Abstract
Myosin-Va (Myo5a) is a motor protein associated with synaptic vesicles (SVs) but the mechanism by which it interacts has not yet been identified. A potential class of binding partners are Rab GTPases and Rab3A is known to associate with SVs and is involved in SV trafficking. We performed experiments to determine whether Rab3A interacts with Myo5a and whether it is required for transport of neuronal vesicles. In vitro motility assays performed with axoplasm from the squid giant axon showed a requirement for a Rab GTPase in Myo5a-dependent vesicle transport. Furthermore, mouse recombinant Myo5a tail revealed that it associated with Rab3A in rat brain synaptosomal preparations in vitro and the association was confirmed by immunofluorescence imaging of primary neurons isolated from the frontal cortex of mouse brains. Synaptosomal Rab3A was retained on recombinant GST-tagged Myo5a tail affinity columns in a GTP-dependent manner. Finally, the direct interaction of Myo5a and Rab3A was determined by sedimentation velocity analytical ultracentrifugation using recombinant mouse Myo5a tail and human Rab3A. When both proteins were incubated in the presence of 1 mm GTPγS, Myo5a tail and Rab3A formed a complex and a direct interaction was observed. Further analysis revealed that GTP-bound Rab3A interacts with both the monomeric and dimeric species of the Myo5a tail. However, the interaction between Myo5a tail and nucleotide-free Rab3A did not occur. Thus, our results show that Myo5a and Rab3A are direct binding partners and interact on SVs and that the Myo5a/Rab3A complex is involved in transport of neuronal vesicles.
Keywords: Actin, G Proteins, Membrane Trafficking, Myosin, Synapses
Introduction
In presynaptic nerve terminals, neurotransmitter molecules are stored in synaptic vesicles (SVs)2 and are released by the fusion of these vesicles with the plasma membrane. This process and the positioning of SVs at the plasma membrane is regulated by SV-associated proteins, including synapsins, synaptotagmin, synaptobrevin, synaptophysin, calmodulin-dependent kinase II (CaMKII), SV2, and the GTPase Rab3A (1). The mechanism by which actin-dependent motors deliver SVs to the plasma membrane for fusion and the proteins involved are not understood. In this study, we investigated the interaction between Myo5a and Rab3A, two known proteins on SVs.
Rab3A belongs to the Ras superfamily of small monomeric GTPases that is implicated in vesicle fusion and regulated secretion, particularly in neurotransmitter release (2). Mammals have four members of the Rab3 family: Rab3A, Rab3B, Rab3C, and Rab3D, which are predominantly expressed in the brain and neuroendocrine tissues (3). Rab3A is localized to SVs in the active or GTP-bound form and dissociates from the vesicle upon GTP hydrolysis or depolarization of the nerve terminal. The inactive or GDP-bound form of Rab3A binds to Rab GDP dissociation inhibitor (RabGDI). When GDP-GTP exchange occurs Rab3A is able to associate with SVs again (4). GTP-bound Rab3A interacts with multiple specific effector proteins, notably Rabphilin3A (5), RIM (6), Rabin3A (7), and synapsin 1 (8). In addition to Rab3A, only Rabphilin3A and synapsin 1 are known to be localized to SVs.
Myosin-Va (Myo5a) is an actin-based molecular motor that is involved in many cellular processes. As with all myosins, the head domain binds to F-actin and hydrolyzes ATP, whereas the tail domain functions in cargo transport and binds to cargo-specific adaptor proteins or associate with lipid membranes (9). In vitro, the globular tail domain of Myo5 can also stabilize the folded and inhibited conformation of Myo5 through interaction with the motor domain (10). Myo5a is highly expressed in neurons (11–12) and has multiple functions that suggest its involvement in synaptic plasticity. Myo5a has been detected in dendrites of Purkinje cells (13–14), presynaptic terminals of photoreceptor cells (15), intact synaptosomes from rat cerebral cortex (16), and is enriched at the postsynaptic density of excitatory synapses (17). Myo5a is responsible for the transport of a variety of organelles in axon terminals and dendritic spines, such as SVs (18–19) and ER-derived vesicles (20). It also is likely to be associated with mitochondria (21–22) and secretory vesicles (23–25). Inhibition of Myo5a activity causes changes in transport of axonal vesicles along actin filaments (26–27).
Rab GTPases have been identified in some cases as link between Myo5 and organelle transport. In melanocytes, Myo5a is recruited to melanosomes by Rab27a (28–29) and its effector melanophilin (30). Previously, direct interactions have been identified between Rab11a and Rab8a and the tails of Myo5a and Myo5b (31–33), as well as between Rab8a and Myo5c tail (12). Most recently, Rab10 has been shown to associate with vertebrate class 5 myosins (34). Prime candidates for such receptors have been identified for neuronal transport, but a Rab GTPase responsible for Myo5a recruitment to SVs has not been determined yet, although Rab3A has been identified on SVs (3, 35).
Here, we show that the globular tail domain of Myo5a binds to the brain-specific GTPase Rab3A. Based on in vitro motility assays using axoplasm from the squid giant axon, we found that RabGDI inhibited Myo5a-based vesicle transport in neurons. Affinity column chromatography revealed that Rab3A in rat brain synaptosomal preparations binds to Myo5a tail domain. Immunofluorescence analysis confirmed our biochemical findings in cultured frontal cortex neurons. In addition, sedimentation velocity analytical ultracentrifugation experiments showed a direct interaction between highly purified Myo5a and GTP-bound Rab3A. Together, our data show that Rab3A and Myo5a can interact directly via its tail domain on synaptic membranes.
EXPERIMENTAL PROCEDURES
Antibodies
The following commercial primary antibodies were used in this study: rabbit polyclonal anti-myosin5a (DIL2, 36), anti-Rab3A and anti-Rabphilin3A (both from StressGen), anti-Rab4A and anti-Rab11 (both from Santa Cruz Biotechnology), anti-GST (GE Healthcare), anti-synapsin 1 (Invitrogen), and mouse monoclonal antibodies directed against Rab3A (Synaptic Systems), PSD95 (ABR), actin (Sigma-Aldrich) and kinesin (Chemicon). Secondary antibodies included anti-mouse IgG and anti-rabbit IgG labeled with horse-radish peroxidase (both from Chemicon), and goat-anti-mouse IgG and goat anti-rabbit IgG labeled with Alexa 488 or Alexa 594 (Molecular Probes).
Preparation of Squid Brain Clarified Extracts and In Vitro Motility Assays
Squid (Loligo pealei) were obtained from the Marine Biological Laboratory in Woods Hole, MA. Squid optic lobes were dissected from live animals and either used fresh or frozen in liquid nitrogen and stored at −80 °C.
A whole-cell extract from squid optic lobes was prepared as described in Brown et al., (27). Briefly, optic lobes were homogenized in 1 ml of TBS-I (50 mm Tris, pH 8.0, 150 mm NaCl, 5 mm Na2EDTA, pH 7.0) and centrifuged at 100,000 × g for 10 min at 4 °C. The supernatant was diluted 1:2 in TBST-I (TBS-I, 1% Triton X-100) and centrifuged again at 100,000 × g for 10 min at 4 °C. The Triton X-100 extracted supernatant is the clarified extract and was used for affinity column chromatography.
