Abstract
Cyclic diguanosine monophosphate (c-di-GMP) is a ubiquitous bacterial second messenger that controls the switch from a single-cell lifestyle to surface-attached, multicellular communities called biofilms. PilZ domain proteins are a family of bacterial c-di-GMP receptors, which control various cellular processes. We have solved the solution structure of the Pseudomonas aeruginosa single-domain PilZ protein PA4608 in complex with c-di-GMP by NMR spectroscopy. Isotope labeling by 13C and 15N of both the ligand and the protein made it possible to define the structure of c-di-GMP in the complex at high precision by a large number of intermolecular and intraligand NOEs and by two intermolecular hydrogen bond scalar couplings. Complex formation induces significant rearrangements of the C- and N-terminal parts of PA4608. c-di-GMP binds as an intercalated, symmetric dimer to one side of the β-barrel, thereby displacing the C-terminal helix of the apo state. The N-terminal RXXXR PilZ domain motif, which is flexible in the apo state, wraps around the ligand and in turn ties the displaced C terminus in a loose manner by a number of hydrophobic contacts. The recognition of the dimeric ligand is achieved by numerous H-bonds and stacking interactions involving residues Arg8, Arg9, Arg10, and Arg13 of the PilZ motif, as well as β-barrel residues Asp35 and Trp77. As a result of the rearrangement of the N and C termini, a highly negative surface is created on one side of the protein complex. We propose that the movement of the termini and the resulting negative surface form the basis for downstream signaling.
Keywords: Cyclic GMP (cGMP), NMR, Prokaryotic Signal Transduction, Protein Folding, Protein Structure, Biofilm, Isotope Labeling, PilZ Domain
Introduction
The second messenger cyclic diguanosine monophosphate (c-di-GMP)5 is widespread in and unique to the bacterial kingdom. Elevated intracellular levels of c-di-GMP generally cause bacteria to change from a motile single-cell state to an adhesive surface-attached multicellular state called biofilm (1–3). Moreover, c-di-GMP controls the virulence of animal and plant pathogens (4–8), progression through the cell cycle (9), antibiotic production (10), and other cellular functions. In several pathogenic bacteria, c-di-GMP has been associated with the regulation of virulence factors and the development of chronic infections. During long term cystic fibrosis lung infections, increased persistence of Pseudomonas aeruginosa correlates with the generation of adaptive colony morphotypes. This includes mucoid colonies and small colony variants, auto-aggregative, hyper-adherent cells whose appearance correlates with poor lung function and persistence of infection. Both morphotypes are strongly linked to elevated levels of c-di-GMP, implicating a central role for this second messenger in chronic P. aeruginosa infections (8, 11–14). These studies emphasize the clinical significance of c-di-GMP and stress the importance for understanding c-di-GMP signaling in the pseudomonads.
c-di-GMP comprises two guanosine monophosphate molecules that are symmetrically connected by 5′-3′ phosphodiester bonds, forming a 12-membered ring (see Fig. 1A). It is synthesized from two molecules of guanosine triphosphate (GTP) by diguanylate cyclases (GGDEF-containing domains) and hydrolyzed into the linear dinucleotide 5′-phosphoguanylyl-(3′-5′)-guanosine by the activity of c-di-GMP-specific phosphodiesterases (EAL-containing domains) (15). The opposing activities of diguanylate cyclases and phosphodiesterases control the intracellular c-di-GMP concentrations, and hence c-di-GMP signaling. Several classes of c-di-GMP effector proteins have been identified so far, including PilZ domain proteins, response regulators, and degenerate GGDEF proteins that have lost catalytic activity (9, 16–19). The PilZ protein family (named after the type IV pilus control protein in P. aeruginosa) represents the best studied class of effectors. Members of this family have been implicated in a diverse range of cellular functions including exopolysaccharide biosynthesis, flagellar motor activity, and virulence gene expression. However, little information is available about the mechanistic details involved in c-di-GMP-mediated PilZ activation of these cellular processes.
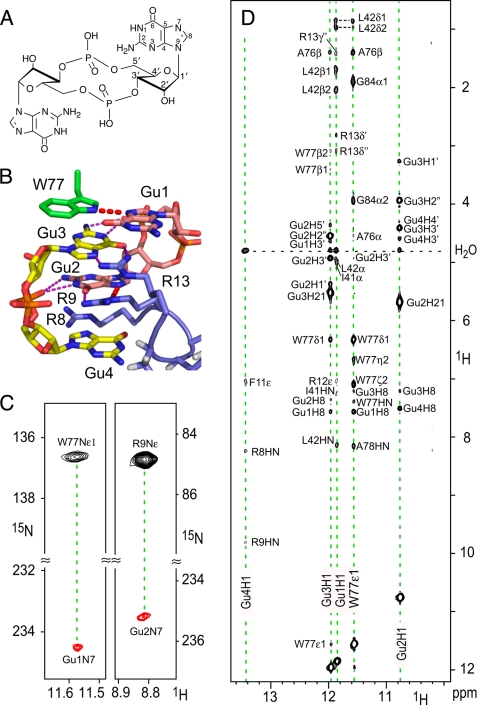
FIGURE 1.
Detection of the dimeric c-di-GMP structure and coordination in the complex with PA4608 by heteronuclear NMR methods. A, the chemical structure c-di-GMP consisting of two GMP moieties linked by O3′-P phosphodiester bonds. B, the intercalated dimeric c-di-GMP as it is bound to PA4608. Because c-di-GMP comprises two guanosine monophosphate molecules that are symmetrically connected by 5′-3′ phosphodiester bonds to a single molecule, we kept the nomenclature of the atoms but numbered the guanosine moieties as guanosines 1 and 2 for the first monomer and guanosines 3 and 4 for the second monomer. For space reasons, guanosines are abbreviated as Gu in the figures. Hydrogen bonds, for which there is experimental evidence, are shown as dashed lines. Two of these result from direct detection via h2JNN couplings (see C) and are shown by thick red dashes. C, extracted regions of two HNN-COSY spectra for direct detection of N–H···N H-bonds (32). The red (negative) signals are cross-peaks resulting from H-bond J-coupling transfer from the H-bond donor nucleus Trp77–15Nϵ1 to the acceptor nucleus Gua1–15N7 (h2JNN = 4.7 ± 0.3 Hz) magnetization and from Arg9–15Nϵ to Gua2–15N7 (h2JNN = 4.8 ± 0.4 Hz), respectively. D, extracted region from the 1H two-dimensional NOESY spectrum showing the NOE contacts from the four imino protons H1 of guanines 1–4 and of Hϵ1 of Trp77. A large number of intermolecular c-di-GMP-to-protein as well as c-di-GMP intradimer contacts are observed that define the c-di-GMP coordination in the complex with PA4608.
