Abstract
The interferon-regulated transcription factor and tumor suppressor protein IRF-1 is predicted to be largely disordered outside of the DNA-binding domain. One of the advantages of intrinsically disordered protein domains is thought to be their ability to take part in multiple, specific but low affinity protein interactions; however, relatively few IRF-1-interacting proteins have been described. The recent identification of a functional binding interface for the E3-ubiquitin ligase CHIP within the major disordered domain of IRF-1 led us to ask whether this region might be employed more widely by regulators of IRF-1 function. Here we describe the use of peptide aptamer-based affinity chromatography coupled with mass spectrometry to define a multiprotein binding interface on IRF-1 (Mf2 domain; amino acids 106–140) and to identify Mf2-binding proteins from A375 cells. Based on their function as known transcriptional regulators, a selection of the Mf2 domain-binding proteins (NPM1, TRIM28, and YB-1) have been validated using in vitro and cell-based assays. Interestingly, although NPM1, TRIM28, and YB-1 all bind to the Mf2 domain, they have differing amino acid specificities, demonstrating the degree of combinatorial diversity and specificity available through linear interaction motifs.
Keywords: Peptides, Protein Domains, Protein Motifs, Protein-Protein Interactions, Transcription Factors, Tumor Suppressor, IRF-1, Intrinsic Disorder, Linear Interaction Motif, Multiprotein Interface
Introduction
Transcription factors and proteins involved in cell signaling pathways appear to have a disproportionately high number of intrinsically disordered (ID)2 domains, suggesting that such regions are of particular importance in their regulation and mode of action (1–5). Current thinking on the role of ID domains suggests that these flexible structures may confer advantages in protein-protein and protein-ligand interactions when compared with more rigid globular domains. For example, it has been proposed that greater plasticity could permit the binding of structurally diverse molecules to the same domain or that they might encourage the kind of highly specific but low affinity interactions that are advantageous in cell signaling. It has also been suggested that ID domains contain a high proportion of the proteome's linear interaction motifs (6, 7). These are protein interaction motifs that may contain 10 or fewer residues. In addition, ID domains appear to facilitate fine control and regulation by post-translational modification (6, 7).
The interferon-regulated transcription factor IRF-1 (interferon regulatory factor-1) is involved in a variety of physiological processes, including the antiviral response and tumor suppression (8, 9). Specifically, IRF-1 regulates the expression of a cohort of genes involved in immunity and/or negative cell growth, either directly, through binding to interferon responsive elements in target genes (10, 11), or indirectly, through its ability to interact with other transcription factors and coactivators (12). For example, IRF-1 binds to an interferon-responsive element in the promoter of target genes like ISG20 and Trail (13, 14). Alternatively, IRF-1 interacts with the transcriptional coactivator p300 through motifs within the transactivation and enhancer domains; this interaction promotes p300-dependent acetylation and activation of p53, leading to an increase in expression of the p53 target gene p21WAF1 (12). Interestingly, the effect of full-length IRF-1 on p300-mediated acetylation of p53 can be recapitulated in trans using peptides containing the p300 binding motifs from IRF-1 (12, 15). This finding suggests that the ability of IRF-1 to interact with other transcriptional regulators is pivotal to its physiological function (12, 15, 16).
Despite the fact that existing evidence suggests that IRF-1 function is highly regulated by post-translational mechanisms (17–20) and that it represents a network hub for radiosensitivity and various human diseases (21–23), the identification of regulatory factors that function through protein-protein interactions with IRF-1 is limited (24, 25). Here we report the application of affinity chromatography using IRF-1-based peptide aptamers to identify proteins that interact with the Mf2 interface from within a major ID domain in the central portion of the IRF-1 protein. This has led us to characterize a multiprotein binding interface employed by IRF-1 regulatory proteins, which interact with discrete but overlapping motifs.
MATERIALS AND METHODS
Reagents
Antibodies were used at 1 μg/ml or as recommended by the supplier and were anti-IRF-1 (BD Biosciences), anti-YB-1 and anti-GAPDH (Abcam), anti-FLAG and anti-GST (Sigma), anti-NPM (Zymed Laboratories Inc.), anti-His (Novagen), anti-Myc (Cancer Research UK), and anti-KAP-1 (Bethyl Laboratories). Secondary antibodies were purchased from Dako Cytomation. Peptide libraries were purchased from Chiron Mimotopes and were synthesized with a biotin tag at the N terminus and an SGSG spacer. Bulk biotin-SGSG-linked peptides were from Genscript and were >93% pure (purification and quality control are described in supplemental Method S1). GST- and FLAG-TRIM28 (KAP-1) were kind gifts from A. Ivanov (26). His-NPM was from F. Carrier (University of Maryland) (27).
Cell Culture, Lysis, and Immunoblots
A375 cells cultured in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% (v/v) fetal bovine serum (Autogen Bioclear) and 1% (v/v) penicillin/streptomycin mix (Invitrogen), were maintained in 10% CO2 at 37 °C. H1299 cells were cultured in RPMI 1640 (Invitrogen) supplemented with serum and antibiotics (as above) and maintained in 5% CO2 at 37 °C. For transient transfection, cells were seeded 24 h before transfection, and DNA (as indicated in the figure legends) was transfected into the cells using Attractene (Qiagen) as described in the manufacturer's handbook. Cell lysis and immunoblotting were as described previously (28).
Peptide Affinity Chromatography
A library of 22 peptide aptamer affinity columns were generated using Mobicol column jackets (MoBiTec) by incubating streptavidin-agarose (50-μl packed volume; binding capacity 24.5 μg biotin/ml) with biotin-peptide (11.5 mm; this was sufficient to saturate all binding sites on the column) for 1 h at room temperature, and peptide-binding proteins from A375 lysate were isolated as described previously (28). For one-dimensional electrophoresis, samples were run out on a 4–12% NuPAGE gel (Invitrogen), and stained with Colloidal Blue (Invitrogen) according to the manufacturer's instructions. For the two-dimensional gel method see supplemental Method S2. Protein bands were excised from the gel using a clean, sharp scalpel and digested with trypsin as described below. Alternately, the NuPAGE gels were transferred to nitrocellulose and immunoblotted as required.
Enzymatic In-gel Digestion
Stained protein bands were excised from the gel, washed with deionized water, cut into small pieces, and destained with freshly prepared 200 mm NH4HCO3 (pH 7.8) in 40% acetonitrile for 20 min at 30 °C, followed by equilibration in 50 mm NH4HCO3 (pH 7.8) in 5% acetonitrile for 30 min at 30 °C. The supernatant was removed, and the gel was dried in a SpeedVac concentrator. Next, the samples were reduced by the addition of 10 mm DTT for 1 h at 60 °C, followed by alkylation with 20 mm iodoacetamide in the dark for 45 min at room temperature. The supernatant was removed, and the gel pieces were washed three times with equilibration buffer and acetonitrile. After vacuum drying, the gel pieces were rehydrated in a cleavage buffer containing equilibration buffer and trypsin (40 ng/μl; Promega; cleaves at the carboxyl side of Lys or Arg except when either is followed by Pro) for 20 min at 4 °C. Finally, 15–60 μl of equilibration buffer was added to cover the gel. The samples were incubated for 18 h at 37 °C. The digestion was stopped by the addition of 5% TFA in acetonitrile, and the aliquot of the resulting peptide mixture was desalted using a GELoader microcolumn (Eppendorf) packed with Poros Oligo R3 material (29). The purified and concentrated peptides were eluted from the microcolumn in several droplets directly onto MALDI plates using 1 μl of α-cyano-4-hydroxycinnamic acid matrix solution (5 mg/ml in 50% acetonitrile, 0.1% TFA).
MALDI Mass Spectrometry
Mass spectra were measured on an Ultraflex III MALDI-TOF/TOF instrument (Bruker Daltonics) equipped with a smartbeamTM solid state laser and LIFTTM technology for MS/MS analysis. Peptide mass fingerprinting spectra were acquired in the positive reflectron mode, in the mass range of 700–4000 Da, and calibrated internally using the monoisotopic [M + H]+ ions of trypsin autoproteolytic fragments (842.5 and 2211.1 Da).
