Abstract
Eubacteria and archaea contain a variety of actin-like proteins (ALPs) that form filaments with surprisingly diverse architectures, assembly dynamics, and cellular functions. Although there is much data supporting differences between ALP families, there is little data regarding conservation of structure and function within these families. We asked whether the filament architecture and biochemical properties of the best-understood prokaryotic actin, ParM from plasmid R1, are conserved in a divergent member of the ParM family from plasmid pB171. Previous work demonstrated that R1 ParM assembles into filaments that are structurally distinct from actin and the other characterized ALPs. They also display three biophysical properties thought to be essential for DNA segregation: 1) rapid spontaneous nucleation, 2) symmetrical elongation, and 3) dynamic instability. We used microscopic and biophysical techniques to compare and contrast the architecture and assembly of these related proteins. Despite being only 41% identical, R1 and pB171 ParMs polymerize into nearly identical filaments with similar assembly dynamics. Conservation of the core assembly properties argues for their importance in ParM-mediated DNA segregation and suggests that divergent DNA-segregating ALPs with different assembly properties operate via different mechanisms.
Keywords: Actin, Bacteria, Electron Microscopy (EM), Microfilaments, Presteady-state Kinetics, Protein Assembly, Bacterial Actin-like Filaments, Plasmid Segregation Proteins, TIRF Microscopy
Introduction
Prokaryotes were long believed to lack cytoskeletons, but recent work demonstrates that eubacteria and archaea use actin-like filaments, tubulin-related polymers, and intermediate filaments to control cellular shape (1), divide (2), establish order in the cytoplasm (3, 4), and move intracellular cargo (5). To understand the evolution of these bacterial cytoskeletal systems, we must understand both their diversity and the structural and functional relationships between them. A recent sequence analysis (6) identified 41 families of actin-like proteins (ALPs)2 in eubacteria and archaea. Seven are known to form filaments, and their functions include controlling cell wall synthesis (MreB) (7), segregating DNA (ParM, AlfA, Alp7A, and pSK41 ParM) (6, 8–10), and aligning organelles (MamK) (3, 4). Five ALPs, MreB, ParM, Ta0583, AlfA, and Psk41 ALP, have been studied in vitro (11–15). Their architectures and dynamics differ significantly from each other and from conventional actin in vitro (11, 14–16). A paradigm emerging from this work is that, unlike the eukaryotic actin cytoskeleton, whose architecture and function are determined by accessory factors, each bacterial actin appears adapted to a specific function, with unique properties that reduce its need for accessory factors.
Given the diversity of the ALPs, we asked whether the biochemical properties we proposed to be important for the cellular function of one actin-like protein, ParM from the R1 plasmid, are conserved across the entire ParM family. R1 ParM is the best understood bacterial actin (17), and it drives plasmid segregation in Gram-negative enteric pathogens by forming a polymerization-based motor (10, 18) that pushes plasmids to opposite poles of rod-shaped cells (19). We previously identified three properties that appear to be essential to the cellular function of ParM and reduce its requirement for accessory factors. These properties are as follows: 1) a stochastic switch between growth and shrinking, called dynamic instability, 2) symmetrical filament elongation, and 3) rapid spontaneous nucleation (18, 20).
Two recently characterized bacterial ALPs assemble into structures that look very different from both ParM and conventional actin filaments. The first, an ALP from plasmid pSK41, was identified initially as a potential member of the ParM family (10). Its sequence similarity to R1 ParM (18%), however, is below the 20% cut-off proposed by Derman et al. (6) for defining ALP families, and its atomic structure appears more closely related to that of an archeal actin, Ta0583, from Thermoplasma acidophilum. Perhaps not surprisingly, pSK41 ALP assembles into filaments with strikingly different architecture and assembly dynamics than R1 ParM; it forms one-strand helical filaments, which are very different from the two-stranded R1 ParM filaments. Nucleation of these filaments is slower than that of R1 ParM and elongation proceeds from a dimeric rather than a trimeric nucleus. Finally, and most interestingly, the pSK41 ALP filaments are not dynamically unstable (15).
The second ALP, AlfA, also is a plasmid-segregating actin with little sequence homology to R1 ParM (15% identity). It also forms unique filaments that bundle spontaneously and lack dynamic instability (14). These findings, especially the differences in polymer assembly dynamics, invite the intriguing conclusion that different ALP families partition plasmid DNA via distinct mechanisms. These results also suggest an important question; how well conserved are the biochemical and biophysical properties of more closely related ALPs, especially as individual ALP families can be more diverse than the entire family of eukaryotic actins?
To address this question, we purified and characterized an actin-like protein encoded by the StbA gene from the Par1 operon of plasmid pB171 from enteropathogenic Escherichia coli (21, 22). R1 ParM and pB171 stbA share 41% identity and 52% similarity. Because this level of conservation is within the cut-off proposed by Derman et al. (6) for prokaryotic ALP families, we will refer to StbA as pB171 ParM. This level of conservation is, however, weak compared with that of eukaryotic actins and is more characteristic of the conservation between conventional actin and the eukaryotic actin-related proteins, which have different activities and cellular functions (23). Using time-resolved light scattering, as well as electron and TIRF microscopy of single filaments, we asked whether the structure and basic biophysical properties of R1 ParM are conserved in pB171 ParM.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
We PCR-amplified the pB171 ParM gene from a mini pB171 plasmid with primers that appended a C-terminal GSKCK tag for later use in maleimide labeling reactions and cloned it into a pET-11a vector (New England Biolabs, Ipswich, MA). We transformed E. coli BL21 cells with the construct, grew them at 37 °C to an optical density of 0.7 at 600 nm, and induced with 0.75 mm isopropyl-β-d-thiogalactopyranoside for 3–5 h. We harvested bacterial pellets via centrifugation and flash froze them in liquid N2. We purified pB171 ParM-GSKCK using the same protocol as for R1 ParM (20) with the following modification; a 0–20% ammonium cut was used to precipitate the pB171 ParM protein out of the clarified bacterial extract as the initial purification step. R1 ParM-GSKCK was expressed and purified as described previously (20).
