Abstract
Animal cells secrete small vesicles, otherwise known as exosomes and microvesicles (EMVs). A short, N-terminal acylation tag can target a highly oligomeric cytoplasmic protein, TyA, into secreted vesicles (Fang, Y., Wu, N., Gan, X., Yan, W., Morell, J. C., and Gould, S. J. (2007) PLoS Biol. 5, 1267–1283). However, it is not clear whether this is true for other membrane anchors or other highly oligomeric, cytoplasmic proteins. We show here that a variety of plasma membrane anchors can target TyA-GFP to sites of vesicle budding and into EMVs, including: (i) a myristoylation tag; (ii) a phosphatidylinositol-(4,5)-bisphosphate (PIP2)-binding domain; (iii), a phosphatidylinositol-(3,4,5)-trisphosphate-binding domain; (iv) a prenylation/palmitoylation tag, and (v) a type-1 plasma membrane protein, CD43. However, the relative budding efficiency induced by these plasma membrane anchors varied over a 10-fold range, from 100% of control (AcylTyA-GFP) for the myristoylation tag and PIP2-binding domain, to one-third or less for the others, respectively. Targeting TyA-GFP to endosome membranes by fusion to a phosphatidylinositol 3-phosphate-binding domain induced only a slight budding of TyA-GFP, ∼2% of control, and no budding was observed when TyA-GFP was targeted to Golgi membranes via a phosphatidylinositol 4-phosphate-binding domain. We also found that a plasma membrane anchor can target two other highly oligomeric, cytoplasmic proteins to EMVs. These observations support the hypothesis that plasma membrane anchors can target highly oligomeric, cytoplasmic proteins to EMVs. Our data also provide additional parallels between EMV biogenesis and retrovirus budding, as the anchors that induced the greatest budding of TyA-GFP are the same as those that mediate retrovirus budding.
Keywords: Endosomes, Lentivirus, Plasma Membrane, Vesicles, Virus Assembly, ELD, EMV, Budding, Exosome, Microvesicle
Introduction
Animal cells release single membrane vesicles that have the same topology as the cell and a variable diameter of ∼50–250 nm (1–8). These vesicles mediate the secretion of a wide variety of proteins, lipids, mRNAs, and micro RNAs, interact with neighboring cells, and can thereby transmit signals, proteins, lipids, and nucleic acids from cell to cell (3, 9–11). Furthermore, the ability of vesicle-derived nucleic acids to alter gene expression in neighboring cells makes it likely that secreted vesicles can fuse with neighboring cells in a nonviral pathway of intercellular vesicle traffic. Not surprisingly, there is increasing evidence that secreted vesicles play important roles in normal physiological processes (e.g. immune signaling (2), development (12, 13) and in human disease (e.g. cancer (14–16), amyloidopathies (17–19), and viral infections (4, 7, 20–22)).
Current models posit that there might be two separate pathways for the formation of small secreted vesicles: microvesicle biogenesis, which involves vesicle budding directly from the plasma membrane (PM)2; and exosome biogenesis, which involves vesicle budding into endosomes to form multivesicular bodies (MVBs), followed by MVB-PM fusion (3, 8). However, differentiating between microvesicles and exosomes is problematic because (i) there is no known physical property or molecular marker that can unambiguously differentiate exosomes from microvesicles (3), and (ii) it is conceptually impossible to know where any particular secreted vesicle was made once it has left the cell. Furthermore, there are several empirical observations that are difficult to reconcile with a strict separation between microvesicle biogenesis and exosome biogenesis. First, it has been shown that exosomal proteins, lipids, and their bound carbohydrates can bud directly from the PM of cells (7, 23–25). Second, MVB biogenesis requires the endosomal sorting complexes required for transport (ESCRT) machinery (26–29), whereas exosome biogenesis does not (7, 30), making it difficult to understand how MVB biogenesis could represent an essential step in exosome biogenesis. Third, it has been reported that MVB-like structures enriched for exosome-like vesicles and exosomal markers are deep invaginations of the plasma membrane and not endosomes at all (31, 32). Given these uncertainties regarding the biogenesis of exosomes and microvesicles, we prefer to refer to small secreted vesicles by the collective acronym of EMV.
One approach to understanding EMV biogenesis is to identify the cis-acting signals that target proteins to sites of EMV budding and into secreted vesicles. We reported previously that the combination of higher order oligomerization and plasma membrane binding are sufficient to target a wide variety of proteins to sites of vesicle budding and into EMVs (7). For example, we found that a simple PM anchor (the N-terminal acylation tag MGCINSKRKD- (33)) targeted a highly oligomeric cytoplasmic protein (the yeast protein TyA) to sites of vesicle budding and into EMVs. In a set of complementary results, we found that inducing the higher order oligomerization of cell surface proteins targeted them to EMVs as well. PM binding and higher order oligomerization also target proteins to HIV particles and are the primary budding information in the HIV Gag protein (7). These data support the idea that retrovirus budding is a form of EMV biogenesis (4). This relationship is also reflected in the sites of EMV and retrovirus budding in different cell types; (i) macrophages bud HIV into MVB-like invaginations of the PM (25, 34), which closely resemble the structures that have been implicated in exosome biogenesis (2, 9, 34); and (ii) T-cells bud both EMVs and HIV particles from discrete regions of the plasma membrane that are enriched for endosomal and exosomal markers and retain the endosomal ability to generate outward budding vesicles (outward indicates a direction away from the cytoplasm) (23).
Here, we explored two facets of EMV cargo biogenesis: (i) whether any other membrane anchors can induce the budding of TyA-GFP and (ii) whether a PM anchor, the N-terminal acylation tag, can induce the budding of any cytoplasmic proteins other than TyA. Our results confirm and extend the hypothesis that PM anchors can induce the budding of highly oligomeric cytoplasmic proteins, raise the possibility that endosomal membrane anchors cannot induce EMV cargo budding, and lend new support to the hypothesis that retrovirus budding is a form of EMV biogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture, Lipid Labeling, Transfection, and Fluorescence Microscopy
A Jurkat T-cell line from the American Type Culture Collection was the original source of the Jurkat T-cell line (a gift from J. Hildreth (23)) used in these experiments. These cells were maintained in AIM-V medium at 37 °C in a humidified chamber with 5% CO2. Cells were labeled with N-Rh-PE by chilling them to 4 °C, incubating them with N-Rh-PE (final concentration, 1 μm) at 4 °C for 60 min, washing the cells three times with PBS, and then incubating the cells in growth medium for 16–20 h at 37 °C. Transfections were by electroporation using a BTX ECM-600 or by nucleofection (Amaxa). For electroporation, we incubated 1.3 × 107 cells with 10 μg DNA in AIM-V medium for 10 min and then electroporated the cells using settings of 300 V, 800 microfarads, and 24 ohms. Nucleofection was performed using the manufacturer's reagents and recommendations (Amaxa) for transfecting Jurkat T-cells. For the production of stable cell lines, we exposed the transfected cells to 1 mg/ml G418 for 2 weeks to generate pools of G418-resistant cell lines and then selected for cells expressing the desired GFP fusion protein by fluorescence-activated cell sorting (Johns Hopkins University FACS facility). Immunofluorescence microscopy was performed at room temperature on a BH2-RFCA microscope (Olympus) equipped with an Olympus S-planApo 60 × 0.40 oil objective and a Sensicam QE (Cooke) digital camera using IPLab 3.6.3 software (Scanalytics, Inc.). Images were converted to tiff files, imported into Adobe Photoshop CS, and assembled into figures using Illustrator CS (Adobe Systems, Inc.). To determine the extent of co-localization between 2xFYVE-TyA-GFP and 2xFYVE-DsRED, the overlap coefficient among 14 separate sets of tiff images was determined using ImageJ software.
