (2, 5).
Cross-talk between the ISR and the other branches of the UPR is mediated through the PERK pathway. Failure to suppress ER stress leads to activation of the ATF6 and IRE1 pathways, which act to alleviate the accumulation of misfolded proteins by up-regulating host factors that increase the capacity of the ER to handle the synthesis of nascent proteins (5, 7). Activation of IRE1 and ATF6 pathways leads to induction of various genes involved in the UPR, such as the chaperone glucose-regulated protein-78/binding immunoglobulin protein (GRP78/BiP), GRP94, and protein-disulfide isomerase (PDI). Under normal conditions, PERK, ATF6, and IRE1 are suppressed via their lumenal domains through interaction with GRP78/BiP, but during ER stress, GRP78 dissociates from these proteins, leading to their activation (8–10). Additionally, in cells under persistent stress conditions, the pro-apoptotic transcription factor CCAT/enhancer-binding protein (CHOP) is induced, which can lead to suppression of the anti-apoptotic protein B cell lymphoma-2 (Bcl-2) (11, 12). Alternatively, CHOP can also function as a pro-survival transcription factor leading to induction of growth arrest and DNA damage-inducible protein (GADD34), a subunit of protein phosphatase 1c (PP1c) that targets the dephosphorylation of phosphorylated eIF2α (eIF2α-P). Thus, depending on the strength, duration, and type of stress encountered by the cell, CHOP can serve as a pro-apoptotic or pro-survival transcription factor (7, 13, 14).
The activation of individual branches and components of the UPR by members of the Flaviviridae family of viruses has been previously reported. Studies with hepatitis C virus (HCV) have demonstrated activation and suppression of the PERK pathway, suggesting a mechanism by which HCV establishes a persistent infection (15). Other reports using the pestivirus bovine viral diarrhea virus have shown that induction of PERK and subsequent eIF2α phosphorylation induces the apoptotic pathway (16). Infection with Japanese encephalitis virus and DENV-2 was shown to induce the IRE-XBP1 pathway and a subset of XBP1-activated genes involved in the ER-associated degradation pathway, protecting cells from virus-induced cytopathic effects (17). In contrast, another group suggested that DENV induction of IRE1-XBP1 and ATF6 pathways contributed to dengue pathogenesis (18), whereas WNV induction of the UPR was shown to lead to CHOP-mediated apoptosis (19). Although these studies demonstrated that viral infection can lead to induction of the individual branches of the UPR and their downstream components, an integrated study of the role the UPR plays during the DENV life cycle and how DENV may modulate the UPR is still lacking.
In this study, we investigate the induction of the three arms of the UPR and determine its effects on the host cell throughout the course of DENV-2 infection, demonstrating that DENV infection leads to induction of the UPR in a time-dependent manner. The induction of the UPR in DENV-infected cells results in the adaptation to ER stress without inducing the apoptotic pathway, consequently prolonging cell viability and the DENV life cycle. By using genetically deficient mouse embryonic fibroblast (MEF) cells, we further demonstrate that certain deficiencies in the UPR signaling pathway (PERK and IRE1) but not others (PKR, XBP1, and ATF6) affect DENV viral output. Together, our results indicate that suppression and induction of various branches of the UPR signaling pathways are modulated during DENV infection in a time-dependent manner and that DENV has evolved to take advantage of the UPR.
EXPERIMENTAL PROCEDURES
Cell Lines and Viruses
Human fibrosarcoma 2fTGH cells (kindly provided by Dr. Stark, Cleveland Clinic Foundation, Cleveland, OH) (20) were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen), 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 10% fetal bovine serum (FBS) (HyClone, Logan, UT) at 37 °C in 5% CO2. Baby hamster kidney cells (BHK-21 clone 15) were grown in minimal essential medium-α (MEM-α; Invitrogen), 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 10% FBS (HyClone). Aedes albopictus (C6/36) cells (American Type Culture Collection, Manassas, VA) were grown in Leibovitz's L-15 medium (Invitrogen) with 100 units/ml of penicillin, 100 μg/ml of streptomycin, 10 mm HEPES, and 10% FBS. PERK+/+ and PERK−/− MEFs (kind gift from D. Ron, New York University School of Medicine, New York), PKR+/+ and PKR−/− MEFs (kind gift from B. R. G. Williams, Cleveland Clinic Foundation, Cleveland, OH) (21), and IRE1α+/+, IREα−/−, XBP1+/+, XBP1−/−, ATF6α+/+, and ATF6α−/− MEFs (kind gift from Randal Kaufman, University of Michigan, Ann Arbor, MI) were grown in DMEM, 100 units/ml of penicillin, 100 μg/ml of streptomycin, 1× non-essential amino acids, 55 μm β-mercaptoethanol, 1× glutamax (Invitrogen), and 10% FBS (HyClone). DENV-2 strain PL046 (kindly provided by H.-Y. Lei, National Cheng Kung University, Tainan, Taiwan) was amplified in C6/36 cells, and plaque-forming units/ml were determined using BHK cells as previously described (22). 2fTGH cells were treated with thapsigargin (Sigma) 3 h (h) post-infection (p.i.) for 1 h, then cells were harvested; in experiments involving cycloheximide (CHX; Sigma), CHX was added at the time of infection and was present throughout the experiment.
Western Blot
Cell lysates were prepared using lysis buffer (1% Triton X-100, 20 mm HEPES, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 10% glycerol) containing complete EDTA-free protease inhibitor (Roche Applied Science) and phosphatase inhibitors (10 mm tetrasodium pyrophosphate, 100 mm sodium fluoride, 17.5 mm β-glycerolphosphate (Sigma), and 1 mm sodium orthovanadate (Santa Cruz Biotechnologies, Santa Cruz, CA)). Cell lysates used in the detection of ATF6 and GADD34 were prepared using RIPA buffer (10 mm Tris-HCl, pH 7.5, 500 mm NaCl, 1 mm EDTA, 0.5% sodium deoxycholate, 0.1% SDS, and 1% Triton X-100) containing complete EDTA-free protease inhibitor and phosphatase inhibitors as above. Equal amounts of protein, as measured by Bradford assay (Bio-Rad), were resolved on 7 or 11% SDS-PAGE gels. Proteins were transferred onto Immobilon-P polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA). Immunoblots were blocked at room temperature for 1 h using blocking buffer (1× phosphate-buffered saline, pH 7.3 (PBS; 137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4·7H2O, 1.4 mm KH2PO4), 5% nonfat dry milk, and 0.1% Tween 20).
Primary antibodies were obtained from the following sources and were diluted in blocking buffer at the following dilutions: anti-eIF2α (1:1,000), anti-phospho-specific eIF2α (1:1,000), anti-eIF2α total (1:1,000), anti-phospho-specific PERK (1:1,000), anti-phospho-specific GCN2 (1:1,000), anti-GCN2-total (1:1,000), anti-Bcl-2 (1:1,000), anti-caspase-3 (1:1,000), anti-caspase-9 (Cell Signaling Technologies, Beverly, MA), anti-ATF6 (1:200), anti-CHOP (1:200), anti-GADD34 (1:200), anti-GRP78 (1:1,000), anti-GRP94 (1:1,000), anti-actin horseradish peroxidase (HRP) (1:1,000) (Santa Cruz Biotechnologies), anti-PDI (1:1,000) (Transduction Laboratories, Lexington, KY), anti-PARP (1:1,000) (Chemicon, Temecula, CA), and mouse monoclonal anti-NS1 7E11 (1:1,000), which recognizes all 4 DENV serotypes (kind gift from R. J. Putnak, Walter Reed Army Institute of Research, Washington, D. C.). A mixture of anti-NS1 monoclonal antibodies previously generated in the laboratory were also used.4 The blots were incubated overnight (a minimum of 12 h for phospho-specific antibodies), washed twice with 1× PBS-T (PBS containing 0.1% Tween 20) at room temperature, and incubated for 1 h with HRP-conjugated goat anti-rabbit or goat anti-mouse at 1:10,000 or 1:5,000, respectively. Quantification was performed using the Chemi-Doc system (Bio-Rad).