Electron Microscopy and Image Analysis
pB171 and R1 ParM were polymerized with 5 mm nucleotide for 5 min and then prepared by negative staining as described (24). Samples were imaged on a Tecnai T12 microscope operating at 120 kV at 62,000× magnification. Images were recorded on a Gatan Ultrascan 4,000 × 4,000 CCD camera, at a pixel size of 1.72 Å. The defocus of each micrograph was determined using the CTFFIND program (25), and the entire micrograph was corrected by phase flipping.
Three-dimensional reconstructions of both pB171 and R1 ParM were performed by iterative helical real space reconstruction, as described (26). A total of 5006 pB171 ParM filament segments 260 Å in length were used in an initial reconstruction. Heterogeneity of the helical symmetry within the dataset was sorted by comparison with a series of references with different helical symmetries, as described for R1 ParM (27). The largest class from this analysis, corresponding to particles with an azimuthal angle of 166.2°, had 1111 helical segments. This class was used in an independent reconstruction, yielding the final structure at a 19 Å resolution. The R1 ParM reconstruction used 4799 helical segments and did not require classification by helical symmetry.
High Speed Sedimentation Assays
We combined various concentrations of pB171 ParM with 10 mm nucleotide in buffer F (100 mm KCl, 30 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 1 mm DTT). For experiments with ATP and GTP, an additional 10 mm MgCl2 was added to the reactions. The reactions were then immediately centrifuged for 15 min at 355,000 × g at 25 °C in a Beckman Coulter TLA 120.1 rotor. The supernatants were resolved on 4–12% precast gradient NuPAGE acrylamide gels (Invitrogen) or on self-cast 13.75% SDS polyacrylamide gels. The gels were stained with SYPRO Red (Invitrogen), scanned with a Typhoon 9400 variable mode imager (GE Healthcare), and quantified using ImageQuant TL software (GE Healthcare). The steady state monomer concentration was estimated as the x-intercept of lines fit to a plot of the calculated amount of protein in the pellet versus the total initial protein.
Etheno-ATP Binding and Nucleotide Competition Assays
Dissociation Rate Constant
Reaction mixtures containing equimolar pB171 ParM and 1,N6-etheno-ATP (Invitrogen) in buffer Q (100 mm KCl, 30 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 1 mm DTT, 200 mm acrylamide) were incubated for 15 min at room temperature and then combined with equal volumes of 10 mm ATP in buffer Q using an SFA-20 rapid mixer (Hi-tech, Bradford-on-Avon, UK). We monitored the fluorescence at 420 nm (excitation, 315 nm) over time with a K2 fluorimeter (ISS, Champagne, IL) and fit exponential decay functions to the data to estimate the dissociation rate constant.
Association Rate Constant
We mixed equal volumes of 1 μm pB171 ParM in with a range of concentrations of etheno-ATP in buffer Q using the rapid mixer and recorded the fluorescence over time. The observed rate constants were estimated by fitting exponential rise functions to the data and plotted versus the etheno-ATP concentrations. The slope of a line fit to the plot estimated the association rate constant (kon) and the y-intercept provided a second estimate of the disassociation rate constant (koff).
Affinity Constants (Kd)
A 1.6 μm ParM-etheno-ATP-buffer Q solution was mixed with equal volumes of a range of concentrations of ATP and GTP. Following a 15-min incubation at room temperature, the fluorescence of the individual reaction mixtures was measured. We fit a four-parameter logistic function to a plot of the percentage of relative binding versus the concentration of competitor nucleotide to estimate the IC50 and converted the IC50 values to a Ki using the online IC50-to-Ki converter tool (BotDB Database (28)).
Bulk Polymerization and Phosphate Release Assays
For the bulk polymerization assays, we rapidly mixed a range of concentrations of pB171 ParM with equal volumes of 10 mm MgCl2-ATP or MgCl2-GTP in buffer F and recorded the right angle light scattering intensity over time with an excitation wavelength of 314 nm. Each trace for a particular concentration is the average of five or more runs performed on the same day. For the assays with varied nucleotide, we rapidly mixed 10 μm pB171 ParM in buffer F with equal volumes of a dilution series of ATP or GTP and recorded the right angle light scattering over time. We measured phosphate release by 5 μm ParM polymerized with ATP or GTP in buffer F using the EnzChek phosphate assay kit (Invitrogen) with an Ultrospec 2100 Pro spectophotometer controlled with SWIFT II software (GE Healthcare). A360 values were converted to inorganic phosphate concentration by using a phosphate standard and parallel right angle light scattering assays were performed on the same day.
pB171 ParM Labeling and TIRF Microscopy
For labeling reactions, monomeric pB171 ParM was combined with Alexa 488-maleimide (Invitrogen) at a 1:1.6 molar ratio in buffer F lacking DTT for 30 min at 4 °C. The reactions were quenched by the addition of 10 mm DTT, and the protein was separated from free dye by gel filtration. The labeling efficiency was 80–100%.