Plasmids
The CFP-Golgi plasmid was obtained from Clontech. All other expression vectors were based on pcDNA3-GFP (23). All amplified regions of all plasmids were sequenced to ensure the absence of any undesired mutations. To generate the plasmid used to express Acyl(G2A)TyA-GFP, we amplified the AcylTyA ORF (7) using a 5′ primer designed to replace the Gly2 codon with an alanine codon and the same 3′ primer used previously to amplify the TyA ORF (7). The resulting product was cleaved with Asp718 and BamHI and cloned between the Asp718 and BamHI sites of pcDNA3-GFP. A similar strategy was used to generate pcDNA3-Acyl(C3A)TyA-GFP, using a 5′ primer designed to replace the Cys3 codon of the Acyl tag with an alanine codon. pcDNA3-HIV Gag-GFP was described previously (7). To generate the Gag(G2A)-GFP expression vector, we amplified the HIV Gag ORF using a 5′ primer designed to replace the Gly2 codon with an alanine codon and the same 3′ primer used originally to amplify the HIV Gag ORF (23). The resulting product was cleaved with Asp718 and BamHI and cloned between the Asp718 and BamHI sites of pcDNA3-GFP to generate pcDNA3-HIV Gag(G2A)-GFP. To generate the plasmid designed to express TyA-GFP-CCKVL, we amplified the GFP ORF using a 5′ primer designed to place a BamHI site in place of the start codon of the GFP ORF and a 3′ primer designed to place codons for the amino acids CCKVL between the last codon of the GFP ORF and a stop codon, followed by an XbaI site. The resulting product was cleaved with BamHI and XbaI and cloned between the BamHI and XbaI sites of pcDNA3-TyA-GFP (7). The same strategy was used to generate pcDNA3-TyA-GFP-CKVL, with the exception that the 3′ primer encoded the amino acids CKVL between the final codon of the GFP ORF and the stop codon.
To generate the plasmids designed to express TyA-GFP-SYN, TyA-GFP-2xFYVE, and TyA-GFP-FAPP, we amplified desired regions of the ORFs of syntenin (codons 101–298 (GenBankTM accession no. AAB97144)), 2xFYVE (a direct repeat of codons 158–221 of human HRS (GenBankTM accession no. BAA23366)), and FAPP (codons 2–100 (GenBankTM accession no. AAH02838)), using primers designed to append an XhoI site upstream of each ORF and a stop codon and an XbaI site downstream of each ORF. The resulting products were cleaved with XhoI and XbaI and cloned between the XhoI and XbaI sites of pcDNA3-TyA-GFP. To generate the plasmid designed to express AKT-TyA-GFP we amplified codons 1 to 116 of the human AKT protein kinase (GenBankTM accession no. P31749) using a 5′ primer that appended a HindIII site and start codon immediately upstream of the AKT ORF and a 3′ primer designed to place an Asp718 site downstream of and in frame with codon 285 of the AKT ORF. The resulting product was cleaved with HindIII and Asp718 and cloned between the HindIII and Asp718 sites of pcDNA3-TyA-GFP. A similar strategy was used to generate the plasmids designed to express CD4-TyA-GFP, CD83-TyA-GFP, CD43-TyA-GFP, CD38-TyA-GFP, and CD69-TyA-GFP with the full ORFs of the CD4 (GenBankTM accession no. AAA16069), CD83 (GenBankTM accession no. CAG33300), CD43 (GenBankTM accession no. AAA51949), CD38 (GenBankTM accession no. BAA18966), and CD69 (GenBankTM accession no. CAA83017) cDNAs inserted between the HindIII and Asp718 sites of pcDNA3-TyA-GFP (7).
To make the plasmid to express GFP-SYN, we excised the XhoI-XbaI fragment containing the SYN ORF from pcDNA3-TyA-GFP-SYN and cloned it between the XhoI and XbaI sites of pcDNA3-GFP, downstream of and in-frame with the GFP ORF. To make a plasmid that would express AKT-GFP we excised the HindIII-Asp718 fragment containing the AKT ORF from pcDNA3-AKT-TyA-GFP and cloned it between the HindIII and Asp718 sites of pcDNA3-GFP, upstream of and in-frame with the GFP ORF. The plasmid pcDNA3-DsRED-2xFYVE was created by inserting the XhoI-XbaI 2xFYVE domain between the XhoI and XbaI sites of pcDNA3-DsRED (7).
To express MusD-GFP, we synthesized a human codon-optimized version of the MusD structural protein ORF (35) and then amplified this new MusD ORF using primers designed to append an Asp718 site immediately upstream of the ORF and a BamHI site in place of the stop codon. We then cleaved the amplified DNA product with these enzymes and inserted it between the Asp718 and BamHI sites of pcDNA3-GFP. To express AcylMusD-GFP, we performed a similar series of steps, with the exception that the 5′ primer contained an Asp718 site and codons designed to express the peptide MGCINSKRKD immediately upstream of the second codon of the MusD ORF (eliminating its normal start codon). The plasmid designed to express AcylSFV-GFP was generated by amplifying the simian foamy virus (SFV) ORF from the plasmid pcDNA3-SFV-GFP (7) using a 5′ primer designed to append an Asp718 site and codons designed to express the peptide MGCINSKRKD immediately upstream of the second codon of the SFV ORF. The product was then cleaved and inserted between the Asp718 and BamHI sites of pcDNA3-GFP.
Collection of EMVs, Preparation of Lysates, and Immunoblot
EMVs were collected 2 days post transfection from N-Rh-PE-labeled Jurkat T-cells (transient expression) or from N-Rh-PE-labeled Jurkat T-cell lines that had been selected previously for the expression of the desired protein. N-Rh-PE labeling was performed 16–20 h before harvesting cells and EMVs. EMVs and viruses were collected as described previously (7, 23). In brief, cells were chilled and collected by spinning the culture at 200 × g for 3 min. (Jurkat T-cells grow in suspension.) EMVs were collected by filtering the supernatant through a 0.22-micron PVDF filter, spinning the supernatant at 10,000 × g for 30 min, spinning the resulting supernatant a second time at 10,000 × g for 30 min, and then spinning the resulting supernatant at 70,000 × g for 60 min to pellet EMVs. The pelleted EMVs were either resuspended in PBS for fluorescence microscopy or lysed in SDS-PAGE loading buffer for immunoblot. Equal ratios of EMV lysates:cell lysates were separated by SDS-PAGE and processed for immunoblot using specific primary antibodies directed against GFP and HRP-conjugated secondary antibodies. Proteins were visualized by chemiluminescent exposure of x-ray film. Films were scanned, converted to tiff files, and the signal for each band was quantified using ImageJ software. Relative budding measurements were based on the resulting band intensities, calculated as (EMV/(EMV + cell)), relative to that of the positive control protein, AcylTyA-GFP. Statistical analyses of the data are presented as the average ± 1 S.D., along with the p value from a Student's t test.