Metabolic Labeling
Cells were washed with DMEM lacking cysteine and methionine (Invitrogen). After washing, cells were incubated in DMEM supplemented with 5% dialyzed fetal calf serum (Invitrogen) for 15 min. Cells were pulse-labeled for 30 min by the addition of 100 μCi/ml of [35S]cysteine-methionine, then harvested and washed twice with ice-cold PBS. Cell lysates were prepared using RIPA buffer as above. Equal amounts of protein were resolved on an 11% SDS-PAGE gel, dried, and exposed on a GE PhosphorScreen (General Electric, Piscataway, NJ). Cycloheximide (20 μm final concentration) was present in all the media including washes. Thapsigargin (1 μm final concentration) was added just prior to labeling the cells. Images were visualized on a GE Typhoon Variable Mode Imager.
Confocal Microscopy
2fTGH cells were seeded at 5 × 104 cells/well in a 12-well plate containing a microscope coverslip in each well and left to adhere overnight. Cells were infected at a m.o.i. of 5 for the indicated times. Coverslips were washed three times with 1× PBS, fixed in 4% formaldehyde for 10 min, and washed three times with 1× PBS. Cells were then permeabilized using 0.5% saponin in PBS for 10 min. Blocking was done using 1% BSA in 0.1% Tween PBS buffer for 1 h. Primary antibodies were diluted 1:50 in blocking buffer for 1 h and obtained from the following sources: anti-GRP78, anti-GRP94, and anti-XBP1 (Santa Cruz Biotechnologies). Anti-DENV E (4G2; ATCC) and anti-NS3 (E1D8)4 antibodies were directly conjugated to Alexa 488 and Alexa 594, respectively. Alexa FluorTM 488 (goat anti-rabbit), Alexa FluorTM 546 (donkey anti-goat), and Alexa FluorTM 647 (goat anti-rabbit) were diluted 1:200 (Molecular Probes, Invitrogen Corporation). Coverslips were mounted using VECTASHIELD® mounting medium with DAPI (Vector Laboratories Burlingame, CA). Images were captured using the ×63 objective on a Zeiss LSM 510 UV/visual Meta laser-scanning confocal microscope (College of Natural Resource's Biological Imaging Facility, University of California at Berkeley, Berkeley, CA).
RESULTS
The ISR and eIF2α Phosphorylation during DENV-2 Infection
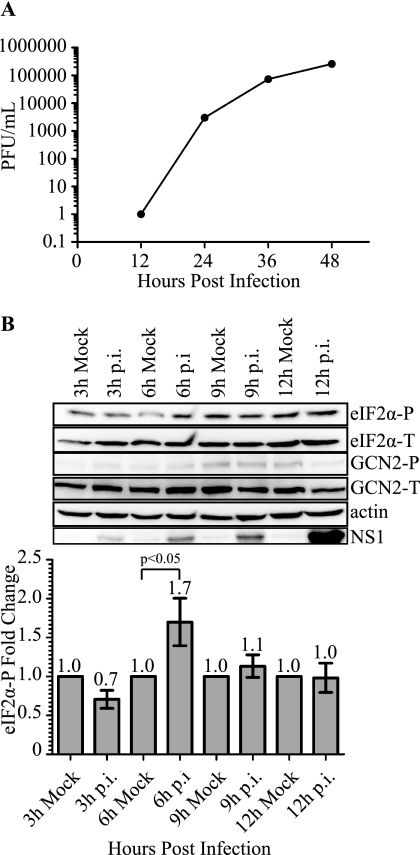
To begin evaluating the role of eIF2α in DENV-2 translation, we first determined the infectivity of the human fibrosarcoma cell line 2fTGH. Cells were infected with DENV-2 (strain PL046) at a m.o.i. of 5, and viral progeny in the supernatant were measured over a 48-h time course. Viral titers were determined by plaque assay, and as expected, viral progeny were increased during infection (Fig. 1A). Using the same infection conditions, we next investigated the phosphorylation state of eIF2α during DENV infection over a 48-h time course. Immunoblot analysis of 2fTGH cell extracts demonstrated a statistically significant increase in levels of phosphorylated eIF2α at 6 h p.i. compared with mock, which then returned to mock levels at later time points (Figs. 1B and 5C, and data not shown).
FIGURE 1.
DENV-2 infectivity of 2fTGH cells and induction of eIF2α phosphorylation during infection. A, DENV-2 viral titers as determined by plaque assay. 2fTGH cells were plated overnight and exposed to DENV-2 infection (PL046) at an m.o.i. of 5. Supernatants were collected at the indicated time points post-infection, and infectious virus titer was determined by plaque assay using BHK21 cells. B, phosphorylation state of eIF2α during DENV-2 infection of 2fTGH cells. Mock and DENV-2-infected 2fTGH cell lysates were collected at the indicated time points, and equal amounts of proteins were assayed by Western blot. Phosphorylated eIF2α was visualized using phosphospecific anti-eIF2α antibodies (eIF2α-P) and quantification of eIF2α-P was normalized to total eIF2α (eIF2α-T). Fold-change was measured within each time point as compared with mock infected controls that were assigned a value of 1. Statistical analysis was performed using the Wilcoxon Rank Sum Test (n = 3). Error bars represent the mean ± S.E. GCN2-P was detected using phosphospecific antibodies to GCN2-P, and total GCN2 was visualized with antibodies directed to GCN2. DENV-2 infection was confirmed by immunoblotting for DENV-2 NS1 protein, and actin was used as a loading control. Shown is an immunoblot representative of 3 independent experiments.
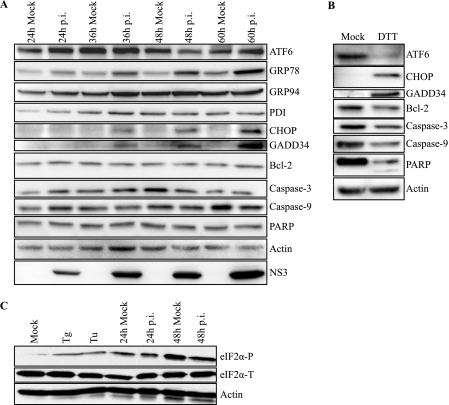
FIGURE 5.
DENV-2 infection induces gene expression of UPR components and activation of the ATF6 pathway. A, immunoblot analysis of components of UPR signaling pathways during a time course of DENV-2 infection. Mock or DENV-2-infected 2fTGH cell lysates were collected at the indicated time points. Lysates were analyzed for the activation and induction of the indicated UPR proteins and markers of apoptosis by immunoblot analysis, as described under “Experimental Procedures.” Actin was used as a loading control, and infection was confirmed by detection of the viral protein NS3. B, positive controls for the induction of UPR proteins and apoptosis markers. Lysates were prepared from 2fTGH cells treated with DTT for 6 h, followed by immunoblot analysis as described under “Experimental Procedures.” C, mock and DENV-2-infected 2fTGH cell lysates were collected at the indicated time points, and equal amounts of proteins were assayed by Western blot. eIF2α-P was visualized and quantified as previously described. Positive controls for eIF2α-P consisted of cells treated with thapsigargin or tunicamycin (Tu) for 30 min. Actin was added as a loading control; the bottom band is the residual HRP activity after probing for eIF2α-P. All data are representative of at least three independent experiments.
We next sought to determine which signaling branch of the ISR pathway mediated eIF2α phosphorylation (eIF2α-P) early during DENV infection. We first investigated the role of PKR in the phosphorylation of eIF2α during DENV infection. Activation of PKR occurs when dsRNA intermediates are produced during viral replication. Recognition of dsRNA by the RNA binding motif of PKR leads to autophosphorylation and subsequent phosphorylation of eIF2α (6). Given that eIF2α-P is observed early during DENV-2 infection, we focused primarily on early time points (3–12 h p.i.). At these time points, we found no difference in PKR phosphorylation in DENV-2-infected 2fTGH cells compared with mock-infected controls, as determined by immunoblot analysis (data not shown). These results are consistent with our previously published reports (23).