To monitor single filament polymerization dynamics, we directly applied 2.7 μl of 25% Alexa 488-labeled pB171 ParM in TIRF buffer (100 mm KCl, 15 mm Tris-HCl, 1 mm MgCl2, 1 mm DTT, 0.8% methylcellulose, 0.5% BSA) and 0.3 μl of 100 mm ATP or AMP-PNP to ethanol-base washed coverslips and performed time-lapse TIRF microscopy using a Nikon Eclipse TE2000-E inverted microscope equipped with an Andor iXon+ EM digital camera and a 40-milliwatt 488/514 argon ion laser. Data were analyzed with ImageJ software (29).
Sequence Alignments and Phylogenetic Analysis
Representative actin and ParM sequence were identified using BLASTP on the NCBI website. The sequences were aligned using the MUSCLE global alignment algorithm (30) and the Jalview alignment editor (31). Phylogenetic analysis was performed with the MEGA4 software (32) using the Neighbor joining (33) and Bootstrap (34) methods.
RESULTS
Comparing Structures of R1 and pB171 ParM Filaments
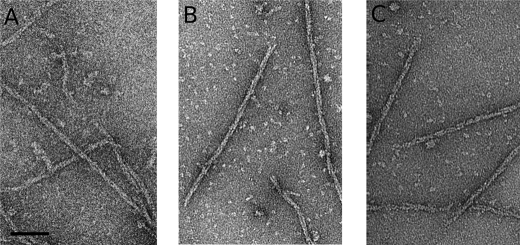
Using electron microscopy, we examined negatively stained pB171 ParM filaments polymerized with AMP-PNP, ATP, and GTP. Under all conditions, we observed well separated, helically wound filaments composed of two strands (Fig. 1, A–C). Filaments formed in AMP-PNP (Fig. 1A) were longer than those formed in either ATP or GTP (Fig. 1, B and C).
FIGURE 1.
pB171 ParM forms filaments in AMP-PNP (A), ATP (B), and GTP (C). pB171 ParM (9.5 μm) was polymerized with 5 mm nucleotide, stained with 0.75% uranyl formate, and visualized by transmission EM. The conditions used were as follows: 100 mm KCl, 30 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 1 mm DTT, 25 °C. Scale bar, 50 nm.
Initial attempts to construct a high resolution model of pB171 ParM filaments in AMP-PNP using iterative helical real space reconstruction (26) failed to converge to a stable solution, even after 60 refinement cycles (supplemental Fig. S1B). In contrast, R1 ParM filaments assembled in AMP-PNP yielded a stable solution after ∼20 iterations (supplemental Fig. S1A). The helical twist of R1 ParM filaments has been shown to be somewhat variable (16, 27), and we interpret the failure of pB171 ParM images to produce a stable reconstruction as evidence that the variation in angles between protomers in these filaments is even higher.
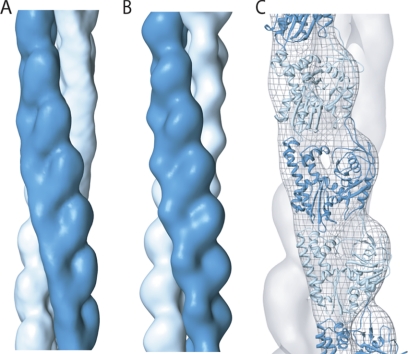
To deal with these heterogeneities, we performed multireference classification of the data set using nine models with different helical symmetries. The largest class, which contained 20% of the entire data set, corresponded to an azimuthal rotation of 166.1° between adjacent protomers. In an independent reconstruction performed using only this class of the data, helical symmetry converged from different initial values to the same solution after ∼10 iterations. Following initial convergence, however, the azimuthal rotations oscillated between 166.0° and 166.25° in subsequent iterations, suggesting some degree of twist heterogeneity even within this class (supplemental Fig. S1C). The final structure of pB171 ParM, with an estimated resolution of 19 Å (supplemental Fig. S1D), closely resembles both the present and previously reported structures of AMP-PNP R1 ParM filaments (Fig. 2A) (27).
FIGURE 2.
EM reconstruction of pB171 ParM filaments. A, the pB171 ParM filament structure, calculated from a subset of filament segments that converged to helical symmetry of 166.1° rotation and 24.2 Å rise per subunit. The map is filtered to 19 Å, the estimated resolution of the reconstruction. Each filament strand is rendered in a different color. B, for comparison, a reconstruction of R1 ParM filaments was calculated, with a refined helical symmetry of 165.4° rotation and 24.5 Å rise per subunit. C, the crystal structure of R1 ParM was manually fit into the EM structure of the pB171 ParM filament, and the fit along a single strand is shown.
We fit the atomic structure of ADP-bound R1 ParM (12) into our pB171 ParM AMP-PNP reconstruction without steric clashes. The inter- and intrastrand contacts between protomers are nearly identical to the model of Galkin et al. (27) (Fig. 2C).
Nucleotide Binding and Sedimentation Assays Demonstrate That pB171 ParM Binds More Tightly to ATP, but Is More Stable in GTP
We determined the distribution of pB171 ParM between monomeric and polymeric states using high speed centrifugation. Similar to other actin-like proteins, assembly of pB171 ParM into filaments required nucleotide triphosphates (either ATP or GTP) and was promoted by MgCl2. ATP-ParM polymerization was reduced in the absence of added MgCl2 and inhibited by 1 mm EDTA. Like other actin-like proteins, pB171 ParM polymerized poorly in the presence of CaCl2. A larger fraction of the pB171 protein pelleted in GTP than ATP in all conditions except buffer containing CaCl2. In this condition, pelleting was identical in ATP and GTP (supplemental Fig. S2, A and B).