RESULTS
A Myristoylation Tag Targets TyA-GFP to Sites of Vesicle Budding and into EMVs
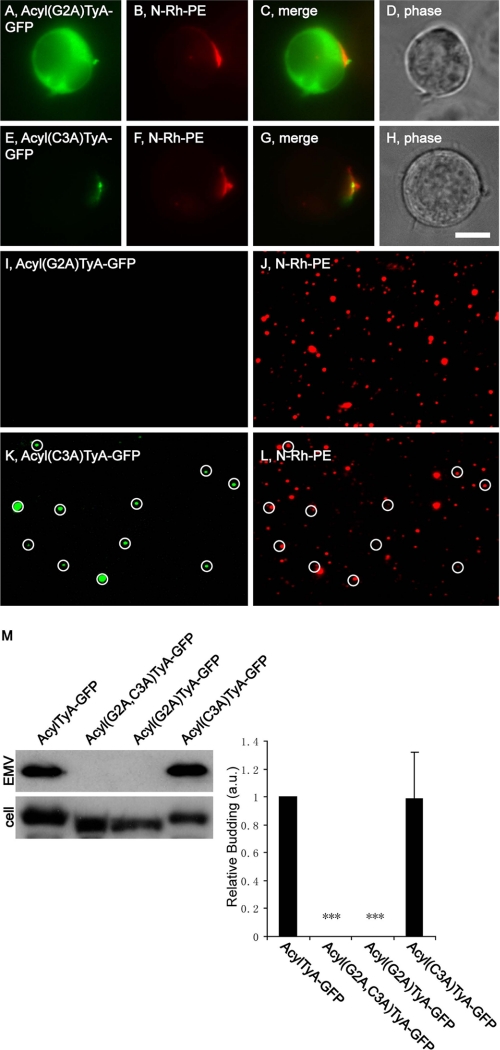
The yeast protein TyA forms highly oligomeric protein complexes in the cytoplasm (36), and a TyA-GFP fusion protein also accumulates in the cytoplasm, at least in a human CD4+ T-cell line (Jurkat) (7). We reported previously that adding a 10-amino acid-long acyl tag, MGCINSKRKD-, to the N terminus of TyA-GFP results in its targeting to the PM, enrichment at endosome-like domains of the plasma membrane, and its secretion from the cell in EMVs (7). This acylation tag can specify the addition of two acyl groups, a myristoyl moiety at Gly2 and a palmitoyl moiety at Cys2 (33). Not surprisingly, we observed previously that mutation of both Gly2 and Cys2 of the tag to alanine residues (G2A and C3A) resulted in a protein, Acyl(G2A,C3A)TyA-GFP, that behaved indistinguishably from TyA-GFP, accumulating in the cell cytoplasm and failing to bud from cells (7).
To determine whether either of the putative acyl attachment sites in the acyl tag were essential for the budding of AcylTyA-GFP, we mutated each to alanine and followed the sorting and secretion of the mutant proteins by Jurkat T-cells. Cells transfected with plasmids designed to encode Acyl(G2A)TyA-GFP or Acyl(C3A)-GFP were incubated for a day, pulse-labeled with N-Rh-PE for 1 h, and incubated an additional day. Cells and EMVs were harvested and processed for both fluorescence microscopy and immunoblot. Cell and EMV lysates were also prepared from Jurkat T-cells expressing AcylTyA-GFP or Acyl(G2A,C3A)TyA-GFP, which served as positive and negative budding controls in immunoblot experiments. We report here that Acyl(G2A)TyA-GFP accumulated in the cytoplasm of Jurkat T-cells (Fig. 1, A–D), was not detected in N-Rh-PE-labeled EMVs by fluorescence microscopy of secreted vesicles (Fig. 1, I and J), and was not detected in secreted vesicles by immunoblot (Fig. 1M) (0 ± 0%; n = 3; p = 0). In contrast, Acyl(C3A)TyA-GFP was targeted to EMV budding sites within the cell (Fig. 1, E–H) and was secreted from the cell in N-Rh-PE-labeled EMVs (Fig. 1, K–M). Furthermore, the extent of secretion was indistinguishable from that of our positive control, AcylTyA-GFP (99 ± 33%; n = 3; p = 0.95). These data indicate that the EMV targeting of AcylTyA-GFP is due primarily to its putative myristoyl attachment site (Gly2), with little if any contribution from its putative palmitoyl attachment site (Cys2). As for whether AcylTyA-GFP or Acyl(C3A)TyA-GFP were actually myristoylated, we have no direct evidence. However, protein myristoylation is expected to cause reduced migration during SDS-PAGE, and we observed that AcylTyA-GFP and Acyl(C3A)TyA-GFP both migrated more slowly than either Acyl(G2A,C3A)TyA-GFP or Acyl(G2A)TyA-GFP (Fig. 1M).
FIGURE 1.
The myristoyl attachment site is required for targeting AcylTyA-GFP to sites of vesicle budding and into EMVs. A–H, fluorescence and phase microscopy images of N-Rh-PE-labeled Jurkat T-cells expressing Acyl(G2A)TyA-GFP (A–D) or Acyl(C3A)TyA-GFP (E–H). Scale bar, 10 μm. I–L, fluorescence microscopy images of EMVs produced by N-Rh-PE-labeled Jurkat T-cells expressing Acyl(G2A)TyA-GFP (I and J) or Acyl(C3A)TyA-GFP (K and L). The white circles show the position of GFP-positive vesicles, some of which also possessed N-Rh-PE fluorescence. M, anti-GFP immunoblot of EMV and cell lysates prepared from Jurkat T-cells expressing AcylTyA-GFP, Acyl(G2A,C3A)TyA-GFP, Acyl(G2A)TyA-GFP, or Acyl(C3A)TyA-GFP. The bar graph to the right shows the average ± 1 S.D. of the relative budding calculated from band intensities (EMV/(EMV + cell)) relative to the AcylTyA-GFP control, which were determined by densitometric analysis of immunoblot films. Results noted with three asterisks have a p value of < 0.0005.