Next, we focused on the role of GCN2 in mediating eIF2α-P during DENV-2 infection at these early time points. The role of GCN2 in controlling viral infection has been previously demonstrated with vesicular stomatitis virus, and infection with Sinbis virus has been shown to lead to eIF2α phosphorylation (22, 24). Given the anti-viral potential of GCN2 and its role in phosphorylating eIF2α, we wanted to determine whether GCN2 also contributed to early phosphorylation of eIF2α during DENV-2 infection. To test this, we infected 2fTGH cells with DENV-2 for 3–12 h and performed an immunoblot analysis of cell extracts. The levels of phosphorylated GCN2 (GCN2-P) showed no increase between DENV-infected cells and their time-matched mock infected controls (Fig. 1B).
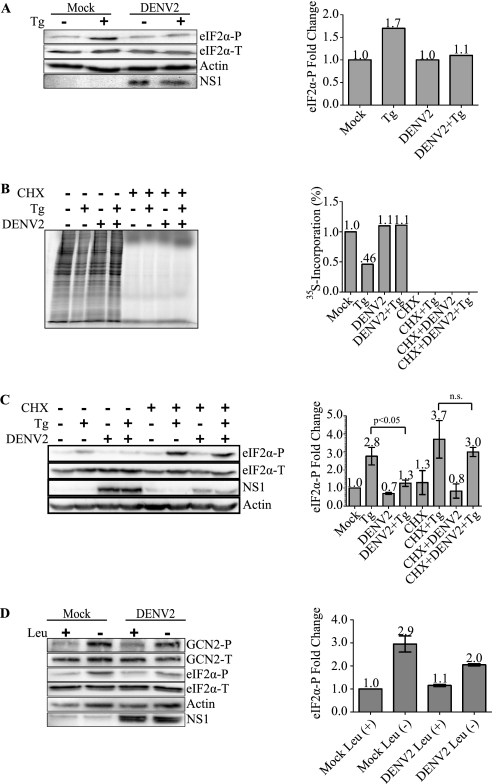
Last, we focused on the PERK pathway and its role in eIF2α phosphorylation. Under conditions of ER stress, the accumulation of unfolded proteins leads to the dissociation of GRP78 from the ER lumenal domain of PERK, which then triggers its activation through dimerization and autophosphorylation (25, 26). We attempted to directly examine the phosphorylation state of PERK using specific antibodies to phosphorylated PERK (PERK-P); however, we were unable to detect PERK-P in 2fTGH cells either infected with DENV-2 or treated with the ER stress inducers thapsigargin (Tg) or tunicamycin (25, 27). This negative result might be due to low levels of endogenous PERK in 2fTGH cells and the difficulty in detecting PERK-P except in a few rare cell lines (8). As an alternative approach, the role of PERK in the phosphorylation of eIF2α early during DENV-2 infection was examined by using the ER stress-inducing drug Tg to activate PERK in both DENV-2-infected and uninfected cells. We infected 2fTGH cells with DENV-2 at an m.o.i. of 5 for 3 h, the earliest time point at which we detect viral protein synthesis, then treated or mock treated the cells with Tg for 1 h. Lysates were analyzed for eIF2α-P by immunoblot. Infected cells displayed similar levels of eIF2α-P as compared with mock infected cells; however, cells that were uninfected and treated with Tg reproducibly showed higher levels of eIF2α-P compared with cells infected with DENV-2 followed by Tg treatment (Fig. 2A).
FIGURE 2.
DENV-2 translation inhibits PERK-mediated eIF2α phosphorylation during ER stress. A, DENV-2 inhibits PERK-mediated eIF2α phosphorylation under ER stress. 2fTGH cells were plated overnight and infected with DENV-2 (PL046) at an m.o.i. of 5. Three hours p.i., cells were mock treated or treated with 1 μm Tg for 1 h. Levels of eIF2α-P and total eIF2α were visualized, quantification of eIF2α-P was normalized to eIF2α-T, and fold-change was compared with mock infected cells assigned a value of 1. DENV-2 NS1 protein expression was detected using a mixture of mouse anti-NS1 monoclonal antibodies. Shown is an immunoblot representative of 3 independent experiments. Aggregate values were statistically analyzed in conjunction with C (lanes 1–4). B, DENV-2 translation inhibits thapsigargin-induced shutdown of protein synthesis. Metabolic [35S]cysteine-methionine incorporation was measured in 2fTGH cells that were mock infected or infected with DENV-2 (PL046) at a m.o.i. of 5 and mock treated or treated with 20 μm CHX at the time of infection. Three hours p.i. cells were mock-treated or treated with 1 μm Tg for 1 h and metabolically labeled with [35S]cysteine-methionine at the time of treatment. Equal amounts of protein were resolved by SDS-PAGE, gels were dried, and 35S incorporation was visualized by autoradiography quantified using ImageQuantTM (GE Healthcare). Values shown are percent incorporation normalized to mock infected control. The gel shown is representative of at least 3 independent experiments. C, viral protein synthesis is necessary to inhibit PERK-mediated eIF2α-P during ER stress. 2fTGH cells were treated as in B. Mock infected and DENV-2-infected 2fTGH cell lysates were harvested, and equal amounts of proteins were assayed by Western blot. eIF2α-P was visualized and quantified as in A. DENV-2 infection was confirmed by immunoblotting for DENV-2 NS1 protein, and actin was used as a loading control. Statistical analyses were performed using the Wilcoxon Rank Sum Test (n = 3), and error bars indicate mean ± S.E.; n.s., not significant. D, DENV-2 translation inhibits GCN2-mediated eIF2α phosphorylation during amino acid deprivation. 2fTGH cells were plated overnight and infected with DENV-2 as in A. Three hours p.i. cells were placed in medium containing or lacking leucine for 3 h. Lysates were collected and analyzed for phosphorylated and total eIF2α, phosphorylated and total GCN2, actin, and NS1 as previously described. Quantification of phosphorylated eIF2α was performed as described in A. Statistical analysis was performed using the Wilcoxon Rank Sum Test (n = 2) and error bars indicate mean ± S.E.
To test more specifically whether active translation of the viral genome was necessary for DENV-2 inhibition of Tg-induced PERK-mediated eIF2α phosphorylation, we used 35S-labeled cysteine-methionine to monitor nascent protein synthesis in 2fTGH cells that were mock infected or infected with DENV-2 for 3 h in the presence or absence of the translation elongation inhibitor CHX followed by a 1-h Tg treatment or mock treatment. 2fTGH cells that were treated with CHX demonstrated complete inhibition of protein synthesis irrespective of infection or addition of Tg (Fig. 2B). Cells that were mock infected or DENV-2-infected demonstrated comparable levels of protein synthesis in the absence of CHX. As expected, 2fTGH cells that were mock infected and treated with Tg displayed a decrease in levels of protein synthesis compared with DENV-2-infected cells treated with Tg. We also analyzed cell lysates for eIF2α phosphorylation by immunoblot. As demonstrated in Fig. 2C (second lane versus fourth lane), 2fTGH cells that were DENV-2 infected and treated with Tg demonstrated a statistically significant reduction in levels of eIF2α phosphorylation compared with mock infected cells treated with Tg. However, cells treated with CHX at the time of DENV-2 infection were no longer able to suppress Tg-induced PERK-mediated eIF2α phosphorylation (sixth lane versus eighth lane), and viral protein synthesis was significantly decreased in these cells as shown by decreased levels of viral protein NS1, further confirming that the CHX was active in inhibiting total protein synthesis and that viral protein synthesis is necessary to inhibit eIF2α-P.