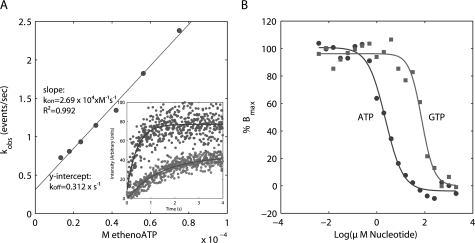
Actins and the prokaryotic ALPs studied to date bind to and polymerize in the presence of ATP and GTP with varying efficiencies (13–15, 27, 35–37). Stopped-flow experiments indicate that pB171 ParM binds the fluorescent ATP analog, 1,N6-etheno-ATP with a rate constant of 25.8 ± 1.1 × 103 s−1 m−1, and the pB171 ParM-etheno-ATP complex disassociated with a rate constant of 0.368 ± 0.142 s−1, corresponding to a Kd of 14.2 ± 5.5 μm (Fig. 3A). Competition binding experiments between etheno-ATP and either ATP or GTP indicated that pB171 ParM has a significantly higher affinity for ATP (Kd = 2.7 ± 1.2 μm) than GTP (Kd = 114.4 ± 33.4 μm) (Fig. 3B). Assuming intracellular ATP and GTP concentrations of 9.4 and 4.9 mm (38), respectively, and ignoring the presence of other nucleotide binding proteins, the measured affinities suggest that, in vivo, 98.8% of pB171 ParM is bound to ATP, and 1.2% is bound to GTP.
FIGURE 3.
pB171 ParM binds preferentially to ATP over GTP. A, pseudo-first order association kinetics of pB171 ParM binding to etheno-ATP. The slope of the line estimates the second order association rate constant and the y intercept estimates the first order disassociation rate constant. The inset shows representative association curves for 75 μm and 13.5 μm 1,N6-etheno-ATP. Buffer conditions were as follows: 100 mm KCl, 30 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 1 mm DTT, 200 mm acrylamide. B, competitive binding experiments of 1,N6-etheno-ATP versus ATP and GTP for pB171 ParM. The data were fit to a four-parameter logistic curve to estimate the IC50. Buffer conditions were the same as above.
For quantitative comparison of cytoskeletal polymers, we will define the following three terms: critical concentration, steady state monomer concentration, and instability ratio. We define a critical concentration only for single-state polymers. Briefly, if polymer assembly is governed by,
where k+ and k− are rate constants for monomer association and dissociation, and [m] and [e] are concentrations of monomer and filament ends, then the critical concentration is defined as the monomer concentration at which polymer neither grows nor shrinks.
 |
Although R1 ParM is normally a two-state (dynamically unstable) polymer, we can convert it into a one-state polymer using point mutants or nonhydrolyzable nucleotide analogs. Under either condition, R1 ParM has a critical concentration (0.6 μm) governed by Equation 1. We will call this the ATP critical concentration (mccATP). R1 ParM is unstable in ADP and has an ADP critical concentration (mccADP) greater than 120 μm. Wild type R1 ParM filaments in the presence of ATP can switch from stable elongation to rapid depolymerization, and the measured steady state monomer concentration under these conditions is 2.3 μm. Because this monomer concentration reflects the behavior of two filament populations, each with a different critical concentration, we will refer to it simply as the steady state monomer concentration (mss) of the polymer. If we assume that the reason mss is greater than mccATP is because at steady state some fraction of filaments (r) have ATP caps at their ends and are governed by Equation 1, whereas the rest (1 − r) have ADP-bound protomers at one or both ends and are catastrophically shortening at a rapid rate (ks), then the steady state monomer concentration is given by Equation 3.
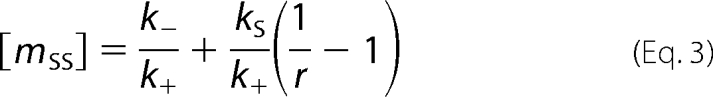 |
Using parameters measured for R1 ParM we calculate that, at steady state, 88% of filaments are stable and 12% are shrinking.
Finally, for polymers whose stability changes upon nucleotide hydrolysis, the ratio of the critical concentration in nucleotide diphosphate (ADP or GDP) over that in nucleotide tri-phosphate (ATP or GTP) is a convenient measure of dynamic instability. We will call this the “instability ratio” of the polymer. For example, actin, which is not generally considered dynamically unstable, has an instability ratio of 1.6 (39). AlfA, which segregates DNA in Bacillus cells, has a similar instability ratio of 4.2 (14). In contrast, dynamically unstable R1 ParM filaments have an instability ratio >160 (20).
Finally, sedimentation assays indicated that pB171 ParM has a lower steady state monomer concentration in GTP (mssGTP = 1.1 ± 0.21 μm) than in ATP (mssATP 1.5 ± 0.12 μm). We find that like R1 ParM, pB171 ParM has an instability ratio >140 (Table 1), in both adenosine and guanosine nucleotides.
TABLE 1.
pB171 ParM steady state monomer concentrations
The steady state monomer concentrations were determined by sedimentation assays.