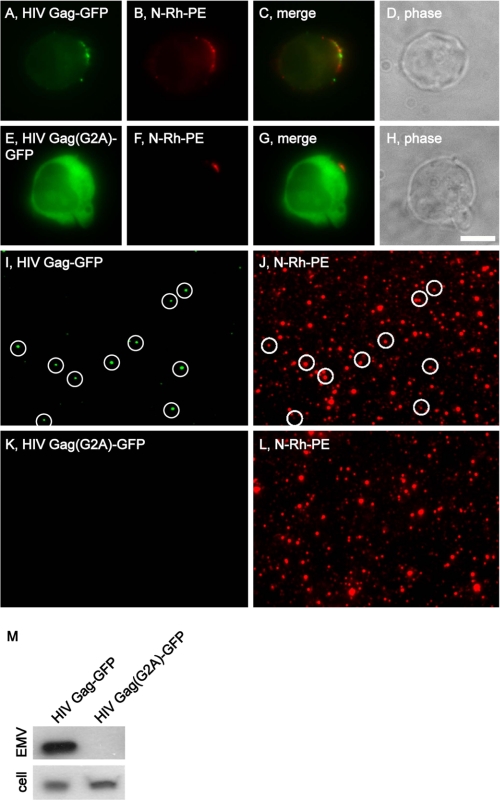
Gag Requires Its Myristoyl Attachment Site for Targeting to Sites of Vesicle Budding and into EMVs
The budding of AcylTyA-GFP and nonbudding of Acyl(G2A)-GFP are reminiscent of an earlier result from the HIV field, that an alanine substitution mutation at the HIV Gag Gly2 codon (HIV Gag(G2A)) eliminates Gag and virus budding (37). To determine whether the G2A mutation has similar effects on the trafficking of HIV Gag to sites of EMV release and into EMVs, we followed the sorting and secretion of HIV Gag-GFP and HIV Gag(G2A)-GFP in N-Rh-PE-labeled Jurkat T-cells. As reported previously (7, 23), Jurkat T-cells target HIV Gag-GFP to endosome-like domains of plasma membrane, where it co-localizes with the classic EMV marker CD63, the ESCRT-associated protein Alix, and as we show again here, N-Rh-PE (Fig. 2, A–D). HIV Gag-GFP was (once again) secreted from the cell in N-Rh-PE-labeled EMVs (Fig. 2, I, J, and M). In contrast, HIV Gag(G2A)TyA-GFP accumulated in the cytoplasm of N-Rh-PE-labeled Jurkat T-cells (Fig. 2, E–H) and was not secreted from the cell in EMVs (Fig. 1, K–M). Thus, the role of myristoylation in the budding of AcylTyA-GFP appears to be similar to that of its role in HIV biogenesis.
FIGURE 2.
The myristoyl attachment site is required for targeting HIV Gag-GFP to sites of vesicle budding and EMVs. A–H, fluorescence and phase microscopy images of N-Rh-PE-labeled Jurkat T-cells expressing HIV Gag-GFP (A–D) or HIV Gag(G2A)-GFP (E–H). Scale bar, 10 μm. I–L, fluorescence microscopy images of EMVs produced by N-Rh-PE-labeled Jurkat T-cells expressing (I and J) HIV Gag-GFP or HIV Gag(G2A)-GFP (K and L). The white circles show the position of GFP-positive vesicles, some of which also possessed N-Rh-PE fluorescence. M, anti-GFP immunoblot of EMV and cell lysates prepared from Jurkat T-cells expressing HIV Gag-GFP or HIV Gag(G2A)-GFP.
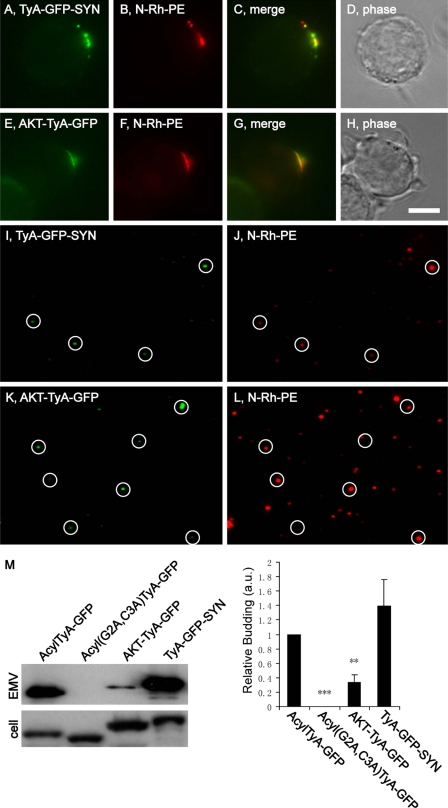
PIP2- and PIP3-binding Domains Target TyA-GFP to Secreted Vesicles
The inner leaflet of the plasma membrane is enriched for PIP2. Moreover, proteins that bind PIP2 can be targeted from the cytoplasm to the inner leaflet of the PM (38). To determine whether such lipid-binding domains can induce the budding of TyA-GFP, we followed the sorting and secretion of a TyA-GFP-SYN fusion protein that contains the PIP2-binding PDZ domains of syntenin (39). N-Rh-PE-labeled Jurkat T-cells trafficked TyA-GFP-SYN to sites of EMV budding that were enriched for N-Rh-PE (Fig. 3, A–D) and secreted TyA-GFP-SYN from the cell in EMVs (Fig. 3, I, J, and M). Moreover, the relative budding of TyA-GFP-SYN was robust, equivalent to that of AcylTyA-GFP (140 ± 37%; n = 3; p = 0.2).
FIGURE 3.
PIP2- and PIP3-binding domains can target TyA-GFP to N-Rh-PE-enriched domains of plasma membrane and into EMVs. A–H, fluorescence and phase microscopy images of N-Rh-PE-labeled Jurkat T-cells expressing TyA-GFP-SYN (A–D) or AKT-TyA-GFP (E–H). Scale bar, 10 μm. I–L, fluorescence microscopy images of EMVs produced by N-Rh-PE-labeled Jurkat T-cells expressing TyA-GFP-SYN (I and J) or AKT-TyA-GFP (K and L). The white circles show the position of GFP-positive vesicles, some of which also possessed N-Rh-PE fluorescence. M, anti-GFP immunoblot of EMV and cell lysates prepared from Jurkat T-cells expressing AcylTyA-GFP, Acyl(G2A,C3A)TyA-GFP, TyA-GFP-SYN, or AKT-TyA-GFP. The bar graph to the right shows the average ± 1 S.D. of the relative budding calculated from band intensities (EMV/(EMV + cell)) relative to the AcylTyA-GFP control, which were determined by densitometric analysis of immunoblot films. Two asterisks reflect a p value of < 0.005; three asterisks reflect a p value of < 0.0005.
Phosphatidylinositol 3,4,5-trisphosphate (PIP3) is also enriched at the plasma membrane and can mediate the PM targeting of proteins that contain PIP3-binding domains (40). One such PIP3-binding module is the PH domain of the AKT protein kinase (41, 42). A fusion protein between the PIP3-binding domain of AKT and TyA-GFP (AKT-TyA-GFP) was targeted to sites of EMV budding and into EMVs (Fig. 3, E–H and K–M). However, the relative budding of AKT-TyA-GFP was only 34 ± 10% (n = 3; p = 0.008) of what we observed for AcylTyA-GFP (Fig. 3M).