Thus far, our data suggest that PERK but not PKR plays a role in early induction and suppression of eIF2α phosphorylation during DENV infection. As our results demonstrated that DENV-2 infection inhibits Tg-induced PERK-mediated eIF2α phosphorylation, we decided to test whether DENV-2 infection could also inhibit GCN2-mediated eIF2α phosphorylation. We tested this by mock infecting or infecting 2fTGH cells with DENV-2 for 3 h followed by 3 h with or without leucine deprivation. Mock infected or DENV-infected cells grown in medium containing leucine displayed similar levels of GCN2-P and eIF2α-P (Fig. 2D). On the other hand, cells that were uninfected and leucine deprived consistently showed higher levels of eIF2α-P compared with cells infected with DENV-2 followed by leucine deprivation (Fig. 2D). However, the levels of GCN2-P in both leucine-deprived mock infected and DENV-infected cells were similar, suggesting that under these conditions, inhibition of eIF2α phosphorylation occurs downstream of GCN2 phosphorylation.
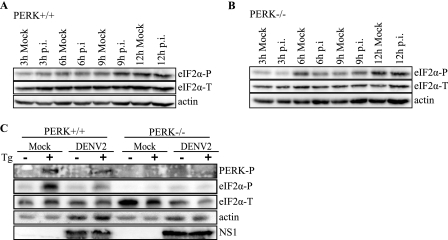
Based on our data in 2fTGH cells, we wanted to further analyze whether PERK was responsible for the transient induction of eIF2α phosphorylation early in DENV-2 infection. For this, we performed similar experiments to those in Fig. 1B and analyzed levels of phosphorylated eIF2α in PERK+/+ or PERK−/− MEF cells mock infected or infected with DENV-2 for 3–12 h. We found that PERK+/+ cells infected with DENV-2 had increased levels of phosphorylated eIF2α 9 h p.i. compared with mock infected controls; however, these levels returned to mock infected levels at later time points (Fig. 3A). In contrast, PERK−/− cells infected with DENV-2 showed no increase in levels of phosphorylated eIF2α as compared with mock infected controls throughout the course of infection (Fig. 3B).
FIGURE 3.
eIF2α phosphorylation during DENV-2 infection in PERK+/+ and PERK−/− cells. A and B, time course of eIF2α phosphorylation in DENV-infected PERK+/+ and PERK−/− MEF cells. Mock and DENV-2-infected (A) PERK+/+ and (B) PERK−/− cell lysates were collected at the indicated time points, and equal amounts of protein were assayed by immunoblot. Levels of eIF2α-P and total eIF2α were visualized as described under “Experimental Procedures.” Actin was included as a loading control. C, DENV-2 inhibits PERK-mediated eIF2α phosphorylation under ER stress. PERK+/+ and PERK−/− cells were plated overnight and infected with DENV-2 (PL046) at an m.o.i. of 5. Three hours p.i. cells were mock-treated or treated with 1 μm Tg for 1 h. Phosphorylated PERK was visualized using phosphospecific PERK antibody, and levels of eIF2α-P and total eIF2α were visualized as described previously. DENV-2 infection was confirmed using a mouse anti-NS1 monoclonal antibody. Actin was used as a loading control.
We next attempted to examine the phosphorylation state of PERK in DENV-infected PERK+/+ MEFs and, as in 2fTGH cells, we were unable to visualize phosphorylated PERK over an infection time course. However, using our alternative approach with the ER stress-inducing drug Tg to activate PERK, we were able to detect PERK-P in both DENV-2-infected and uninfected PERK+/+ cells; as expected, no PERK-P was observed in PERK−/− cells (Fig. 3C). PERK+/+ and PERK−/− cells were infected with DENV-2 at an m.o.i. of 5 for 3 h, then treated or mock treated with Tg for 1 h. Lysates were then analyzed by immunoblot. PERK+/+ cells that were uninfected and treated with Tg demonstrated higher levels of eIF2α-P compared with cells infected with DENV-2 followed by Tg treatment (Fig. 3C), similar to our results in 2fTGH cells (Fig. 2, A and C). No difference in eIF2α phosphorylation was observed in PERK−/− cells regardless of treatment conditions.
Previous studies have demonstrated the selective increase in translation of ATF4 under conditions that induce robust and persistent eIF2α phosphorylation (28). Consistent with the eIF2α data presented here, we find that expression of ATF4 at these early time points is not induced (data not shown), suggesting it is not expressed or is transiently expressed and too faint to detect. Together, these results demonstrate that DENV-2 infection induces eIF2α phosphorylation early during infection in 2fTGH and PERK+/+ cells but that this effect is rapidly reversed. Our 2fTGH data shows that translation of the viral genome is necessary for inhibition of Tg-induced PERK-mediated eIF2α phosphorylation. Additionally, although DENV-2 is able to inhibit GCN2-mediated eIF2α under conditions of amino acid deprivation, the MEF results suggest that PERK and not GCN2 is responsible for the phosphorylation of eIF2α because the transient induction of eIF2α phosphorylation was only observed in PERK+/+ cells infected with DENV-2 and not in DENV-2-infected PERK−/− cells.
Induction of the IRE1-XBP1 and ATF6 Pathways in Mid and Late DENV Infection
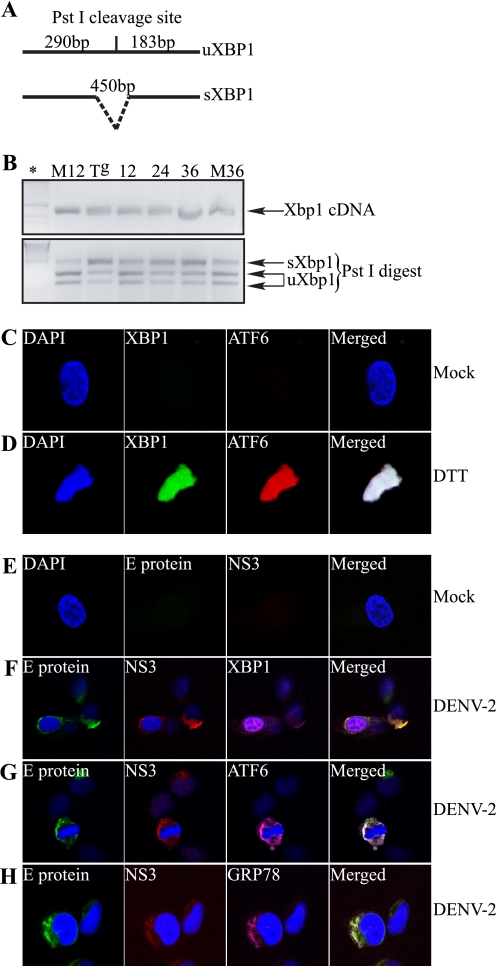
Persistent ER stress that is not resolved through attenuation of global protein synthesis induces the cell to activate either the IRE1-XBP1 or ATF6 pathways or both to resolve cellular stress. After showing that DENV infection modulates the ISR early during infection, we wanted to determine whether and when during DENV-2 infection the IRE1-XBP1 and ATF6 pathways are induced. We first focused on the IRE1-XBP1 pathway. Activation of IRE1 causes post-transcriptional cleavage of the Xbp1 mRNA (uXbp1) that produces the spliced form of Xbp1 mRNA (sXbp1), which encodes the transcriptionally active form of XBP1. 2fTGH cells were mock-infected or infected with DENV-2 over a 36-h time course. Cells were harvested and total RNA was extracted at the indicated time points. Using a standard RT-PCR protocol coupled with restriction enzyme digestion for detection of the sXbp1 and uXbp1 mRNA forms (Fig. 4A), we found that the IRE1-XBP1 pathway was induced in DENV-2-infected cells. Starting at 12 h p.i., 2fTGH cells infected with DENV-2 began to accumulate the sXbp1 mRNA, and levels of this mRNA continued to increase through 36 h p.i. (Fig. 4B). To further confirm the synthesis and nuclear translocation of the transcriptionally active XBP1 protein, we used laser scanning confocal microscopy (LSCM). Guided by our RT-PCR data, we grew 2fTGH cells on coverslips and mock infected (Fig. 4E) or infected cells with DENV-2 at an m.o.i. of 5 (Fig. 4, F and G) for 24 h, at which time cells were fixed, stained using conjugated mouse monoclonal antibodies against DENV-2 E and NS3 proteins, and mounted on slides for processing. As positive controls for the induction of the IRE1-XBP1 and ATF6 pathways, we treated uninfected cells with the stress-inducing agent dithiothreitol (DTT) for 6 h (Fig. 4D). Confocal microscopy demonstrated that at 24 h p.i., XBP1 was synthesized and targeted to the nucleus of DENV-2-infected cells, consistent with nuclear localization observed in DTT-treated cells (Fig. 4, D and F).