Several structures of PilZ domain proteins have been solved in their apo and holo forms. One is the two-domain Vibrio cholerae protein VCA0042. A comparison of apo VCA0042 with its holo structure (20) shows that binding of a single c-di-GMP molecule induces a conformational change in the loop connecting the C-terminal PilZ domain to the N-terminal YcgR-N domain. This brings the two domains into close proximity, thereby forming a new allosteric interaction surface with c-di-GMP at their mutual interface. It has been shown recently that YcgR, a homolog of VCA0042 in Escherichia coli, is able to bind to the flagellar motor in its c-di-GMP-bound form (21). Although VCA0042 appears to bind only one molecule of c-di-GMP and does not alter its quaternary structure upon ligand binding, PP4397 from Pseudomonas putida binding of two molecules of c-di-GMP in the junction of its YcgR-N and PilZ domains results in a dimer-monomer transition (22). This suggested that different PilZ domain proteins exhibit distinct binding stoichiometries and mechanistic principles.
All structures of c-di-GMP protein complexes solved until today involve multidomain proteins (for review, see Ref. 15). In these structures, the binding of c-di-GMP within a hinge-like region induces an allosteric rearrangement of the neighboring domains via fixation to the ligand. It was proposed that this c-di-GMP-induced domain immobilization represents a general mechanism for signal transduction. In contrast to these multidomain proteins, the PilZ domain PA4608 from P. aeruginosa is a single domain c-di-GMP-binding protein. The apo solution structure of PA4608 was solved by Ramelot et al. (23), who also confirmed binding of c-di-GMP by chemical shift analysis. NMR chemical shift mapping was then used to localize the ligand binding site of PA4608 to one side of the protein surface and to demonstrate that c-di-GMP binds to PA4608 as a dimer (16). Furthermore, these analyses demonstrated that PA4608 remains monomeric after ligand binding (16).
Here we report the structure of the complex of PA4608 with c-di-GMP solved by advanced solution NMR techniques. Uniform 13C, 15N isotope labeling of both c-di-GMP and PA4608 allowed the observation of a large number of intermolecular and intraligand NOE contacts, 1DHN and 1DCH ligand and protein residual dipolar couplings (RDCs), and intermolecular hydrogen bond scalar couplings. Both termini of the protein undergo large structural changes upon ligand binding while retaining a certain amount of flexibility. In particular, the C-terminal helix is displaced by the ligand, thereby opening one side of the β-barrel as a binding site, and the N terminus containing the RXXXR PilZ domain motif wraps around the ligand and in turn ties the C-terminal helix in a loose conformation induced by a number of hydrophobic contacts. The rearranged termini expose a highly negatively charged surface on one side of the complex to a possible effector protein. This induced folding of the flexible N terminus containing the PilZ signature RXXXR and the resulting clustering of surface charges may present a general mechanism for c-di-GMP readout by PilZ domains.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Overexpression, 13C, 15N labeling, and purification of PA4608 were achieved as described previously (16). In the protein construct used, the PA4608 sequence is preceded by a hexahistidine tag with the sequence MGSSHHHHHHSSGLVPRGSH. c-di-GMP was produced enzymatically from GTP by using a non-feedback-inhibited mutant of the diguanylate cyclase DgcA named DgcA0646, in which the inhibition site RESD is replaced by GRDC (24, 25). Uniformly 13C- and 15N-labeled c-di-GMP was produced in the same manner but starting with 13C-, 15N-labeled GTP (Spectra Stable Isotopes) as a precursor.
NMR Samples
Uniformly 13C,15N- and 15N-labeled samples of apo PA4608 (0.8–1.2 mm) were prepared in 250 mm NaCl, 10 mm Tris-HCl at pH 6.7, 0.01% NaN3 (w/v), and 5% (v/v) D2O as sample volumes of 200–400 μl. For complex formation, c-di-GMP was titrated to the apo PA4608 samples from a 20 mm stock solution, and the fraction of apo- and ligand-bound protein was monitored by 1H-15N heteronuclear single quantum correlation spectra. The titration was stopped at two equivalents of c-di-GMP, i.e. when the protein was saturated. For preparation of D2O samples, the H2O samples were lyophilized and redissolved in D2O twice. Non-isotropic samples of complexed (apo) protein were prepared by adding 18 mg/ml (26 mg/ml) filamentous phage Pf1 (Asla Biotech).
NMR Spectroscopy and Resonance Assignments
NMR spectra were recorded at 293 K on Bruker DRX 600 and DRX 800 NMR spectrometers equipped with TXI and TCI probe heads, respectively. For protein assignment, structure information, and assessment of backbone dynamics by 15N relaxation, standard two-dimensional and three-dimensional NMR spectra were acquired similar to the ones described (26). c-di-GMP ribose resonances were connected by three-dimensional HCCH-COSY and -TOCSY spectra recorded on 13C/15N-labeled c-di-GMP bound to 13C/15N-labeled PA4608. Ribose moieties and guanine H1/N1 and H8/C8 resonances were assigned via their NOE contacts. Details of the acquired spectra are indicated in the supplemental Table S1. NMR data were processed using the NMRPipe suite of programs (27). Spectra were displayed and analyzed with the programs SPARKY (28) and PIPP (29).
Structure Calculations
Structure calculations were performed with the program XPLOR-NIH (30) using two consecutive simulated annealing protocols (31). In a first step, calculations started from an extended strand with c-di-GMP bound to the protein via the measured H-bond between Hϵ1 of Trp77 and N7 of Gua1, using intraprotein restraints only; the second step included all restraints. c-di-GMP molecules were defined as two-residue, circular strands of RNA. The 20-residue His tag and linker were not included in structure calculations. A total of 200 structures were calculated, and the 20 lowest energy structures were selected for deposition (Protein Data Bank (PDB) code 2L74). The structural statistics are given in Table 1.
TABLE 1.
Assignment and structure statistics
The statistics comprise an ensemble of the final 20 simulated annealing structures. Individual structures were fitted to each other using residues in the β-sheets of the barrel: 19–24, 27–37, 40–44, 57–62, 68–79, and 82–90. The numbers of the various constraints are given in parentheses.