In vitro motility assays from squid axoplasm were performed as previously described in Brown et al. (27). Squid giant axons were dissected from freshly caught squid, placed in filtered seawater, and then finely dissected in Ca2+-free seawater. Immediately following the extrusion of axoplasm, 20 μl of TAMD buffer (25 mm Tris, 5 mm ATP, 5 mm MgCl2, 2 mm DTT, pH 7.5) supplemented with 1 μm rhodamine-phalloidin (Sigma-Aldrich) was added to each slide. Vesicle movement was monitored by video-enhanced contrast differential interference contrast (VEC-DIC) microscopy, and the presence of actin filaments was confirmed by fluorescence microscopy. For motility inhibition experiments, freshly dissected axons were cut in half; one half was extruded onto a coverslip in TAMD buffer containing GST-RabGDI at different concentrations and the other half in TAMD buffer containing GST as a control.
The level of motile activity (number of vesicles moving/field/minute) on actin filaments was determined by counting the total number of moving vesicles on tracks invisible by VEC-DIC microscopy. For each preparation, three random fields (22 × 25 μm) at a distance of 20–60 μm from the bulk axoplasm were selected for analysis. The number of vesicles moving in each field during a period of 3 min was counted and averaged for the three fields. A vesicle was counted each time it showed continuous directed movement on an actin track. If a vesicle stopped and then resumed directed movement in the same or another direction during the observation period, it was counted as a new vesicle.
Isolation of Synaptosomes
Rat brains (Sprague-Dawley rats, 7–8-week-old) from Pel-Freez Biologicals and C57BL/6J Myo5ad-l20J/d-l20J (null mutant) mouse brains from McLaughlin Research Institute Animal Resource Center were obtained frozen and stored at −20 °C until use.
According to the protocols from Carlin et al., (37) and Jordan et al., (38), synaptosomal fractions containing pre- and postsynaptic elements were isolated from adult rat brains and null mutant mouse brains. Briefly, brains were homogenized with a motorized Teflon-glass homogenizer (10 strokes) using 10 g brains/40 ml solution A (0.32 m sucrose, 1 mm NaHCO3, 1 mm MgCl2, 0.5 mm CaCl2, and 0.2 mg/ml CompleteTM protease inhibitor mixture (Roche Applied Science) and diluted homogenates were then centrifuged at 1400 × g for 10 min. The supernatant solution (S1) was saved, and the pellet was resuspended in solution A and subjected to centrifugation (710 × g for 10 min). The resulting supernates (S2) were pooled and subjected to a second centrifugation (710 × g for 10 min). Supernates (S3) were spun again at 30,000 × g for 20 min to obtain a crude fraction and the pellet (P4) was resuspended in solution B (0.32 m sucrose, 1 mm NaHCO3). This homogenate was layered on top of a three-step sucrose gradient (0.85 m, 1.0 m, 1.2 m) and centrifuged at 82,500 × g for 2 h. The crude synaptosomal fraction was collected at the 1.0 m-1.2 m sucrose interface, resuspended in 4 volumes of solution B and centrifuged at 48,200 × g for 20 min. The supernatant (S5) was discarded, and the resulting pellet was resuspended in TBS-I. Aliquots were frozen in liquid nitrogen and stored at −80 °C. All steps during the preparation were carried out on ice or at 4 °C.
Affinity Column Chromatography
GST-Myo5a tail affinity column chromatography was performed as described in Brown et al., (27). Briefly, purified GST-Myo5a tail bacterial lysate was loaded onto a GSTrap column and washed with PBS. Synaptosomal fractions prepared from rat and null Myo5a mutant mouse brains were treated with 0.2 mg/ml complete protease inhibitor mixture and 1% Triton X-100, filtered through a syringe filter unit (Millipore) and applied to the GST-Myo5a tail affinity column. The GSTrap column (GE Healthcare) was washed again with TBS-I and bound proteins were eluted with 50 mm Tris, pH 8.0, and 10 mm glutathione. Fractions were collected and analyzed by SDS-PAGE and Western blots. For GST-RabGDI affinity isolations, a plasmid containing the full-length cDNA for Drosophila GST-RabGDI was expressed in Escherichia coli (BL21). The 75 kDa GST-tagged protein was purified on a GST affinity column and was used in pull-down experiments as stated above.
The effect of the following reagents on affinity column chromatography was tested: 1 mm GDP, 1 mm GTP, 1 mm GTPγS, 2 μm staurosporine (all from Sigma-Aldrich), 20 nm okadaic acid, and 2 μm CaMKIIi (both from Calbiochem). Triton X-100 treated synaptosomes were incubated in the presence of GDP, GTP, and GTPγS and protein kinase/phosphatase inhibitors for 1 h at room temperature and were then applied to GST-Myo5a tail or GST-RabGDI affinity columns.
Cell Culture and Immunocytochemistry
Primary frontal cortex neurons from embryonic day 15 Shoe/-NRMI or null Myo5a mutant mice were prepared as described previously (39). Briefly, frontal cortices were dissociated by papain enzymatic digestion, and then mechanically by transfer pipettes, and plated on glass coverslips coated with poly-l-ornithine at 1.0 × 106 cells/ml. Neurons were prepared for immunofluorescence microscopy at 6–12 days in vitro. Cells were washed with PBS, fixed with 4% paraformaldehyde in 4% sucrose containing PBS for 20 min, treated with 50 mm ammonium chloride in PBS for 10 min, and permeabilized with 0.2% Triton X-100 in PBS for 5 min. Primary (1:100) and secondary (1:300) antibodies were diluted in 0.2% gelatin in PBS and incubated successively for 30–60 min at room temperature. Coverslips were mounted in DABCO (Sigma-Aldrich) and observed by fluorescence microscopy.
Protein Expression and Purification
All recombinant proteins were individually expressed in E. coli (Rosetta II, Novagen) and purified as described previously (40). Cells were harvested, resuspended in lysis buffer (50 mm Tris, pH 7.3; 300 mm NaCl; 5 mm dithiothreitol; 0.2 mg/ml complete protease inhibitor mixture), followed by lysis with a microfluidizer cell disrupter and clarification by centrifugation. The clarified supernatants containing the GST-Myo5a tail protein were passed over a glutathione-Sepharose column (GSTrapTM FF column, GE Healthcare) and GST-Myo5a tail was eluted with a gradient of reduced glutathione. Fractions containing GST-Myo5a tail were combined, treated with thrombin, and dialyzed with three changes against PBS containing 5 mm dithiothreitol. Myo5a tail was further purified over a glutathione-Sepharose column followed by gel filtration chromatography. The GFP-Rab3A construct (41) was PCR-subcloned into the pHis parallel vector (42), and purified by nickel affinity chromatography (HisTrap column, GE Healthcare) followed by dialysis and TEV protease treatment, and further purification over a HisTrap column. Finally, all proteins were further purified using a gel filtration column (Superdex 200, GE Healthcare) pre-equilibrated with 20 mm Tris (pH 7.5), 300 mm NaCl, 1 mm TCEP, and 1 μm ZnCl2.
Analytical Ultracentrifugation
Analytical ultracentrifugation experiments were carried out as previously described (43). A ProteomeLabTM XL-A analytical ultracentrifuge (Beckman Coulter) equipped with absorbance optics and an eight-hole An-50 Ti analytical rotor was used for sedimentation experiments that were carried out at 10 °C and 50,000 rpm (200,000 × g) using 3-mm two-sector charcoal-filled Epon centerpieces with quartz windows. For each protein sample, 300 scans were collected in a period of ∼11 h. Sedimentation boundaries were analyzed by the continuous sedimentation coefficient distribution (c(s)) method using the program SEDFIT (44).