FIGURE 4.
DENV-2 induces Xbp1 mRNA splicing and nuclear localization, leading to GRP78 up-regulation in 2fTGH cells. A, schematic representation of the RT-PCR product for uXbp1 and sXbp1, indicating the presence or absence of the PstI restriction site. B, RT-PCR analysis of uXbp1 and sXbp1 during DENV-2 infection. 2fTGH cells were mock infected (M) or infected with DENV-2 at an m.o.i. of 5 for the indicated times (in hours). The positive control consisted of 2fTGH cells that were treated with 1 μm Tg for 12 h (Tg). Total mRNA was extracted, and 1 μg of total mRNA was amplified by RT-PCR with primers targeting Xbp1. Xbp1 cDNA was subjected to PstI digestion, and products were resolved on a 2% agarose gel. uXbp1 or sXbp1 following PstI digestion are shown. *, molecular weight marker. C–H, confocal microscopy visualization of the induction of components of the UPR signaling pathways in DENV-2-infected 2fTGH cells. C and D, intracellular staining using anti-XBP1 (green) and anti-ATF6 (red) antibodies followed by secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 546, respectively, in mock treated (C) or DTT-treated (D) 2fTGH cells. E, intracellular staining of mock infected 2fTGH cells with anti-E (green) or anti-NS3 (red) mouse monoclonal antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 594, respectively. F–H, DENV-2-infected 2fTGH cells were stained intracellularly for the activation and nuclear localization of XBP1 (F) and ATF6 (G) and GRP78 up-regulation (H). Cells were infected with DENV-2 for 24 h. Intracellular staining was carried out using anti-XBP1, anti-ATF6, or anti-GRP78 antibodies followed by secondary antibodies conjugated to Alexa Fluor 647 (magenta). Additionally, the nucleus was visualized using DAPI (blue), and viral proteins were visualized using anti-E or anti-NS3 monoclonal antibodies directly conjugated to Alexa Fluor 488 (green) or Alexa Fluor 594 (red), respectively. All images were captured using a Zeiss 510 Laser Scanning microscope. All data shown are representative of at least three experiments.
The continued burden and accumulation of unfolded viral proteins in the ER, in addition to activating the PERK and IRE1-XBP1 pathways, can also lead to activation of the ATF6 pathway. Activation of the ATF6 pathway targets ATF6 from the ER to the Golgi, where it is proteolytically cleaved, and its cytosolic DNA-binding portion targeted to the nucleus to activate gene expression. When we used LSCM to determine whether ATF6 is also activated and localized to the nucleus after DENV-2 infection, we found that at 24 h p.i., it remained inactivated and colocalized to the perinuclear area with DENV-2 proteins E and NS3 (Fig. 4G), unlike DTT-treated cells where activated ATF6 was found in the nucleus (Fig. 4D). Additional controls showing staining of XBP1 and ATF6 in wild-type and XBP−/− and ATF6α−/− cells are shown in supplemental Fig. S1.
One consequence of activating either the IRE1-XBP1 or ATF6 pathway is up-regulation of genes involved in protein folding, such as the chaperone and master regulator GRP78 (29, 30). We demonstrated that DENV-2 infection of 2fTGH cells for 24 h leads to induction of the IRE1-XBP1 pathway but not the ATF6 pathway. We next examined GRP78 levels and found that at 24 h p.i., GRP78 is also up-regulated, suggesting that IRE1-XBP1 is responsible for the induction of GRP78 (Fig. 4H). In addition, GRP78 colocalizes with DENV proteins E and NS3. Interestingly, however, when we used LSCM to analyze the up-regulation of GRP94, also an ER resident chaperone, we did not find it to be up-regulated at 24 h p.i. (data not shown).
We wanted to further confirm that up-regulation of GRP78, but not GRP94, was mediated by induction of the IRE1-XBP1 pathway and not the ATF6 pathway. To do this, we infected 2fTGH cells with DENV-2 over a 60-h time course. Cell lysates were collected at the indicated time points and analyzed by immunoblot. Using ATF6-specific antibodies that recognize the intact 90-kDa form of the ATF6 protein (pATF6), we demonstrated that proteolytic processing of pATF6 only occurs transiently at 48 h p.i. in DENV-2-infected cells compared with mock-infected cells (Fig. 5A). As a positive control, we showed that the ATF6 pathway was functional by using DTT, a strong reducing agent and potent ER stress inducer (Fig. 5B). We next probed for the expression of GRP78, GRP94, and PDI; all ER-resident chaperone proteins (16). We observed that levels of GRP78 were elevated compared with mock-infected controls at 24–60 h p.i. (Fig. 5A). However, consistent with our LSCM results (data not shown), we did not detect an increase in either GRP94 or PDI protein levels (Fig. 5A). Together, these data suggest that activation of the IRE1-XBP1 pathway leads to selective up-regulation of GRP78, based on the similar kinetics of IRE1-XBP1 activation and GRP78 expression.
The activation IRE1-XBP1 or ATF6 in cells that are exposed to prolonged ER stress can also lead to the induction and expression of the pro-survival or pro-apoptotic protein CHOP. Based on our activation data of IRE1-XBP1 (Fig. 4, A and B) and ATF6 (Figs. 4D and 5A) pathways at different time points post infection, we wanted to determine whether induction of CHOP expression correlated with the induction of either pathway. To assess this, we probed cell lysates from DENV-2-infected or mock infected cells for CHOP expression 24–60 h p.i. We found that CHOP expression began at 36 h p.i. in DENV-2-infected cells but not in mock infected cells (Fig. 5A). We also showed that induction of CHOP leads to activation of its downstream target GADD34 in DENV-2-infected cells starting at 36 h p.i. and continuing to 60 h p.i. (Fig. 5A). The stress-inducing agent DTT was used as a positive control for the induction of CHOP and GADD34 during ER stress (Fig. 5B). Expression of GADD34 has been shown to serve as a negative feedback loop to aid PP1c in reversing eIF2α phosphorylation and suppressing the ISR (31). Consistent with this observation, we do not observe higher levels of eIF2α-P in DENV-2-infected cells compared with their mock-infected counterparts at 24 or 48 h p.i. (Fig. 5C). Together, these data suggest that induction of CHOP and GADD34 is mediated by the IRE1-XBP1 pathway rather than the ATF6 pathway, as the timing of their induction correlated with activation of the IRE1-XBP1 pathway but not the ATF6 pathway. Furthermore, this illustrates how DENV virus is able to suppress eIF2α-P early in infection, via a yet undetermined mechanism, whereas also eliciting the induction of genes involved in the feedback mechanism, GADD34, which targets eIF2α for dephosphorylation later in the course of infection.
Additionally, we found that induction of CHOP during DENV-2 infection did not lead to the apoptosis-signaling cascade (Fig. 5A). Previous reports have demonstrated that expression of CHOP can lead to decreased levels of the anti-apoptotic protein Bcl-2 (19); however, we did not detect changes in Bcl-2 expression in DENV-2-infected cells compared with mock-infected cells (Fig. 5A). It has also been demonstrated that ER stress activates pro-apoptotic factors pro-caspase-9, pro-caspase-3, and poly(ADP-ribose) polymerase (PARP) via proteolytic processing (32–34), as evidenced in positive controls showing cleavage of these proteins after pharmacological induction of ER stress by DTT (Fig. 5B). However, we found that levels of intact pro-caspase-9, pro-caspase-3, and PARP were equivalent in mock-infected cells compared with DENV-2-infected cells throughout the time course of infection (24–60 h), further indicating that apoptosis is not taking place (Fig. 5A). Lack of apoptosis was further confirmed by propidium iodide and annexin V staining of DENV-infected 2fTGH cells 48 h after infection (data not shown).
