Abstract
Nephronophthisis is the most common genetic cause of end-stage renal failure during childhood and adolescence. Genetic studies have identified disease-causing mutations in at least 11 different genes (NPHP1–11), but the function of the corresponding nephrocystin proteins remains poorly understood. The two evolutionarily conserved proteins nephrocystin-1 (NPHP1) and nephrocystin-4 (NPHP4) interact and localize to cilia in kidney, retina, and brain characterizing nephronophthisis and associated pathologies as result of a ciliopathy. Here we show that NPHP4, but not truncating patient mutations, negatively regulates tyrosine phosphorylation of NPHP1. NPHP4 counteracts Pyk2-mediated phosphorylation of three defined tyrosine residues of NPHP1 thereby controlling binding of NPHP1 to the trans-Golgi sorting protein PACS-1. Knockdown of NPHP4 resulted in an accumulation of NPHP1 in trans-Golgi vesicles of ciliated retinal epithelial cells. These data strongly suggest that NPHP4 acts upstream of NPHP1 in a common pathway and support the concept of a role for nephrocystin proteins in intracellular vesicular transport.
Keywords: Centrosome, Epithelium, Golgi, Protein-tyrosine Kinase (Tyrosine Kinase), Vesicles, Pyk2, Cilium, Cystic Kidney Disease, Nephrocystin
Introduction
Nephronophthisis (NPH)3 is a genetically heterogeneous renal cystic disease with an autosomal recessive mode of transmission (for review, see Refs. 1, 2). NPH is the most common cause of hereditary renal failure in children and young adults. Mutations in 11 different genes have been identified over the last years (1–3). Most mutations affect the NPHP1 gene, which encodes for nephrocystin-1 (NPHP1) (4, 5). NPHP1 interacts with the gene products of NPHP2 (6), NPHP3 (7), and NPHP4 (8), three other genes involved in the development of NPH. Mutations in NPHP2 and NPHP3 present differently from NPHP1 and NPHP4 mutations, e.g. in age of disease onset. It has recently been shown that NPHP1 and NPHP4 localize to primary cilia of renal tubular epithelial cells (6, 9, 10).
NPHP1 and NPHP4 are conserved in the nematode Caenorhabditis elegans (11–13), where the homologs of NPHP1 and NPHP4 are localized to the transition zone of sensory cilia of ciliated neurons. nph-1 and nph-4 mutant worms show subtle structural ciliary defects (14). However, double mutants display much stronger functional ciliary impairment (11, 12). Moreover, NPH-4 has been shown to be required for the correct localization of NPH-1 in these neurons in C. elegans (12). In mammalian cells NPHP1 and NPHP4 interact with the 116-kDa cytoplasmic protein-tyrosine kinase Pyk2 (10, 15), which is activated by a variety of stimuli that increase intracellular calcium (16–19). Pyk2 appears to play an important role in the integration of environmental stimuli and the polarized organization of cytoskeletal components in cell migration (20). Interestingly, the interaction with NPHP1 increases Pyk2 activity (15).
Here, we report that NPHP4 negatively regulates Pyk2-induced tyrosine phosphorylation of NPHP1 by controlling the NPHP1/Pyk2 interaction. Phosphorylation at three defined tyrosine residues increases binding of NPHP1 to the trans-Golgi sorting protein PACS-1 (phosphofurin acidic cluster sorting protein 1). By counteracting this process NPHP4 controls subcellular localization of NPHP1 in human ciliated epithelial cells. Several patient mutations of NPHP4 lost their ability to affect tyrosine phosphorylation of NPHP1 which supports a critical function for NPHP4 and Pyk2 in controlling NPHP1 and the NPH protein complex.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
NPHP1 and Pyk2 constructs have previously been described (15, 21). Full-length NPHP3 and NPHP4 were cloned from a human kidney cDNA library. HA-tagged Pyk2 constructs and Src cDNA were kindly provided by Dr. I. Dikic (University of Frankfurt, Germany) and Dr. J. Brugge (Harvard Medical School, Boston). Site-directed mutagenesis was performed using a modified QuikChange Site-Directed Mutagenesis kit (Stratagene). All plasmids were verified by automated DNA sequencing. Antibodies were obtained from Sigma (anti-FLAG, anti-acetylated tubulin), Santa Cruz (anti-myc, anti-HA, anti-src, pY99), BD Transduction (anti-Pyk2, anti-PY 4G10), Serotec (anti-V5), and Abcam (anti-Pericentrin).
Generation and Purification of NPHP1-specific Monoclonal Antibodies
Bacterially expressed and affinity-purified His-tagged NPHP112–205 was used to immunize mice following a standard immunization protocol (9). Fusions resulted in the generation of more than 30 specific monoclonal antibodies producing hybridome clones. Antibodies were screened with immunofluorescent stainings, immunoblotting, and immunoprecipitation. Protein G columns were used to concentrate the NPHP1-specific antibodies. Specificity was verified again by using bacterially expressed recombinant proteins and cell lysates from transfected cells.
Cell Culture and Transfections
HEK293T cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). For transfection experiments, cells were grown until 60–80% confluence and transfected with plasmid DNA using a modified calcium phosphate method as described previously (15). hTERT-RPE1 were cultured in a 1:1 mixture of DMEM and Ham's F12 medium supplemented with 10% FBS, 2 mm l-glutamine, and sodium bicarbonate (2.6 g/liter). Cilia formation was induced by serum depletion over 48 h. For siRNA transfection experiments cells were grown until 60–80% confluence and transfected with siRNA to a final concentration of 20 nm using Oligofectamine (Invitrogen). Cy3-labeled control siRNA was transfected in parallel and served as transfection control. siRNAs targeting NPHP4 were directed against the following sequences: CTCGTTATCGCTGTTGCTCAA (siRNA 1), CAGCCGCTTTGTCCATCTCAA (siRNA 2), AAGCAACGAGATGGTGCTACA (siRNA 3), and CAGATCTCGGGTCATCTCAAA (siRNA 4). Control siRNA strands were purchased from Biomers and had the sequences 5′-GUGACACGUUCGGAGAATTAC-3′ and 5′-AATTCTCCGAACGUGUCACGU-3′. For the transfection of cDNA into hTERT-RPE1, GeneJuice (Merck) was used. For the inhibition of tyrosine phosphatases peroxovanadate was prepared as described (22). Cultured cells were incubated for 15 min with peroxovanadate (final concentration 0.5 mm).