Quantitative Analysis
Images for quantitative analysis were saved as PICT files from high-resolution scanned scientific imaging films (Kodak). Image Gauge software (Fujifilm Inc.) was used to measure pixel intensities (grayscale intensities) of proteins detected by Western blot analysis. Measurements of proteins bound to GST-Myo5a tail or GST-RabGDI were individually normalized against the background of each individual film and renormalized against the signal obtained with the same proteins in synaptosomes. The resulting individual, normalized signal intensities were averaged for each experiment and analyzed by a two-tailed t test.
RESULTS
RabGDI Inhibits Myo5a-based Vesicle Motility
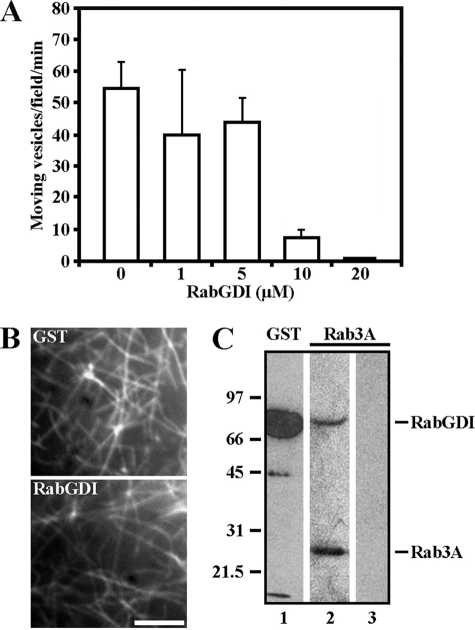
In vitro motility assays were performed to determine whether a Rab GTPase is involved in neuronal Myo5a-based vesicle transport (27). Axoplasm from the squid giant axon was treated with bacterially expressed Rab GDP dissociation inhibitor (RabGDI). In axoplasm preparations on glass coverslips, actin spontaneously assembled into filaments, forming a network on the coverslip surface. For these experiments, each axon was divided in half; one half was incubated in buffer containing RabGDI at concentrations between 1 and 20 μm RabGDI and the other half in buffer containing GST (Fig. 1A). GST, which had no effect, was used as a control at identical concentrations as RabGDI and referred to as 0 μm (supplemental video S1). The axoplasmic actin filament network was stable and did not change in the presence of GST-RabGDI during observation time (Fig. 1B). Vesicle movement was observed in the presence of ATP and motile activity was measured as the number of moving vesicles/field/min. RabGDI inhibited the motile activity in a concentration-dependent manner (Fig. 1A), and motile activity was inhibited by 99% in the presence of 20 μm RabGDI (Fig. 1A, supplemental video S2).
FIGURE 1.
Myo5a-dependent neuronal vesicle motility requires a Rab GTPase. A, vesicle motility in squid axoplasm was inhibited by 99% at 20 μm GST-RabGDI compared with controls (0 μm). All data are mean ± S.D. of three independent experiments. B, 20 μm GST-RabGDI did not affect actin filament network formation as visualized by fluorescence microscopy after incubation with 0.5 μm rhodamine-phalloidin. Bar, 5 μm. C, GST-RabGDI was used in an affinity column to isolate Rab GTPases from squid brain. Western blots of eluted fractions probed with antibodies to GST (lane 1) and Rab3A (lane 2) revealed the presence of Rab3A. Rab3A was not present if no RabGDI was applied to affinity column (lane 3).
We then used GST-RabGDI as a matrix for affinity chromatography to isolate GDP-bound Rab GTPases. Clarified squid brain homogenate prepared from squid optic lobes was saturated with 1 mm GDP and applied to an immobilized RabGDI affinity column. Bound proteins were eluted with glutathione, then analyzed by SDS-PAGE and Western blots. As a control, GDP-loaded brain homogenate was applied to a GSTrap column to which RabGDI was omitted. The presence of the 75 kDa GST-RabGDI fusion protein was confirmed by immunoblotting (Fig. 1C, lane 1). Among the proteins specifically eluted from RabGDI affinity column, we identified Rab3A (Fig. 1C, lane 2). These data demonstrated that Myo5-based neuronal vesicle transport (27) depends on an active Rab GTPase and that native Rab3A is present in brain homogenates. Thus, our data are consistent with findings that Rab3A is expressed in squid brain and is present on SVs (35).
Rab3A/Rabphilin3A Complex Is a Novel Myo5a Binding Partner
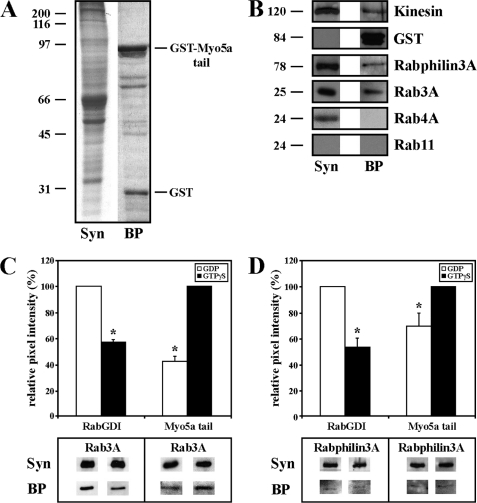
To identify the Rab GTPase that interacts with Myo5a at the synapse, the synaptosomal fraction isolated from rat brains rather than whole-brain homogenate was used in affinity isolation experiments. The synaptosomal fraction (supplemental Fig. S1) was loaded onto GST-tagged Myo5a tail affinity columns prepared as described (27) and the bound proteins (BP) were eluted with glutathione (Fig. 2A). We tested the ability of the GST-tagged fragment of the globular tail domain of mouse Myo5a to pull-down native kinesin from synaptosomes to verify its functionality (45). As expected, kinesin was identified in eluates obtained from GST-Myo5a tail affinity columns (Fig. 2B). Because of the knowledge that Rab3A is a major presynaptic protein, we tested proteins specifically eluted from GST-Myo5a affinity columns for the presence of Rab3A, which was confirmed by immunoblotting (Fig. 2B). We also identified the putative Rab3A effector protein Rabphilin3A in chromatographic eluates. These data suggested that the recombinant Myo5a tail forms a complex with native Rab3A and Rabphilin3A. In control experiments, the synaptosomal fraction was loaded onto a GSTrap column to which only GST was applied. No binding of synaptosomal proteins to affinity columns was detected (supplemental Fig. S2). Rab4A and Rab11, Rabs shown to be involved in membrane trafficking and sorting of endocytosed material between different cell compartments, were not present in the eluates as revealed by Western blots. However, Rab11 has been shown to bind to another member of the myosin-V family, Myo5b, in neurons (46).
FIGURE 2.
Rab3A is a novel Myo5a binding partner. A, synaptosomal fraction (Syn) and bound proteins (BP) eluted from immobilized GST-Myo5a tail affinity column were analyzed by SDS-PAGE stained with Coomassie. GST-Myo5a tail (84 kDa) and GST (25 kDa) are indicated. B, Western blots of the synaptosomal fraction (Syn) and bound proteins (BP) eluted from GST-Myo5a tail affinity column are shown. Blots were probed for proteins as indicated. Rab3A was detected in the eluted fraction. C-D, Rab3A and Rabphilin3A bind to Myo5a tail in a GTP-dependent manner. Inactive GDP-bound Rab3A (C, left panel) and Rabphilin3A (D, left panel) bind to RabGDI. Active GTP-bound Rab3A (C, right panel) and Rabphilin3A (D, right panel) bind to Myo5a tail in the presence of GTPγS. Signal intensities of Rab3A and Rabphilin3A were individually normalized against the signal obtained with these proteins in synaptosomes. All data are mean ± S.D. of three independent experiments. Statistically significant differences at p < 0.001 are indicated by asterisks (*).