Effects of Genetic Deficiencies in UPR Components on DENV-2 Viral Output
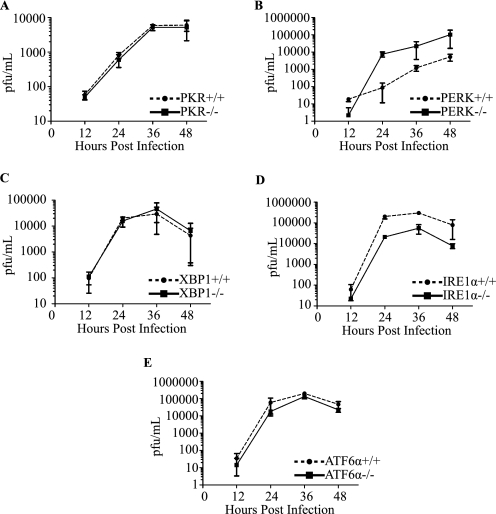
We have demonstrated that DENV-2 infection leads to the induction of proximal transducers and downstream targets of the UPR without leading to apoptosis. Although several reports have demonstrated that flavivirus infection activates the UPR (18, 19), the effect of genetically knocking out individual components of the UPR on DENV-2 production has not been addressed. We began by testing components that affect the ISR arm of the UPR. For this, we obtained congenic PKR+/+ or PKR−/− MEFs and assessed viral titers over a 48-h course of infection. Induction of PKR by the dsRNA replication intermediates of DENV could lead to an induction of eIF2α-P and attenuation of protein synthesis, resulting in reduction or elimination of viral replication. However, we did not observe any effect on DENV-2 growth kinetics in PKR−/− compared with PKR+/+ cells, consistent with our previous observations (Fig. 6A and data not shown) (16, 19).
FIGURE 6.
Effects of UPR mutants on DENV-2 virus production. Congenic MEF cells deficient in PKR (A), PERK (B), IRE1α (C), XBP1 (D), and ATF6α (E) were infected for the indicated times, and viral production was measured by plaque assay as previously described (see “Experimental Procedures”). Error bars indicate mean ± S.D.; n = 3.
Next, we examined the role of PERK in controlling DENV infection. Using congenic MEF cells that were PERK+/+ or PERK−/−, we assessed viral titers as above. PERK−/− cells reproducibly displayed 1–2 logs higher viral titers as compared with their PERK+/+ counterparts (Fig. 6B). These data indicate that PERK plays an antiviral role in regulating the synthesis of DENV-2 viral proteins, and that the loss of PERK results in an increase in virion production. This result is consistent with our previous observation that DENV-2-infected cells are able to suppress Tg-induced PERK-mediated eIF2α-P and further suggest that PERK is the kinase responsible for phosphorylation of eIF2α early in DENV-2 infection.
Failure to suppress ER stress early on leads to activation of the IRE1-XBP1 and ATF6 pathways. Given the activation kinetics of the IRE1-XBP1 and ATF6 pathways in 2fTGH cells, we first tested the effect of mutations in the IRE1-XBP1 pathway on DENV-2 viral progeny output. Using both IRE1α−/− and XBP−/− congenic MEF cell lines, we hypothesized that mutations in either component of the IRE-XBP1 pathway would lead to a decrease in viral progeny output given its role in inducing GRP78 (Fig. 4A). IRE1α−/− cells reproducibly displayed 10-fold lower viral titers as compared with their IRE1α+/+ counterparts (Fig. 6C). However, when XBP1+/+ and XBP1−/− MEF cells were infected with DENV-2, we observed no difference in viral progeny output (Fig. 6D), suggesting redundancy in this part of the UPR pathway. Last, we tested the ATF6 signaling component of the UPR to determine whether mutations in this pathway had any adverse effects on DENV-2 viral production. Congenic ATF6α+/+ or ATF6α−/− MEF cells were infected with DENV-2, supernatants were collected, and viral titers were determined over a 48-h course of infection. No differences were observed in viral titers in ATF6α+/+ cells compared with ATF6α−/− cells (Fig. 6E).
DISCUSSION
In this study, we investigated the activation and modulation of the three arms of the UPR during DENV-2 infection. We demonstrate that induction of the UPR in DENV-2-infected 2fTGH cells occurs in a time-dependent manner that does not lead to the induction of apoptosis. Very early during DENV-2 infection, we observed a transient up-regulation in the phosphorylation of eIF2α, indicating the activation and suppression of the ISR pathway. We found that PKR, a component of the ISR, did not play a role in controlling DENV-2 infection, in agreement with previous reports (23). This is likely due to exclusion of PKR from DENV replication sites, where dsRNA intermediates are shielded in virally induced ER vesicle compartments (35, 36). Instead, we showed that PERK, another component of the ISR, plays an antiviral role, as viral titers are increased in PERK−/− cells. Through the use of immunoblot analysis and metabolic labeling, we demonstrated that DENV-2-infected cells are able to suppress PERK-mediated eIF2α phosphorylation, suggesting that PERK is responsible for phosphorylation of eIF2α early in infection.
The induction and subsequent suppression of PERK-mediated eIF2α phosphorylation during DENV-2 infection are consistent with previous reports regarding HCV, also a member of the Flaviviridae family. HCV encodes the E2 protein, which binds to PERK and inhibits PERK-mediated eIF2α phosphorylation by acting as a pseudosubstrate (15). We postulate that DENV may encode a viral protein that functions similarly to the E2 protein of HCV, as we do not detect activation of downstream components of the PERK pathway early during infection. The involvement of DENV proteins is supported by the fact that suppression of PERK-mediated eIF2α phosphorylation is lost when viral protein synthesis is inhibited. Although we do show that DENV-2 infection can also inhibit GCN2-mediated eIF2α phosphorylation, we do not believe that the conditions needed to induce this pathway (i.e. amino acid starvation) are present in a human DENV infection, nor did we observe induction of GCN2 phosphorylation after DENV infection in vitro. Overall, our results indicate that PERK but not PKR is an important component of the UPR involved in controlling viral infection.
Additionally, we find that mid-course during DENV infection (24–36 h), the IRE1-XBP1 pathway is activated, as demonstrated by the presence of the sXbp1 mRNA and nuclear localization of XBP1, in agreement with previous reports for Japanese encephalitis virus and DENV (17). Activation of the IRE1-XBP1 pathway occurs during the phase of the viral life cycle in which protein synthesis and viral particle formation are taking place, causing both ER stress and phospholipid depletion. The up-regulation of downstream genes that reduce ER stress, such as GRP78 (Figs. 4C and 5A), is consistent with previously published reports for bovine viral diarrhea virus (16), WNV (19), and Japanese encephalitis virus (32); however, contrary to these reports, we do not observe changes in expression levels of GRP94 or PDI proteins, both of which are also involved in maintaining ER homeostasis.