Immunoprecipitation
Immunoprecipitations were performed as described (15). Briefly, HEK293T cells were transiently transfected by the calcium phosphate method. After incubation for 24 h, cells were washed twice and lysed in a 1% Triton X-100 lysis buffer. After centrifugation (15,000 × g, 15 min, 4 °C) and ultracentrifugation (100,000 × g, 30 min, 4 °C) cell lysates containing equal amounts of total protein were incubated for 1 h at 4 °C with the appropriate antibody followed by incubation with 30 μl of protein G-Sepharose beads (GE Healthcare) for ∼1 h. For FLAG tags the lysates were incubated at 4 °C with anti-FLAG (M2) agarose beads (Sigma) for ∼1 h. For evaluation of tyrosine phosphorylation the ultracentrifuge step was omitted, and incubation of the lysate with antibodies and beads was reduced to 30 min. The beads were washed extensively with lysis buffer, and bound proteins were resolved by 10% SDS-PAGE and visualized with enhanced chemiluminescence after incubation of the blots with the respective antibodies. Experiments were repeated at least three times with identical results.
32P Labeling and Two-dimensional Mapping of Phosphorylation Sites
32P labeling of cells (1–2 mCi/ml for 6–8 h), solubilization, and immunoprecipitation of NPHP1 were done as described previously (23, 24). All experiments were performed in 6-well plates. After separation of the immunoprecipitated proteins by 10% SDS-PAGE, proteins were transferred onto nitrocellulose membranes. Radiolabeled proteins were detected by PhosphorImager (BAS2000, Fuji) analysis, and tryptic digests were performed as described previously with minor modifications (23). Briefly, membrane pieces containing the 32P-labeled proteins were cut out and blocked with 0.5% polyvinylpyrrolidone 40 in 0.6% acetic acid, for 30 min at 37 °C. Following extensive washes with water, membrane-bound NPHP1 was cleaved in situ with sequencing grade trypsin in 50 mm (NH4)HCO3 for 12 h at 37 °C. Released tryptic peptides were vacuum-dried and oxidized with performic acid for 1 h on ice. Reactions were stopped by dilution with 20% (v/v) ammonia solution. Thereafter, samples were frozen and vacuum-dried, and a second digest was performed for 12 h at 37 °C. Following vacuum drying, samples were dissolved in electrophoresis buffer (formic acid:acetic acid:water, 46:156:1790 (v/v/v)), and phosphopeptides were separated by electrophoresis on cellulose thin layer plates in a first dimension (2000 V, 40 min, electrophoresis buffer) and ascending chromatography in a second dimension (15 h, isobutyric acid, 1-butyl alcohol, pyridine, acetic acid, water, 1250:38:96:58:558 (v/v/v/v/v)). Phosphopeptides were detected by PhosphorImager analysis and eluted from the cellulose matrix with 20% (v/v) acetonitrile in a sonicated water bath for 15 min. Part of the extract (25–100 cpm) was hydrolyzed with 6 m HCl for 1 h at 110 °C and subjected to a phosphoamino acid analysis. The second fraction (50–500 cpm) was sequenced by Edman degradation using a solid phase sequencer (ABI 477). Twenty sequencing cycles were collected, dried, and analyzed for their content of 32P radioactivity using a PhosphorImager. Data obtained from Edman degradation were used to predict phosphorylation sites. The prediction was verified by in vitro mutagenesis of corresponding phosphoacceptor sites.
Immunofluorescence Staining of hTERT-RPE1
hTERT-RPE1 were seeded on coverslips and cultured as described above. Cilia formation was induced by serum depletion over 48 h (25). After washing with ice-cold PBS, cells were fixed using methanol at −20 °C for 8 min. Cells were washed three times with PBS, and double immunofluorescence staining was performed sequentially with the antibodies as indicated in the absence of Triton X-100. After washing, slides were incubated with the secondary antibody, Cy2-labeled anti-mouse, and Cy3-labeled anti-rabbit IgG, washed again, mounted in ProLong Gold antifade reagent with DAPI (Invitrogen), and subjected to immunofluorescence microscopy with a microscope equipped with the apotome system (Axiovert 200; Carl Zeiss MicroImaging) and the software Axiovision (Carl Zeiss MicroImaging). The appropriate controls were performed without the first and/or second primary antibodies.
RESULTS
NPHP4 Modulates Pyk2-induced Tyrosine Phosphorylation of NPHP1
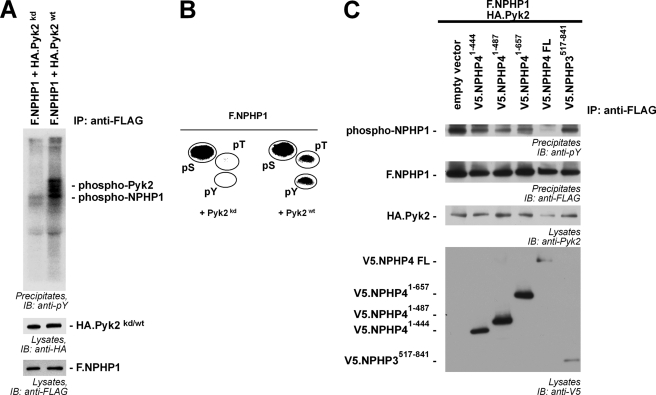
To test whether Pyk2 not only interacted with but also phosphorylated NPHP1, we co-expressed FLAG-tagged NPHP1 (F.NPHP1) and wild-type Pyk2 (Pyk2wt) or a kinase-dead mutant (Pyk2kd) in transiently transfected HEK293T cells. After serum starvation of the cells for 4 h, the cells were lysed, and NPHP1 was precipitated with anti-FLAG antibody. As described previously, hyperphosphorylated Pyk2 interacted with NPHP1 (15). Moreover, wild-type Pyk2 but not the kinase-dead mutant of the kinase induced strong tyrosine phosphorylation of NPHP1 (Fig. 1A). To analyze Pyk2-mediated phosphorylation of NPHP1 in more detail, we radioactively labeled NPHP1-expressing cells with 32P in vivo. NPHP1 was precipitated, and the proteins of the precipitate were separated by SDS-PAGE and blotted on nitrocellulose membranes. Precipitated NPHP1 was cut out of the membrane, extracted, and phosphoamino acid analysis was performed revealing a strong basal level of serine phosphorylation. In addition to this basal level of phosphorylation, Pyk2-dependent phosphorylation of threonine and tyrosine residues could be detected (Fig. 1B). Surprisingly, co-expression of NPHP4 inhibited the Pyk2-induced phosphorylation of NPHP1 (Fig. 1C), whereas NPHP4 itself was only weakly tyrosine-phosphorylated in presence of Pyk2 (supplemental Fig. 1). To assess the pathophysiological relevance of this finding further we included three known NPHP4 patient mutations in the analysis (26). Disease-causing NPHP4 mutations showed a strongly reduced ability to affect NPHP1 phosphorylation, whereas a truncation of NPHP3, another NPH disease gene product, was nearly without effect (Fig. 1C).