We next determined whether Myo5a tail association with Rab3A is GTP-dependent. Synaptosomal fractions preincubated with either 1 mm GDP alone or 1 mm GTP in the presence of 1 mm GTPγS were loaded onto Myo5a tail affinity columns. Bound proteins were eluted and their levels in eluates were measured biochemically by Western blots and normalized against background intensities and signal intensities obtained with proteins in synaptosomes. Incubation of the synaptosomal fraction with GDP increased the amount of both Rab3A and Rabphilin3A bound to RabGDI (Fig. 2C, left panel, Fig. 2D, left panel, respectively). Activation of Rab3A with GTP in the presence of GTPγS increased binding affinity of Rab3A (Fig. 2C, right panel) and Rabphilin3A (Fig. 2D, right panel) to Myo5a tail. In conclusion, these data indicate that Myo5a tail associates with Rab3A in a complex with Rabphilin3A in a GTP-dependent manner.
Protein Kinase Activity Regulates Association of Myo5a with Rab3A
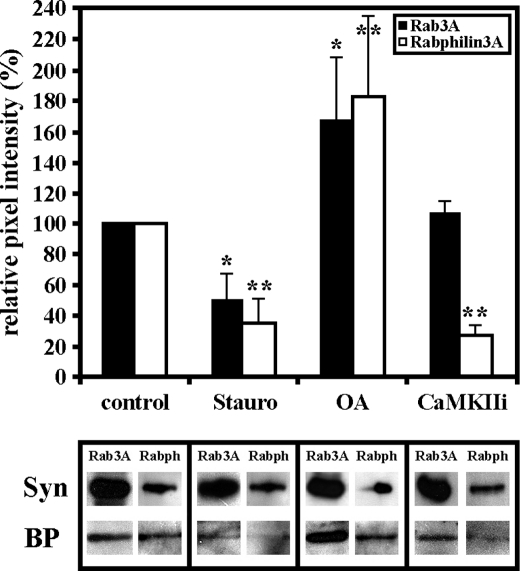
We next tested for a requirement for protein phosphorylation for Myo5a tail association with Rab3A. Synaptosomes were first treated with 2 μm staurosporine, a broad-spectrum inhibitor of protein kinases, and analyzed by Myo5a tail affinity chromatography. This treatment inhibited Rab3A binding to the Myo5 tail by 50% as revealed by Western blots of eluted fractions (Fig. 3). Treatment with 20 nm okadaic acid, an inhibitor of protein phosphatase 1 and 2A, increased the binding activity of Rab3A to Myo5a tail by 90% (Fig. 3). Surprisingly, the addition of 2 μm CaMKII inhibitory peptide did not affect the association of Rab3A to the Myo5a tail (Fig. 3). The addition of protein kinase inhibitors had no significant effect on Myo5a tail as determined by quantitative Western blot analysis (data not shown). These results suggest that CaMKII as well as other protein kinases may be involved in regulating the interaction of Myo5a and Rab3A on SVs.
FIGURE 3.
Protein kinase activity regulates the association of Myo5a with Rab3A. Synaptosomal fractions were preincubated with 2 μm staurosporine (Stauro), 2 μm CaMKII inhibitory peptide (CaMKIIi), or 20 nm okadaic acid (OA) and then passed over a GST-Myo5a tail affinity column. Western blots of the total amount of Rab3A or Rabphillin3A (Syn) and bound proteins (BP) eluted from GST-Myo5a tail affinity column are shown. 2 μm staurosporine (Stauro) decreased binding of Rab3A and Rabphilin3A to GST-Myo5a column. Inhibition of CaMKII activity did not affect the binding affinity of Rab3A to Myo5a tail but reduced Rabphilin3A binding. Inhibition of protein phosphatases 1 and 2A increased binding affinity of both Rab3A and Rabphilin3A to Myo5a tail (OA). Signal intensities of Rab3A and Rabphilin3A were individually normalized against the signal obtained with these proteins in synaptosomes. All data are mean ± S.D. of three independent experiments. Statistically significant differences compared with the controls are indicated by *, p < 0.001.
The Tail of Myo5a Binds to Rab3A in Dilute-lethal (Myo5ad-l/d-l/Myo5ad-l/d-l) Synaptosomes
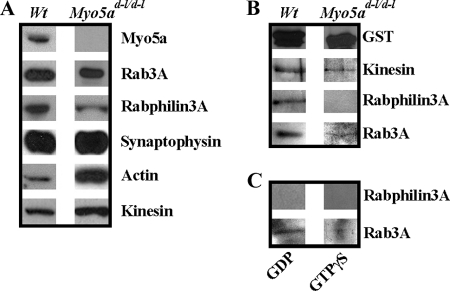
To exclude the possibility that the interaction between Myo5a and Rab3A is through a tail/tail interaction between recombinant and native Myo5a in the synaptosomal fraction, we used a synaptosomal fraction isolated from dilute-lethal homozygous-null mutant mouse brains (Myo5ad-l/d-l/Myo5ad-l/d-l) lacking Myo5a. These dilute-lethal null mutant mice exhibit impaired melanosome transport and pronounced neurological symptoms (11). First, we compared the protein composition of wild-type and null mutant synaptosomes by Western blots. Biochemical characterization revealed that both brain fractions were similar, except for the expected absence of Myo5a in null mutant synaptosomes (Fig. 4A). Surprisingly, the detected amounts of actin and kinesin were significantly higher in null mutant preparations contrasted with wild-type synaptosomes (Fig. 4A).
FIGURE 4.
Myo5a is absent from Myo5ad-l/d-l synaptosomes as detected by Western blot analysis. A, comparison of major proteins present in rat wild type synaptosomes (Wt) and mouse dilute-lethal synaptosomes (Myo5ad-l/d-l). Western blots revealed the presence of Rab3A, Rabphilin3A, and synaptophysin in both synaptosomal preparations, whereas Myo5a is absent in Myo5ad-l/d-l synaptosomes. B, Western blots of bound proteins (BP) eluted from GST-Myo5a tail affinity columns probed for proteins as indicated. C, Rab3A from Myo5ad-l/d-l-null mutant synaptosomes binds to Myo5a tail in a GTP-dependent manner. Inactivation of Rab3A in the presence of GDP resulted in binding of Rab3A (C, left panel) to RabGDI. Activation of Rab3A with GTP in the presence of GTPγS decreased binding affinity of Rab3A to RabGDI (C, right panel). Rabphilin3A could not be detected by Western blot analysis.
We next performed affinity isolation experiments using the GST-Myo5a tail. In null mutant chromatographic eluates we detected not only kinesin but also Rab3A, whereas Rabphilin3A was not detectable (Fig. 4B). The association of Myo5a tail and Rab3A from null mutant synaptosomes also was GTP-dependent as revealed by RabGDI column affinity chromatography (Fig. 4C). In conclusion, our data show that Rab3A from Myo5a-deficient null mutant synaptosomes interacts with the recombinant Myo5a tail and that Rab3A from mutant mice behaves similarly to Rab3A from wild-type synaptosomes.