Another target of the IRE1-XBP1 pathway is CHOP, a transcriptional factor that can activate both pro-survival and pro-apoptotic proteins (11–13, 37). One of these pro-survival proteins is GADD34, a phosphatase regulatory subunit that forms a complex with PP1c and targets dephosphorylation of eIF2α-P, ensuring protein synthesis during later phases of the ISR (2, 31). We find that activation of the IRE1-XBP1 pathway correlates with expression of CHOP and GADD34 and that at these time points we detect similar levels of eIF2α-P in DENV-2-infected cells and time-matched mock infected controls. This contrasts with previous reports, where eIF2α-P was detected late in DENV and WNV infection (18, 19). These differences with respect to DENV infection might be due in part to the mock infected controls chosen for comparison. In the previous report, only one mock infected control at a single time point was used for comparison to DENV-infected cells at multiple time points (18), whereas we included a mock infected control at every time point to account for levels of eIF2α-P over time. For example, we found that a mock infected control plated for 12 h displayed a lower level of eIF2α-P than the mock infected controls at 24 and 48 h. Alternatively, the differences we observe compared with DENV (18) and WNV (19) could be attributed to the different cell types used. Furthermore, infection with bovine viral diarrhea virus and WNV has been reported to induce CHOP expression late in infection, leading to reduced levels of Bcl-2 and cleavage of PARP. However, we found that the induction of CHOP during DENV-2 infection of 2fTGH cells does not lead to decreased expression of Bcl-2 or proteolytic cleavage of pro-caspase-9, pro-caspase-3, or PARP. The observation that late eIF2α phosphorylation or induction of apoptotic markers late in DENV-2 infection are not present in our system may be due to the increased expression of GRP78 and GADD34, which prevents CHOP-mediated induction of the apoptotic signaling cascade and is consistent with previous reports (31, 37, 38).
A distinctive morphological characteristic of flavivirus-infected cells is the proliferation and hypertrophy of the ER (1). These changes to the ER can lead to activation of IRE1-XBP1 and ATF6 pathways to expand the ER and also to compensate for phospholipid depletion during viral particle production (5, 39). Although we did not directly examine endomembrane proliferation or phospholipid biosynthesis after DENV infection, we tested whether deletion of IRE1 or XBP1 had any effect on viral production. We found that XBP1−/− and XBP1+/+ MEF cells yielded similar numbers of viral progeny, even though earlier experiments demonstrated XBP1 activation (Fig. 4). These results suggest that redundant pathways might compensate for mutations in this arm of the UPR, as previously reported for WNV (19). When IREα−/− MEF cells were infected with DENV-2, we found viral titers to be 10-fold lower than in congenic IREα+/+ MEFs. Given that IRE1 has two functions, one as an endoribonuclease involved in the IRE1-XBP1 pathway and a second function as a kinase involved in the IRE1-JNK signaling pathway (40), it is possible that signaling through this latter pathway may be important during DENV infection, with IRE1 coupling ER stress with the activation of JNK, a mitogen-activated protein kinase (MAPK). Previous reports have demonstrated the activation of two MAPKs, p38 and ERK1/2, in DENV-infected endothelial cells, leading to the up-regulation of protease-activated receptor type-1 and tissue factor receptor via phosphorylation and suggesting an involvement in DENV pathogenesis (41). Interestingly, a recent report showed a link between cholesterol and JNK signaling and demonstrated its importance for DENV replication in infected macrophages (42). In addition, several studies on WNV have reported the cellular redistribution of cholesterol and its requirement for viral entry and replication (43, 44). Finally, using DENV subgenomic luciferase replicon-expressing cell lines, treatment with various anti-cholesterol pharmacological drugs were shown to affect replication (45). These findings, along with those presented in this study, lead us to hypothesize that signaling through the ER via IRE1-JNK pathway might be important during the DENV life cycle.
We have demonstrated that activation of the ATF6 pathway is transient and occurs late in DENV-2 infection. The observation that the ATF6 pathway is activated is in agreement with previous studies for WNV and DENV; however, here we report that this activation is transient and is preceded by activation of the IRE1-XBP1 pathway. Umareddy et al. (18) reported that induction of the ATF6 pathway led to up-regulation of uXbp1 mRNA in DENV-infected cells; however, these studies did not address the contribution of XBP1 to its own up-regulation (46). We also speculated that activation of ATF6 up-regulates the synthesis of uXbp1, thus restoring the depleted message and allowing for a sustained induction of the UPR as long as IRE1 remains active. However, when we infected ATF6α−/− MEFs with DENV, we found no difference in viral titers, suggesting that ATF6 is dispensable or that XBP1 transactivation of itself is sufficient.
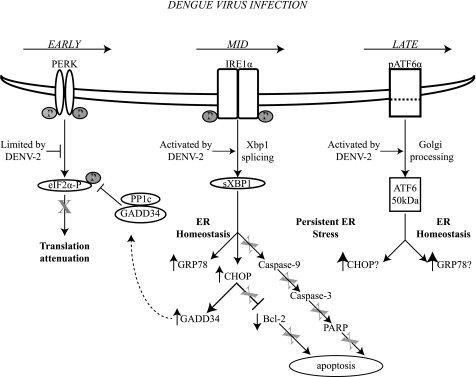
Here we have demonstrated that DENV infection of 2fTGH cells leads to sequential activation of the individual branches of the UPR, allowing DENV to adapt the cell to the stress of infection and preventing the activation of pro-apoptotic events. Our proposed model (Fig. 7) shows that translation of the viral genome early in infection triggers the ISR, specifically activating the PERK pathway. PERK activation and subsequent eIF2α phosphorylation exposure is short-lived, as DENV soon suppresses this pathway. This transient activation, however, allows for resetting of the cellular translational program, liberating ribosomes and translational factors that accumulate as free subunits, of which the viral RNA can take advantage (2). Accumulation of viral proteins and the formation of mature viral particles then trigger the IRE-XBP1 pathway, which serves various functions. One is to induce the expression of the chaperone protein GRP78 to allow translocation of nascent viral polyproteins into the lumen of the ER, and another is to activate the downstream transcription factor CHOP that subsequently triggers the expression of GADD34. Expression of GADD34 targets PP1c to dephosphorylate eIF2α-P, allowing for the continued synthesis of viral and cellular proteins. This is further supported by experiments performed using the drug salubrinal, a specific GADD34/PP1c inhibitor, which demonstrated that salubrinal treatment of A549 cells infected with DENV2 resulted in decreased viral output throughout the course of infection (18). Finally, overexpression of GRP78 prevents CHOP-mediated induction of the apoptotic signaling cascade, consistent with previous reports. Further accumulation of viral proteins and the proliferation of the ER induce ATF6. Activation of the ATF6 pathway enhances the UPR response, presumably by activating the transcription of uXbp1 and allowing for a sustained amount of uXbp1 precursor to be spliced into its active form.
FIGURE 7.
Model of DENV modulation of the unfolded protein response. Schematic diagram illustrating the effects of DENV-2 infection on the UPR according to the results presented in this study, demonstrating the time-dependent activation of the three branches of the UPR and showing that induction of the UPR does not lead to the induction of apoptosis. Arrows indicate activation or induction by DENV-2, and T-lines and X indicate inhibition as a result of DENV infection.
By adapting its viral life cycle to the UPR, DENV is able to manipulate the sequence of events to activate and suppress the three different branches of the UPR in a time-dependent manner to prevent premature apoptosis and prolong the viral life cycle. Our results also have broader implications with respect to understanding how the cell regulates activation of the individual branches of the UPR in response to distinct environmental stressors. Last, our model system will allow further investigation of the role that UPR plays in lipid biosynthesis and ER expansion during a DENV infection.
Acknowledgments
We thank George R. Stark for 2fTGH human fibrosarcoma cells; Bryan R. G. Williams for PKR+/+ and PKR−/− MEFs; David Ron for PERK+/+ and PERK−/− MEFs; and Randal Kaufman for IRE1α+/+, IRE1α−/−, XBP1+/+, XBP1−/−, ATF6α+/+, and ATF6α−/− MEFs. We are grateful to Jennifer Kyle, Sondra and Milt Schlesinger, Alex Costa, Peter Friebe, and Suman Paranjape for helpful discussion and editorial comments in the preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AI052324 (to E. H.) and a supplemental National Institutes of Health grant (to J. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
P. R. Beatty and E. Harris, unpublished results.
- DENV-2
- dengue virus-2
- UPR
- unfolded protein response
- LSCM
- laser scanning confocal microscopy
- m.o.i.