FIGURE 1.
NPHP4 controls Pyk2-induced tyrosine phosphorylation of NPHP1. A, Pyk2 induces tyrosine phosphorylation of NPHP1. F.NPHP1 was precipitated (IP), and tyrosine phosphorylation of precipitated proteins was detected with an anti-phosphotyrosine antibody (pY99, upper panel). Strong tyrosine phosphorylation of precipitated NPHP1 and co-precipitating Pyk2 was dependent on Pyk2 kinase activity (kd, kinase-dead mutant). IB, immunoblotting. B, phosphoamino acid analysis of precipitated NPHP1 after in vivo labeling of HEK293T cells with 32P. Phosphorylated amino acids were identified by co-migration with standard nonradioactive phosphoamino acids, ninhydrin staining (indicated by circles), and autoradiography. C, wild-type NPHP4 (WT NPHP4) but not various patient mutations abolishes Pyk2-induced tyrosine phosphorylation of NPHP1. F.NPHP1 was precipitated, and tyrosine phosphorylation was evaluated with an anti-phosphotyrosine antibody. All NPHP4 constructs reduced tyrosine phosphorylation of NPHP1 compared with the empty vector control, but not to the degree of WT NPHP4. NPHP3517–841 served as control protein. All experiments were performed at least three times with identical results.
NPHP4 Interferes with the Interaction of Pyk2 and NPHP1
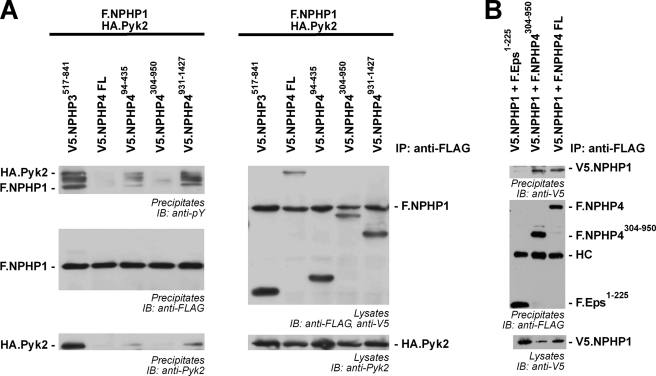
Next, we addressed the mechanism by which NPHP4 reduces Pyk2-induced tyrosine phosphorylation of NPHP1. V5-tagged versions of NPHP4 were co-expressed with F.NPHP1 and Pyk2, and F.NPHP1 was precipitated and analyzed for tyrosine phosphorylation. Although all truncations of NPHP4 weakened the phosphorylation of NPHP1 and attenuated co-precipitation of Pyk2, full-length NPHP4 and NPHP4304–950 abolished tyrosine phosphorylation and were both sufficient to abrogate co-precipitation of Pyk2. These findings suggest that NPHP4 interferes with the binding of Pyk2 to its substrate NPHP1, thereby decreasing NPHP1-induced activation of Pyk2 and phosphorylation of NPHP1 (Fig. 2A). To investigate whether this NPHP4 truncation interacts with NPHP1, F.NPHP4 or F.NPHP4304–950 and V5-tagged NPHP1 were co-expressed and FLAG-tagged proteins were precipitated. NPHP1 co-precipitated with both wild-type NPHP4 and the truncation (Fig. 2B). Interestingly, NPHP4304–950 contains a proline-rich region. The interaction of NPHP1 and Pyk2 has been mapped to the SH3 domain of NPHP1 and the proline-rich region of Pyk2 (15). Therefore, it may be tempting to speculate that the two proline-rich regions of Pyk2 and NPHP4 compete for the binding to NPHP1.
FIGURE 2.
NPHP4304–950 is sufficient to control Pyk2-induced tyrosine phosphorylation of NPHP1 by interfering with the NPHP1/Pyk2 interaction. A, NPHP4304–950 controls tyrosine phosphorylation of NPHP1 and interferes with the NPHP1/Pyk2 interaction. F.NPHP1 was precipitated, and tyrosine phosphorylation of precipitated proteins was detected with an anti-phosphotyrosine antibody in the presence of either a control protein, WT NPHP4, or different NPHP4 truncations. Precipitates were also stained for co-precipitating Pyk2. WT NPHP4 and NPHP4304–950 decrease the tyrosine phosphorylation of NPHP1 (upper panel) and abolish the NPHP1/Pyk2 interaction (lower panel). B, NPHP1 co-precipitates with NPHP4304–950. FLAG-tagged proteins were precipitated, and co-precipitating V5.NPHP1 was evaluated. V5.NPHP1 specifically co-precipitated with both WT NPHP4 and the NPHP4304–950 truncation. The immunoprecipitation experiments were performed four times.