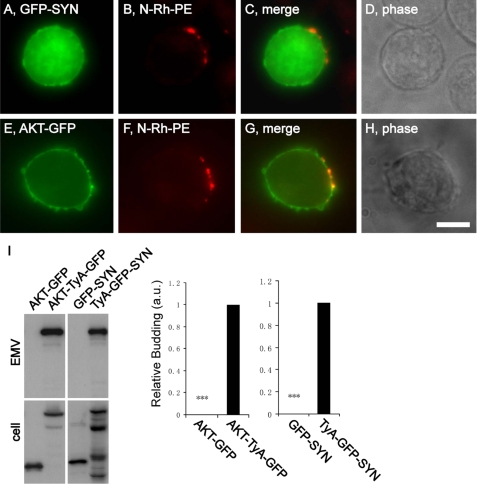
The localization of TyA-GFP-SYN and AKT-TyA-GFP to sites of vesicle budding might reflect an enrichment of PIP2 and PIP3 at these regions of the PM, rather than the oligomerization-induced targeting of PM proteins to these destinations. To address this issue, we determined the subcellular distribution of PIP2 and PIP3 reporter proteins in N-Rh-PE-labeled Jurkat T-cells. GFP-SYN and AKT-GFP are expected to bind to PIP2 and PIP3, respectively, and were localized throughout the PM (Fig. 4, A–H). The PIP2 reporter, GFP-SYN, never displayed any enrichment at sites of budding, and although we occasionally detected a slight enrichment of AKT-GFP at these sites, this occurred in only a minor proportion of the cells. As for whether AKT-GFP and GFP-SYN were secreted from the cell in EMVs, immunoblot analysis of EMV and cell lysates indicated that their secretion was extremely low relative to that of AKT-TyA-GFP and TyA-GFP-SYN, respectively (Fig. 4I).
FIGURE 4.
PIP2- and PIP3-binding domains do not target GFP to sites of vesicle budding or to EMVs. A–H, fluorescence and phase microscopy images of N-Rh-PE-labeled Jurkat T-cells expressing GFP-SYN (A–D) or AKT-GFP (E–H). Scale bar, 10 μm. I, anti-GFP immunoblot of EMV and cell lysates prepared from Jurkat T-cells expressing AKT-GFP, AKT-TyA-GFP, GFP-SYN, or TyA-GFP-SYN. The bar graph to the right shows the average ± 1 S.D. of the relative budding calculated from band intensities (EMV/(EMV + cell)) relative to the AcylTyA-GFP control, which were determined by densitometric analysis of immunoblot films. Three asterisks reflect a p value of < 0.0005.
A C-terminal Prenylation and Palmitoylation Tag Targets TyA to EMVs
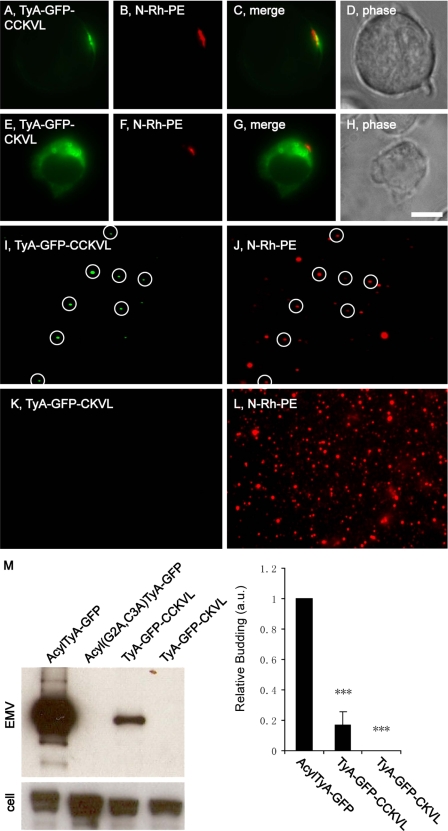
The robust budding of the C-terminally anchored TyA-GFP-SYN protein raised the possibility that a C-terminally positioned lipid anchor tag might also be able to induce the budding of TyA-GFP. To examine this possibility, we followed the sorting and secretion of a TyA-GFP variant carrying the amino acids CCKVL at its C terminus, a dual prenylation/palmitoylation tag; CCKVL are the C-terminal five amino acids of RhoB and can specify prenylation at Cys(−4) and palmitoylation at Cys(−5) (43). Jurkat T-cells trafficked TyA-GFP-CCKVL to sites of vesicle budding and secreted it from the cell in EMVs (Fig. 5, A-D, I, J, and M). However, the co-localization of TyA-GFP-CCKVL with N-Rh-PE appeared to be less robust than what we observed for AcylTyA-GFP and perhaps even less than we observed for AKT-TyA-GFP. As for the budding of TyA-GFP-CCKVL, this was much reduced, only 17 ± 9% (n = 3; p = 0.004) relative to control. TyA-GFP-CKVL, which is predicted to cause C-terminal prenylation but lacks the palmitoyl attachment site, accumulated on the surface of cytoplasmic vesicles of unknown identity rather than at plasma membrane domains enriched for other EMV cargoes (Fig. 5, E–H). In addition, we found no evidence that TyA-GFP-CKVL was secreted from the cell in EMVs (Fig. 5, K–M).
FIGURE 5.
A C-terminal prenylation/palmitoylation tag targets TyA-GFP to EMVs. A–H, fluorescence and phase microscopy images of N-Rh-PE-labeled Jurkat T-cells expressing TyA-GFP-CCKVL (A–D) or TyA-GFP-CKVL (E–H). Scale bar, 10 μm. I–L, fluorescence microscopy images of EMVs produced by N-Rh-PE-labeled Jurkat T-cells expressing TyA-GFP-CCKVL (I and J) or TyA-GFP-CKVL (K and L). The white circles show the position of GFP-positive vesicles, some of which also possessed N-Rh-PE fluorescence. M, anti-GFP immunoblot of EMV and cell lysates prepared from Jurkat T-cells expressing AcylTyA-GFP, Acyl(G2A,C3A)TyA-GFP, TyA-GFP-CCKVL, or TyA-GFP-CKVL. The bar graph to the right shows the average ± 1 S.D. of the relative budding calculated from band intensities (EMV/(EMV + cell)) relative to the AcylTyA-GFP control, which were determined by densitometric analysis of immunoblot films. Three asterisks reflect a p value of < 0.0005.