| Assignment statistics | |
| Completeness of resonance assignments (%) | |
| Protein, backbonea (461/502) | 91.8 |
| Protein, all atomsb (1195/1342) | 89.0 |
| c-di-GMPc (58/68) | 85.0 |
| Structure statistics | |
| r.m.s.d. value from experimental distance restraints (Å) | |
| All (1951) | 0.036 ± 0.001 |
| Protein, intraresidue (i = j) (163) | 0.017 ± 0.002 |
| Protein, sequential (|i − j| = 1) (598) | 0.031 ± 0.002 |
| Protein, medium range (1 < |i − j| ≤ 5) (348) | 0.029 ± 0.003 |
| Protein, long range (|i − j| >5) (625) | 0.042 ± 0.002 |
| Protein to ligandd (103) | 0.058 ± 0.006 |
| Within ligand (59) | 0.021 ± 0.005 |
| Hydrogen bondse (55) | 0.033 ± 0.002 |
| r.m.s.d. values from RDCs (Hz) | |
| Proteinf (152) | 0.14 ± 0.01 |
| Ligandg (15) | 0.15 ± 0.03 |
| r.m.s.d. values from TALOS torsion angle restraintsh (°) (169) | 0.88 ± 0.05 |
| r.m.s.d. values from experimental dihedral restraintsi (°) (16) | 0.16 ± 0.17 |
| Deviations from idealized covalent geometryj | |
| Bonds (Å) | 0.003 ± 0.0001 |
| Angles (°) | 0.65 ± 0.01 |
| Impropers (°) | 0.57 ± 0.01 |
| Coordinate precisionk (Å) | |
| Protein backbone non-hydrogen atoms | 0.20 ± 0.05 |
| Protein all non-hydrogen atoms | 0.88 ± 0.07 |
| c-di-GMP non-hydrogen atoms | 0.58 ± 0.18 |
| c-di-GMP non-hydrogen atomsl | 0.38 ± 0.14 |
| PROCHECK quality indicatorsm | |
| In most favored regions of Ramachandran plot (%) | 86.9 |
| In additional allowed regions of Ramachandran plot (%) | 11.7 |
| In generously allowed regions of Ramachandran plot (%) | 1.3 |
| In disallowed regions of Ramachandran plot (%) | 0.1 |
| χ1 pooled S.D. (°) | 22.9 |
| χ2 pooled S.D. (°) | 22.5 |
a Considering 1HN, 15N (except Pro), 13Cα, and 1Hα resonances of residues Met1–Asp125. No resonances were assigned for residues Asp108 through Arg114. The first numbers in parentheses are the assigned resonances, and the second numbers are all occurring resonances.
b Considering routinely assignable 1H, 15N, and 13C resonances of residues Met1–Asp125, excluding N-terminal and Lys amino groups, Arg guanidino groups, hydroxyl protons of Ser, Thr, Tyr, thiol protons of Cys, carboxyl resonances of Asp and Glu, non-protonated aromatic carbons, and Pro 15N. 1H belonging to the same methyl group and Phe, Tyr 1Hδ, 1Hϵ are counted as one.
c Considering H1′, C1′, H2′, C2′, H3′, C3′, H4′, C4′, H5′, H5″, C5′, H1, N1, N2, N7, H8, and C8. Two additional H21 resonances were observed due to hydrogen bonds.
d Derived from two-dimensional, isotope-filtered two-dimensional, and three-dimensional 13C and 15N-edited NOESY spectra.
e There are 49 hydrogen bonds within the protein, 4 within the ligand, and 2 directly measured between protein and ligand. For each backbone hydrogen bond constraint, there are two distance restraints: rNH-O, 1.8–2.5Å, and rN-O, 2.8–3.5Å.
f These comprise 147 RDCs from N-HN and Cα-Hα, and 5 RDCs from aromatic rings.
g 11 RDCs are from the ribose moieties and four of the guanine rings.
h Backbone torsion angle restraints were derived from 13Cα and 13Cβ chemical shifts using the program TALOS+ (43).
i χ1 (15) and χ2 (5) torsion angles were obtained from HN-Hβ ROE distances and 3JHαHβ, 3JNCγ, and 3JC'Cγ couplings.
j The improper torsion restraints maintain planarity and chirality.
k The coordinate precision is the average r.m.s. difference between the individual simulated annealing structures and the mean coordinates. The calculation includes all residues that do not exhibit large amplitude internal motions as evidenced from the 15N relaxation experiments. These consist of all residues from 16 to 103 with the exception of 18, 24, 25, 38, 39, 47–53, and 64–67. The latter are predominantly located in loops.
l From an optimal superposition of all non-hydrogen atoms of c-di-GMP.
m These values are calculated with the program PROCHECK-NMR (38). Values are reported for all non-mobile residues, i.e. Glu7–Phe11, Arg13–Gly107, and Leu116–Ser121, excluding glycines and prolines.
RESULTS AND DISCUSSION
NMR Spectroscopy, Resonance Assignments, and Structure Determination
Standard triple-resonance NMR spectra recorded on PA4608 saturated with c-di-GMP were used to assign backbone and side chain resonances of PA4608. Resonance assignments were obtained for about 92% of the protein backbone in the PA4608·c-di-GMP complex. Missing backbone assignments comprise parts of the N-terminal His tag sequence as well as residues His12 and Asp108–Arg114. In almost all of these cases, resonances were not visible in the 1H-15N heteronuclear single quantum correlation spectra, presumably due to chemical exchange broadening resulting from flexibility on the microsecond to millisecond time scale.
The chemical shifts of c-di-GMP bound to PA4608 are very distinct from free c-di-GMP. In particular, four different imino H1 proton resonances are observed, indicative of four guanosines in an asymmetric environment. This is consistent with the binding of one c-di-GMP dimer to one protein monomer. Of these 4 guanosine residues, all one-bond 1H1/15N1 and 1H/13C resonance pairs could be assigned with the exception of Gua1-H5" and Gua4-H5" by using information from three-dimensional HCCH-TOCSY/-COSY and 15N- or 13C-edited NOESY experiments. For both Gua2 and Gua3, one of the two N2-bound protons could also be clearly observed. In free guanosines, these proton resonances are usually strongly broadened from the rotation around the C2–N2 bond. The observation of the resonances together with the measured NOEs indicates H-bond formation between the intercalated dimers in the following way (Fig. 1B): Gua1-O1P···H1–N1–Gua3, Gua1-O1P···H21–N2–Gua3, Gua4-O1P···H1–N1–Gua2, and Gua4-O1P···H21–N2–Gua2. Two additional H-bonds between c-di-GMP and the protein, Gua1-N7···Hϵ1–Nϵ1-Trp77 and Gua2-N7···Hϵ—Nϵ-Arg9, were detectable by the HNN-COSY experiment (32) (Fig. 1C) and are indicated in Fig. 1B as red dashed lines. The position of the c-di-GMP dimer on the protein is well defined by 103 intermolecular NOEs. A small section of the two-dimensional NOESY spectrum (Fig. 1C) shows some of these NOEs from the four imino protons of the c-di-GMP dimer and the Trp77 Hϵ1 proton, indicating contacts between the ligand and protein residues Arg8, Arg9, Phe11, Arg13, Arg12, Ile41, Leu42, Ala76, and Trp77. The c-di-GMP dimer itself is well defined by 59 intramolecular NOEs and a further 15 1DCH RDCs. Structural restraints of the protein comprise 1734 intramolecular NOEs and 152 1DHN and 1DCH RDCs.
Based on these restraints, a well defined structure of the PA4608·c-di-GMP complex was calculated using conventional simulated annealing protocols. Its quality is reflected in a heavy atom coordinate r.m.s.d. of 0.20 Å for the backbone in the ordered region of the protein and of 0.38 Å for the c-di-GMP dimer. Further structural statistics are given in Table 1.