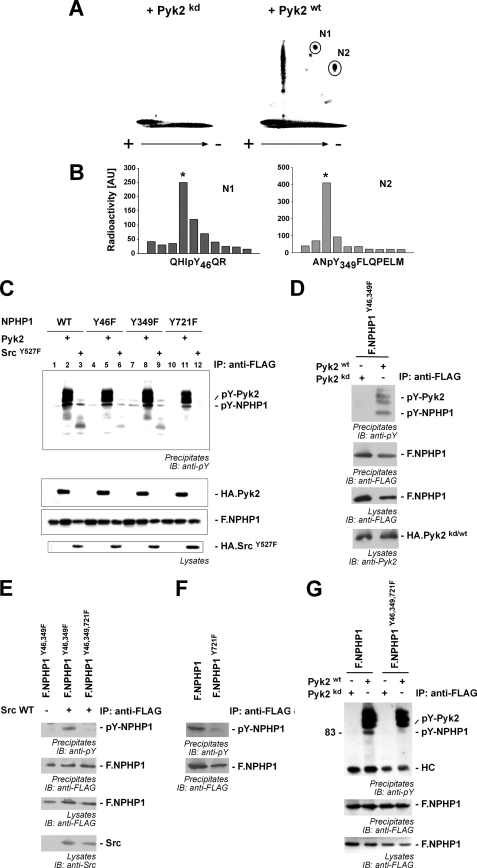
Pyk2 Induces Tyrosine Phosphorylation of NPHP1 at Tyr-46, Tyr-349, and Tyr-721
To identify the phosphorylated tyrosine residues of NPHP1 in vivo, we repeated the 32P labeling experiments and subjected precipitated NPHP1 to a complete tryptic digest. Phosphoamino acid analysis of precipitated NPHP1 revealed several major phosphopeptides (Fig. 3A). Total hydrolysis and phosphoamino acid analysis performed on these spots identified two spots (N1 and N2) as containing exclusively phosphotyrosine, but no phosphorylated serine or threonine residues (data not shown). The major fraction of these peptides was used for solid phase Edman degradation. Comparison with the fragments resulting from in silico tryptic digest identified the peptide fragments, indicating that tyrosines 46 and 349 on NPHP1 were phosphorylated in vivo (Fig. 3B). A third site at position 721 on NPHP1 was predicted to be phosphorylated by Src kinase by Scansite and NetPhos programs. Interestingly, the nonreceptor tyrosine kinase Src has been shown to be activated by Pyk2 kinase (21). Tyrosine-to-phenylalanine mutation of single residues did not demonstrate an effect on Pyk2-induced phosphorylation of NPHP1 (Fig. 3C). Src-induced tyrosine phosphorylation could be prevented by mutating the tyrosine residue at position 721 to phenylalanine (Fig. 3C, lane 12) suggesting that Src kinase phosphorylates NPHP1 at tyrosine 721. This is supported by the finding that Pyk2 induced tyrosine phosphorylation of the NPHP1 Y46F/Y349F double mutant (Fig. 3D). This can be explained by activation of endogenous c-Src kinase by Pyk2 and subsequent phosphorylation of the NPHP1 tyrosine 721. In line with this concept, c-Src induced tyrosine phosphorylation of the NPHP1 Y46F/Y349F double mutant but not of the NPHP1 Y46F/Y349F/Y721F triple mutant (Fig. 3E). In addition, the NPHP1 Y721F mutant showed reduced endogenous tyrosine phosphorylation of NPHP1 (Fig. 3F). The combined mutation of the three tyrosine residues abolished the Pyk2-dependent anti-phosphotyrosine signal (Fig. 3G) completely.
FIGURE 3.
Pyk2 induces NPHP1 phosphorylation of tyrosines 46, 349, and 721. A, transiently transfected HEK293T cells were labeled with 32P and lysed, and F.NPHP1 was immunoprecipitated. Precipitated NPHP1 was separated by SDS-PAGE, transferred to nitrocellulose membranes, and digested in situ with trypsin, and the peptides were separated on TLC plates by high voltage electrophoresis and ascending chromatography. + and − indicate voltage during electrophoresis. Separated phosphopeptides were visualized by autoradiography. Two Pyk2-dependent phosphopeptide fragments could be identified as phosphotyrosine-containing peptides (N1 and N2, marked with circles). B, phosphopeptides N1 and N2 were analyzed by Edman degradation which identified the position of the phosphorylated tyrosine residue in the peptide. C, tyrosine residues 46, 349, and 721 in NPHP1 were mutated to phenylalanine residues, and FLAG-tagged versions of these single mutants were co-expressed with empty vector (lanes 1, 4, 7, and 10), HA-tagged Pyk2 (lanes 2, 5, 8, and 11) or SrcY527F, a constitutively active mutant of c-Src (lanes 3, 6, 9, and 12). NPHP1 was precipitated and analyzed for tyrosine phosphorylation (upper panel). D, combined mutation of tyrosine residues 46 and 349 does not abrogate Pyk2-induced phosphorylation. Double mutant NPHP1 Y46F/Y349F was precipitated (IP), and tyrosine phosphorylation was monitored in presence of wild-type Pyk2 or a kinase-dead mutant. IB, immunoblotting. E, wild-type (WT) c-Src induces phosphorylation of the NPHP1 Y46F/Y349F double mutant, but not of the NPHP1 Y46F/Y349F/Y721F triple mutant. NPHP1 proteins were precipitated, and tyrosine phosphorylation was monitored. F, tyrosine residue 721 is endogenously phosphorylated. HEK293T cells were transfected and incubated for 15 min with peroxovanadate to inhibit tyrosine dephosphorylation. After cell lysis, NPHP1 proteins were precipitated and stained for phosphotyrosine. Mutation of Tyr-721 attenuated tyrosine phosphorylation of NPHP1. G, combined mutation of the mapped tyrosine residues abrogates tyrosine phosphorylation of NPHP1. NPHP1 proteins were precipitated, and tyrosine phosphorylation was monitored, revealing tyrosine phosphorylation of WT NPHP1 but not of the triple mutant. All immunoprecipitation experiments were performed at least three times.