| Nucleotide | Steady-state monomer concentration |
|---|---|
| μm | |
| ATP | 1.5 ± 0.12 |
| ADP | >114 |
| AMP-PNP | 0.5 ± 0.04 |
| GTP | 1.1 ± 0.21 |
| GDP | >114 |
| GMP-PNP | 0.77 ± 0.1 |
Rapid Nucleation and Nucleotide Hydrolysis by pB171 ParM
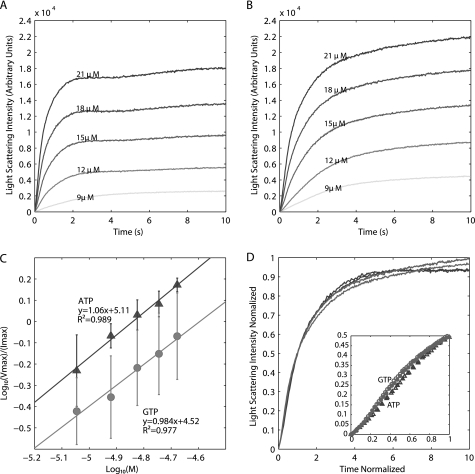
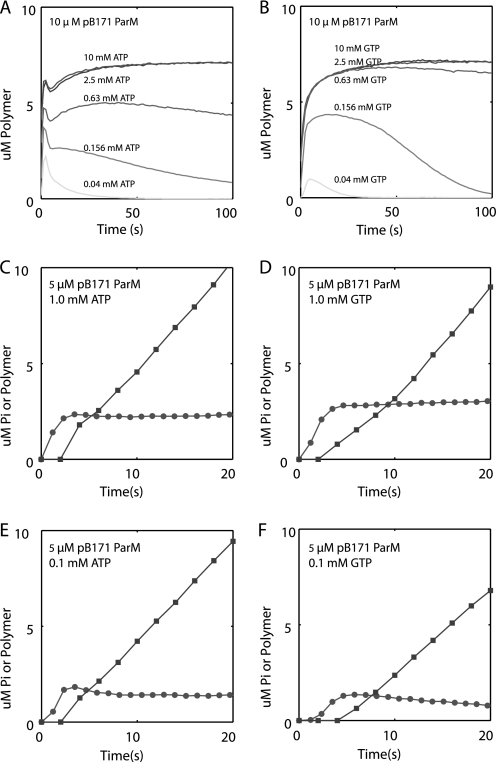
For linear helical polymers such as actin and R1 ParM, filament assembly is governed by several parameters that include nucleus size, nucleation rate, elongation rate, stability of the polymer in subsaturating nucleotide, and rates of nucleotide hydrolysis and phosphate release. Using right angle light scattering, pB171 ParM rapidly assembles in the presence of ATP, suggesting that nucleation is fast. The ATP-pB171 ParM assembly curves have three phases: an initial increase in polymer, followed by a brief decrease, and then a slower approach to steady state (Fig. 4A). pB171 ParM also rapidly assembled in GTP (Fig. 4B). However, the traces lacked the middle phase observed in ATP.
FIGURE 4.
Light scattering assays demonstrate that pB171 ParM nucleates and assembles rapidly in ATP and GTP. A and B, rapid assembly of pB171 ParM in 10 mm MgCl2-ATP (A) and 10 mm MgCl2-GTP (B) monitored by light scattering. Buffer conditions for all experiments in this figure were as follows: 100 mm KCl, 30 mm Tris-HCl, 1 mm MgCl2, 1 mm DTT. C, determination of the nucleus size and relative rates of nucleation in ATP and GTP. The log of the maximal rate of assembly (Vmax) normalized by the maximum light scattering intensity was plotted versus the log concentration of pB171 ParM. The nucleus size (n) is estimated as two times the slope of the linear fit, and the x-intercept is proportional to the square root of the nucleation rate constant times the elongation rate constant. The error bars are the S.D. of calculated values from three separate experiments. D, normalized intensity (I(t)/Imax) plotted versus normalized time (t/t½) for the highest two concentrations of pB171 ParM in ATP (dark gray) and GTP (light gray). Inset, earliest time points (triangles, ATP; circles, GTP).
To estimate the nucleus size and nucleation rate, we plotted the intensity-normalized maximum polymerization rate versus the protein concentration on a log-log plot and fitted a line to the transformed data (14, 20, 40). Using this method, we estimate that the size of the nucleus, the last unstable intermediate in the filament assembly pathway, for pB171 ParM filaments is a dimer in ATP and GTP (Fig. 4C). In contrast, assembly of actin and R1 ParM begins with creation of a trimeric nucleus (20, 41). Overall, our analysis indicates that pB171 ParM filaments assemble spontaneously much more quickly than actin filaments.
We also used the method of Flyvbjerg et al. (42) to estimate the nucleus size from early time points of our light scattering data. We normalized the amplitudes and times of light scattering curves collected at different concentrations of pB171 ParM and observed that all of the ATP data collapsed on to one curve. The GTP data collapsed onto a similar curve, indicating that pB171 ParM assembles via the same mechanism in ATP and GTP (Fig. 4D) but with different rate constants. To estimate the nucleus size, we plotted the normalized data for the earliest time points on log-log plots. The slope for the earliest time points reflects the number of kinetic steps in nucleation (42). This analysis indicated that nucleus formation occurs in one step in both ATP and GTP, further indicating that the pB171 ParM nucleus is a dimer (supplemental Fig. S3).
To determine the stability of pB171 ParM in limiting concentrations of nucleotide, we polymerized 10 μm ParM in the presence of varying concentrations of ATP and GTP. Following an initial rapid polymerization, pB171 ParM filaments depolymerized when assembled in limiting concentrations of ATP and GTP (Fig. 5, A and B). Consistent with our measurements of nucleotide affinity, the initial rates of pB171 ParM polymerization were more sensitive to limiting concentrations of GTP than ATP (supplemental Fig. S4, E and F). These data also suggest that pB171 ParM filaments are more stable in GTP than ATP, because the rate of decay in 40 μm ATP was faster than the rate of decay in 40 μm GTP (supplemental Fig. S4G). Similar results were obtained for pB171 ParM polymerized with various concentrations of MgCl2-ATP and MgCl2-GTP (supplemental Fig. S4, A–D).
FIGURE 5.