Myo5a Associates with Rab3A in Neuronal Cultures
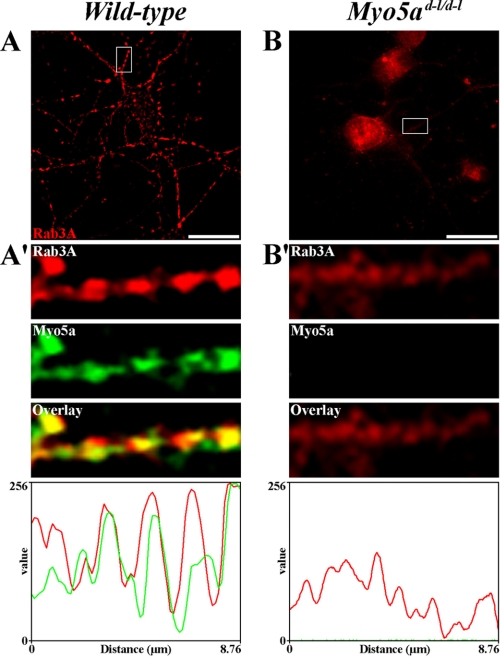
To investigate the association and functional relevance between Myo5a and Rab3A in vivo, we cultured frontal cortex neurons that were isolated from wild-type and null mutant mouse brains. Using double immunofluorescence microscopy, we analyzed whether Myo5a is codistributed with Rab3A in these neuronal cultures. Both proteins exhibited a punctate staining pattern in the soma and neurites and strong colocalization was detected between Myo5a and Rab3A in neurites of wild-type neurons by linescan analysis (Fig. 5A and 5A′). Next, the distribution of Rab3A was assessed in null mutant neurons and compared with that of wild-type neurons. Null mutant neurons grew and extended neurites normally in culture and Rab3A had a staining pattern that was comparable to that in wild-type neurons in the absence of Myo5a (Fig. 5B). No colocalization was detected between Myo5a and Rab3A immunoreactivity in null mutant neurons (Fig. 5B′).
FIGURE 5.
The association of Myo5a with Rab3A is altered in mouse dilute-lethal (Myo5ad-l/d-l) neurons. A–B, indirect immunofluorescent localization of Myo5a (green) and Rab3A (red) in 12 DIV wild-type (A) and 6 DIV dilute-lethal Myo5ad-l/d-l (B) mouse frontal cortex neurons. A′–B′, insets show a high magnification of a dendritic segment where colocalization between Myo5a and Rab3A was detectable in wild-type (A′) but absent in dilute-lethal Myo5ad-l/d-l neurons (B′). Fluorescence intensity was measured for Myo5a (green) and Rab3A (red) along the dendrites shown in the magnified insets (A′–B′). Bars, 20 μm.
To determine whether areas of Myo5a and Rab3A colocalization corresponded to SVs, we analyzed the distribution of Rab3A and the SV marker synapsin 1 in wild-type and null mutant mouse neurons (supplemental Fig. S3). We found that Rab3A and synapsin 1 colocalized in neurites of wild-type neurons (supplemental Fig. S3A) and that both proteins showed partial colocalization in some areas of null mutant neurons (supplemental Fig. S3B).
Myo5a Tail Interacts Directly with Rab3A
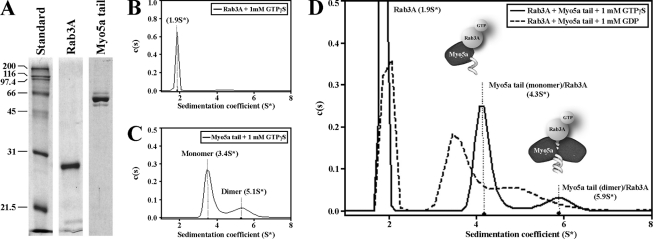
To determine if Rab3A interacts directly with Myo5a tail, sedimentation velocity analytical ultracentrifugation (AUC) was performed on highly purified recombinant Myo5a tail and Rab3A (Fig. 6A). Sedimentation velocity profiles for the purified samples were fitted to a distribution of Lamm equation solutions to determine the diffusion-free sedimentation coefficient distribution (c(s)) for each protein (Fig. 6, B and C and Table 1) (44). The sedimentation velocity profile for 2.6 μm Rab3A showed a mostly monodisperse distribution in solution that sedimented with an s value of 1.90 ± 0.3 and an experimentally determined molecular mass of 23.5 kDa (Fig. 6B and Table 1). In contrast, 0.51 μm Myo5a tail sedimented as two distinct species with s values of 3.4 ± 1.5 and 5.1 ± 0.8 as displayed in the sedimentation velocity profile (Fig. 6C). Modeling of the sedimentation coefficients identified two distinct peaks representing monomeric (61.5%, 3.4 ± 1.5) and dimeric (38.5%, 5.1 ± 0.8) species of the Myo5a tail (Fig. 6C and Table 1). The molecular masses calculated from the sedimentation coefficients were 52.9 kDa for the monomeric and 96.8 kDa for the dimeric species of Myo5a tail very similar to that of the theoretical molecular masses for the monomer and dimer (54.7 kDa and 109.4 kDa) (Table 1). To characterize the interaction between Rab3A and Myo5a tail, 2.6 μm Rab3A and 0.51 μm Myo5a tail were mixed and sedimentation coefficients were determined in the presence of GDP or GTPγS. When both proteins were mixed in the presence of 1 mm GTPγS, a direct interaction was observed and the protein-protein complex sedimented with s values of 4.3 ± 1.3 and 5.9 ± 0.6 (Fig. 6D and Table 1). These results suggest a direct interaction between Myo5a tail and GTP-bound Rab3A. Further analysis revealed the presence of excess free Rab3A (67%) and that GTP-bound Rab3A interacts with both the monomeric (27.5%) and dimeric (5.5%) species of the Myo5a tail (Fig. 6D). The experimentally obtained molecular masses were 73.8 kDa for a complex between GTP-bound Rab3A and monomeric Myo5a tail, and 135 kDa for a complex containing GTP-bound Rab3A and dimeric Myo5a tail (Table 1). These masses are similar to theoretical molecular masses for a 1:1 complex between Rab3A and the Myo5a tail monomer at 4.3S*, and a 1:2 complex between Rab3A and the Myo5a tail dimer at 5.9S*. However, Myo5a tail and Rab3A sedimented as separate species in the presence of 1 mm GDP (Fig. 6D).
FIGURE 6.
Purification and characterization of the protein-protein interaction between Myo5a tail and Rab3A. A, Myo5a tail and Rab3A were analyzed by SDS-PAGE gel electrophoresis stained with Coomassie after protein purification by affinity and gel filtration chromatography. B–D, examples of diffusion-free sedimentation coefficient distributions (c(s)) derived from sedimentation velocity data of individual Rab3A (2.6 μm, B) and Myo5a tail (0.5 μm, C), and Rab3A plus Myo5a tail together (D). Separate monomer and dimer peaks were observed in the Myo5a tail sample (C). D, GTP-bound Rab3A was found in association with monomeric and dimeric Myo5a tail. The diagrams illustrate possible protein-protein interactions between Myo5a tail regions and GTP-bound Rab3A. All analytical ultracentrifugation experiments were performed in the presence of 1 mm GTPγS or 1 mm GDP.
TABLE 1.
Summary of sedimentation coefficients derived from sedimentation velocity analysis of individual Rab3A and Myo5a tail and the Rab3A/Myo5a tail complex
| Protein | S*a | S20,Wb | f/f0c | Calculated mass | Theoretical mass |
|---|---|---|---|---|---|
| kDa | kDa | ||||
| Rab3A + 1 mm GTPγS | 1.9 ± 0.3 | 2.1 | 1.4 | 23.5 | 24.9 |
| Myo5a tail + 1 mm GTPγS | 3.4 ± 1.5 | 3.6 | 1.3, | 52.9 | 54.7 |
| 5.1 ± 0.8 | 5.4 | 1.3 | 96.8 | 109.4 | |
| Rab3A/Myo5a tail + 1 mm GTPγS | 4.3 ± 1.3 | 4.6 | 1.3 | 73.8 | 79.6 |
| 5.9 ± 0.6 | 6.3 | 1.5 | 135 | 134.3 |
a S*, experimental sedimentation coefficient determined at 10 °C (± S.E. from two or three independent experiments).
b S20,W, standard sedimentation coefficient after correcting for water at 20°C.
c f/f0, ratio of the experimental frictional coefficient to the theoretical frictional coefficient for a perfect sphere of the same molecular mass.