Tyrosine Phosphorylation of NPHP1 Modulates Binding of the Transport Protein PACS-1
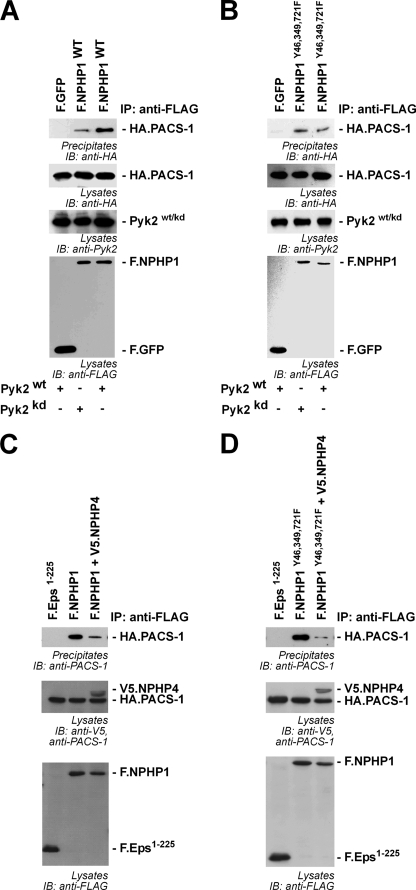
We have previously shown that NPHP1 localizes to cilia of kidney epithelial cells (6). Localization to cilia requires binding of PACS-1 (9). PACS-1 has been identified as a sorting connector, which retrieves membrane-associated proteins to the trans-Golgi Network (27, 28). These processes are regulated through casein kinase 2-dependent protein interactions of cargo proteins with PACS-1 (9), a mechanism that has been confirmed for the ciliary membrane protein CNGB1b (cyclic nucleotide-gated channel β1) very recently (29). To test whether tyrosine phosphorylation of NPHP1 may influence binding of PACS-1, HEK 293T cells were transiently transfected with HA-tagged PACS-1 and FLAG-tagged NPHP1 in the presence of wild-type or a kinase-dead mutant of Pyk2. Co-expression of wild-type Pyk2, but not of the kinase-dead mutant, facilitated the interaction of PACS-1 with NPHP1 (Fig. 4A). Mutation of the Pyk2-dependent tyrosine phosphorylation sites in NPHP1 (Tyr-46, Tyr-349, and Tyr-721) to phenylalanines abrogated the Pyk2-mediated effect on PACS-1 binding, suggesting that this effect was intrinsic to NPHP1 and a result of Pyk2-dependent tyrosine phosphorylation of NPHP1 (Fig. 4B) rather than an indirect effect of Pyk2 overexpression. Based on our findings that NPHP4 reduces Pyk2-induced NPHP1 phosphorylation, we speculated that NPHP4 might also influence the interaction of NPHP1 with PACS-1. Strikingly, overexpression of NPHP4 substantially attenuated the interaction between NPHP1 and PACS-1 (Fig. 4C). Still, this NPHP4 effect was also seen in the NPHP1 Y46F/Y349F/Y721F triple mutant, suggesting that NPHP4 affects NPHP1 in multiple ways (Fig. 4D).
FIGURE 4.
Tyrosine phosphorylation of NPHP1 facilitates interaction with PACS-1. A, FLAG-tagged proteins were precipitated (IP), and co-precipitating HA.PACS-1 was analyzed (upper panels). IB, immunoblotting. The expression of WT Pyk2 enhances the amount of co-precipitating PACS-1 with WT NPHP1 compared with the presence of the kinase-dead (kd) variant of Pyk2. B, this effect was lost after mutation of the three phosphorylated tyrosines to phenylalanines, showing the importance of NPHP1 phosphorylation. C, the presence of NPHP4 significantly weakened the interaction of NPHP1 with PACS-1. D, this was also the case for the NPHP1 Y46F/Y349F/Y721F triple mutant. All experiments were performed at least three times.
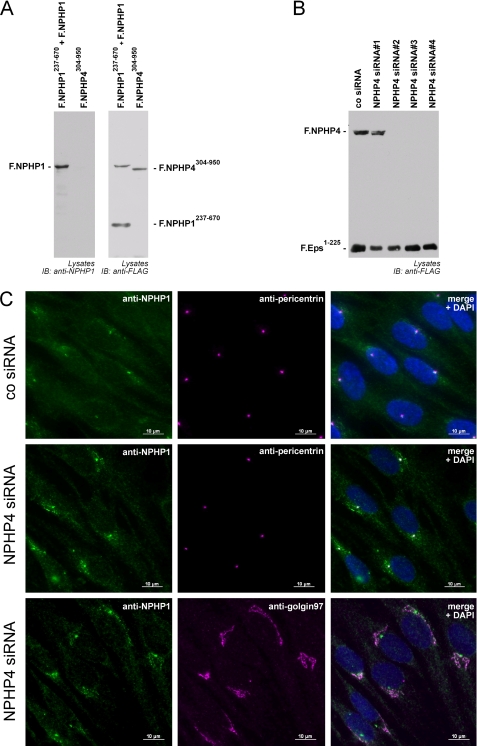
NPHP4 Controls Subcellular Localization of NPHP1
To test for a role of NPHP4 in controlling NPHP1 localization, we generated novel monoclonal antibodies against the N terminus of NPHP1. The monoclonal antibody detected specifically and exclusively NPHP1 but none of the control proteins (Fig. 5A). Several siRNAs targeting human NPHP4 were tested by co-transfection with FLAG-tagged NPHP4 and a FLAG-tagged control protein. Three of four siRNAs showed efficient knockdown activity against NPHP4 (Fig. 5B). To assess a role for NPHP4 in controlling ciliogenesis and/or NPHP1 trafficking, retinal pigment epithelial cells were transfected with siRNA directed against NPHP4 or with control siRNA. Ciliogenesis was induced by serum withdrawal for 48 h. NPHP1 localization was evaluated using the novel monoclonal antibody alone (data not shown) or in combination with a polyclonal antiserum directed against the basal body and centrosomal marker protein pericentrin (Fig. 5C). Although cells transfected with a negative control siRNA showed a predominant staining of NPHP1 at the ciliary base, transfection of NPHP4 siRNA led to a redistribution of NPHP1 to vesicle-like structures surrounding the basal body (Fig. 5C, upper two panels). To clarify this staining pattern further, we performed co-stainings of NPHP1 with an antiserum against the trans-Golgi protein Golgin-97. These stainings revealed a co-localization of NPHP1 with Golgin-97 (Fig. 5C, bottom panel), suggesting a role of NPHP4 in the regulation of bidirectional NPHP1 trafficking between trans-Golgi and the ciliary base. Interestingly, overexpression of NPHP4 had no obvious effect on NPHP1 localization (supplemental Fig. 2). Taken together, these data indicate that NPHP4 acts upstream of NPHP1 and regulates the localization of NPHP1 at the ciliary base.