For peptide mass fingerprinting data base searching, peak lists in XLM data format were created using flexAnalysis software (version 3.0, Bruker Daltonics) with the SNAP peak detection algorithm. No smoothing was applied, and the maximal number of assigned peaks was set to 50. After peak labeling, all known contaminant signals were removed. The peak lists were searched using an in-house MASCOT search engine against a SwissProt 2010_07 database subset of human proteins (518,415 entries) with the following search settings: peptide tolerance of 20 ppm, missed cleavage site value set to 2, variable carbamidomethylation of cysteine, oxidation on methionine, and protein N-terminal acetylation. No restrictions on protein molecular weight and pI value were applied. Proteins with molecular weight search scores over the threshold 56 calculated for the used settings were considered as identified. If the score was lower or only slightly higher than the threshold value, the identity of the protein candidate was confirmed by MS/MS analysis. In addition to the above-mentioned MASCOT settings, fragment mass tolerance of 0.6 Da and instrument type MALDI-TOF/TOF was applied for searching of MS/MS spectra.
Affinity Chromatography Using GST-IRF-1
GST-IRF-1, GST-AGR2, and GST alone were expressed in BL21-AI (Invitrogen) cells and purified using glutathione-Sepharose (GE Healthcare) according to the manufacturer's instructions. However, the protein was not eluted; instead, binding assays were carried out as described previously (28).
Protein Interaction Assay
GST-IRF-1 or GST alone (250 ng) were coated onto a white 96-well plate in 0.1 m NaHCO3 buffer (pH 8.6) overnight at 4 °C. Non-reactive sites were blocked using PBS containing 3% (w/v) BSA. His-NPM (0–64 ng) was added in Reaction Buffer (25 mm Hepes, pH 7.5, 50 mm KCl, 10 mm MgCl2, 5% (v/v) glycerol, 0.1% (v/v) Tween 20, 2 mg/ml BSA) for 1 h at room temperature. After washing extensively with PBS supplemented with 0.1% (v/v) Tween 20 (PBST) between steps, binding was measured using anti-His and HRP-tagged anti-mouse antibodies, and luminescence was quantified using a luminometer (Labsystems, Fluoroskan Ascent FL). Alternatively, His-NPM (100 ng) (or TRIM28; see below) was coated onto a microtiter plate as described above. After blocking, the wells were incubated with a titration of GST alone, GST-IRF-1 WT, or GST-IRF-1 Δ106–140 (0–25 ng) in Reaction Buffer, and binding was detected using anti-GST and HRP-tagged anti-mouse antibodies, with extensive washing in between as above. Luminescence was quantified using a luminometer (Labsystems, Fluoroskan Ascent FL).
For TRIM28-IRF-1 protein interaction assays, untagged IRF-1, expressed and purified using the PUREsystem Classic II (Post Genome Institute, Tokyo, Japan), was coated onto a white 96-well plate as above. TRIM28 (0–128 ng), purified as GST-TRIM28 using glutathione-Sepharose and subsequently cleaved on the column using thrombin, was added in the mobile phase. It should be noted that TRIM28 itself contains a thrombin cleavage site and was therefore processed into two fragments during cleavage, both of which were present in the mobile phase. Binding was detected using anti-TRIM28 and HRP-tagged anti-rabbit antibodies.
For peptide-protein interaction assays (direct peptide binding assay), streptavidin (1 μg/well) was coated onto a microtiter plate overnight at 37 °C and then incubated with biotin-tagged peptide to saturate the wells (∼60 pmol). Wells were washed and blocked as above and incubated with a titration of His-NPM (0–250 ng) in Reaction Buffer as above. Binding was detected using anti-His and HRP-tagged anti-mouse antibodies, and luminescence was quantified using a luminometer as described above.
Peptide Competition Assay
GST-IRF-1 (100 ng) was coated onto a white 96-well plate in 0.1 m NaHCO3 buffer (pH 8.6) overnight at 4 °C. Non-reactive sites were blocked using PBS containing 3% BSA. Peptide (0–1.25 μm or 0–5 μm as indicated) or an equal amount of carrier alone was added to His-NPM (100 ng) (or TRIM28) in Reaction Buffer (as above) and incubated for 15 min at room temperature. The mix was then added to the 96-well plate coated with GST-IRF-1 and incubated for a further 1 h at room temperature. Following extensive washing in PBST, binding was detected using anti-His and HRP-tagged anti-mouse antibodies (or anti-TRIM28 and anti-rabbit antibodies), and luminescence was measured as above.
Immunoprecipitation
A375 cells were harvested and lysed with 20 mm Tris-HCl, pH 7.4, 1 mm EDTA, 1% Nonidet P-40, 0.15 m NaCl, 10% glycerol, 0.1 mm sodium molybdate, 1 mm pefabloc, 1 mm sodium orthovanadate, and 1 mm DTT. The lysate (4 mg of total protein) was incubated (150 min at 4 °C) with anti-IRF-1 antibody (1 μg/reaction) chemically cross-linked to protein G-Sepharose using DMP (30). In the bead control, lysate was incubated with untreated protein G-Sepharose. The beads were washed two times in Wash Buffer containing 0.5 m NaCl followed by two washes in Wash Buffer only (25 mm HEPES, pH 7.5, 1 mm EDTA, 0.1% Nonidet P-40, 10% glycerol, 0.2 mg/ml benzamidine, 10 mm β-glycerophosphate, 0.5 mm sodium orthovanadate, and 50 mm NaF). Bound proteins were analyzed following the addition of sample buffer using 10% SDS-PAGE/immunoblot.
Complex Pull-downs
OneSTREP-tagged IRF-1 (or empty vector) alone or plus FLAG-TRIM28 was transfected into A375 cells (2 × 100-mm plates/IP condition, transfected according to the manufacturer's instructions). Post-transfection (24 h), cells were harvested and lysed in Triton Lysis Buffer (50 mm HEPES, pH 7.5, 0.2% (v/v) Triton X-100, 150 mm NaCl, 10 mm NaF, 2 mm DTT, 0.1 mm EDTA, protease inhibitor mix). Following this, tagged complexes were isolated using either Streptactin macroprep (IBA), or FLAG-agarose beads (Sigma) according to the manufacturer's instructions. Eluates were analyzed by SDS-PAGE/immunoblot using anti-IRF-1, anti-YB-1, and anti-FLAG antibodies. In the case of OneStrep-IRF-1·NPM complex formation, A375 cells were transiently transfected with 0.5 μg of OneStrep-IRF-1 or empty vector and scraped into ice-cold PBS 24 h post-transfection. Cells were lysed as described above (see “Immunoprecipitation”), following which OneStrep-tagged complexes were isolated using Streptactin macroprep essentially according to the manufacturer's instructions except using the wash conditions described under “Immunoprecipitation.”
Dual Luciferase Reporter Assay
Luciferase reporter assays were carried out in H1299 cells as described previously (31) using 120 ng of either p125-luc IFNβ (which contains the human IFN-β promoter region −125 to +19) or a control plasmid p55-luc IFNβ (which lacks the interferon-stimulated response element or ISRE; promoter region −55 to +19), TLR3-Luc (hTLR3–588) or hTLR3IRF (a mutant lacking the ISRE), TRAIL (pTRL3 or a mutant minus the ISRE/IRFE, pTRL3m6), and IL-7 (−609-Luc or a mutant −609-mtIRF-E-Luc, which is missing the ISRE) (28, 31). Reporter activity was determined 24 h post-transfection using luciferase luminescence as a read-out.
RESULTS
The Use of Peptide Aptamer Affinity Chromatography
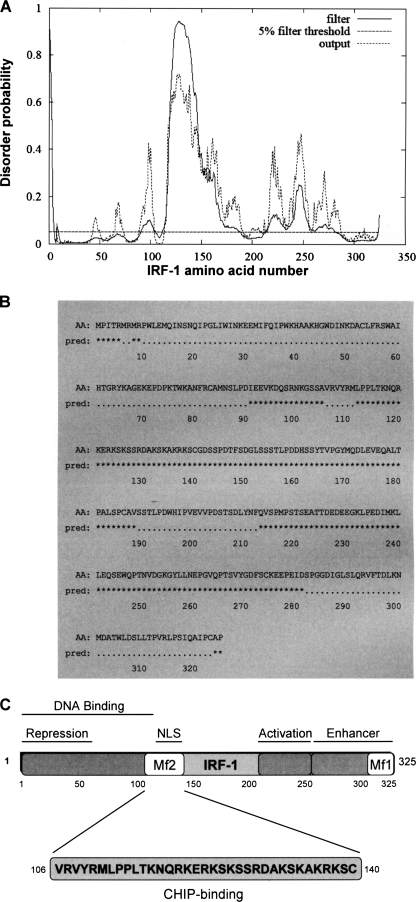
Using data generated by the DISOPRED2 Disorder Prediction Server (32), we estimate that >30% of IRF-1 structure is intrinsically disordered (Fig. 1, A and B), identifying IRF-1 as a member of a class of proteins that are described as highly unstructured (3, 7). We recently discovered that the E3-ubiquitin ligase CHIP binds to a site (amino acids 106–140) within the major disordered domain of IRF-1 (Fig. 1C), facilitating ubiquitination of IRF-1 under specific stress conditions (33). Because disordered domains appear to favor multiple protein interactions, we speculated that the major disordered domain in IRF-1 may bind to other components of the IRF-1 regulatory network.