Light scattering and phosphate release assays demonstrate that pB171 ParM is unstable in limiting concentrations of nucleotide and rapidly hydrolyzes nucleotide and releases phosphate. A and B, assembly of pB171 ParM in various concentrations of ATP (A) and GTP (B). C–F, phosphate release assays. The amount of phosphate released by 5 μm pB171 ParM polymerized in 1 mm ATP (C), 1 mm GTP (D), 0.1 mm ATP (E), or 0.1 mm GTP (F) are plotted versus time (closed squares). Parallel assembly reactions were monitored with light scattering using the same stock protein solutions (closed circles). Buffer conditions for all experiments in this figure are the same as those used for the experiment in Fig. 4.
Phosphate release assays indicated that phosphate production lagged behind polymerization in low (0.1 mm) and high (1 mm) concentrations of ATP and GTP (Fig. 5, C–F) Surprisingly, we observed similar rates of phosphate production in 1 mm ATP (0.577 ± 0.001 μm/s) and 1 mm GTP (0.593 ± 0.006 μm/s) at steady state. Consistent with the difference in measured affinities, steady state pB171 ParM phosphate production was greater in ATP (0.513 ± 0.017 μm/s) than GTP (0.445 ± 0.09 μm/s) at 0.1 mm nucleotide concentrations.
TIRF Microscopy of Individual Filaments Confirm That pB171 ParM Elongates Symmetrically, Is Dynamically Unstable, and That Nucleotide Hydrolysis Is Required for Dynamic Instability
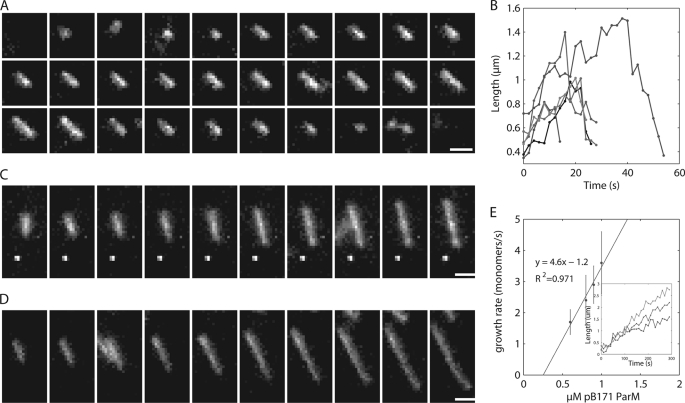
Using time-lapse TIRF microscopy to monitor individual filaments of 25% Alexa 488 labeled ParM polymerized in ATP, we observed many short individual filaments that diffused rapidly, binding only transiently to the coverglass before detaching. However, all filaments that remained attached to the coverslip surface for an extended period elongated symmetrically from both ends prior to undergoing catastrophic depolymerization, which appeared to occur primarily from one end. No rescue events were observed; all filaments that we tracked either detached from the slide or completely depolymerized. We measured the rates of filament elongation (10.6 +/4.9 monomers/s) and depolymerization following catastrophe (22.9 ± 9.4 monomers/s) (Fig. 6, A and B), which are similar to those we measured previously for R1 ParM (20).
FIGURE 6.
Time-lapse TIRF microscopy observing individual filaments demonstrate that pB171 ParM is dynamically unstable when polymerized in ATP and appears to elongate symmetrically. A, montage of an individual pB171 ParM filament in ATP. 25% Alexa 488-labeled 2.8 μm B171 ParM was polymerized in the presence of 10 mm ATP and imaged via TIRF microscopy every 2 s. Buffer conditions were as follows: 100 mm KCl, 30 mm KCl, 1 mm MgCl2, 1 mm DTT, 0.8% methylcellulose, 0.5% BSA. Scale bar, 1 μm. B, length versus time for six representative filaments polymerized in ATP. C and D, montage of individual filaments polymerized in nonhydrolyzable AMP-PNP. 25% Alexa 488-labeled 0.8 μm or 1.0 μm pB171 ParM was polymerized in the presence of 10 mm AMP-PNP. The time interval is 20 s between each frame. Buffer conditions are the same as in ATP. Scale bar, 1 μm. E, rate of elongation of AMP-ParM. The rate of elongation was measured at various concentrations of pB171 ParM in AMP-PNP and plotted versus the μm pB171 ParM. The line fit to the data represents the equation: rate of filament elongation = kon x (μm protein)-koff. Inset shows three representative filaments growth over time from 0.6, 0.8, and 1.0 μm pB171 concentrations.
To determine whether nucleotide hydrolysis regulates dynamic instability in pB171 ParM, we performed TIRF microscopy on various concentrations of pB171 ParM polymerized in AMP-PNP. At all concentrations tested, the AMP-PNP filaments appeared to elongate from both ends and attain lengths much greater than ATP filaments (Fig. 6, C and D). Although filament fragmentation occurred, we observed no examples of catastrophe. We occasionally observed two types of bundling behaviors that appeared to be length dependent: 1) lateral binding of a smaller filament to the center of a longer filament and 2) the collision of two long filament ends that lead to filament zippering. We measured an elongation rate constant in AMP-PNP of 2.3 monomers × s−1 μm−1 per filament end and a noncatastrophic depolymerization rate constant of 0.6 monomers per filament end per second (Fig. 6E). Using these parameters, together with the steady state monomer concentrations in ATP and AMP-PNP, we calculate from Equation 2 that, at steady state, 89% of the pB171 ParM filaments are stable and growing, whereas 11% are unstable and shrinking when polymerized in ATP. Comparing this steady state behavior to R1 ParM indicates that the two polymers maintain a similar balance between nucleation, growth, and catastrophe.