DISCUSSION
Neurotransmission, the process by which neurons communicate with their target cells, requires a balance between endocytosis and exocytosis of SVs. The release of neurotransmitter relies on SV trafficking, which involves the coordinated activity of many proteins including small GTP-binding proteins. Previous studies have established that Rab3A, a highly abundant small GTPase on brain SVs in the presynaptic nerve terminal, controls the Ca2+-mediated release of neurotransmitter (47–48). However, it is not well understood whether and how SVs are actively transported between different compartments in the presynaptic nerve terminal. In this study we confirmed that Myo5a tail associates with SVs in the rat brain synaptosomal fraction and we showed for the first time that the globular tail domain of recombinant Myo5a forms a complex with Rab3A in a GTP-dependent manner. We further demonstrated that purified Rab3A in its GTP-bound form interacts directly with both monomeric and dimeric species of Myo5a tail. Therefore, we conclude that Rab3A is the receptor for Myo5a on SVs.
The findings in this report add to the established roles for Rab3A. It is well known that Rab3A regulates SV trafficking including targeting, fusion, and repriming of SVs after exocytosis, and regulates the dynamics of the actin cytoskeleton (49). The cycling of Rab3A between the active GTP-bound or inactive GDP-bound form is associated with changes in the trafficking cycle of SVs at presynaptic nerve terminals. In its GTP-bound form Rab3A is largely associated with SVs at the synapse, while GTP-hydrolysis has been linked to Ca2+-dependent exocytosis resulting in the dissociation of Rab3A from SVs (50–52). After fusion with the plasma membrane, SVs are retrieved through endocytosis followed by reassociation of Rab3A with SVs, a process that is facilitated by RabGDI (5, 53). Stimulation of synaptic terminals by chemical depolarization resulted in the redistribution of SVs from the readily releasable pool toward the active zone (54). This process is completely blocked in RAB3A knock-out mice indicating an active role for Rab3A in SV trafficking (55).
Our results help to answer one of the key questions of myosin driven vesicle transport in neurons, i.e. how the motor binds to vesicles. It has been shown that Myo5a functions in axonal vesicle transport on actin filaments (20). Even though Myo5a has been implicated in the regulation of secretory vesicle trafficking its interaction with Rab3A had not been established previously. Studies of Myo5a in different cell types shows that this motor is involved in the capture of vesicles and their actin-based transport, as well as in late steps in exocytosis (56). Thus, our results support the idea that GTP-bound Rab3A is involved in Myo5a-dependent vesicle transport.
Distinct members of the Rab family are often used by unconventional myosin motors to attach to their cargo, to regulate the motor activity, and to mediate vesicle transport along actin filaments (57–58). Therefore, the formation of the Myo5a/Rab3A complexes is likely to be a critical step that is required for the correct binding, translocation, and delivery of cargoes to the plasma membrane. The Rab27a/Myo5a interaction was the first evidence for the association of a Rab GTPase and a myosin. Rab27a is present at the surface of mature melanosomes and recruits Myo5a via Melanophilin/Slac-2, which regulates its motor activity (29–30, 59–60). Therefore, melanophilin is required as a linker protein that targets Myo5a to melanosomes. Rab3A has multiple binding partners and therefore multiple functions. A number of Rab effector proteins including Rabphilin3A contain a Rab-binding domain and a linker domain, which can interact with the target membrane. As shown in earlier studies, Rab3A recruits Rabphilin3A to SVs before docking to the plasma membrane and neurotransmitter release (5). In this study, we showed that Myo5a forms a complex with Rab3A and Rabphilin3A on SVs. However, in contrast to Myo5a attachment to melanosomes, we found that the globular tail domain of Myo5a interacts directly with GTP-bound Rab3A on SVs and that Rabphilin3A may not be required for binding of Rab3A to the Myo5a tail. Furthermore, Rabphilin3A is not needed for the regulatory functions of Rab3A in neurotransmitter release as demonstrated in Rabphilin knock-out mice (61).
Similarly, Myo5b associates with Rab11a in a GTP-dependent manner and regulates a step in plasma membrane recycling. This interaction is facilitated by the Rab11 family-interacting protein-2 (Rab11-FIP-2), which links the Myo5b tail to Rab11a and regulates endosome movement (31, 62). Moreover, Rab11 was shown to interact with Myo5b and regulate AMPA receptor trafficking (46). Myo5c, the third member of the vertebrate class 5 myosins, has been shown to interact with the transferrin receptor and Rab8a, which regulates the biosynthetic pathway between the Golgi apparatus and the plasma membrane (12). In addition, Roland et al. (32) demonstrated the interaction of Myo5b with Rab8a in compartments containing EHD protein family members in the absence of Rab11a (32). More recently, the same group used dominant-negative Myo5 tail constructs to confirm the interaction of all three vertebrate class 5 myosins with Rab8a and Rab10 (34). Furthermore, they showed that multiple Rab GTPases could regulate all class 5 myosins.
Although Myo5a has been shown to bind to SVs that contain Rab3 those results did not demonstrate a direct binding between both proteins (18). When we reconstituted the protein-protein interaction with highly purified recombinant mouse Myo5a tail and human Rab3A, our functional results showed a direct interaction between the AF-6/cno-homology tail region of Myo5a and Rab3A. As Myo5a tail did not bind to nucleotide-free Rab3A, the data indicated that this interaction is strongly GTP-dependent. We also found that GTP-bound Rab3A binds at a 1:1 stochiometry to the monomeric Myo5a tail in the monomeric form, and at 1:2 stochiometry to the Myo5a tail in the dimeric form. At present, the exact nature of the protein-protein interaction in the monomeric and dimeric forms of Myo5a is not clear. We speculate that binding of Rab3A to monomeric Myo5a is favored, which could be explained by the lower Kd value for GTP-bound Rab3A and Myo5a tail monomer interaction as compared with GTP-bound Rab3A and the Myo5a tail dimer interaction. Future studies that distinguish among these possibilities will be required to understand this protein-protein interaction in neuronal cells.
Protein-protein interactions are important key events for the correct targeting of proteins and the precise timing of exocytosis in nerve terminals. In a recent study coming from Giovedi et al., (8), synapsin 1 has been identified as a binding partner of SV-associated Rab3A. Synapsin 1 was originally identified as a phosphoprotein that cross-links the reserve pool of SV away from the active zone (63). When synapsin 1 becomes phosphorylated by CaMKII, its affinity for SVs and Rab3A is greatly reduced which causes the release of SVs from the actin cytoskeleton (8, 64). However, studies of Myo5a in synaptosomes isolated from rat hippocampi showed that Myo5a did not bind directly to synapsin 1 (18). These data were interpreted to suggest that Myo5a could bind to SVs that are mobilized after release from synapsin/actin interactions. In additional cross-linking experiments, the same authors identified the synaptobrevin II/synaptophysin complex as a preliminary binding partner for Myo5a on SVs (18).