FIGURE 1.
The Mf2 region of IRF-1 lacks a defined structure and is highly disordered. A, analysis of the IRF-1 protein sequence for disorder probability using DISOPRED2 (available on the World Wide Web) (35) with a false positive rate of 5%. The “filter” curve represents the output from DISOPRED2, and the “output” curve shows the output from a linear SVM classifier (DISOPREDsvm). The output from DISOPREDsvm is included to indicate shorter, low confidence predictions of disorder. B, amino acid sequence (AA) of IRF-1 with predicted (pred) disordered residues (*) obtained by DISOPRED2 analysis shown. C, schematic showing the location of the highly disordered Mf2 region in relation to other defined domains within the IRF-1 protein. Mf1 is a multifunctional regulatory domain (28).
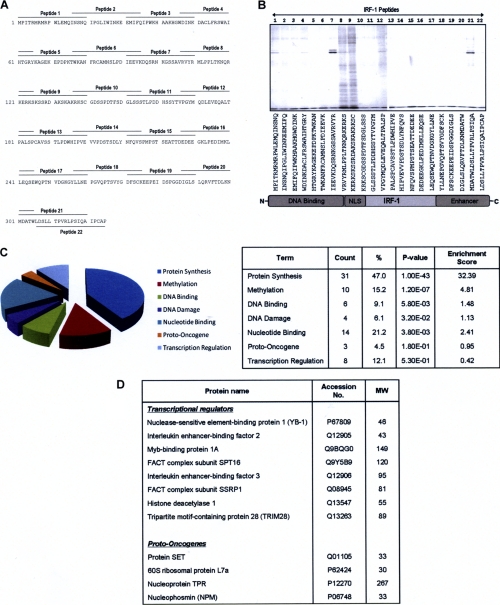
Rather than selecting just the CHIP-binding domain for analysis (Fig. 1) (33), we decided to take a less biased approach by determining the binding profile for aptamers that spanned the entire sequence of IRF-1 (Fig. 2A). Affinity columns bearing overlapping IRF-1-based peptides were generated and used to screen for interacting proteins from A375 cell lysates (supplementary Fig. S3A). When total bound protein was analyzed (Fig. 2B), it was striking that only a limited number of columns (e.g. columns 8, 9, 12, and 21) gave a diversity of interacting factors relative to background (Fig. 2B) and to a column bearing a biotinylated control peptide (supplemental Fig. S3B). In addition, the binding proteins did not necessarily correspond to proteins that were highly represented in the crude lysate (supplemental Fig. S3B). Consistent with the hypothesis that the major unstructured domain of IRF-1 (Leu112–Val188; Fig. 1B) is likely to be a hot spot for regulatory interactions and post-translational events, tens of protein bands were eluted from two overlapping peptides from the major ID domain (peptides 8 and 9; Fig. 2, A and B). These two peptides also contain the CHIP-docking site (33), suggesting that they may form a multiprotein binding interface on IRF-1. We have named this the Mf2 (multifunctional 2) domain (Fig. 1C).
FIGURE 2.
Development of a biochemical screen to identify novel IRF-1-binding proteins. A, overlapping peptides (20 amino acids long with an N-terminal biotin tag and SGSG spacer and 5-amino acid overlap) spanning the entire length of the IRF-1 protein that were used to generate peptide aptamer affinity columns. B, eluates from the peptide aptamer affinity chromatography columns were analyzed on 4–12% gradient gels, and protein bands were detected using colloidal blue stain. C, pie chart showing the IRF-1-binding proteins identified by mass spectrometry that were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (34, 35) and classified by Uniprot keyword into functional biological categories. The table shown lists the number of identified proteins in each functional category; the percentage with respect to the total number of identified proteins (%); the p value, indicating significance; and the enrichment score, showing the enrichment of each category in the IRF-1 column eluates when compared with its occurrence in the human proteome. D, table expanding on the output from DAVID showing proteins classified as transcriptional regulators and proto-oncogenes.
Preliminary analyses, where a range of protein bands from the Mf2 columns were selected, demonstrated that the amount of individual proteins was sufficient for identification by mass spectrometry. A comprehensive analysis of the proteins bound to columns 8 and 9 was therefore carried out (supplemental Fig. S3B) based on the identification of peptide profiles from the mass spectrum of digested proteins (peptide mass fingerprinting) and confirmation of protein identity by fragmentation of one or more selected peptides from the MS spectrum in the case of poor peptide mass fingerprinting identification. We used this approach for identification of samples from both one- and two-dimensional gels (supplemental Fig. S3C and Method S2), and the measurement was performed by MALDI TOF/TOF mass spectrometry (supplemental Table T1). The identified proteins were then analyzed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID, version 6.7) (34,35) to group them into functional biological categories (Fig. 2C).
Because we are particularly interested in the identification of IRF-1-interacting proteins that may regulate its transactivation or tumor suppressor activity, we focused on proteins that fell into the categories of transcriptional regulators and proto-oncogenes (Fig. 2D). From this list, we picked three proteins that were of particular interest with respect to potential post-translational regulation of IRF-1 (i.e. known transcriptional regulators that also had a link to cancer) (NPM, YB-1, and TRIM28) and carried out a more detailed validation. Interestingly, although a number of novel Mf2 domain interactions were identified, we did not identify CHIP by this method. This could be due to the low levels of CHIP expressed in A375 cells, or it may reflect the fact that IRF-1·CHIP complexes form preferentially in cells exposed to heat or heavy metal stress (33). Thus, CHIP binding to the Mf2 domain of IRF-1 in unstressed cells may be negatively regulated by other CHIP-interacting proteins or by post-translational modification.
Validation of NPM Binding to IRF-1
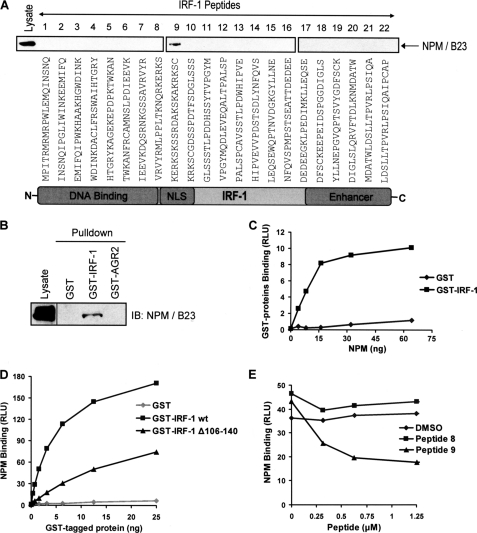
One of the Mf2-interacting proteins identified above, NPM (supplemental Table T1), is a nuclear phosphoprotein and chaperone that has been implicated in the regulation of a number of transcription factors with oncogene or tumor suppressor activity and can itself act as an oncogene (36–40). Although previously identified as a potential regulator of IRF-1 (25), the link between IRF-1 and NPM has not been characterized at the molecular level. To address this, we first validated binding of NPM to the Mf2 domain peptides using immunoblot analysis (Fig. 3A). NPM bound primarily to peptide 9, although in line with the MS analysis (supplemental Table T1), weak binding of NPM to peptide 8 was also detected on longer exposures (data not shown). Thus, NPM predominantly binds to peptide 9 of the Mf2 domain, forming a stable isolatable complex.
FIGURE 3.