FIGURE 5.
NPHP4 controls the subcellular localization of NPHP1 in human ciliated retinal pigment epithelial cells. A, characterization of a novel murine monoclonal antibody detecting the N terminus of NPHP1. FLAG-tagged NPHP1 and NPHP1237–670 or NPHP4304–950 were expressed in HEK293T cells. Immunoblotting (IB) with an anti-FLAG antibody revealed expression of the indicated proteins. The novel NPHP1 mAb detected full-length NPHP1 but neither NPHP4304–950 nor NPHP1237–670. B, characterization of NPHP4 siRNA. Plasmids for F.NPHP4 and F.Eps1–225 (epidermal growth factor receptor pathway substrate 15-like 1) and the indicated siRNAs were co-transfected in HEK293T cells. NPHP4 siRNAs 2, 3, and 4 specifically reduced NPHP4 expression. C, NPHP4 knockdown leads to an accumulation of NPHP1 in the trans-Golgi network. hTERT-RPE1 were transfected with either control siRNA or siRNA directed against NPHP4. After methanol fixation immunofluorescence staining was performed with the indicated antibodies in the absence of Triton X-100. Transfection of NPHP4 siRNA led to an increased NPHP1 signal co-localizing with a staining of the trans-Golgi marker Golgin-97. All RNAi experiments were performed three times.
DISCUSSION
Mutations affecting the transition zone protein NPHP1 are responsible for >25% of all cases of nephronophthisis, making NPHP1 the most frequently affected single gene in this disease (26). The protein has been shown to form a complex with NPHP4 (8) and other proteins such as NPHP2 (30), NPHP3 (7), and NPHP9.4 NPHP4 also interacts with NPHP8 (31). The domain architecture, subcellular localization, and interaction with signaling molecules suggest that NPHP1 and the NPH protein complex are involved in sensory pathways that transmit extracellular signals such as mechanical stress, osmotic or acidic stimuli, or chemosensation to the interior of the cell (32, 33). This hypothesis is supported by a recent study that showed that the C. elegans homologs of NPHP1 and NPHP4 are expressed in ciliated sensory neurons. nph-1;nph-4 double, but not single-mutant males are response-defective (11), indicating that NPH-1 and NPH-4 play important and redundant roles in facilitating ciliary sensory signal transduction. Recently, NPHP1 and NPHP4 have been shown to interact with the tight junction and polarity proteins PALS1, PATJ, and Par6 in mammalian cells and to be involved in epithelial tight junction formation and the regulation of epithelial morphogenesis (34). The NPHP1/4 protein complex includes further signaling and scaffolding proteins such as the tyrosine kinase Pyk2, the adaptor protein p130Cas, the GTPase regulator RPGRIP1, and others (7, 10, 15, 35). However, the function of this protein complex as well as its regulation at the molecular level have remained elusive.
In the present study we demonstrate a direct interrelation between the two evolutionarily conserved nephrocystins, NPHP1 and NPHP4, and the nonreceptor tyrosine kinase Pyk2. We demonstrate that NPHP4 acts upstream of NPHP1 regulating the tyrosine phosphorylation and subcellular localization of NPHP1 and the interaction with Pyk2 kinase. We have previously demonstrated that the trans-Golgi sorting protein PACS-1 is required for the localization of NPHP1 at the transition zone at the ciliary base (9). Our new data now link Pyk2 and NPHP4 to this regulation, as illustrated schematically in supplemental Fig. 3 and demonstrate that in addition to a casein kinase 2-dependent mechanism also tyrosine phosphorylation of NPHP1 enhances the interaction with PACS-1. At this point, the exact molecular mechanism still remains unclear. Because the three tyrosine phosphorylation sites are distributed all along the protein (Tyr-46, Tyr-349, Tyr-721) one can speculate that the addition of negatively charged phosphate groups may induce conformational changes of NPHP1, thus enhancing either the affinity to PACS-1 or the accessibility for casein kinase 2 to the serine residues. To clarify this issue, protein structure determination of full-length NPHP1 and further studies whether PACS-1 binding is regulated by sequential phosphorylation events will be needed. Irrespective of the underlying mechanism NPHP1 binding to the trans-Golgi protein, PACS-1 seems to be tightly regulated by NPHP4, casein kinase 2, and Pyk2.
Interestingly, our data reveal that the presence of NPHP4 attenuates the binding of NPHP1 to PACS-1 without impairing the ciliary localization of NPHP1. In contrast, the knockdown of NPHP4 clearly affected the subcellular localization NPHP1 in human RPE cells. This is in accordance with findings in C. elegans, where NPH-4 has been shown to be required for the correct subcellular localization of NPH-1 in ciliated neurons (12). In mammalian cells NPHP1 and NPHP4 are enclosed in one protein complex, and the association of NPHP1 and NPHP4 seems to be very tight.4 Therefore, NPHP4 appears to be the favorite binding partner of NPHP1 and may serve as a molecular anchor to keep NPHP1 at a defined subcellular localization, at the ciliary base. This pool of NPHP4 associated NPHP1 would not be within reach for PACS-1 or Pyk2 in a noteworthy amount. However, after the release of NPHP1 from NPHP4 binding, which in our experiments is mimicked by the knockdown of NPHP4, both Pyk2 and PACS-1 could act to recover NPHP1 to the TGN. A role for PACS-1 in TGN retrieval has previously been described (28, 36–38). This model is in accordance with our previous findings, which revealed a vesicular distribution of NPHP1 after addition of a dominant negative PACS-1 mutation (9).