Ligand-induced Changes in the Backbone Structure of PA4608
As compared with apo PA4608, the complexed protein shows strong chemical shift changes in the N-terminal region around residue Arg9, in the C-terminal region around residue Val120, as well as in the central β-barrel around residue Val36 (16). An analysis of the secondary structure (Fig. 2) and backbone dynamics (see below) shows that structural changes mainly occur in the N- and C-terminal regions, whereas the central β-barrel is largely unaffected. Ligand binding induces a transition of the N-terminal region from an unfolded state to an ordered structure and a strong destabilization of the C-terminal region in particular between residues Asp108 and Arg114. For residues Glu110 to Arg114, no resonances can be detected at all due to exchange broadening, indicating the disappearance of the apo 310 helix γ1 (Glu109–Glu113). Residues Glu115 up to the C terminus are again observable but correspond to an α-helix (α2) between residues Ala117 and Ser121 rather than to the apo 310 helix γ2 (Leu116–Leu118).
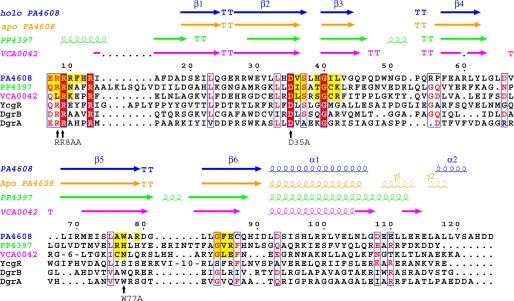
FIGURE 2.
Sequence-structure alignment of various c-di-GMP binding PilZ domains generated with MultAlin (42) and further adjusted manually. Arrows represent β-sheets, and coils represent helices of holo PA4608 (this work), apo PA4608 (PDB ID 1YWU (23)), holo PP4397 (PDB ID 3KYF (22)), and holo VCA0042 (PDB ID 2RDE (20)). Yellow residues represent van der Waals contacts with c-di-GMP. The structures of the c-di-GMP-binding proteins DgrA and DgrB from C. crescentus and YcgR from S. typhimurium are unknown. The signature motifs of the PilZ domain (15) RXXXR and DXSXXG are almost fully conserved (solid red) in all six PilZ domains. The black arrows show DgrA residues that are important for c-di-GMP binding and in vivo function as identified by point mutations (16).
Fig. 2 shows a sequence and secondary structure alignment of PA4608 with the other PilZ domains, for which structures complexed with c-di-GMP have been solved, i.e. P. putida PP4397 (22) and V. cholerae VCA0042 (20). In addition, Caulobacter crescentus DgrA and DgrB and Salmonella typhimurium YcgR have been included because they have similar nanomolar affinities for c-di-GMP as PA4608, and key residues for ligand binding are identified by point mutations (16). Despite a rather low sequence similarity, the β-barrel and helix α1 secondary structure elements of the solved structures are largely conserved. Most importantly, in all aligned proteins, the PilZ domain signature motifs RXXXR and DXSXXG are completely conserved apart from a single amino acid replacement (S/A) for DgrA. Residues that have van der Waals contacts to the ligand in the solved holo structures (yellow) are also largely conserved. These comprise the two RXXXR and DXSXXG motifs and residues around Trp77 (PA4608) and Gly84 (PA4608), which are located on adjacent β-strands. Together with the DVSLHG40 region, the latter form the surface of the β-barrel that is in direct contact with the ligand. Not surprisingly, mutations of the conserved residues Arg8, Arg9, Asp35, or Trp77 (PA4608 numbering) strongly reduce or abolish c-di-GMP binding in DgrA (16).
Description of the PA4608·c-Di-GMP Complex Structure
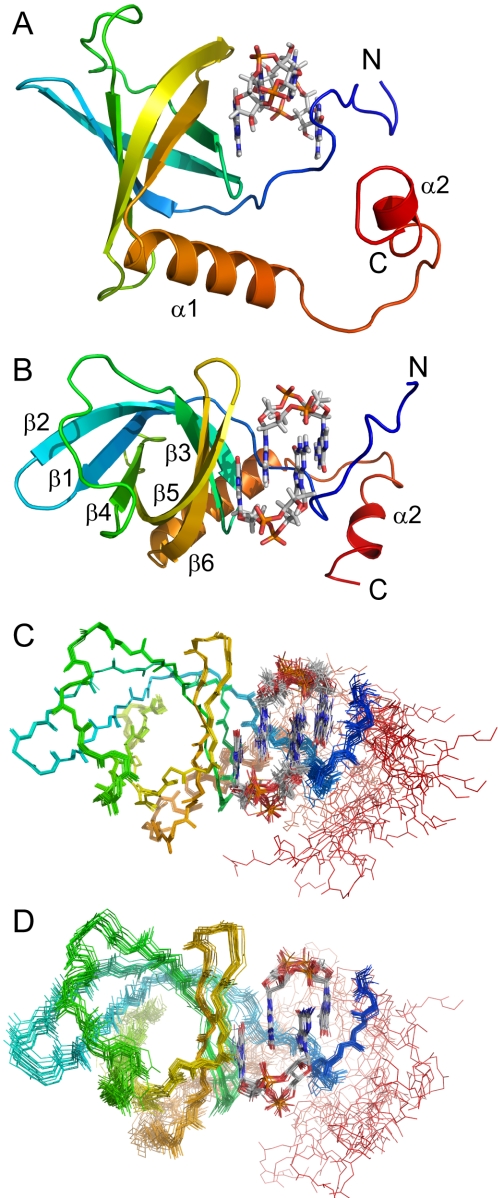
The core structure of PA4608 in complex with c-di-GMP consists of a six-stranded antiparallel β-barrel with an elongated α-helix across one barrel opening (Fig. 3, A and B). The antiparallel β-strands are in the order 1-2-3-6-5-4 with 1 and 4 closing the barrel in an antiparallel manner. The intercalated c-di-GMP dimer is located at one side of the barrel. It makes contacts with residues of the barrel strands β5, β6, β3, and β2 as well as with side chains from the N-terminal part of the sequence, which is unstructured in the apo state. This N-terminal end wraps around the external side of the intercalated c-di-GMP dimer. The face of the N-terminal region, which points away from the ligand, makes contacts to the C-terminal region, thereby also anchoring helix α2 toward the ligand. Fig. 3, C and D, show bundle representations of the 20 lowest energy structures in a best fit superposition of the barrel backbone atoms and the ligand atoms, respectively. The high coordinate precision of the protein core, c-di-GMP, and the N terminus is evident. In contrast, the C-terminal region is less well defined, but clearly not random.
FIGURE 3.
Structure of PA4608 with bound c-di-GMP. A, ribbon representation of the lowest energy structure of holo PA4608 with c-di-GMP in stick representation. B, the same structure rotated by 90° around the y axis. C, overlay of the 20 lowest energy structures in the same orientation and color coding as in B. The first 4 disordered residues are omitted. Structures are superimposed as a best fit of β-barrel residues 19–24, 27–37, 40–44, 57–62, 68–79, and 82–90. D, same as C except that structures are superimposed as a best fit of the c-di-GMP dimer.