NPM binds to the Mf2 region of IRF-1. A, eluates from the peptide aptamer affinity chromatography columns (see supplemental Fig. S3A) were analyzed on 4–12% gradient gels. NPM binding was determined by immunoblot developed using anti-NPM. B, immunoblot showing the binding of NPM from A375 lysates to recombinant GST, GST-IRF-1, and GST-AGR2 immobilized on glutathione-Sepharose. The immunoblot was developed using anti-NPM. The lysate was analyzed on the same gel, but the lanes were not contiguous and the data are representative of at least two independent experiments. C, GST or GST-IRF-1 was coated onto a microtiter plate and incubated with a titration (0–64 ng) of His-NPM. Binding was detected using an anti-His antibody and enhanced chemiluminescence. The amount of protein (ng) against binding, expressed as relative light units (RLU), is shown. The results are representative of two separate experiments. D, recombinant His-NPM was coated on a microtiter plate and incubated with a titration (0–25 ng) of GST alone, GST-IRF-1 WT, or GST-IRF-1 Δ106–140. Binding was detected as above except using anti-GST. E, GST-IRF-1 was coated onto a microtiter plate, following which a fixed amount of His-NPM preincubated with a titration (0–1.25 μm) of peptide 8, peptide 9, or the DMSO carrier, was added to the plate. Binding was detected as in C. Peptide concentration against binding, expressed as relative light units, is shown. The data are representative of two independent experiments.
In order to establish if NPM could bind to full-length IRF-1, a column of GST-IRF-1 immobilized on glutathione-Sepharose beads was loaded with A375 cell lysate and washed extensively. Bound proteins were analyzed by SDS-PAGE/immunoblot following incubation of the beads with SDS sample buffer. NPM bound to the IRF-1 containing column but not to GST alone or GST-AGR2 control columns (Fig. 3B). The interaction between IRF-1 and NPM was demonstrated to be direct by carrying out a protein interaction assay using purified components. GST or GST-IRF-1 was coated onto a microtiter well and incubated with His-NPM, expressed and purified from E. coli, which was in the mobile phase. This showed that IRF-1, but not GST, could interact directly with NPM in the absence of other cellular factors (Fig. 3C). To determine whether the binding of NPM to full-length IRF-1 was through the Mf2 domain, the following approaches were taken. First, a mutant IRF-1 protein was used in which the Mf2 domain (amino acids 106–140; GST-IRF-1 Δ106–140) had been deleted (33). Binding of NPM to the Mf2 domain deletion mutant was significantly impaired when compared with binding to wild-type IRF-1 (Fig. 3D). Second, the ability of peptide 9 to diminish NPM binding by competing with full-length IRF-1 was measured. When NPM and IRF-1 levels were kept constant and peptide 9 was titrated into the assay, it competed with full-length IRF-1 for binding to NPM (Fig. 3E), whereas, consistent with the weak binding detected in Fig. 3A, peptide 8 had no significant effect on NPM binding when compared with a DMSO control. The data presented in this section suggest that the region represented by peptide 9 (amino acids 121–140) is the major interface for NPM on IRF-1.
NPM Binding to a Short Linear Motif
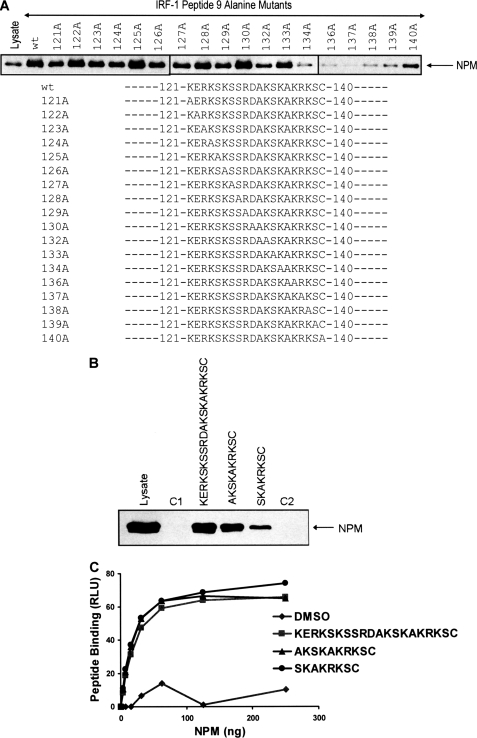
Recent studies have suggested that ID domains frequently house linear interaction motifs that can be composed of 10 or fewer amino acids. To determine whether the Mf2 domain contained a linear interaction motif that bound to NPM, we defined residues critical for binding using a library of peptide 9 derivatives in which each amino acid had been sequentially mutated to Ala (Fig. 4A). The library was used to generate individual affinity chromatography columns, and the ability of NPM from A375 cell extracts to bind the mutant peptides was determined. This showed that a core motif comprising Lys134–Ser139 (KAKRKS) of IRF-1 contained residues that were essential for stable NPM binding, with mutation of Arg137 being sufficient to essentially abolish the interaction. It should be noted that some other residues, such as Lys132, also appeared to make a contribution to binding, although they were not as critical as the core motif. Based on the results of the Ala scan, two shorter peptides 131AKSKAKRKSC140 and 133SKAKRKSC140 were generated, both of which contained the core KAKRKS residues and one of which also contained Lys132. When asked to bind NPM from cell lysates, whereas the 131AKSKAKRKSC140 peptide retained capacity similar to that of the WT peptide, the shorter 133SKAKRKSC140 peptide showed reduced binding, suggesting that it had lost some of its affinity for NPM (Fig. 4B). However, 133SKAKRKSC140 was as efficient as the WT and 131AKSKAKRKSC140 peptides in binding to recombinant NPM (Fig. 4C). This suggests that either Lys132 is more important for binding to endogenous NPM than the recombinant protein or that the shorter peptide loses some specificity and can therefore bind to a greater range of cellular proteins, some of which would then compete with NPM for binding. However, the fact that the 133SKAKRKSC140 peptide retains significant activity for NPM binding suggests that the core residues are both required and sufficient for the interaction.
FIGURE 4.
NPM binds specifically to a basic linear interaction motif within the Mf2 region of IRF-1. A, eluates from peptide aptamer affinity chromatography columns based on peptide 9, where each amino acid had been sequentially mutated to alanine, were analyzed on 4–12% gradient gels, and NPM was detected by immunoblot using anti-NPM. B, affinity columns were generated using the indicated peptides. C1 (IRF-1 peptide 4) and C2 (IRF-1 peptide 21) were used as controls for nonspecific binding, and NPM binding was analyzed as in A. C, a fixed amount of the indicated peptides (or a carrier alone control; DMSO) was immobilized on a streptavidin-coated microtiter plate and incubated with a titration (0–250 ng) of His-NPM. Binding was detected using an anti-His antibody and enhanced chemiluminescence. The amount of protein (ng) against binding, expressed as relative light units (RLU) is shown.
YB-1 and TRIM28 Bind to the ID Region of IRF-1
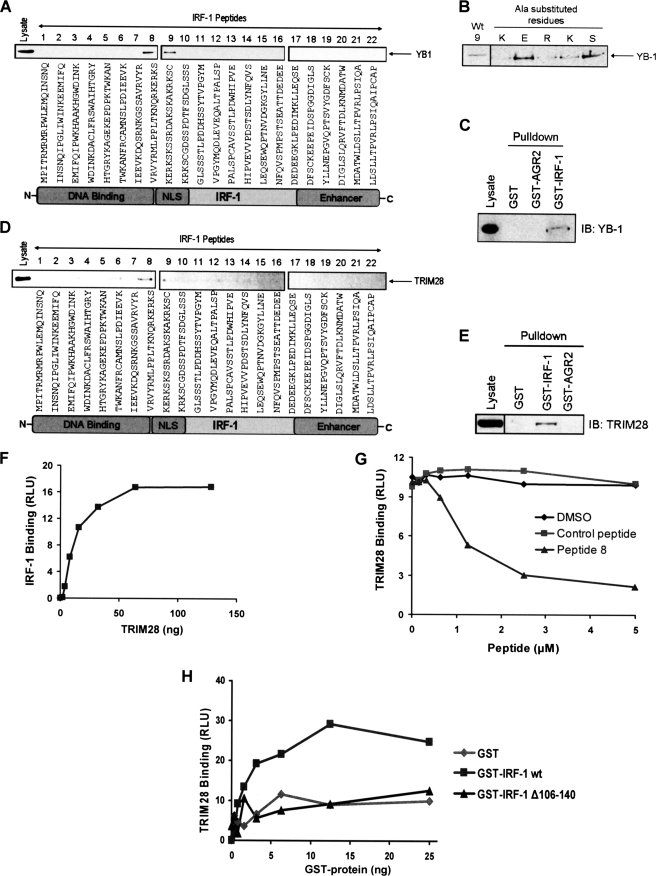
Having validated NPM, a previously identified regulator of IRF-1 (25), as a direct binding partner that interacts with a short linear motif from the Mf2 domain of IRF-1, we moved on to look at two potentially novel IRF-1-interacting factors. The two proteins YB-1 and TRIM28 (also known as KAP1) were chosen from the list of Mf2 binding proteins identified by MS analysis (supplemental Table T1) on the basis that both of these proteins are associated with the regulation of gene transcription (41–43). First, we screened the IRF-1 peptide series for binding to YB-1, a Y-box protein that is overexpressed in various cancer types and has been reported to regulate both transcription and translation. We found that, unlike NPM, which bound preferentially to peptide 9 of the Mf2 domain, cellular YB-1 interacted equally well with both peptides 8 and 9 (Fig. 5A). This suggests that although both YB-1 and NPM can bind to the Mf2 domain, there is a difference in the exact interface they employ for binding. The specificity of YB-1 binding was demonstrated using a series of peptides based on peptide 9, where residues that overlapped (KERKS) with the second YB-1-interacting aptamer, peptide 8, were sequentially mutated to Ala. This showed that although substitution of the Ser and Glu residues had no impact on binding, the binding was specific because loss of a single basic amino acid was sufficient to significantly reduce the interaction (Fig. 5B) despite the fact that the net charge of the alanine mutant peptides was still highly basic. In order to verify that YB-1 could bind to the Mf2 domain when in the context of full-length IRF-1, binding of YB-1 to GST-IRF-1 immobilized on glutathione-Sepharose was assessed. Fig. 5C shows that when A375 cell lysate was passed down the GST-IRF-1 column, YB-1 bound specifically to IRF-1 because it was not present in the eluate from either a GST alone column or a control column containing GST-AGR2. Thus, endogenous cellular YB-1 can bind to full-length immobilized IRF-1 protein.