DISCUSSION
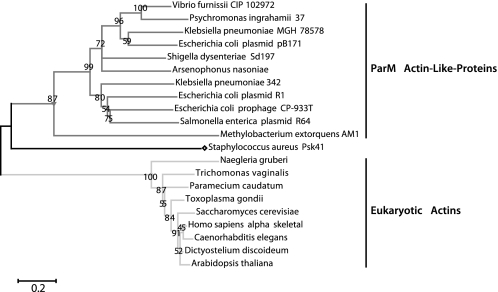
Eukaryotic actins are highly conserved, probably due to the large number of conserved binding partners that regulate their assembly and function. Across metazoan species, for example, actin sequences are ∼98% identical. The primary sequences of protozoan actins are more variable but the level of sequence conservation is still much greater than that observed in bacterial actin families. Bacterial actin families are, in fact, much less well defined than eukaryotic actins. The majority of known bacterial actins have been identified by genome searches and the homology cut-off proposed by Derman et al. for defining families (6) is more or less arbitrary. We were interested in determining whether one particular clade of bacterial actins, the Alp3 or ParM group, represents a bona fide family with conserved structure and activity. As Fig. 7 demonstrates, the ParM family is quite divergent in comparison with the eukaryotic actins.
FIGURE 7.
Evolutionary relationships demonstrate that the ParM family is more divergent than eukaryotic actins. The tree represents a bootstrap consensus tree inferred from 1000 replicate trees generated using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches. The branch lengths are proportional to the relative evolutionary distance, and the scale bar is in units of the number of amino acid substitutions per site. The GI numbers for the sequences and the sequence alignments used to generate this phylogram are provided in the supplemental material. Evolutionary analyses were conducted in MEGA4.
The ParM protein, encoded by the R1 plasmid is, to date, the most well characterized bacterial ALP (17). It polymerizes into left-handed, double-stranded, helical filaments that nucleate rapidly, elongate symmetrically, and are dynamically unstable (20). Here, we show that ParM from the Par1 locus of pB171, although only 41% identical to R1 ParM, polymerizes via a similar pathway into filaments that have a remarkably similar structure. Using electron microscopy, we found that pB171 ParM monomers build filaments that are more similar to R1 ParM than to actin or any other characterized actin-like protein (MreB, AlfA, or the pSK41 ALP). Light scattering assays demonstrated that pB171 ParM filaments rapidly and spontaneously form filaments in the absence of nucleation factors in both ATP and GTP. Sedimentation assays demonstrated that pB171 ParM does not polymerize in ADP or GDP, and light scattering assays revealed that pB171 ParM filaments rapidly depolymerize in limiting amounts of ATP and GTP prior to complete nucleotide hydrolysis. This is consistent with our previous observation that substoichiometric ratios of ADP to ATP (∼20%) destabilize R1 ParM filaments, regardless of the total ATP concentration (20). For both R1 and pB171 ParM, we attribute this effect to nucleotide exchange on the terminal subunit of the filament, a phenomenon initially observed for actin filaments (43). Finally, TIRF microscopy of labeled pB171 ParM revealed both symmetrical filament elongation and dynamic instability.
In addition to the basic similarities, we also note three minor differences between the biochemical and biophysical properties of ParM proteins from plasmids R1 and pB171. First, the structure of pB171 ParM filaments is more heterogeneous. This is shown mainly as variability in the degree of helical twist of the strands that compose the filament. We hypothesize that this reflects weaker lateral interactions between the strands that permit them to either rotate or slip more freely. Second, we find that pB171 ParM is slightly more stable in GTP than ATP, as evidenced by the lower steady state monomer concentration. Increased stability in GTP may reflect an increase in the rate of polymerization, a decrease in the rate of depolymerization of stable or unstable filament ends or a decrease in the propensity of GTP filaments to undergo catastrophe due to a reduced rate of nucleotide hydrolysis or phosphate release. Although our analysis did not identify the mechanism of the increased stability in GTP, our phosphate release assays suggests that pB171 ParM filaments hydrolyze GTP at similar rates as ATP. Third, the size of the pB171 ParM filament nucleus is a dimer in ATP and GTP, whereas the apparent size of the R1 ParM nucleus is a trimer (20, 27). This probably reflects slight differences in one of the monomer-monomer interfaces. Depending on buffer conditions, the nucleus size of conventional actin ranges from two to four subunits (41). Osawa and Kansai (44) predicted that all linear helical polymers would generally assemble from nuclei in this size range.
Popp et al. (15) recently characterized the structure and assembly of a divergent ALP from plasmid pSK41, identified previously by Møller-Jensen et al. (10) as a possible member of the ParM family. This ALP is, however, only 18% similar to R1 ParM and falls below the proposed cut-off for definition of ALP families. As noted above, however, this sequence similarity criterion is fairly arbitrary and a more rigorous definition of families requires combining sequence similarity with structural and functional information. Filaments formed by the pSK41 ALP protein lack many of the longitudinal monomer-monomer contacts that define the two long-pitch helices found in actin and ParM and these filaments are best described as single-stranded. In addition, they are not dynamically unstable, and they do not elongate symmetrically. Finally, and most intriguingly, the atomic structure of the pSK41 ALP is more similar to the archeal actin Ta0583 from T. acidophilum than to R1 ParM. Popp et al. argue that the pSK41 protein represents an evolutionary intermediate between a chromosomally encoded ALP and the plasmid-encoded ParM-family proteins and suggested that it promotes plasmid partitioning via a very different mechanism. Our investigation and that of Popp et al. highlight the importance of careful structural and functional studies in defining families of bacterial actin-like proteins.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and additional information.
- ALP
- actin-like protein
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- TIRF
- total internal reflection fluorescence.