CD43 Weakly Targets TyA-GFP to EMVs
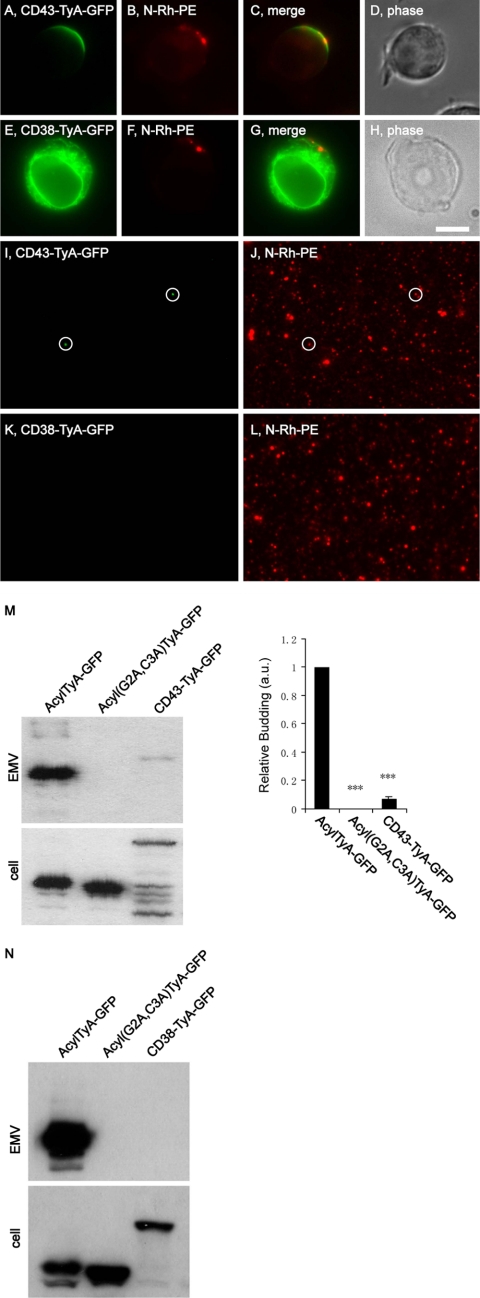
The PM anchors assayed above all allow for direct, cytoplasm-to-PM trafficking of the cargo protein. We next tested whether the classic pathway of integral PM protein biosynthesis could deliver proteins to sites of budding and into EMVs. One approach to this issue was to follow the sorting and secretion of CD43-TyA-GFP, a fusion protein between CD43, a type-1 integral membrane protein, and TyA-GFP, with the TyA-GFP sequences added to the cytoplasmic C terminus of CD43. When expressed in N-Rh-PE-labeled Jurkat T-cells, CD43-TyA-GFP was trafficked to the plasma membrane where it showed significant overlap with N-Rh-PE (Fig. 6, A–D). However, the distribution of CD43-TyA-GFP was always broader than that of N-Rh-PE, extending laterally from the sides of the endosome-like domains of PM, similar to what we observed for TyA-GFP-CCKVL. Moreover, the budding of CD43-TyA-GFP was weak, as CD43-TyA-GFP fluorescence was detected in only a few secreted EMVs (Fig. 6, I and J), and immunoblot analysis showed only a modest signal in EMVs (6.6 ± 2% (n = 3; p = 0.0002) relative to AcylTyA-GFP (Fig. 6M)). Complicating the interpretation of these results was the observation that CD43-TyA-GFP was subject to extensive proteolysis, both in cell lysates and in purified EMVs. In fact, we could not detect full-length CD43-TyA-GFP in EMV lysates; only a smaller protein was detected. This anti-GFP reactive protein had a higher Mr than AcylTyA-GFP, presumably due to cleavage of CD43-TyA-GFP somewhere within the CD43 portion of the fusion protein.
FIGURE 6.
Fusions of TyA-GFP to integral plasma membrane proteins. A–H, fluorescence and phase microscopy images of N-Rh-PE-labeled Jurkat T-cells expressing CD43-TyA-GFP (A–D) or CD38-TyA-GFP (E–H). Scale bar, 10 μm. I–L, fluorescence microscopy images of EMVs produced by N-Rh-PE-labeled Jurkat T-cells expressing CD43-TyA-GFP (I and J) or CD38-TyA-GFP (K and L). The white circles show the position of GFP-positive vesicles, some of which also possessed N-Rh-PE fluorescence. M, anti-GFP immunoblot of EMV and cell lysates prepared from Jurkat T-cells expressing AcylTyA-GFP, Acyl(G2A,C3A)TyA-GFP, or CD43-TyA-GFP. The bar graph to the right shows the average ± 1 S.D. of the relative budding calculated from band intensities (EMV/(EMV + cell)) relative to the AcylTyA-GFP control, which were determined by densitometric analysis of immunoblot films. Three asterisks reflect a p value of < 0.0005. N, anti-GFP immunoblot of EMV and cell lysates prepared from Jurkat T-cells expressing AcylTyA-GFP, Acyl(G2A,C3A)TyA-GFP, or CD38-TyA-GFP.
Although the budding of CD43-TyA-GFP was low relative to our positive control, it was better than what we observed for other fusions between TyA-GFP and integral PM proteins. For example, we could not detect the expression of fusion proteins between TyA-GFP and the type-1 integral PM proteins CD4 and CD83 (data not shown), perhaps because they are eliminated by the endoplasmic reticulum-associated degradation pathway (44). In the case of CD38-TyA-GFP, a fusion of TyA-GFP to the C terminus of the type-2 integral PM protein CD38, the protein appeared to be retained in the ER (Fig. 6, E–H) and showed no evidence for budding (Fig. 6, K, L, and N). Similar results were observed for a fusion between the type-2 PM protein CD69 and TyA-GFP (data not shown). In summary, it appears that the classic secretory pathway can support the biogenesis of a genetically encoded, highly oligomeric EMV cargo, but perhaps only for a select subset of integral PM proteins.
Targeting TyA-GFP to Endosome Membranes Induces only Very Weak Budding
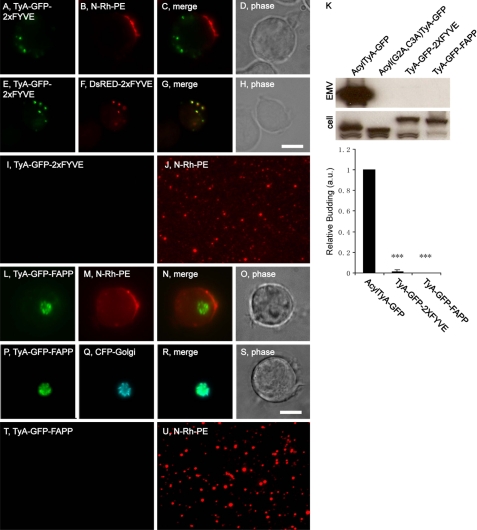
The classic model of exosome biogenesis is that proteins are targeted to endosome membranes, bud into endosomes, and are released later upon endosome-PM fusion (3). If this model is correct, it should be possible to induce the budding of TyA-GFP by targeting it to endosome membranes. To test this hypothesis, we took advantage of the fact that endosome membranes are enriched for the lipid phosphatidylinositol 3-phosphate (PI3P). The FYVE domain from the ESCRT-0 protein Hrs binds specifically to PI3P, and two tandem repeats of the HRS domain (2xFYVE) efficiently target heterologous proteins to the outer leaflet of PI3P-rich endosome membranes (45). More specifically, we transfected Jurkat T-cells with a plasmid designed to express a TyA-GFP-2xFYVE fusion protein, pulse-labeled them with N-Rh-PE a day later, and incubated them an additional day. Cells and EMVs were then harvested and examined by both fluorescence microscopy and immunoblot. We also co-transfected Jurkat T-cells with plasmids designed to express TyA-GFP-2xFYVE and DsRED-2xFYVE (a marker for PI3P-rich membranes), incubated the cells for 2 days, and examined them by fluorescence microscopy. These experiments revealed that Jurkat T-cells did not localize TyA-GFP-2xFYVE to endosome-like domains of plasma membrane (Fig. 7, A–D) but instead targeted it to PI3P-rich endosomes (Fig. 7, E–H). Even though the HRS protein and other ESCRT proteins are implicated in outward vesicle budding during MVB biogenesis, these cells failed to bud TyA-GFP-2xFYVE at robust levels. This is reflected in the fact that the vast majority of N-Rh-PE-positive vesicles lacked GFP fluorescence (Fig. 7, I and J) and that immunoblot analysis of EMV fractions detected only the faintest of signals, visible only upon prolonged exposure of the EMV blot (Fig. 7K). Nevertheless, a specific signal was detected, 1.4 ± 1.5% of control (n = 3; p = 8 × 10−5), and we did, on rare occasions, observe an N-Rh-PE-positive vesicle with GFP fluorescence (data not shown). Thus, while targeting TyA-GFP to PI3P-rich endosomes was not an efficient mechanism for inducing its budding, it did induce a slight amount of TyA-GFP budding.