FIGURE 5.
YB-1 and TRIM28 bind to the highly disordered Mf2 region of IRF-1. A, eluates from the peptide aptamer affinity chromatography columns (see supplemental Fig. S3A) were analyzed on 4–12% gradient gels. YB-1 binding was determined by immunoblot developed using anti-YB-1. B, affinity columns were generated using peptides based on peptide 9, where the residues that overlap with peptide 8 (KERKS) were sequentially mutated to Ala. YB-1 binding was analyzed as in A. C, immunoblot showing the binding of YB-1 from A375 lysate to recombinant GST, GST-IRF-1, and GST-AGR2 immobilized on glutathione-Sepharose. The immunoblot was developed using anti-YB-1 polyclonal antibody. The data are representative of at least two independent experiments. D, as in A except that TRIM28 binding was determined by immunoblot developed using anti-TRIM28. E, as in C, GST, GST-IRF-1, and GST-AGR2 were immobilized on glutathione-Sepharose, following which A375 lysate was incubated with the immobilized proteins for 1 h. After extensive washing, bound proteins were eluted using sample buffer and analyzed by SDS-PAGE/immunoblot using anti-TRIM28. F, a fixed amount of untagged IRF-1 synthesized using the PUREsystem classic II protein synthesis kit was coated onto a microtiter plate and incubated with a titration (0–128 ng) of recombinant TRIM28. Binding was detected using anti-TRIM28 and enhanced chemiluminescence. The amount of protein (ng) against binding, expressed as relative light units (RLU) is shown. G, GST-IRF-1 was coated onto a microtiter plate, following which a fixed amount of recombinant TRIM28 preincubated with a titration (0–5 μm) of peptide 8, a control peptide (IRF-1 peptide 4), or the DMSO carrier was added to the plate. Binding was detected as in F. Peptide concentration against binding, expressed as relative light units, is shown. H, recombinant TRIM28 was coated on a microtiter plate and incubated with a titration (0–25 ng) of GST alone, GST-IRF-1 WT, or GST-IRF-1 Δ106–140. Binding was detected and expressed as in F except using anti-GST.
We moved on to assess the possible interaction of TRIM28 with IRF-1. TRIM28 is a transcriptional corepressor that normally functions in concert with other repressors to regulate gene expression (44, 45) and that has recently been associated with metastasis of primary breast cancers (46). When we characterized the interaction of endogenous cellular TRIM28 with the IRF-1 peptide aptamer series, it bound preferentially to peptide 8 from within the Mf2 region, although weaker but consistent binding to peptide 9 and to a couple of other N-terminal domain peptides was also detected, suggesting that it forms a complex interface with IRF-1 (Fig. 5D). This hypothesis was supported by the fact that cellular TRIM28 could bind recombinant GST-IRF-1 immobilized on glutathione-Sepharose with relatively high affinity to form a stable isolatable complex (Fig. 5E). To verify that the interaction between TRIM28 and IRF-1 was direct and that it did not require the presence of other cellular proteins, we used purified proteins and demonstrated binding of TRIM28 to IRF-1 when it was immobilized on a microtiter plate, after cleaving off the GST tag (Fig. 5F). Although data presented in Fig. 5D suggested that the interface between IRF-1 and TRIM28 may be complex, the Mf2 domain is shown to be integral to the interaction because peptide 8 can compete very effectively with full-length IRF-1 for binding to TRIM28 when compared with a control peptide or with the peptide carrier (Fig. 5G; DMSO). Additionally, deletion of the Mf2 domain from IRF-1 reduces TRIM28 binding to background levels (Fig. 5H), suggesting that the Mf2 domain contains the primary binding interface for TRIM28 on IRF-1.
Cellular IRF-1 Complexes
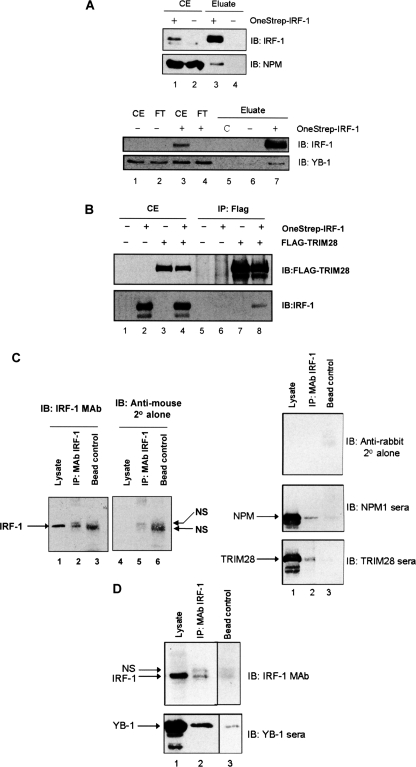
Finally, we carried out protein complex capture and immunoprecipitation assays in order to establish whether any of the binding proteins characterized above could be isolated in IRF-1 complexes from cells. First, we determined the ability of OneStrep-tagged IRF-1 to interact with endogenous NPM and YB-1 following capture of the IRF-1 complexes using Streptactin beads (Fig. 6A). Both endogenous NPM and YB-1 were found in the OneStrep-IRF-1 pull-down and not in the OneStrep control, suggesting that IRF-1·NPM and IRF-1·YB-1 complex formation occurs in A375 cells (Fig. 6A) (for NPM, top, compare lanes 3 and 4; for YB-1, bottom, compare lanes 6 and 7). For the IRF-1-TRIM28 interaction, OneStrep-IRF-1 was co-transfected with FLAG-tagged TRIM28. TRIM28 was then immunoprecipitated using an anti-FLAG monoclonal antibody. Under these conditions, IRF-1 co-precipitates with the TRIM28 immunocomplex (Fig. 6B, compare lane 8 with lane 6). The results of the experiments described in this section suggest that IRF-1 can be found in cellular complexes with all three of the Mf2-binding proteins investigated.
FIGURE 6.
Both endogenous and exogenously expressed IRF-1 can associate with NPM, YB-1, and TRIM28 in A375 cells. A, immunoblot (IB) of OneStrep-IRF-1 isolated using Streptactin from A375 cells that had been transiently transfected with OneStrep-IRF-1 or empty vector as indicated. The immunoblots were probed for IRF-1 and NPM (top panels). A duplicate experiment was performed, and immunoblots were probed for IRF-1 and YB-1 (bottom panels). Crude cell extract (CE) and flow-through (FT) from the Streptactin column are shown. C, a beads only control. B, immunoblot of FLAG-TRIM28 immunoprecipitated (IP) with anti-FLAG antibody from A375 cells transiently transfected with FLAG-TRIM28, OneStrep-IRF-1, or empty vector, as indicated. The immunoblots were probed for IRF-1 and FLAG. C, A375 cell lysate (4 mg of total protein/condition) was incubated with protein-G beads alone (Bead control) or with protein G that had been cross-linked to anti-IRF-1 mAb (IP: mAb IRF-1). Following extensive washing, bound proteins were analyzed by 10% SDS-PAGE/immunoblot together with the load (Lysate). The left panels show the membrane developed initially with anti-mouse secondary antibody (2°) alone (middle panel); the same membrane was then probed with anti-IRF-1 followed by the anti-mouse secondary antibody to show specific IRF-1 bands (far left panel). Right panels, the membrane was probed with anti-rabbit secondary antibody alone (top) and then sequentially with NPM polyclonal sera and TRIM28 polyclonal sera. The data are representative of two separate experiments. D, a separate experiment was carried out as in C and probed with either anti-IRF-1 mAb or polyclonal YB-1 sera. NS, a nonspecific band picked up by the secondary antibody.