- multiplicity of infection
- ISR
- integrated stress response
- IRE1
- inositol-requiring protein-1
- ATF6
- activating transcription factor-6
- PKR
- protein kinase R
- PERK
- PKR-like endoplasmic reticulum kinase
- GCN2
- general control non-derepressible-2 kinase
- PDI
- protein-disulfide isomerase
- GRP78-BiP
- glucose-regulated protein-78/binding immunoglobulin protein
- dsRNA
- double-stranded RNA
- ER
- endoplasmic reticulum
- CHOP
- CCAT/enhancer-binding protein
- HCV
- hepatitis C virus
- MEF
- mouse embryonic fibroblast
- p.i.
- post-infection
- CHX
- cycloheximide
- Tg
- thapsigargin
- PARP
- poly(ADP-ribose) polymerase
- PP1
- protein phosphatase 1
- WNV
- West Nile virus.
REFERENCES
- 1. Lindenbach B. D., Rice C. M., Thiel H. J. (2007) in Fields Virology (Knipe D. M., Howley P. M. eds) 5th Ed., pp. 1101–1152, Lippincott, Williams & Wilkins, Philadelphia [Google Scholar]
- 2. Ron D., Harding H. P. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonemberg N., Hershey J. W. B. eds) pp. 345–368, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 3. Schröder M., Kaufman R. J. (2005) Mutat. Res. 569, 29–63 [DOI] [PubMed] [Google Scholar]
- 4. Shen J., Prywes R. (2005) Methods 35, 382–389 [DOI] [PubMed] [Google Scholar]
- 5. Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 6. Dever T. E., Dar A. C., Sicheri F. (2007) in Translational Control in Biology and Medicine (Mathews M. B., Sonemberg N., Hershey J. W. B. eds) pp. 319–344, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 7. Malhotra J. D., Kaufman R. J. (2007) Semin. Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. (2000) Nat. Cell Biol. 2, 326–332 [DOI] [PubMed] [Google Scholar]
- 9. Shen J., Chen X., Hendershot L., Prywes R. (2002) Dev. Cell 3, 99–111 [DOI] [PubMed] [Google Scholar]
- 10. Lee A. S. (2005) Methods 35, 373–381 [DOI] [PubMed] [Google Scholar]
- 11. McCullough K. D., Martindale J. L., Klotz L. O., Aw T. Y., Holbrook N. J. (2001) Mol. Cell. Biol. 21, 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oyadomari S., Mori M. (2004) Cell Death Differ. 11, 381–389 [DOI] [PubMed] [Google Scholar]
- 13. Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. (2006) PLoS Biol. 4, e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hetz C. A. (2007) Antioxid. Redox Signal. 9, 2345–2355 [DOI] [PubMed] [Google Scholar]
- 15. Pavio N., Romano P. R., Graczyk T. M., Feinstone S. M., Taylor D. R. (2003) J. Virol. 77, 3578–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jordan R., Wang L., Graczyk T. M., Block T. M., Romano P. R. (2002) J. Virol. 76, 9588–9599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu C. Y., Hsu Y. W., Liao C. L., Lin Y. L. (2006) J. Virol. 80, 11868–11880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Umareddy I., Pluquet O., Wang Q. Y., Vasudevan S. G., Chevet E., Gu F. (2007) Virol. J. 4, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Medigeshi G. R., Lancaster A. M., Hirsch A. J., Briese T., Lipkin W. I., Defilippis V., Früh K., Mason P. W., Nikolich-Zugich J., Nelson J. A. (2007) J. Virol. 81, 10849–10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pellegrini S., John J., Shearer M., Kerr I. M., Stark G. R. (1989) Mol. Cell. Biol. 9, 4605–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang Y. L., Reis L. F., Pavlovic J., Aguzzi A., Schäfer R., Kumar A., Williams B. R., Aguet M., Weissmann C. (1995) EMBO J. 14, 6095–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berlanga J. J., Ventoso I., Harding H. P., Deng J., Ron D., Sonenberg N., Carrasco L., de Haro C. (2006) EMBO J. 25, 1730–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Diamond M. S., Harris E. (2001) Virology 289, 297–311 [DOI] [PubMed] [Google Scholar]
- 24. Krishnamoorthy J., Mounir Z., Raven J. F., Koromilas A. E. (2008) Cell Cycle 7, 2346–2351 [DOI] [PubMed] [Google Scholar]
- 25. Harding H. P., Zhang Y., Ron D. (1999) Nature 397, 271–274 [DOI] [PubMed] [Google Scholar]
- 26. Wek R. C., Cavener D. R. (2007) Antioxid. Redox Signal. 9, 2357–2371 [DOI] [PubMed] [Google Scholar]
- 27. Shi Y., Vattem K. M., Sood R., An J., Liang J., Stramm L., Wek R. C. (1998) Mol. Cell. Biol. 18, 7499–7509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 29. Yoshida H., Haze K., Yanagi H., Yura T., Mori K. (1998) J. Biol. Chem. 273, 33741–33749 [DOI] [PubMed] [Google Scholar]
- 30. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 31. Novoa I., Zeng H., Harding H. P., Ron D. (2001) J. Cell Biol. 153, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su H. L., Liao C. L., Lin Y. L. (2002) J. Virol. 76, 4162–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferri K. F., Kroemer G. (2001) Nat. Cell Biol. 3, E255–E263 [DOI] [PubMed] [Google Scholar]
- 34. Rao R. V., Castro-Obregon S., Frankowski H., Schuler M., Stoka V., del Rio G., Bredesen D. E., Ellerby H. M. (2002) J. Biol. Chem. 277, 21836–21842 [DOI] [PubMed] [Google Scholar]
- 35. Uchil P. D., Satchidanandam V. (2003) J. Biol. Chem. 278, 24388–24398 [DOI] [PubMed] [Google Scholar]
- 36. Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C. K., Walther P., Fuller S. D., Antony C., Krijnse-Locker J., Bartenschlager R. (2009) Cell Host Microbe 5, 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X. Z., Lawson B., Brewer J. W., Zinszner H., Sanjay A., Mi L. J., Boorstein R., Kreibich G., Hendershot L. M., Ron D. (1996) Mol. Cell. Biol. 16, 4273–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rao R. V., Hermel E., Castro-Obregon S., del Rio G., Ellerby L. M., Ellerby H. M., Bredesen D. E. (2001) J. Biol. Chem. 276, 33869–33874 [DOI] [PubMed] [Google Scholar]
- 39. Bommiasamy H., Back S. H., Fagone P., Lee K., Meshinchi S., Vink E., Sriburi R., Frank M., Jackowski S., Kaufman R. J., Brewer J. W. (2009) J. Cell Sci. 122, 1626–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P., Ron D. (2000) Science 287, 664–666 [DOI] [PubMed] [Google Scholar]
- 41. Huerta-Zepeda A., Cabello-Gutiérrez C., Cime-Castillo J., Monroy-Martínez V., Manjarrez-Zavala M. E., Gutiérrez-Rodríguez M., Izaguirre R., Ruiz-Ordaz B. H. (2008) Thromb. Haemost. 99, 936–943 [DOI] [PubMed] [Google Scholar]
- 42. Ceballos-Olvera I., Chávez-Salinas S., Medina F., Ludert J. E., del Angel R. M. (2010) Virology 396, 30–36 [DOI] [PubMed] [Google Scholar]
- 43. Mackenzie J. M., Khromykh A. A., Parton R. G. (2007) Cell Host Microbe 2, 229–239 [DOI] [PubMed] [Google Scholar]
- 44. Medigeshi G. R., Hirsch A. J., Streblow D. N., Nikolich-Zugich J., Nelson J. A. (2008) J. Virol. 82, 5212–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rothwell C., Lebreton A., Young Ng C., Lim J. Y., Liu W., Vasudevan S., Labow M., Gu F., Gaither L. A. (2009) Virology 389, 8–19 [DOI] [PubMed] [Google Scholar]
- 46. Yoshida H., Matsui T., Hosokawa N., Kaufman R. J., Nagata K., Mori K. (2003) Dev. Cell 4, 265–271 [DOI] [PubMed] [Google Scholar]