Furthermore, this is the first study to link nonreceptor tyrosine kinase activity with the function and/or localization of ciliary proteins in mammalian cells. Using the biflagellate algae Chlamydomonas reinhardtii as model system, Wang and Snell have shown that flagellar adhesion results in activation of an unknown tyrosine kinase (39). This activity was inhibited by the tyrosine kinase inhibitor genistein, which also inhibits adhesion-dependent cellular programs (for review, Refs. 40, 41). Although the identity of the Chlamydomonas kinase remains unknown, it is tempting to speculate that Pyk2 may represent one of the mammalian orthologs that are activated in response to ciliary sensing. Pyk2 is a calcium-responsive tyrosine kinase (17). Ciliary calcium signaling in response to mechanical stimuli has been linked to the transient receptor potential channel TRPP2/polycystin-2 that forms part of a pressure sensing protein complex (42, 43). Ciliary signal transduction might therefore induce calcium transients that are transmitted to the calcium-responsive tyrosine kinase Pyk2 to regulate NPHP1 protein interactions. Other factors activating Pyk2 are increased salt concentration and an acidic pH, so one could speculate whether Pyk2 links NPH protein function to these physiological stimuli.
Taken together, our data suggest a critical role for NPHP4 in dynamically regulating the Pyk2-induced tyrosine phosphorylation of NPHP1 and its targeting to either trans-Golgi network or the ciliary base. Based on the calcium dependence of Pyk2, one can envision a signaling cascade triggered by urine flow and ciliary bending or chemical extracellular stimuli that increases intraciliary or intracellular calcium, activation of Pyk2, phosphorylation of nephrocystin, and subsequent interaction with PACS-1. In this cascade NPHP4 would be a negative regulator by antagonizing Pyk2-induced phosphorylation and decreasing the interaction of NPHP1/PACS-1. This role of NPHP4 in keeping NPHP1 at the ciliary base can now explain the role for NPH-4 in controlling NPH-1 localization in C. elegans (12).
Acknowledgments
We thank Stefanie Keller, Bettina Maar, Ruth Herzog, Manuela Hochberger, Christina Engel, Charlotte Meyer, and Petra Dämisch for excellent technical assistance and members of the laboratories for helpful discussions. We thank Dr. Ivan Dikic for providing cDNAs.
This work was supported by German Research Foundation Grants BE 2212, SCHE 1562, SFB 635, and SFB 832.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
M. C. Liebau, unpublished observation.
- NPH
- nephronophthisis
- hTERT-RPE1
- retinal pigment epithelial cells immortalized by using human telomerase reverse transcriptase
- F.NPHP
- FLAG-tagged NPHP
- NPH-1
- nephronophthisis homolog 1 in C. elegans
- NPH-4
- nephronophthisis homolog 4 in C. elegans
- NPHP
- nephronophthisis protein/nephrocystin
- NPHP1
- nephrocystin-1
- NPHP4
- nephrocystin-4
- PACS-1
- phosphofurin acidic cluster sorting protein 1
- Pyk2
- protein-tyrosine kinase 2β.
REFERENCES
- 1. Hildebrandt F., Attanasio M., Otto E. (2009) J. Am. Soc. Nephrol. 20, 23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Salomon R., Saunier S., Niaudet P. (2009) Pediatr. Nephrol. 24, 2333–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Otto E. A., Tory K., Attanasio M., Zhou W., Chaki M., Paruchuri Y., Wise E. L., Wolf M. T., Utsch B., Becker C., Nürnberg G., Nürnberg P., Nayir A., Saunier S., Antignac C., Hildebrandt F. (2009) J. Med. Genet. 46, 663–670 [DOI] [PubMed] [Google Scholar]
- 4. Hildebrandt F., Otto E., Rensing C., Nothwang H. G., Vollmer M., Adolphs J., Hanusch H., Brandis M. (1997) Nat. Genet. 17, 149–153 [DOI] [PubMed] [Google Scholar]
- 5. Saunier S., Calado J., Heilig R., Silbermann F., Benessy F., Morin G., Konrad M., Broyer M., Gubler M. C., Weissenbach J., Antignac C. (1997) Hum. Mol. Genet. 6, 2317–2323 [DOI] [PubMed] [Google Scholar]
- 6. Otto E. A., Schermer B., Obara T., O'Toole J. F., Hiller K. S., Mueller A. M., Ruf R. G., Hoefele J., Beekmann F., Landau D., Foreman J. W., Goodship J. A., Strachan T., Kispert A., Wolf M. T., Gagnadoux M. F., Nivet H., Antignac C., Walz G., Drummond I. A., Benzing T., Hildebrandt F. (2003) Nat. Genet. 34, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olbrich H., Fliegauf M., Hoefele J., Kispert A., Otto E., Volz A., Wolf M. T., Sasmaz G., Trauer U., Reinhardt R., Sudbrak R., Antignac C., Gretz N., Walz G., Schermer B., Benzing T., Hildebrandt F., Omran H. (2003) Nat. Genet. 34, 455–459 [DOI] [PubMed] [Google Scholar]
- 8. Mollet G., Salomon R., Gribouval O., Silbermann F., Bacq D., Landthaler G., Milford D., Nayir A., Rizzoni G., Antignac C., Saunier S. (2002) Nat. Genet. 32, 300–305 [DOI] [PubMed] [Google Scholar]
- 9. Schermer B., Höpker K., Omran H., Ghenoiu C., Fliegauf M., Fekete A., Horvath J., Köttgen M., Hackl M., Zschiedrich S., Huber T. B., Kramer-Zucker A., Zentgraf H., Blaukat A., Walz G., Benzing T. (2005) EMBO J. 24, 4415–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mollet G., Silbermann F., Delous M., Salomon R., Antignac C., Saunier S. (2005) Hum. Mol. Genet. 14, 645–656 [DOI] [PubMed] [Google Scholar]