FIGURE 7.
PI3P-binding and phosphatidylinositol 4-phosphate-binding domains do not induce robust budding of TyA-GFP. A–D, fluorescence and phase microscopy images of N-Rh-PE-labeled Jurkat T-cells expressing TyA-GFP-2xFYVE. E–H, fluorescence and phase microscopy images of Jurkat T-cells expressing TyA-GFP-2xFYVE and DsRED-2xFYVE. Scale bar, 10 μm. I and J, fluorescence microscopy images of EMVs produced by N-Rh-PE-labeled Jurkat T-cells expressing TyA-GFP-2xFYVE. K, anti-GFP immunoblot of EMV and cell lysates prepared from Jurkat T-cells expressing AcylTyA-GFP, Acyl(G2A,C3A)TyA-GFP, TyA-GFP-2xFYVE, or TyA-GFP-FAPP. The bar graph below the immunoblot shows the average ± 1 S.D. of the relative budding calculated from band intensities (EMV/(EMV + cell)) relative to the AcylTyA-GFP control, which were determined by densitometric analysis of immunoblot films. Three asterisks reflect a p value of < 0.0005. L–O, fluorescence and phase microscopy images of N-Rh-PE-labeled Jurkat T-cells expressing TyA-GFP-FAPP. P–S, fluorescence and phase microscopy images of Jurkat T-cells expressing TyA-GFP-FAPP and CFP-Golgi. Scale bar, 10 μm. T and U, fluorescence microscopy images of EMVs produced by N-Rh-PE-labeled Jurkat T-cells expressing TyA-GFP-FAPP.
Targeting TyA-GFP to Golgi Membranes Does Not Induce Its Budding
We also tested whether targeting TyA-GFP to Golgi membranes could induce its budding. The Golgi apparatus is enriched for phosphatidylinositol 4-phosphate and phosphatidylinositol 4-phosphate-binding domains can target proteins to the Golgi membrane, including the PH domain of FAPP (46–48). Jurkat T-cells were transfected with a plasmid designed to express a fusion between TyA-GFP and the phosphatidylinositol 4-phosphate-binding PH domain of FAPP, TyA-GFP-FAPP, pulse-labeled with N-Rh-PE a day later, and then incubated one additional day. Cells and EMVs were harvested and assayed by fluorescence microscopy and immunoblot. In addition, Jurkat T-cells were co-transfected with plasmids designed to express TyA-GFP-FAPP and a Golgi-targeted form of CFP, incubated for 2 days, and then processed for fluorescence microscopy. These experiments revealed that Jurkat T-cells did not traffic TyA-GFP-FAPP to N-Rh-PE-rich domains of plasma membrane (Fig. 7, L–O), but instead targeted it to Golgi membranes, where it co-localized with CFP-Golgi (Fig. 7, P–S). Furthermore, we were unable to detect TyA-GFP-FAPP in secreted vesicles, either by immunoblot of EMVs (Fig. 7K) or by fluorescence microscopy of secreted vesicles (Fig. 7, T and U).
Acylation Tag Targets Other Highly Oligomeric Cytoplasmic Proteins to EMVs
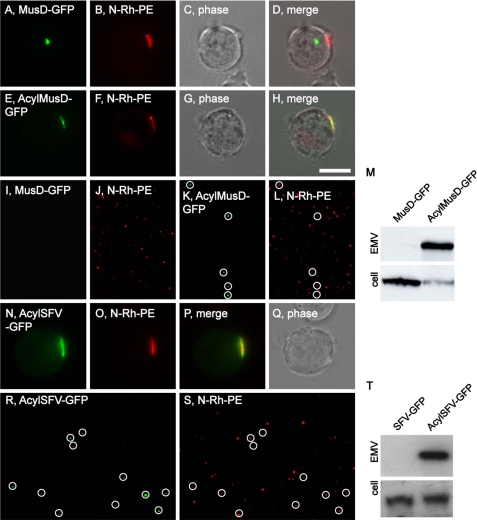
Although multiple PM anchors targeted TyA-GFP into secreted vesicles, it is not known whether PM anchors can target any other proteins to sites of vesicle budding and into EMVs. To explore this issue, we appended the 10-amino acid-long acyl tag (MGCINSKRKD-) to the N terminus of MusD-GFP. (MusD is the structural protein of the MusD LTR retrotransposon, does not bud, and accumulates in cytoplasmic retrotransposon assembly particles (35).) Jurkat T-cells were transfected with plasmids designed to express MusD-GFP or AcylMusD-GFP, incubated for a day, pulse-labeled with N-Rh-PE, incubated for another day, and then cells and EMVs were examined by fluorescence microscopy and immunoblot. Jurkat T-cells accumulated MusD-GFP in a perinuclear, cytoplasmic structure that was clearly distinct from the plasma membrane domains associated with vesicle budding (Fig. 8, A–D). In contrast, AcylMusD-GFP co-localized with N-Rh-PE at EMV budding sites (Fig. 8, E–H). Fluorescence microscopy of purified EMVs revealed that MusD-GFP was not secreted in vesicles, whereas AcylMusD-GFP fluorescence could be detected in many N-Rh-PE-containing EMVs (Fig. 8, I–L). This difference was corroborated by immunoblot analysis (Fig. 8M), which showed robust budding of AcylMusD-GFP and little or no budding of MusD-GFP.
FIGURE 8.
The 10-amino acid-long acylation tag targets MusD and SFV to sites of vesicle budding and into EMVs. A–H, fluorescence and phase microscopy images of N-Rh-PE-labeled Jurkat T-cells expressing MusD-GFP (A–D) or AcylMusD-GFP (E–H). Scale bar, 10 μm. I–L, fluorescence microscopy images of EMVs produced by N-Rh-PE-labeled Jurkat T-cells expressing MusD-GFP (I and J) or AcylMusD-GFP (K and L). The white circles show the position of GFP-positive vesicles, some of which also possessed N-Rh-PE fluorescence. M, anti-GFP immunoblot of EMV and cell lysates prepared from Jurkat T-cells expressing MusD-GFP or AcylMusD-GFP. N–Q, fluorescence and phase microscopy images of N-Rh-PE-labeled Jurkat T-cells expressing AcylSFV-GFP. R and S, fluorescence microscopy images of EMVs produced by N-Rh-PE-labeled Jurkat T-cells expressing AcylSFV-GFP. The white circles show the position of GFP-positive vesicles, some of which also possessed N-Rh-PE fluorescence. T, anti-GFP immunoblot of EMV and cell lysates prepared from Jurkat T-cells expressing SFV-GFP or AcylSFV-GFP.