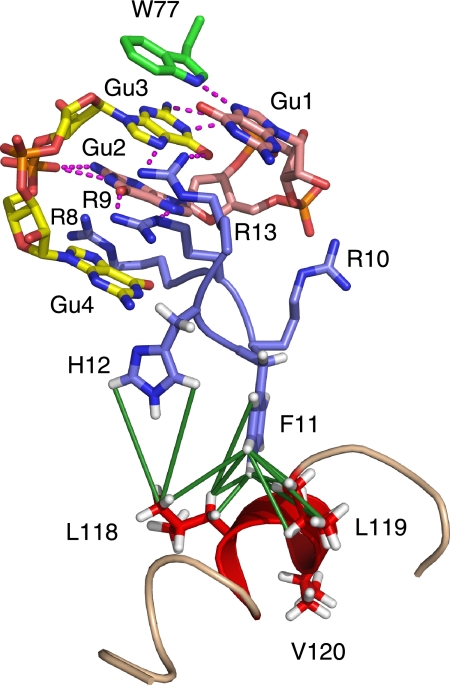
Details of the recognition of c-di-GMP dimer by PA4608 are shown in Fig. 4. The main residues involved are Trp77 and Arg8 through Arg13. Trp77 recognizes the side of the Gua1 base by the h2JNN-detected H-bond from its indole Nϵ1 donor to the Gua1 N7 acceptor (Fig. 1). This locates the Gua1 base in the same plane as the aromatic ring of Trp77. A further H-bond may be possible from the Gua1 H1 hydrogen to the side chain of Asp35 but is not clearly detectable due to low definition of the Asp35 side chain (not shown). The Gua1 base also packs against the side chain of Arg13. The pyrimidine ring of Gua3 stacks below the pyrrole ring of Trp77. The N7 and O6 atoms of Gua3 are recognized by two further H-bonds from the Arg13 Nη1 and Nη2 atoms. Besides the structural proximity, evidence for these H-bonds comes from the observation of resonances and NOE contacts from the Nη1 and Nη2 atoms of Arg13. Usually these proton resonances are not detectable in free arginine side chains due to chemical exchange broadening from the rotation around the Cζ–Nη1 and Cζ–Nη2 bonds. The imino N1 and the amino N2 groups of Gua3 are in H-bonding distance to the Gua1 phosphate group of the first c-di-GMP monomer.
FIGURE 4.
Structural details of the N- and C-terminal coordination of PA4608. Parts of the N-terminal region (blue) comprising residues Arg8, Arg9, Arg10, Phe11, His12, and Arg13 and parts of the C-terminal region comprising helix α2 (red) are shown. The green lines represent the 15 NOE contacts observed between the side chains of Phe11 and His12 and the side chains of Leu118 and Leu119.
The pyrimidine ring of Gua2 stacks below the pyrimidine ring of Gua3. Its imino N1 and the amino N2 groups are in H-bonding distance to the Gua4 phosphate group of the second c-di-GMP monomer. The N7 and O6 atoms of Gua2 are recognized by H-bonds from the Nϵ and the Nη2 atoms of Arg9, respectively. The Arg9-Nϵ–Hϵ···N7–Gua2 H-bond is directly detected by h2JNN couplings (Fig. 1), whereas again the close proximity and the detection of both Arg9 Hη21 and Hη22 protons indicate the presence of the Arg9-Nη2-Hη21···O6-Gua2 H-bond. Finally, the base of the last guanosine Gua4 is packing against the side chain of Arg9 and is turned away from the stacked Trp77/Gua3/Gua2 ring system. Nevertheless, all four guanine bases and the indole ring of Trp77 are nearly coplanar.
The observed H-bond interactions of the N7 and O6 acceptors of Gua3 and Gua2 with the Nη and Nϵ donors of Arg13 and Arg9, respectively, are a very common mode of recognition of guanosines by arginines in protein-DNA complexes (33). A further common recognition motif is identified in the favorable cation-π interactions (34, 35) that can be assumed for the packing of Gua1 against Arg13 and Gua4 against Arg9. The high degree of conservation of Arg9 and Arg13 (Fig. 2) is fully consistent with this observed key role in c-di-GMP dimer recognition. The side chains of Arg8 and Arg10 are not as well defined in the structure, and hence do not allow the identification of non-ambiguous H-bonds. However, the proximity of Arg8 to the guanine ring of Gua4 and of Arg10 to the negatively charged phosphate of Gua2 suggests that Arg8 and Arg10 are also involved in c-di-GMP recognition.
Residues Phe11 and His12 are in the center of the RXXXR13 c-di-GMP recognition motif. Their backbone 1H-15N resonances are very weak (Phe11) or not observed at all (His12), indicating flexibility on the microsecond to millisecond time scale. Nevertheless, the side chains of Phe11 and His12 could be assigned, and 15 unambiguous NOEs are detected from these residues to the side chains of Leu118 and Leu119 in the C-terminal helix α2 (Fig. 4). Despite the apparently high structural flexibility in this region, these hydrophobic contacts clearly dock the C-terminal helix α2 onto the top of the N-terminal c-di-GMP recognition site. Thus, the RXXXR signature motif couples ligand binding directly to the reorientation of the C-terminal part of the protein.
Comparison of the PA4608 c-Di-GMP Binding Mode to Other Known Structures
Besides PA4608, PilZ structures have been solved in both apo and holo forms for two additional proteins, PP4397 (22) and VCA0042 (20). PP4397 consists of a C-terminal PilZ and an N-terminal YcgR domain. It dimerizes in its apo form via interactions between the RXXXR signature motif of one protomer and the PilZ domain of the other protomer. Binding of an intercalated dimer of c-di-GMP to the RXXXR motif in the linker region between the two PP4397 domains induces the separation of the protein dimer into monomers.
Fig. 5, A and B, show a comparison of the c-di-GMP dimer recognition in PA4608 and PP4397. The similarity of the c-di-GMP coordination is very strong but may not be surprising based on the high sequence conservation of the recognition motifs. Thus, (i) Arg123 and Arg127 in PP4397 recognize the bases of Gua2 and Gua3 by the same H-bonds as Arg9 and Arg13 in PA4608; (ii) the side chain of His201 in PP4397 replaces the Trp77 side chain in PA4608 to recognize Gua1 by H-bonds; and (iii) Arg122 in PP4397 has well defined H-bonds to Gua4 that correspond to H-bonds from Arg8 in PA4608 but are not well detected in the NMR structure due to the low definition of the side chain. Further similarities are (iv) the H-bond recognition of the Gua1 imino group by the side chain of Asp157 in PP4397 and (v) the H-bond recognition of Gua2 phosphate by Asn124 in PP4397, which are replaced by Asp35 and Arg10 in PA4608, respectively.
FIGURE 5.
Comparison of c-di-GMP dimer recognition in the stand-alone PilZ protein PA4608, the YcgR-PilZ protein PP4397, and the diguanylate cyclase PleD. A, lowest energy NMR structure of PA4608 (green) coordinating the intercalated two c-di-GMP molecules (pink and yellow). H-bonds are shown as gold dashed lines. B, PilZ (green) and YcgR-N (magenta) domains of PP4397 from P. putida (PDB ID 3KYF (22)) with bound c-di-GMP dimer. The bottom part gives an overview how the c-di-GMP dimer fits into the junction between the YcgR-N (magenta) and PilZ (green, gold) domains of PP4397. C, binding of c-di-GMP dimer to the allosteric inhibition site of the diguanylate cyclase PleD (PDB ID 1W25 (36)). Residues Arg390, Asp362, and Arg359 are from the GGDEF domain (blue), and Arg148, Arg178, and Gly174 are from the D2 adaptor domain (green) of PleD.