We next determined whether endogenous IRF-1 complexes contained any of the Mf2-binding proteins. Because IRF-1 runs close to the antibody heavy chain on SDS-PAGE, we avoided the majority of heavy chain contamination by first cross-linking the IRF-1 mAb to protein G beads. The IRF-1 mAb cross-linked beads were incubated with A375 cell lysate, and bound proteins were analyzed by immunoblot. Fig. 6C (lanes 4–6) shows the background banding pattern when the membrane was incubated with anti-mouse secondary antibody (2°) alone; this illustrates nonspecific background bands picked up in the beads plus lysate control lane and the IP lane (NS). However, when the IRF-1 primary antibody was applied to the same membrane followed by the secondary antisera, a specific IRF-1 band was detected in the IP lane (lane 2) that corresponded to the IRF-1 band from whole cell lysate (lane 1). A duplicate membrane showed little background from the anti-rabbit secondary antibody (right, top); however, when NPM (middle) or TRIM28 (bottom) antisera were used, both proteins were shown to co-immunoprecipitate with IRF-1, and background binding in the bead control was negligible (compare lane 2 with lane 3). In a separate experiment (Fig. 7D), although YB-1 gave some background binding to the bead alone control (bottom; lane 3) a significant increase in YB-1 protein was readily detected when the IRF-1 mAb was present (lane 2), demonstrating complex formation between these two proteins. Thus, a portion of the endogenous IRF-1 protein can be found in complexes with NPM, YB-1, and TRIM28.
FIGURE 7.
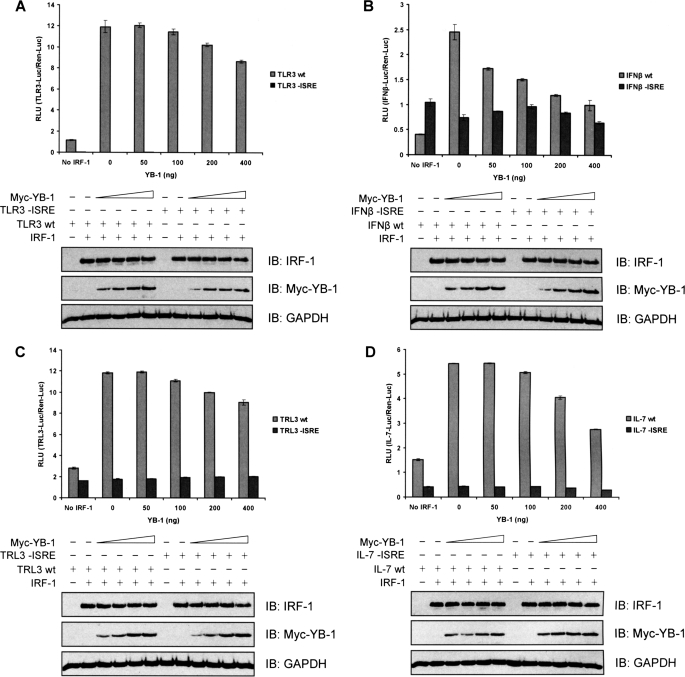
YB-1 represses IRF-1-dependent gene activation. A, H1299 cells were co-transfected with a TLR3-Luc reporter plasmid (WT or mutant lacking the ISRE; 140 ng), a control Renilla-Luc plasmid (60 ng), pcDNA3-IRF-1 (100 ng), and a titration of Myc-YB-1 (0–400 ng), as indicated. DNA amounts were normalized across samples using pcDNA3 empty vector. Post-transfection (24 h), the cells were harvested, and dual luciferase reporter assays were performed. Results were normalized by expressing firefly luciferase/Renilla luciferase activity in relative light units (RLU) as the mean ± S.D. (error bars). YB-1, IRF-1, and GAPDH (loading control) protein levels were detected by SDS-PAGE/immunoblot. B–D, as in A except that a reporter plasmid containing firefly luciferase regulated by the IFNβ promoter (WT or mutant lacking the ISRE; B) or the TRAIL promoter (TRL3 WT or mutant without the ISRE; C) or the IL-7 promoter (WT or mutant minus the ISRE; D) were used instead of the TLR3 reporter construct.
YB-1 Can Repress IRF-1-dependent Transcription
We were keen to establish whether binding of either of the novel Mf2 domain-interacting proteins, TRIM28 or YB-1, had consequences in terms of IRF-1 activity. Because TRIM28 has been reported to function as part of a multiprotein repressor complex (47), we decided to concentrate on YB-1 because it is, by itself, sufficient to modulate the activity of some transcription factors (66, 67). Reporter assays were carried out using a range of IRF-1-responsive gene promoters linked to luciferase. To ensure that we measured IRF-1-dependent transcription, H1299 cells (which have low expression of endogenous IRF-1) were used, and the transcriptional activity of exogenous IRF-1 was determined. We also established that reporter constructs where the IRF-1-responsive element had been mutated, or deleted, were not stimulated by transfected IRF-1 (Fig. 7, A–D). Interestingly, although YB-1 was able to repress IRF-1-dependent transcription from all of the promoter constructs tested, there was a significant difference in their apparent sensitivity. Thus, YB-1 inhibited IRF-1 activity by up to 25–30% (relative to the background; i.e. in the absence of exogenous IRF-1) when its activity was determined using either the TLR3 (Fig. 7A) or TRAIL (TRL3; Fig. 7C) constructs. However, IRF-1 activity against the IFN-β (Fig. 7B) and IL-7 (Fig. 7D) promoters was reduced by as much as 60–80%. In addition, the kinetics of YB-1-mediated repression varied, with IRF-1 activity against the IFN-β reporter showing significant inhibition at the lowest amount of YB-1 used (50 ng), whereas effects on IRF-1 activity assayed using the other reporters required higher amounts of YB-1 (200–400 ng). In all cases, no effect of YB-1 was seen on the activity of promoters that did not contain a functional IRF-1 response element. The data presented in Fig. 7 suggest that binding of YB-1 to the Mf2 domain of IRF-1 can be exploited to modulate the function of IRF-1 as a regulator of gene expression.
DISCUSSION
It is now well accepted that ID proteins or domains are highly represented in the eukaryotic proteome and are particularly prevalent in the proteins involved in cell signaling and the control of gene transcription (1, 48, 49). Thus, more than half of all eukaryotic proteins have unstructured domains, whereas it is estimated that 25–30% of proteins can be found in a mostly disordered state. The challenge for experimentalists is therefore to design approaches to validate ID domains and define their functions in the absence of conventional structural data. Because it is vital to identify the interactome of a protein or domain in order to understand its role in any cellular process, defining ID-binding factors for a given protein or protein domain must be a priority. Here we used IRF-1-based aptamers to ask whether the Mf2 interface from the major intrinsically disordered domain of IRF-1 was a multiprotein binding site for regulators of IRF-1 function.
Exogenous IRF-1 is expressed at low levels in cells, making it difficult to use conventional approaches, such as TAP-tagging linked to mass spectrometry, to identify regulators. In fact, we can find no reports of these approaches being applied successfully to the identification of IRF-1-interacting proteins. On the other hand, full-length immobilized His- or GST-tagged IRF-1 has been used as an affinity matrix to identify interacting proteins from metabolically labeled K-562 cells (25) or from Jurkat T cells (50). These studies identified NPM (25) and CKII (50), respectively, as IRF-1-interacting factors and potentially relevant physiological regulators. In the current study, we used peptide aptamers based on the sequence of IRF-1 to generate affinity matrices because we reasoned that this approach would favor the identification of (i) relatively low affinity interactions as high concentration of the “bait” peptide could be achieved and (ii) short linear interaction motifs prevalent in ID domains. In addition to extending our knowledge about the interactions between NPM (25) and CKII (50) with IRF-1 by defining them as Mf2 domain-binding proteins, we also identified a number of other potential IRF-1-interacting proteins (supplemental Table T1). A portion of the proteins identified as binding to the Mf2 domain are known to be involved in protein synthesis and more precisely are ribosome-associated proteins; however, many of these factors have previously been identified as components of the NPM interactome (51–54) and may therefore bind IRF-1 indirectly. We therefore narrowed down our proteins of interest by leaving out those that fell into the category of protein synthesis when analyzed by DAVID (Fig. 2C). Instead, we focused on proteins with a link to transcriptional regulation that have also been implicated in cancer.