REFERENCES
- 1. Jones L. J., Carballido-López R., Errington J. (2001) Cell 104, 913–922 [DOI] [PubMed] [Google Scholar]
- 2. Erickson H. P. (1995) Cell 80, 367–370 [DOI] [PubMed] [Google Scholar]
- 3. Komeili A., Li Z., Newman D. K., Jensen G. J. (2006) Science 311, 242–245 [DOI] [PubMed] [Google Scholar]
- 4. Pradel N., Santini C. L., Bernadac A., Fukumori Y., Wu L. F. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17485–17489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Møller-Jensen J., Borch J., Dam M., Jensen R. B., Roepstorff P., Gerdes K. (2003) Mol. Cell 12, 1477–1487 [DOI] [PubMed] [Google Scholar]
- 6. Derman A. I., Becker E. C., Truong B. D., Fujioka A., Tucey T. M., Erb M. L., Patterson P. C., Pogliano J. (2009) Mol. Microbiol. 73, 534–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daniel R. A., Errington J. (2003) Cell 113, 767–776 [DOI] [PubMed] [Google Scholar]
- 8. Becker E., Herrera N. C., Gunderson F. Q., Derman A. I., Dance A. L., Sims J., Larsen R. A., Pogliano J. (2006) EMBO J. 25, 5919–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schumacher M. A. (2008) Biochem. J. 412, 1–18 [DOI] [PubMed] [Google Scholar]
- 10. Møller-Jensen J., Jensen R. B., Löwe J., Gerdes K. (2002) EMBO J. 21, 3119–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van den Ent F., Amos L. A., Löwe J. (2001) Nature 413, 39–44 [DOI] [PubMed] [Google Scholar]
- 12. van den Ent F., Møller-Jensen J., Amos L. A., Gerdes K., Löwe J. (2002) EMBO J. 21, 6935–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hara F., Yamashiro K., Nemoto N., Ohta Y., Yokobori S., Yasunaga T., Hisanaga S., Yamagishi A. (2007) J. Bacteriol. 189, 2039–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Polka J. K., Kollman J. M., Agard D. A., Mullins R. D. (2009) J. Bacteriol. 191, 6219–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Popp D., Xu W., Narita A., Brzoska A. J., Skurray R. A., Firth N., Goshdastider U., Maéda Y., Robinson R. C., Schumacher M. A. (2010) J. Biol. Chem. 285, 10130–10140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Orlova A., Garner E. C., Galkin V. E., Heuser J., Mullins R. D., Egelman E. H. (2007) Nat. Struct. Mol. Biol. 14, 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Needleman D. J. (2008) Curr. Biol. 18, R212–214 [DOI] [PubMed] [Google Scholar]
- 18. Garner E. C., Campbell C. S., Weibel D. B., Mullins R. D. (2007) Science 315, 1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Campbell C. S., Mullins R. D. (2007) J. Cell Biol. 179, 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garner E. C., Campbell C. S., Mullins R. D. (2004) Science 306, 1021–1025 [DOI] [PubMed] [Google Scholar]
- 21. Tobe T., Hayashi T., Han C. G., Schoolnik G. K., Ohtsubo E., Sasakawa C. (1999) Infect. Immun. 67, 5455–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ebersbach G., Gerdes K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 15078–15083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dion V., Shimada K., Gasser S. M. (2010) Curr. Opin. Cell Biol. 22, 383–391 [DOI] [PubMed] [Google Scholar]
- 24. Ohi M., Li Y., Cheng Y., Walz T. (2004) Biol. Proced. Online 6, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mindell J. A., Grigorieff N. (2003) J. Struct. Biol. 142, 334–347 [DOI] [PubMed] [Google Scholar]
- 26. Egelman E. H. (2000) Ultramicroscopy 85, 225–234 [DOI] [PubMed] [Google Scholar]
- 27. Galkin V. E., Orlova A., Rivera C., Mullins R. D., Egelman E. H. (2009) Structure 17, 1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cer R., Mudunuri U., Stephens R., Lebeda F. (2009) Nucleic Acids Res. 37, W441–W445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abramoff M., Magelhaes P. (2004) Biophotonics International 11, 36–42 [Google Scholar]
- 30. Edgar R. C. (2004) Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 33. Saitou N., Nei M. (1987) Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 34. Felsenstein J. (1985) Evolution 39, 783–791 [DOI] [PubMed] [Google Scholar]
- 35. Wen K. K., Yao X., Rubenstein P. A. (2002) J. Biol. Chem. 277, 41101–41109 [DOI] [PubMed] [Google Scholar]
- 36. Esue O., Wirtz D., Tseng Y. (2006) J. Bacteriol. 188, 968–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Popp D., Narita A., Oda T., Fujisawa T., Matsuo H., Nitanai Y., Iwasa M., Maeda K., Onishi H., Maéda Y. (2008) EMBO J. 27, 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bennett B. D., Kimball E. H., Gao M., Osterhout R., Van Dien S. J., Rabinowitz J. D. (2009) Nat. Chem. Biol. 5, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pollard T. D. (1986) J. Cell Biol. 103, 2747–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nishida E., Sakai H. (1983) J. Biochem. 93, 1011–1020 [DOI] [PubMed] [Google Scholar]
- 41. Cooper J. A., Buhle E. L., Jr., Walker S. B., Tsong T. Y., Pollard T. D. (1983) Biochemistry 22, 2193–2202 [DOI] [PubMed] [Google Scholar]
- 42. Flyvbjerg H., Jobs E., Leibler S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5975–5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Teubner A., Wegner A. (1998) Biochemistry 37, 7532–7538 [DOI] [PubMed] [Google Scholar]
- 44. Oosawa F., Kasai M. (1962) J. Mol. Biol. 4, 10–21 [DOI] [PubMed] [Google Scholar]