VCA0042 also consists of a C-terminal PilZ and an N-terminal YcgR domain, dimerizes via the YcgR domain, and binds c-di-GMP in the linker region between the two domains. However, in contrast to PP4397, the bound c-di-GMP is in monomeric form. It was recently proposed that the replacement of the arginine, which precedes the RXXXR motif, by a leucine (Fig. 2) is responsible for the recognition of a c-di-GMP monomer instead of a dimer (20, 22). It is interesting to note that the respective residues in PA4608 (Arg8) and PP4397 (Arg122) are both arginines and recognize the base of Gua4 in the second c-di-GMP monomer by hydrogen bonds, consistent with the binding of dimeric c-di-GMP to both PA4608 and PP4397.
PleD, a multidomain diguanylate cyclase from C. crescentus, is a member of an additional class of c-di-GMP-binding proteins (36, 37). In this protein family, the c-di-GMP binding site constitutes an allosteric site responsible for tight feedback inhibition of the enzymatic activity (24). Similar to PA4608 and PP4397, PleD binds a dimer of c-di-GMP. Fig. 5C shows the dimer coordination in the same orientation as for PA4608 in Fig. 5A. Again, the specific recognition is mainly achieved by arginines. Thus, Arg390, Arg359, and Arg178 assume the role of the base-recognizing Arg8, Arg9, and Arg13 in PA4608. However, these side chains originate obviously from very different regions of the primary sequence and even from different domains of the protein. In addition, no equivalent of the recognition of Gua1 by the aromatic residue Trp77 (His201) in PA4608 (PP4397) is present in the PleD structure. Thus, although the principle of base recognition by arginine fingers is similar, the overall architecture of the c-di-GMP binding sites is distinct for the two protein families.
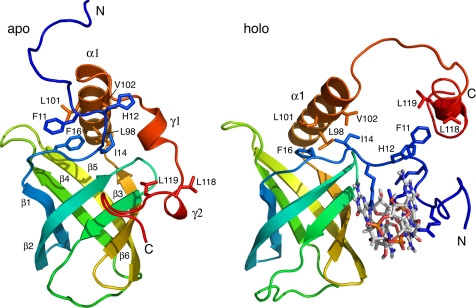
Comparison of PA4608 Apo and Holo Forms
Pronounced changes in the chemical shifts between the apo and holo form of PA4608 indicate significant structural rearrangements in the N- and C-terminal regions upon ligand binding. Fig. 6 shows a comparison between the two states. In the apo form, the c-di-GMP binding side of the β-barrel comprising strands β2, β3, β5, and β6 is partially covered by the C-terminal 310 helices γ1 and γ2. Binding of c-di-GMP to this site pushes the C-terminal region away from the β-barrel and causes the unfolding of helix γ1. In turn, the N-terminal region, which is flexible in the apo state, buries the outer side of c-di-GMP with the residues RRRFHR13 of the recognition motif. The end of the C-terminal region comprising the α-helix α2 covers this part of the N terminus by hydrophobic contacts in a lid-like fashion.
FIGURE 6.
Comparison of overall structure of apo and holo PA4608. A ribbon representation highlighting the ligand-induced reorientation of the N- (blue) and C-terminal (red) parts of the protein is shown. The orientation of the barrel is the same for both structures. Residues involved in contacts between the N terminus and the C terminus and in anchoring helix α1 are shown in stick representation.
During this process, the β-barrel remains basically unchanged. This is reflected by an r.m.s.d. of 0.9 Å between the heavy backbone atom coordinates of the barrel in the apo and the holo structures. However, a certain rearrangement of helix α1 connecting the barrel to the C-terminal part is evident in Fig. 6. It corresponds to a rotation of 11° of the helix axis away from the barrel. The helix is structurally well defined with all its residues recognized as α-helical by PROCHECK-NMR (38) for all 20 lowest energy structures. Its change in orientation was confirmed experimentally from RDCs of the complex and the apo state, which were additionally acquired (supplemental Fig. S1 and Table S2). In the region of helix α1, the RDCs of the apo and holo form agree well with the apo and holo structures, respectively, but deviate for an interchange of structures. The difference in helix orientation is apparently caused by the hydrophobic interactions between helix residues Leu98, Leu101, and Val102 and the N-terminal residues Ile14 and Phe16 in the apo state and rearrangement of the N- and C-terminal regions, which pull the helix away from the barrel in the complexed state.
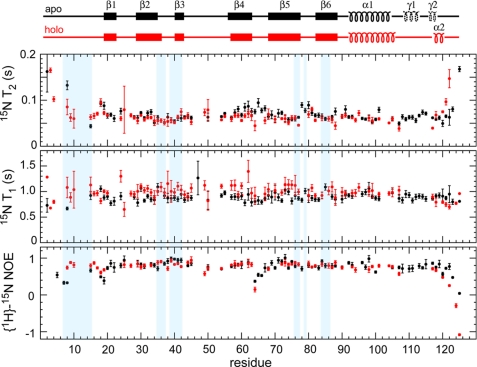
The changes in the protein backbone dynamics between the apo and holo form were also characterized by 15N relaxation experiments carried out at 293 K (Fig. 7). For most residues, the 15N T1 and T2 relaxation times as well as the heteronuclear {1H}-15N NOEs are very uniform and correspond to a well folded structure. Moreover, they are also almost identical between the apo and holo forms. An analysis by the program TENSOR (16, 39) yields isotropic rotational correlation times (τc) of 11.3 ns (12.3 ns) for the apo (holo) form, which are in reasonable agreement with values expected for monomeric PA4608 in its apo form (16.7 kDa) and bound to a dimer of c-di-GMP (18.1 kDa). Also, in agreement with the monomeric state of the complex, no NOEs were detected that would indicate intermolecular dimer contacts. Thus, PA4608 is monomeric in the apo and holo form.
FIGURE 7.
15N T2, T1, and heteronuclear {1H}-15N NOE relaxation data for apo (black) and holo (red) PA4608 as a function of residue number. The secondary structures of apo and holo PA4608 are shown on top. Residues highlighted in pale blue are in van der Waals contact with the ligand. Solid coils represent α-helices, and broken coils represent 310 helices γ1 and γ2.