Although, as noted above, NPM had previously been identified as a potential regulator of IRF-1-mediated gene expression (25), the interaction between these two proteins has not been characterized in detail. Here we show that NPM binds directly to IRF-1 through a short linear motif in the Mf2 domain where amino acids 133–140 are required for efficient binding and are sufficient to form a stable interaction, competing for binding with full-length IRF-1 to NPM (Figs. 3 and 4). Interestingly, the NPM binding motif lies within the nuclear localization sequence (NLS) of IRF-1 (amino acids 116–139) (55), and previous studies have demonstrated binding of NPM to the NLS of several viral proteins, including SV40 T-antigen (56, 57). Classical NLS sequences are characterized by clusters of basic residues and therefore fit well into the context of ID domains that are favored by the presence of polar hydrophilic amino acids (58, 59). The location of NLS motifs within inherently flexible ID regions is advantageous because it allows modification of local structures in response to different binding proteins and facilitates competitive binding to multiple targets. For example, the disordered NLS in NF-κB is exposed when the protein is both free and in the DNA bound state, and this appears to facilitate its interaction with either importin-α or members of the IκB family that can inhibit its nuclear import (59). Thus, some of the proteins that bind to the Mf2 domain of IRF-1 may be expected to regulate its nuclear import. Indeed, NPM has been shown to stimulate nuclear import of some of its client proteins (57, 60).
Of the other proteins identified in the IRF-1 aptamer screen, we chose to concentrate on two transcriptional regulators YB-1 and TRIM28 based on their link to cancer development (46, 61–64). TRIM28 has been shown to function, as part of a multiprotein complex, to repress gene expression (47, 65). YB-1 can interact with various transcription factors to either repress or activate gene expression (66, 67). In the current study, we found that YB-1 could repress IRF-1 transcriptional activity as determined using reporter assays. Interestingly, unlike NPM, which appears to repress IRF-1 activity to a similar extent independent of the target promoter (25), there was a significant difference in the efficacy of YB-1 as an inhibitor of IRF-1 dependent on the reporter construct used. It will therefore be of interest to determine whether YB-1 is a signal-specific regulator of IRF-1 and if its ability to bind to and inhibit IRF-1 is linked to the oncogenic activity of YB-1 (68, 69).
In conclusion, one of the main aims of research in the life sciences is to understand how signal transduction and functional pathways operate by “discovering” novel binding proteins. The use of mass spectrometry to identify “stable” protein·protein complexes has proved to be a paradigm-shifting tool. Indeed, this approach has expanded our view of the scope and diversity of an interactome for a given protein. For example, 284 proteins have been identified in an ERK interactome screen (70), and 292 proteins have been identified as binding partners for 14-3-3ζ (71). We report here on the use of peptide aptamers to identify a multiprotein binding interface for a range of cellular proteins that can bind to the tumor suppressor protein IRF-1. This has highlighted a potential role for the Mf2 domain in the negative regulation of IRF-1 transcription by known transcriptional regulators like NPM (25), YB-1 (67), and TRIM28 (64, 65). Thus, in addition to laying the foundations for further studies to characterize the physiological role of YB-1 and TRIM28 as regulators of IRF-1 function, this study suggests that the Mf2 interactome will be modulated in response to IRF-1-activating signals. We will therefore consider expanding on this approach to define the dynamic Mf2 domain interactome.
Acknowledgments
We thank Mirjam Eckert, Jennifer Fraser, Judith Nicholson, and Emmanuelle Pion for helpful discussion and constructive comments.
This work was supported in part by Czech Science Foundation Grant P304/10/0868; Ministry of Defense, Czech Republic, Grant FVZ0000604; Ministry of Education, Youth, and Sports of the Czech Republic Grant LC07017; Institutional Research Concept AV0Z50200510; and Regional Center for Applied Molecular Oncology Grant CZ.1.05/2.1.00/03.0101 and Cancer Research UK Programme Grant C377/A6355.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods S1, S2, and S4; Fig. S3, A–C; and Table T1.
- ID
- intrinsically disordered
- ISRE
- interferon-stimulated response element
- IP
- immunoprecipitation
- NLS
- nuclear localization sequence
- CHIP
- carboxy terminus of Hsp 70 binding protein.
REFERENCES
- 1. Garza A. S., Ahmad N., Kumar R. (2009) Life Sci. 84, 189–193 [DOI] [PubMed] [Google Scholar]
- 2. Miller M. (2009) Curr. Protein Pept. Sci. 10, 244–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gsponer J., Babu M. M. (2009) Prog. Biophys. Mol. Biol. 99, 94–103 [DOI] [PubMed] [Google Scholar]
- 4. Uversky V. N., Oldfield C. J., Dunker A. K. (2008) Annu. Rev. Biophys. 37, 215–246 [DOI] [PubMed] [Google Scholar]
- 5. Iakoucheva L. M., Brown C. J., Lawson J. D., Obradović Z., Dunker A. K. (2002) J. Mol. Biol. 323, 573–584 [DOI] [PubMed] [Google Scholar]
- 6. Collins M. O., Yu L., Campuzano I., Grant S. G., Choudhary J. S. (2008) Mol. Cell Proteomics 7, 1331–1348 [DOI] [PubMed] [Google Scholar]
- 7. Gsponer J., Futschik M. E., Teichmann S. A., Babu M. M. (2008) Science 322, 1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Romeo G., Fiorucci G., Chiantore M. V., Percario Z. A., Vannucchi S., Affabris E. (2002) J. Interferon Cytokine Res. 22, 39–47 [DOI] [PubMed] [Google Scholar]
- 9. Kröger A., Köster M., Schroeder K., Hauser H., Mueller P. P. (2002) J. Interferon Cytokine Res. 22, 5–14 [DOI] [PubMed] [Google Scholar]
- 10. Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. (2001) Annu. Rev. Immunol. 19, 623–655 [DOI] [PubMed] [Google Scholar]
- 11. Dror N., Alter-Koltunoff M., Azriel A., Amariglio N., Jacob-Hirsch J., Zeligson S., Morgenstern A., Tamura T., Hauser H., Rechavi G., Ozato K., Levi B. Z. (2007) Mol. Immunol. 44, 338–346 [DOI] [PubMed] [Google Scholar]
- 12. Dornan D., Eckert M., Wallace M., Shimizu H., Ramsay E., Hupp T. R., Ball K. L. (2004) Mol. Cell Biol. 24, 10083–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gongora C., Degols G., Espert L., Hua T. D., Mechti N. (2000) Nucleic Acids Res. 28, 2333–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clarke N., Jimenez-Lara A. M., Voltz E., Gronemeyer H. (2004) EMBO J. 23, 3051–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Senger K., Merika M., Agalioti T., Yie J., Escalante C. R., Chen G., Aggarwal A. K., Thanos D. (2000) Mol. Cell 6, 931–937 [DOI] [PubMed] [Google Scholar]
- 16. Masumi A., Wang I. M., Lefebvre B., Yang X. J., Nakatani Y., Ozato K. (1999) Mol. Cell Biol. 19, 1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park J., Kim K., Lee E. J., Seo Y. J., Lim S. N., Park K., Rho S. B., Lee S. H., Lee J. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17028–17033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marsili G., Remoli A. L., Sgarbanti M., Battistini A. (2004) Ann. N.Y. Acad. Sci. 1030, 636–643 [DOI] [PubMed] [Google Scholar]