- 11. Jauregui A. R., Barr M. M. (2005) Exp. Cell Res. 305, 333–342 [DOI] [PubMed] [Google Scholar]
- 12. Winkelbauer M. E., Schafer J. C., Haycraft C. J., Swoboda P., Yoder B. K. (2005) J. Cell Sci. 118, 5575–5587 [DOI] [PubMed] [Google Scholar]
- 13. Wolf M. T., Lee J., Panther F., Otto E. A., Guan K. L., Hildebrandt F. (2005) J Am. Soc. Nephrol. 16, 676–687 [DOI] [PubMed] [Google Scholar]
- 14. Jauregui A. R., Nguyen K. C., Hall D. H., Barr M. M. (2008) J. Cell Biol. 180, 973–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Benzing T., Gerke P., Höpker K., Hildebrandt F., Kim E., Walz G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9784–9789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dikic I., Tokiwa G., Lev S., Courtneidge S. A., Schlessinger J. (1996) Nature 383, 547–550 [DOI] [PubMed] [Google Scholar]
- 17. Lev S., Moreno H., Martinez R., Canoll P., Peles E., Musacchio J. M., Plowman G. D., Rudy B., Schlessinger J. (1995) Nature 376, 737–745 [DOI] [PubMed] [Google Scholar]
- 18. Astier A., Avraham H., Manie S. N., Groopman J., Canty T., Avraham S., Freedman A. S. (1997) J. Biol. Chem. 272, 228–232 [DOI] [PubMed] [Google Scholar]
- 19. Tokiwa G., Dikic I., Lev S., Schlessinger J. (1996) Science 273, 792–794 [DOI] [PubMed] [Google Scholar]
- 20. Okigaki M., Davis C., Falasca M., Harroch S., Felsenfeld D. P., Sheetz M. P., Schlessinger J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10740–10745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blaukat A., Ivankovic-Dikic I., Grönroos E., Dolfi F., Tokiwa G., Vuori K., Dikic I. (1999) J. Biol. Chem. 274, 14893–14901 [DOI] [PubMed] [Google Scholar]
- 22. Ruff S. J., Chen K., Cohen S. (1997) J. Biol. Chem. 272, 1263–1267 [DOI] [PubMed] [Google Scholar]
- 23. Blaukat A., Pizard A., Breit A., Wernstedt C., Alhenc-Gelas F., Muller-Esterl W., Dikic I. (2001) J. Biol. Chem. 276, 40431–40440 [DOI] [PubMed] [Google Scholar]
- 24. Kruljac-Letunic A., Moelleken J., Kallin A., Wieland F., Blaukat A. (2003) J. Biol. Chem. 278, 29560–29570 [DOI] [PubMed] [Google Scholar]
- 25. Pugacheva E. N., Jablonski S. A., Hartman T. R., Henske E. P., Golemis E. A. (2007) Cell 129, 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoefele J., Sudbrak R., Reinhardt R., Lehrack S., Hennig S., Imm A., Muerb U., Utsch B., Attanasio M., O'Toole J. F., Otto E., Hildebrandt F. (2005) Hum. Mutat. 25, 411. [DOI] [PubMed] [Google Scholar]
- 27. Crump C. M., Xiang Y., Thomas L., Gu F., Austin C., Tooze S. A., Thomas G. (2001) EMBO J. 20, 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wan L., Molloy S. S., Thomas L., Liu G., Xiang Y., Rybak S. L., Thomas G. (1998) Cell 94, 205–216 [DOI] [PubMed] [Google Scholar]
- 29. Jenkins P. M., Zhang L., Thomas G., Martens J. R. (2009) J. Neurosci. 29, 10541–10551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Otto E., Hoefele J., Ruf R., Mueller A. M., Hiller K. S., Wolf M. T., Schuermann M. J., Becker A., Birkenhäger R., Sudbrak R., Hennies H. C., Nürnberg P., Hildebrandt F. (2002) Am. J. Hum. Genet. 71, 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arts H. H., Doherty D., van Beersum S. E., Parisi M. A., Letteboer S. J., Gorden N. T., Peters T. A., Märker T., Voesenek K., Kartono A., Ozyurek H., Farin F. M., Kroes H. Y., Wolfrum U., Brunner H. G., Cremers F. P., Glass I. A., Knoers N. V., Roepman R. (2007) Nat. Genet. 39, 882–888 [DOI] [PubMed] [Google Scholar]
- 32. Igarashi P., Somlo S. (2002) J. Am. Soc. Nephrol. 13, 2384–2398 [DOI] [PubMed] [Google Scholar]
- 33. Watnick T., Germino G. (2003) Nat. Genet. 34, 355–356 [DOI] [PubMed] [Google Scholar]
- 34. Delous M., Hellman N. E., Gaudé H. M., Silbermann F., Le Bivic A., Salomon R., Antignac C., Saunier S. (2009) Hum. Mol. Genet. 18, 4711–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roepman R., Letteboer S. J., Arts H. H., van Beersum S. E., Lu X., Krieger E., Ferreira P. A., Cremers F. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18520–18525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hinners I., Wendler F., Fei H., Thomas L., Thomas G., Tooze S. A. (2003) EMBO Rep. 4, 1182–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scott G. K., Fei H., Thomas L., Medigeshi G. R., Thomas G. (2006) EMBO J. 25, 4423–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scott G. K., Gu F., Crump C. M., Thomas L., Wan L., Xiang Y., Thomas G. (2003) EMBO J. 22, 6234–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Q., Snell W. J. (2003) J. Biol. Chem. 278, 32936–32942 [DOI] [PubMed] [Google Scholar]
- 40. Pan J., Wang Q., Snell W. J. (2005) Lab. Invest. 85, 452–463 [DOI] [PubMed] [Google Scholar]
- 41. Snell W. J., Pan J., Wang Q. (2004) Cell 117, 693–697 [DOI] [PubMed] [Google Scholar]
- 42. Nauli S. M., Alenghat F. J., Luo Y., Williams E., Vassilev P., Li X., Elia A. E., Lu W., Brown E. M., Quinn S. J., Ingber D. E., Zhou J. (2003) Nat. Genet. 33, 129–137 [DOI] [PubMed] [Google Scholar]
- 43. Sharif-Naeini R., Folgering J. H., Bichet D., Duprat F., Lauritzen I., Arhatte M., Jodar M., Dedman A., Chatelain F. C., Schulte U., Retailleau K., Loufrani L., Patel A., Sachs F., Delmas P., Peters D. J., Honoré E. (2009) Cell 139, 587–596 [DOI] [PubMed] [Google Scholar]