The relaxation data also give evidence for changes in dynamics upon complex formation for certain parts of the sequence. Thus, the N-terminal residues around Arg8 have low {1H}-15N NOE and large T2 values corresponding to large amplitude motions on the nanosecond time scale for the apo state, whereas these values become comparable with folded residues in the rest of the sequence in the holo state. This is consistent with folding upon ligand binding of the N terminus. In contrast, the C-terminal end around residue Ala122 has the opposite behavior, indicating that the very C-terminal end becomes flexible upon ligand binding. Nevertheless, the adjacent helix α2 of the complexed state appears relatively stable because residues Ala117 and Leu118 of this helix have 15N relaxation parameters that correspond to a folded structure. Consistent with this stability of helix α2, all its 5 residues (Ala117 to Ser121) are found in α-helical conformations in 90% of the lowest energy structures. In the apo state, the region of residues Asp108 to Leu116, which contains the 310 helix γ1, has relaxation parameters of a well folded protein. In contrast, resonances from these residues are strongly broadened or not detectable at all in the holo state, which must be caused by micro- to millisecond dynamics of the protein backbone. Thus, this region undergoes an unfolding transition upon ligand binding.
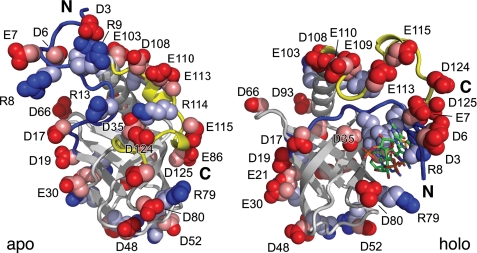
Clustering of PA4608 Surface Charges upon c-di-GMP Binding
The conformational changes in PA4608 induced by c-di-GMP binding lead to a severe rearrangement of surface charges (Fig. 8). Besides the four positive arginines (Arg8, Arg9, Arg10, Arg13), the N-terminal region contains 3 negative residues (Asp3, Asp6, Glu7). The coordination of the ligand by the four arginines causes their side chains to point inward. The concomitant transition to a folded structure of this region reorients the side chains of Asp3, Asp6, and Glu7 toward the outside and creates a negative, surface-exposed cluster (Fig. 8). Likewise, the C terminus contains 7 negative residues centered on the 310 helix γ1 (Asp108, Glu109, Glu110, Glu113, Glu115) and at the very C terminus (Asp124, Asp125). The ligand-induced rearrangement of this region positions these negative charges on top of the negative cluster around residue Glu7. Together with further negative residues of the β-barrel (Asp17, Asp19, Glu21, Glu30, Asp35, Asp48, Asp80) and helix α1 (Glu103), which do not move upon ligand binding, the combined structural changes create one face of the protein, which is strongly negatively charged and devoid of any positive charges. It is attractive to speculate that this newly generated molecular surface constitutes the readout of this small signaling protein by providing a highly charged interaction surface for high affinity regulatory interactions with downstream target proteins.
FIGURE 8.
Changes in surface charge distribution induced by c-di-GMP binding to PA4608. Apo (left) and holo (right) structures are shown in the same orientation as in Fig. 6. Charged residues (Arg, Lys, Asp, and Glu) are indicated by blue and red space-filling models of the side chains with interior atoms painted in fainter colors. The protein backbone is indicated in ribbon representation with N-terminal and C-terminal regions in blue and yellow, respectively.
Although the PilZ domain associates with a plethora of known signaling, catalytic, or transport domains (40), stand-alone or single domain PilZ proteins represent the largest subclass of this protein family (see NCBI/CDART (Conserved Domain Architecture Retrieval Tool)). It is interesting to note that some of the surface-exposed charged side chains that were found to cluster on one side of the c-di-GMP-bound form of PA4608 show conservation within the subclass of stand-alone PilZ domain proteins. In particular, positions corresponding to Glu7 and to a lesser extent Glu115 have conserved negative charges within this family but not within the YcgRN-PilZ family represented by the VCA0042 and PP4397 proteins (supplemental Fig. S2). Consistent with this view, the C terminus of PilZ domains varies substantially between these two PilZ protein families (supplemental Fig. S2). Although the PilZ domains of YcgR homologs have rather conserved sequences of defined length, the C termini of single domain PilZ proteins share little sequence homology, vary substantially in length, and are usually strongly charged. This indicates a high degree of modularity within the C termini of single domain PilZ proteins and suggests that the clustering of key C-terminal residues engaged in potential electrostatic interactions might be a conserved principle for many of these c-di-GMP-binding proteins.
The motional freedom of the C-terminal region of the complexed form of PA4608 together with the abundant negative surface charges may indicate that more than one downstream effector protein with a positively charged surface site exists. Patterns of spatially clustered surface charges that have co-evolved with their respective interaction partners may provide binding specificity for a large variety of protein-protein interactions. This emphasizes the potential of this large family of proteins for functional diversification as signal transducer. Given the diverse cellular functions controlled by c-di-GMP in bacteria, rapid adaptation of this sensing module to different signaling pathways could explain the wide distribution and evolutionary success of stand-alone PilZ domain proteins.
Despite the similar binding of c-di-GMP by the PilZ domains in PA4608, VCA0042, and PP4397 (Fig. 5), the three proteins show fundamental differences in how the ligand binding translates into a molecular readout. Although PA4608 is a monomer in its apo- and ligand-bound form, both VCA0042 and PP4397 form dimers. Ligand binding to YcgR homologs induces large conformational changes within the dimer (VCA0042 (20)) or causes a dimer-to-monomer transition (PP4397 (22)). In both structures, the RXXXR binding motif, also called the c-di-GMP switch (20), forms a hinge-like connector between the N-terminal YcgR and the C-terminal PilZ domain. In VCA0042, the switch is flexible and disordered in the apo structure but undergoes conformational changes upon ligand binding that cause a rotation of the entire PilZ domain and make it switch back toward the N-terminal YcgR domain (20). In contrast, in apo PP4397, the switch forms a short helix with residues within and next to the switch participating in dimerization. Upon c-di-GMP binding, this helix unwinds and wraps around the ligand, thereby contributing to the dimer-to-monomer transition. Thus, in all three proteins, the c-di-GMP switch region is critically involved in signal transduction by making extensive contacts both to c-di-GMP and to the flanking domains (VCA0042 and PP4397) or the C terminus of the same PilZ protomer (PA4608). Despite the different structural context, the central role of the c-di-GMP switch in all three PilZ proteins seems to be to “decode” ligand binding and to transfer this information to flanking regions within the same domain or interacting domains. Although this translates into repositioning of the two domains in the YcgR homologs, it amounts to a striking clustering of charged residues on one side of the PilZ domain protein PA4608. This stresses the versatility and evolutionary flexibility of the PilZ c-di-GMP binding module, which allows for rapid functional expansion in cellular signaling pathways.
During the preparation of this manuscript, Shin et al. (41) published a structural model of PA4608 bound to c-di-GMP based on solution NMR data. The overall structural arrangement is similar. However, in contrast to our work, the position and coordination of the ligand are not well defined because the ligand and the intermolecular interactions were not assigned. Furthermore, NOEs indicating the coordination of the C terminus to the N terminus were not assigned or detected.
Acknowledgments
We thank Matthias Christen, Beat Christen and Regula Aregger for help in producing isotope-labeled c-di-GMP and PA4608, Eric Hajjar for sequence analysis of PilZ domain proteins, as well as Tilman Schirmer for stimulating and helpful discussions.