- 19. Watanabe N., Sakakibara J., Hovanessian A. G., Taniguchi T., Fujita T. (1991) Nucleic Acids Res. 19, 4421–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pion E., Narayan V., Eckert M., Ball K. L. (2009) Cell. Signal. 21, 1479–1487 [DOI] [PubMed] [Google Scholar]
- 21. Eschrich S., Zhang H., Zhao H., Boulware D., Lee J. H., Bloom G., Torres-Roca J. F. (2009) Int. J. Radiat. Oncol. Biol. Phys. 75, 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dezso Z., Nikolsky Y., Nikolskaya T., Miller J., Cherba D., Webb C., Bugrim A. (2009) BMC Syst. Biol. 3, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lovegrove F. E., Gharib S. A., Patel S. N., Hawkes C. A., Kain K. C., Liles W. C. (2007) Am. J. Pathol. 171, 1894–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakagawa K., Yokosawa H. (2002) FEBS Lett. 530, 204–208 [DOI] [PubMed] [Google Scholar]
- 25. Kondo T., Minamino N., Nagamura-Inoue T., Matsumoto M., Taniguchi T., Tanaka N. (1997) Oncogene 15, 1275–1281 [DOI] [PubMed] [Google Scholar]
- 26. Ivanov A. V., Peng H., Yurchenko V., Yap K. L., Negorev D. G., Schultz D. C., Psulkowski E., Fredericks W. J., White D. E., Maul G. G., Sadofsky M. J., Zhou M. M., Rauscher F. J., 3rd (2007) Mol. Cell 28, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang C., Maiguel D. A., Carrier F. (2002) Nucleic Acids Res. 30, 2251–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Narayan V., Eckert M., Zylicz A., Zylicz M., Ball K. L. (2009) J. Biol. Chem. 284, 25889–25899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gobom J., Nordhoff E., Mirgorodskaya E., Ekman R., Roepstorff P. (1999) J. Mass Spectrom. 34, 105–116 [DOI] [PubMed] [Google Scholar]
- 30. Harlow E., Lane D. (1999) Using Antibodies: A Laboratory Manual, p. 323, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Möller A., Pion E., Narayan V., Ball K. L. (2010) J. Biol. Chem. 285, 38348–38361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 33. Narayan V., Pion E., Landré V., Müller P., Ball K. L. (2011) J. Biol. Chem. 286, 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang da W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 35. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 36. Yung B. Y. (2007) Chang Gung Med. J. 30, 285–293 [PubMed] [Google Scholar]
- 37. Meani N., Alcalay M. (2009) Expert Rev. Anticancer Ther. 9, 1283–1294 [DOI] [PubMed] [Google Scholar]
- 38. Colombo E., Marine J. C., Danovi D., Falini B., Pelicci P. G. (2002) Nat. Cell Biol. 4, 529–533 [DOI] [PubMed] [Google Scholar]
- 39. Bertwistle D., Sugimoto M., Sherr C. J. (2004) Mol. Cell Biol. 24, 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brady S. N., Yu Y., Maggi L. B., Jr., Weber J. D. (2004) Mol. Cell Biol. 24, 9327–9338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kohno K., Izumi H., Uchiumi T., Ashizuka M., Kuwano M. (2003) BioEssays 25, 691–698 [DOI] [PubMed] [Google Scholar]
- 42. Alter M. D., Hen R. (2008) Neuron 60, 733–735 [DOI] [PubMed] [Google Scholar]
- 43. Lorenz P., Koczan D., Thiesen H. J. (2001) Biol. Chem. 382, 637–644 [DOI] [PubMed] [Google Scholar]
- 44. Lee Y. K., Thomas S. N., Yang A. J., Ann D. K. (2007) J. Biol. Chem. 282, 1595–1606 [DOI] [PubMed] [Google Scholar]
- 45. Liao G., Huang J., Fixman E. D., Hayward S. D. (2005) J. Virol. 79, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ho J., Kong J. W., Choong L. Y., Loh M. C., Toy W., Chong P. K., Wong C. H., Wong C. Y., Shah N., Lim Y. P. (2009) J. Proteome Res. 8, 583–594 [DOI] [PubMed] [Google Scholar]
- 47. Briers S., Crawford C., Bickmore W. A., Sutherland H. G. (2009) J. Cell Sci. 122, 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sigalov A. B. (2010) Mol. Biosyst. 6, 451–461 [DOI] [PubMed] [Google Scholar]
- 49. Mittag T., Kay L. E., Forman-Kay J. D. (2010) J. Mol. Recognit. 23, 105–116 [DOI] [PubMed] [Google Scholar]
- 50. Lin R., Hiscott J. (1999) Mol. Cell Biochem. 191, 169–180 [PubMed] [Google Scholar]
- 51. Piñol-Roma S. (1999) Mol. Biol. Cell 10, 77–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lindström M. S., Zhang Y. (2008) J. Biol. Chem. 283, 15568–15576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maggi L. B., Jr., Kuchenruether M., Dadey D. Y., Schwope R. M., Grisendi S., Townsend R. R., Pandolfi P. P., Weber J. D. (2008) Mol. Cell Biol. 28, 7050–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu Y., Maggi L. B., Jr., Brady S. N., Apicelli A. J., Dai M. S., Lu H., Weber J. D. (2006) Mol. Cell Biol. 26, 3798–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schaper F., Kirchhoff S., Posern G., Köster M., Oumard A., Sharf R., Levi B. Z., Hauser H. (1998) Biochem. J. 335, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Szebeni A., Herrera J. E., Olson M. O. (1995) Biochemistry 34, 8037–8042 [DOI] [PubMed] [Google Scholar]
- 57. Szebeni A., Mehrotra B., Baumann A., Adam S. A., Wingfield P. T., Olson M. O. (1997) Biochemistry 36, 3941–3949 [DOI] [PubMed] [Google Scholar]
- 58. Bäuerle M., Doenecke D., Albig W. (2002) J. Biol. Chem. 277, 32480–32489 [DOI] [PubMed] [Google Scholar]
- 59. Lätzer J., Papoian G. A., Prentiss M. C., Komives E. A., Wolynes P. G. (2007) J. Mol. Biol. 367, 262–274 [DOI] [PubMed] [Google Scholar]
- 60. Gao H., Jin S., Song Y., Fu M., Wang M., Liu Z., Wu M., Zhan Q. (2005) J. Biol. Chem. 280, 10988–10996 [DOI] [PubMed] [Google Scholar]
- 61. Wang C., Ivanov A., Chen L., Fredericks W. J., Seto E., Rauscher F. J., 3rd, Chen J. (2005) EMBO J. 24, 3279–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Braithwaite A. W., Del Sal G., Lu X. (2006) Cell Death Differ. 13, 984–993 [DOI] [PubMed] [Google Scholar]
- 63. Wilkinson M. F., Shyu A. B. (2001) BioEssays 23, 775–787 [DOI] [PubMed] [Google Scholar]
- 64. Chatterjee M., Rancso C., Stühmer T., Eckstein N., Andrulis M., Gerecke C., Lorentz H., Royer H. D., Bargou R. C. (2008) Blood 111, 3714–3722 [DOI] [PubMed] [Google Scholar]
- 65. Groner A. C., Meylan S., Ciuffi A., Zangger N., Ambrosini G., Dénervaud N., Bucher P., Trono D. (2010) PLoS Genet. 6, e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Okamoto T., Izumi H., Imamura T., Takano H., Ise T., Uchiumi T., Kuwano M., Kohno K. (2000) Oncogene 19, 6194–6202 [DOI] [PubMed] [Google Scholar]
- 67. Higashi K., Inagaki Y., Fujimori K., Nakao A., Kaneko H., Nakatsuka I. (2003) J. Biol. Chem. 278, 43470–43479 [DOI] [PubMed] [Google Scholar]
- 68. Fujii T., Kawahara A., Basaki Y., Hattori S., Nakashima K., Nakano K., Shirouzu K., Kohno K., Yanagawa T., Yamana H., Nishio K., Ono M., Kuwano M., Kage M. (2008) Cancer Res. 68, 1504–1512 [DOI] [PubMed] [Google Scholar]
- 69. Basaki Y., Hosoi F., Oda Y., Fotovati A., Maruyama Y., Oie S., Ono M., Izumi H., Kohno K., Sakai K., Shimoyama T., Nishio K., Kuwano M. (2007) Oncogene 26, 2736–2746 [DOI] [PubMed] [Google Scholar]
- 70. von Kriegsheim A., Baiocchi D., Birtwistle M., Sumpton D., Bienvenut W., Morrice N., Yamada K., Lamond A., Kalna G., Orton R., Gilbert D., Kolch W. (2009) Nat. Cell Biol. 11, 1458–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ge F., Li W. L., Bi L. J., Tao S. C., Zhang Z. P., Zhang X. E. (2010) J. Proteome Res. 9, 5848–5858 [DOI] [PubMed] [Google Scholar]