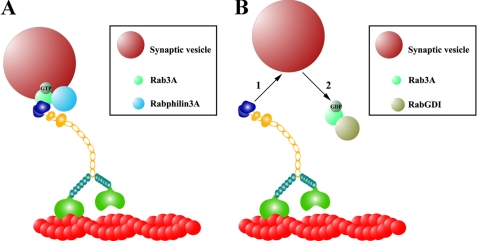
Our data provide evidence for a Myo5a tail association with Rab3A on SVs in vitro and are consistent with the model presented in Fig. 7. We suggest that GTP-bound Rab3A binds directly to the AF-6/cno-homology tail region of Myo5a and associates with Rabphilin3A on SVs (Fig. 7A). After GTP hydrolysis caused by Ca2+-induced exocytosis the GDP-bound form Rab3A is then released from the Myo5a tail, where it then binds to RabGDI to undergo GDP/GTP exchange (Fig. 7B). After release from RabGDI, the GTP-bound or active form of Rab3A can associate with SVs again and bind to Myo5a or putative effector proteins such as Rabphilin 3A (5). What could be a possible function for Myo5a in SV transport and synaptic transmission? It has been shown that Myo5a is important in recycling and replenishing of synaptic or neurosecretory vesicles (18, 23, 26, 65). Furthermore, the subcelluar localization of Myo5a in neurons suggests several roles in synaptic physiology as shown for photoreceptor cells in the retina (66). Our findings support the idea that Myo5a is an important factor for Rab3A-mediated synaptic transmission including SV availability, because neurotransmitter release is regulated by multiple steps, which includes the release of SVs from the actin cytoskeleton, docking to the presynaptic plasma membrane followed by exocytosis.
FIGURE 7.
Proposed model for the Myo5a/Rab3A interaction on SVs. This model integrates the SV abundant membrane proteins Rab3A and Rabphilin3A and illustrates a possible interaction of the Myo5a tail-domain with the SV. A, active or GTP-bound form of Rab3A builds a complex with Rabphilin3A on the surface of SV that binds directly to the globular tail domain of Myo5a. B, Rab3A dissociates from Myo5a tail and Rabphilin3A after GTP hydrolysis (1) and binds to RabGDI in its inactive or GDP-bound form (2) until GDP/GTP exchange.
In conclusion, we have shown that the tail domain of Myo5a forms a complex with Rab3A on SVs in vitro. Biochemical and kinetic studies demonstrated a direct GTP-dependent interaction between Myo5a and Rab3A. A further characterization of Myo5a binding to SVs and the identification of additional binding partners will increase our understanding of Myo5a-mediated transport of SVs.
Acknowledgments
We thank Carl DeSelm (Washington University, St. Louis, MO) and Jeremiah Brown (Dartmouth Medical School, Hanover, NH) for experimental design and technical assistance with preparation of squid brain extracts and in vitro motility assays. We thank Bärbel Redlich (University of Rostock, Rostock, Germany) and Sadie Bartholomew (Department of Biochemistry, Stanford University, Stanford, CA) for help with culture and immunostaining of mouse frontal cortex neurons and Venkatasubramanian Dharmarajan (Syracuse University, Syracuse, NY) for technical assistance with protein purification. We thank Romano Regazzi (University of Lausanne, Lausanne, Switzerland) for the GFP-Rab3A construct.
This work was supported, in whole or in part, by National Institutes of Health Grants CA140522 (to M. S. C.), GM066901, and TW007220 (to J. A. M. and D. W. P.) and NSF Grant MCB-0517303 (to G. M. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental videos S1–S2 and Figs. S1–S3.
- SV
- synaptic vesicle
- Myo5a
- myosin-Va
- CaMKII
- calmodulin-dependent kinase II
- GDI
- GDP dissociation inhibitor
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
REFERENCES
- 1. Jahn R., Südhof T. C. (1994) Annu. Rev. Neurosci. 17, 219–246 [DOI] [PubMed] [Google Scholar]
- 2. Geppert M., Bolshakov V. Y., Siegelbaum S. A., Takei K., De Camilli P., Hammer R. E., Südhof T. C. (1994) Nature 369, 493–497 [DOI] [PubMed] [Google Scholar]
- 3. Schlüter O. M., Khvotchev M., Jahn R., Südhof T. C. (2002) J. Biol. Chem. 277, 40919–40929 [DOI] [PubMed] [Google Scholar]
- 4. Fischer von Mollard G., Stahl B., Li C., Südhof T. C., Jahn R. (1994) Trends Biochem. 19, 164–169 [DOI] [PubMed] [Google Scholar]
- 5. Stahl B., Chou J. H., Li C., Südhof T. C., Jahn R. (1996) EMBO J. 15, 1799–1809 [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y., Okamoto M., Schmitz F., Hofmann K., Südhof T. C. (1997) Nature 388, 593–598 [DOI] [PubMed] [Google Scholar]
- 7. Brondyk W. H., McKiernan C. J., Fortner K. A., Stabila P., Holz R. W., Macara I. G. (1995) Mol. Cell. Biol. 15, 1137–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giovedì S., Vaccaro P., Valtorta F., Darchen F., Greengard P., Cesareni G., Benfenati F. (2004) J. Biol. Chem. 279, 43760–43768 [DOI] [PubMed] [Google Scholar]
- 9. Desnos C., Huet S., Darchen F. (2007) Biol. Cell 99, 411–423 [DOI] [PubMed] [Google Scholar]
- 10. Trybus K. M. (2008) Cell. Mol. Life Sci. 65, 1378–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mercer J. A., Seperack P. K., Strobel M. C., Copeland N. G., Jenkins N. A. (1991) Nature 349, 709–713 [DOI] [PubMed] [Google Scholar]
- 12. Rodriguez O. C., Cheney R. E. (2002) J. Cell Sci. 115, 991–1004 [DOI] [PubMed] [Google Scholar]
- 13. Dekker-Ohno K., Hayasaka S., Takagishi Y., Oda S., Wakasugi N., Mikoshiba K., Inouye M., Yamamura H. (1996) Brain Res. 714, 226–230 [DOI] [PubMed] [Google Scholar]
- 14. Takagishi Y., Oda S., Hayasaka S., Dekker-Ohno K., Shikata T., Inouye M., Yamamura H. (1996) Neurosci. Lett. 215, 169–172 [DOI] [PubMed] [Google Scholar]
- 15. Schlamp C. L., Williams D. S. (1996) Exp. Eye Res. 63, 613–619 [DOI] [PubMed] [Google Scholar]
- 16. Mani F., Espreafico E. M., Larson R. E. (1994) Braz. J. Med. Biol. Res. 27, 2639–2643 [PubMed] [Google Scholar]
- 17. Walikonis R. S., Jensen O. N., Mann M., Provance D. W., Jr., Mercer J. A., Kennedy M. B. (2000) J. Neurosci. 20, 4069–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prekeris R., Terrian D. M. (1997) J. Cell Biol. 137, 1589–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Evans L. L., Lee A. J., Bridgman P. C., Mooseker M. S. (1998) J. Cell Sci. 111, 2055–2066 [DOI] [PubMed] [Google Scholar]
- 20. Tabb J. S., Molyneaux B. J., Cohen D. L., Kuznetsov S. A., Langford G. M. (1998) J. Cell Sci. 111, 3221–3234 [DOI] [PubMed] [Google Scholar]
- 21. Morris R. L., Hollenbeck P. J. (1995) J. Cell Biol. 131, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pathak D., Sepp K. J., Hollenbeck P. J. (2010) J. Neurosci. 30, 8984–8992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosé S. D., Lejen T., Casaletti L., Larson R. E., Pene T. D., Trifaró J. M. (2003) J. Neurochem. 85, 287–298 [DOI] [PubMed] [Google Scholar]