This work was supported by Swiss National Science Foundation Grants 31-109712 and 31-132857 (to S. G.) and 31-108186 (to U. J.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (code 2L74) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2, Tables S1 and S2, and a sequence alignment file.
- c-di-GMP
- cyclic diguanosine monophosphate
- RDC
- residual dipolar coupling
- r.m.s.
- root mean square
- r.m.s.d.
- root mean square deviation
- NOE
- nuclear Overhauser effect
- ROE
- rotating frame Overhauser effect.
REFERENCES
- 1. Jenal U., Malone J. (2006) Annu. Rev. Genet. 40, 385–407 [DOI] [PubMed] [Google Scholar]
- 2. Hengge R. (2009) Nat. Rev. Microbiol. 7, 263–273 [DOI] [PubMed] [Google Scholar]
- 3. Yan H., Chen W. (2010) Chem. Soc. Rev. 39, 2914–2924 [DOI] [PubMed] [Google Scholar]
- 4. Cotter P. A., Stibitz S. (2007) Curr. Opin. Microbiol. 10, 17–23 [DOI] [PubMed] [Google Scholar]
- 5. Dow J. M., Fouhy Y., Lucey J. F., Ryan R. P. (2006) Mol. Plant Microbe Interact. 19, 1378–1384 [DOI] [PubMed] [Google Scholar]
- 6. Ryan R. P., Fouhy Y., Lucey J. F., Jiang B. L., He Y. Q., Feng J. X., Tang J. L., Dow J. M. (2007) Mol. Microbiol. 63, 429–442 [DOI] [PubMed] [Google Scholar]
- 7. Tamayo R., Pratt J. T., Camilli A. (2007) Annu. Rev. Microbiol. 61, 131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malone J. G., Jaeger T., Spangler C., Ritz D., Spang A., Arrieumerlou C., Kaever V., Landmann R., Jenal U. (2010) PLoS Pathog 6, e1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duerig A., Abel S., Folcher M., Nicollier M., Schwede T., Amiot N., Giese B., Jenal U. (2009) Genes Dev. 23, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fineran P. C., Williamson N. R., Lilley K. S., Salmond G. P. C. (2007) J. Bacteriol. 189, 7653–7662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Merighi M., Lee V. T., Hyodo M., Hayakawa Y., Lory S. (2007) Mol. Microbiol. 65, 876–895 [DOI] [PubMed] [Google Scholar]
- 12. Drenkard E., Ausubel F. M. (2002) Nature 416, 740–743 [DOI] [PubMed] [Google Scholar]
- 13. Meissner A., Wild V., Simm R., Rohde M., Erck C., Bredenbruch F., Morr M., Römling U., Häussler S. (2007) Environ. Microbiol. 9, 2475–2485 [DOI] [PubMed] [Google Scholar]
- 14. Hickman J. W., Tifrea D. F., Harwood C. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schirmer T., Jenal U. (2009) Nat. Rev. Microbiol. 7, 724–735 [DOI] [PubMed] [Google Scholar]
- 16. Christen M., Christen B., Allan M. G., Folcher M., Jenö P., Grzesiek S., Jenal U. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee V. T., Matewish J. M., Kessler J. L., Hyodo M., Hayakawa Y., Lory S. (2007) Mol. Microbiol. 65, 1474–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hickman J. W., Harwood C. S. (2008) Mol. Microbiol. 69, 376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krasteva P. V., Fong J. C., Shikuma N. J., Beyhan S., Navarro M. V., Yildiz F. H., Sondermann H. (2010) Science 327, 866–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benach J., Swaminathan S. S., Tamayo R., Handelman S. K., Folta-Stogniew E., Ramos J. E., Forouhar F., Neely H., Seetharaman J., Camilli A., Hunt J. F. (2007) EMBO J. 26, 5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boehm A., Kaiser M., Li H., Spangler C., Kasper C. A., Ackermann M., Kaever V., Sourjik V., Roth V., Jenal U. (2010) Cell 141, 107–116 [DOI] [PubMed] [Google Scholar]
- 22. Ko J., Ryu K. S., Kim H., Shin J. S., Lee J. O., Cheong C., Choi B. S. (2010) J. Mol. Biol. 398, 97–110 [DOI] [PubMed] [Google Scholar]
- 23. Ramelot T. A., Yee A., Cort J. R., Semesi A., Arrowsmith C. H., Kennedy M. A. (2007) Proteins 66, 266–271 [DOI] [PubMed] [Google Scholar]
- 24. Christen B., Christen M., Paul R., Schmid F., Folcher M., Jenoe P., Meuwly M., Jenal U. (2006) J. Biol. Chem. 281, 32015–32024 [DOI] [PubMed] [Google Scholar]
- 25. Christen M., Christen B., Folcher M., Schauerte A., Jenal U. (2005) J. Biol. Chem. 280, 30829–30837 [DOI] [PubMed] [Google Scholar]
- 26. Grzesiek S., Bax A., Hu J. S., Kaufman J., Palmer I., Stahl S. J., Tjandra N., Wingfield P. T. (1997) Protein Sci. 6, 1248–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 28. Goddard T., Kneller D. (2007) SPARKY 3. 114, University of California, San Francisco [Google Scholar]
- 29. Garrett D., Powers R., Gronenborn A., Clore G. (1991) J. Magn. Reson. 95, 214–220 [DOI] [PubMed] [Google Scholar]
- 30. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 31. Nilges M., Clore G. M., Gronenborn A. M. (1988) FEBS Lett. 229, 317–324 [DOI] [PubMed] [Google Scholar]
- 32. Dingley A. J., Nisius L., Cordier F., Grzesiek S. (2008) Nat. Protoc. 3, 242–248 [DOI] [PubMed] [Google Scholar]
- 33. Luscombe N. M., Laskowski R. A., Thornton J. M. (2001) Nucleic Acids Res. 29, 2860–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dougherty D. A. (1996) Science 271, 163–168 [DOI] [PubMed] [Google Scholar]
- 35. Plevin M. J., Bryce D. L., Boisbouvier J. (2010) Nat. Chem. 2, 466–471 [DOI] [PubMed] [Google Scholar]
- 36. Chan C., Paul R., Samoray D., Amiot N. C., Giese B., Jenal U., Schirmer T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17084–17089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wassmann P., Chan C., Paul R., Beck A., Heerklotz H., Jenal U., Schirmer T. (2007) Structure 15, 915–927 [DOI] [PubMed] [Google Scholar]
- 38. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 39. Dosset P., Hus J. C., Blackledge M., Marion D. (2000) J. Biomol. NMR 16, 23–28 [DOI] [PubMed] [Google Scholar]
- 40. Amikam D., Galperin M. Y. (2006) Bioinformatics 22, 3–6 [DOI] [PubMed] [Google Scholar]
- 41. Shin J. S., Ryu K. S., Ko J., Lee A., Choi B. S. (2011) Protein Sci. 20, 270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Corpet F. (1988) Nucleic Acids Res. 16, 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]