- 24. Rudolf R., Kögel T., Kuznetsov S. A., Salm T., Schlicker O., Hellwig A., Hammer J. A., 3rd, Gerdes H. H. (2003) J. Cell Sci. 116, 1339–1348 [DOI] [PubMed] [Google Scholar]
- 25. Desnos C., Huet S., Fanget I., Chapuis C., Böttiger C., Racine V., Sibarita J. B., Henry J. P., Darchen F. (2007) J. Neurosci. 27, 10636–10645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bridgman P. C. (1999) J. Cell Biol. 146, 1045–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brown J. R., Stafford P., Langford G. M. (2004) J. Neurobiol. 58, 175–188 [DOI] [PubMed] [Google Scholar]
- 28. Bahadoran P., Aberdam E., Mantoux F., Buscà R., Bille K., Yalman N., de Saint-Basile G., Casaroli-Marano R., Ortonne J. P., Balloti R. (2001) J. Cell Biol. 152, 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu X. S., Rao K., Zhang H., Wang F., Sellers J. R., Matesic L. E., Copeland N. G., Jenkins N. A., Hammer J. A., 3rd (2002) Nat. Cell Biol. 4, 271–278 [DOI] [PubMed] [Google Scholar]
- 30. Provance D. W., James T. L., Mercer J. A. (2002) Traffic 3, 124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lapierre L. A., Kumar R., Hales C. M., Navarre J., Bhartur S. G., Burnette J. O., Provance D. W., Jr., Mercer J. A., Bähler M., Goldenring J. R. (2001) Mol. Biol. Cell 12, 1843–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roland J. T., Kenworthy A. K., Peranen J., Caplan S., Goldenring J. R. (2007) Mol. Biol. Cell 18, 2828–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Correia S. S., Bassani S., Brown T. C., Lisé M. F., Backos D. S., El-Husseini A., Passafora M., Esteban J. A. (2008) Nat. Neurosci. 11, 457–466 [DOI] [PubMed] [Google Scholar]
- 34. Roland J. T., Lapierre L. A., Goldenring J. R. (2009) J. Biol. Chem. 284, 1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chin G. J., Goldman S. A. (1992) Brain Res. 571, 89–96 [DOI] [PubMed] [Google Scholar]
- 36. Wu X., Bowers B., Wei Q., Kocher B., Hammer J. A., 3rd (1997) J. Cell Sci. 110, 847–859 [DOI] [PubMed] [Google Scholar]
- 37. Carlin R. K., Grab D. J., Cohen R. S., Siekevitz P. (1980) J. Cell Biol. 86, 831–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jordan B. A, Fernholz B. D., Boussac M., Xu C., Grigorean G., Ziff E. B., Neubert T. A. (2004) Mol. Cell. Proteomics 3.9, 857–871 [DOI] [PubMed] [Google Scholar]
- 39. Gramowski A., Jügelt K., Stüwe S., Schulze R., McGregor G. P., Wartenberg-Demand A., Loock J., Schröder O., Weiss D. G. (2006) Eur. J. Neurosci. 24, 455–465 [DOI] [PubMed] [Google Scholar]
- 40. Patel A., Vought V. E., Dharmarajan V., Cosgrove M. S. (2008) J. Biol. Chem. 283, 32162–32175 [DOI] [PubMed] [Google Scholar]
- 41. Waselle L., Coppola T., Fukuda M., Iezzi M., El-Amraoui A., Petit C., Regazzi R. (2003) Mol. Biol. Cell 14, 4103–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sheffield P., Garrard S., Derewenda Z. (1999) Protein Expr Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 43. Patel A., Dharmarajan V., Vought V. E., Cosgrove M. S.(2009) J. Biol. Chem. 284, 24242–24256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang J. D., Brady S. T., Richards B. W., Stenoien D., Resau J. H., Copeland N. G., Jenkins N. A. (1999) Nature 397, 267–270 [DOI] [PubMed] [Google Scholar]
- 46. Lisé M. F., Wong T. P., Trinh A., Hines R. M., Liu L., Kang R., Hines D. J., Lu J., Goldenring J. R., Wang Y. T., El-Husseini A. (2006) J. Biol. Chem. 281, 3669–3678 [DOI] [PubMed] [Google Scholar]
- 47. Fischer von Mollard G., Mignery G. A., Baumert M., Perin M. S., Hanson T. J., Burger P. M., Jahn R., Südhof T. C. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 1988–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Geppert M., Goda Y., Stevens C. F., Südhof T. C. (1997) Nature 387, 810–814 [DOI] [PubMed] [Google Scholar]
- 49. Jahn R., Lang T., Südhof T. C. (2003) Cell 112, 519–533 [DOI] [PubMed] [Google Scholar]
- 50. Fischer von Mollard G., Südhof T. C., Jahn R. (1991) Nature 349, 79–81 [DOI] [PubMed] [Google Scholar]
- 51. Stahl B., von Mollard G. F., Walch-Solimena C., Jahn R. (1994) J. Biol. Chem. 269, 24770–24776 [PubMed] [Google Scholar]
- 52. Star E. N., Newton A. J., Murthy V. N. (2005) J. Physiol. 569, 103–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chou J. H., Jahn R. (2000) J. Biol. Chem. 275, 9433–9440 [DOI] [PubMed] [Google Scholar]
- 54. Leenders A. G., Lopes, da Silva F. H., Ghijsen W. E., Verhage M. (2001) Mol. Biol. Cell 12, 3095–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schlüter O. M., Schmitz F., Jahn R., Rosenmund C., Südhof T. C. (2004) J. Neurosci. 24, 6629–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Eichler T. W., Kögel T., Bukoreshtliev N. V., Gerdes H. H. (2006) Biochem. Soc. Trans. 34, 671–674 [DOI] [PubMed] [Google Scholar]
- 57. Jordens I., Marsman M., Kuijl C., Neefjes J. (2005) Traffic 6, 1070–1077 [DOI] [PubMed] [Google Scholar]
- 58. Grosshans B. L., Ortiz D., Novick P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11821–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Matestic L. E., Yip R., Reuss A. E., Swing D. A., O'Sullivan T. N., Fletcher C. F., Copeland N. G., Jenkins N. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10238–10243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li X. D., Ikebe R., Ikebe M. (2005) J. Biol. Chem. 280, 17815–17822 [DOI] [PubMed] [Google Scholar]
- 61. Schlüter O. M., Schnell E., Verhage M., Tzonopoulos T., Nicoll R. A., Janz R., Malenka R. C., Geppert M., Südhof T. C. (1999) J. Neurosci. 19, 5834–5846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hales C. M., Vaerman J. P., Goldenring J. R. (2002) J. Biol. Chem. 277, 50415–50421 [DOI] [PubMed] [Google Scholar]
- 63. Landis D. M., Hall A. K., Weinstein L. A., Reese T. S. (1988) Neuron 1, 201–209 [DOI] [PubMed] [Google Scholar]
- 64. Turner K. M., Burgoyne R. D., Morgan A. (1999) Trends Neurosci. 22, 459–464 [DOI] [PubMed] [Google Scholar]
- 65. Schnell E., Nicoll R. A. (2001) J. Neurophysiol. 85, 1498–1501 [DOI] [PubMed] [Google Scholar]
- 66. Libby R. T., Lillo C., Kitamoto J., Williams D. S., Steel K. P. (2004) J. Cell Sci. 117, 4509–4515 [DOI] [PubMed] [Google Scholar]