We also tested whether the acylation tag could induce the budding of the SFV Gag protein (7). Foamy viruses such as SFV are the closest relatives to the true retroviruses (orthoretroviridae) but differ from the orthoretroviruses in several ways, two of which are that their Gag proteins do not bud from cells and that virus budding is mediated instead by their ENV proteins (49). It is for this reason that we previously used SFV Gag-GFP as a negative, nonbudding control protein in a study of whether orthoretroviral Gag proteins are targeted to sites of vesicle secretion and bud from the cell in EMVs (they are (7)). Here, we followed the sorting and secretion of AcylSFV-GFP by N-Rh-PE-labeled Jurkat T-cells. In contrast to SFV-GFP, which accumulates in the cytoplasm of Jurkat T-cells (7), AcylSFV-Gag-GFP was targeted to the PM where it co-localized with N-Rh-PE (Fig. 8, N–Q). Furthermore, fluorescence microscopy of EMVs released by these cells demonstrated that AcylSFV-GFP was secreted from the cell in EMVs (Fig. 8, R and S). Immunoblot analysis of cell and EMV lysates confirmed the specific budding of AcylSFV-GFP (Fig. 8T).
DISCUSSION
There is increasing evidence that the secretion of EMVs is a conserved process of animal cells with important implications for multiple physiological processes and several human diseases (3). However, our understanding of EMV biogenesis is rudimentary, and little is known about the signals that target proteins to sites of EMV budding and into EMVs. The data presented in this report expand our understanding of EMV biogenesis by showing that several different types of membrane anchors can target highly oligomeric cytoplasmic proteins to sites of vesicle secretion and EMVs.
Implications for EMV Biogenesis
The membrane anchors used in our experiments were diverse (Fig. 9). The membrane anchors that induced the most efficient budding of TyA-GFP were (i) a myristoylation tag and (ii) a PIP2-binding domain. Both of these modifications are known to target proteins to the plasma membrane, and thus, their ability to target TyA-GFP to sites of EMV budding and into secreted vesicles is consistent with the hypothesis that PM anchors can target highly oligomeric cytoplasmic proteins to EMVs (7). In addition, the robust budding of TyA-GFP-SYN demonstrates that C-terminally positioned PM anchors can function as well as N-terminal PM anchors in targeting cargoes to sites of vesicle budding and into EMVs.
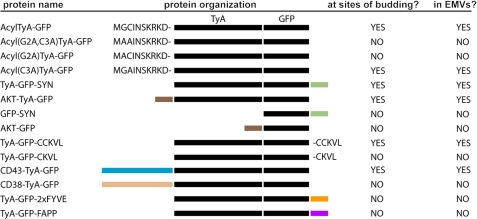
FIGURE 9.
Line diagram of TyA-GFP fusion proteins, their linear organization, sorting to sites of budding, and secretion from the cell in EMVs.
The levels of budding that we observed for AKT-TyA-GFP, TyA-GFP-CCKVL, and CD43-TyA-GFP were all significantly less, ∼34%, ∼17%, and ∼7% of control, respectively. We do not know why these three PM anchors failed to induce a more robust budding of TyA-GFP. These cargoes all displayed a distribution that was somewhat smoother and broader than that of the bona fide EMV cargo lipid within the cell, another indication that they are relatively poor cargoes. In the case of the PIP3-binding domain of AKT, the reduced budding might reflect the relative abundance of the two target lipids in the inner leaflet of the plasma membrane. As for TyA-GFP-CCKVL, its intermediate level of budding (∼15% of control) could reflect substoichiometric palmitoylation, as this protein requires the putative palmitoyl attachment site for budding (TyA-GFP-CKVL did not bud), and protein palmitoylation is known to be reversible and often substoichiometric (50). In regard to the CD43-TyA-GFP fusion, this was the only one of five different fusions between TyA-GFP and an integral plasma membrane protein that was reliably expressed and delivered to the plasma membrane, with the others either being retained in the ER or expressed at something less than our level(s) of detection. In this context, it could be that the low level of CD43-TyA-GFP budding (∼7% of control) reflects difficulties in the transport of this protein from the ER and through the classical secretory pathway.
Perhaps the most unexpected observation in this report is the failure of a PI3P-binding domain to induce the vesicular secretion of TyA-GFP. Exosomes are thought to arise by budding at endosome membranes by the same mechanisms as bona fide MVBs (3), a process that is initiated by the PI3P-binding HRS protein (26). As such, we expected that targeting TyA-GFP to PI3P-rich membranes by the FYVE domains of HRS should induce a level of budding similar to that of the most active PM anchor. However, this was not the case, as the budding of TyA-GFP-2xFYVE was barely detectable (only ∼2% of control). TyA-GFP-2xFYVE fusion was expressed relatively well, was highly fluorescent, and was properly targeted to PI3P-rich endosome membranes, indicating that it was probably not subject to gross defects in folding, solubility, etc. Furthermore, the general organization of TyA-GFP-2xFYVE is the same as TyA-GFP-SYN, which budded extremely well from Jurkat T-cells. These considerations raise the possibility that protein targeting to endosome membranes might play less of a role in EMV biogenesis than believed currently. However, additional experiments are required to determine whether TyA-GFP-2xFYVE accumulates at the limiting membrane of endosomes or whether it buds into endosomes, but the internal vesicles are delivered to the lysosome rather than the extracellular milieu. In addition, it is unclear whether this particular result is specific for TyA-GFP or is seen for other highly oligomeric cytoplasmic proteins targeted to the endosome membrane. As for budding into the Golgi, we found that a Golgi-localized form of TyA-GFP budded even less than TyA-GFP-2xFYVE, indicating that Golgi membranes also do not support EMV biogenesis.
Relevance to Retrovirus Budding
Previous studies have noted a number of similarities between EMV biogenesis and retrovirus budding, including the molecular composition of the released particles, sites of budding in different cell types, and the targeting signals that deliver proteins to EMVs and retrovirus particles (7, 23, 24, 51). These and other observations support the hypothesis that retrovirus budding is, at its core, a form of EMV biogenesis (4). The data presented here, which demonstrate that the best EMV sorting signals for TyA-GFP were a myristoylation tag and a PIP2-binding domain, lends even more support to this hypothesis, for these are the same membrane anchors that mediate the budding of retroviral Gag proteins and infectious retroviruses (52–54). The exosome/microvesicle model of retroviral biogenesis also posits that the acquisition of EMV sorting information is a critical step in the evolution of retroviruses from an LTR retrotransposon (4). The data presented here indicate that mutations that conferred an N-terminal myristoylation site or PIP2-binding activity on a retrotransposon structural protein would also be sufficient to induce the budding of a retrotransposon from the cell. If this occurs in an organism that exchanges EMVs, such mutations might also be sufficient to mediate its transformation from a retrotransposon to an infectious agent.
Acknowledgments
We thank Dan Raben and Michael Caterina for comments and suggestions during the course of this study.
The work was supported, in whole or in part, by National Institutes of Health Grant DK45787.
- PM
- plasma membrane
- EMV
- exosome/microvesicle
- ER
- endoplasmic reticulum
- ESCRT
- endosomal sorting complexes required for transport
- MVB
- multivesicular body
- N-Rh-PE
- N-rhodamine-phosphatidylethanolamine
- SFV
- simian foamy virus
- PI3P
- phosphatidylinositol 3-phosphate
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PIP3
- phosphatidylinositol 3,4,5-trisphosphate.
REFERENCES
- 1. Schorey J. S., Bhatnagar S. (2008) Traffic 9, 871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Théry C., Ostrowski M., Segura E. (2009) Nat. Rev. Immunol. 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 3. Simons M., Raposo G. (2009) Curr. Opin. Cell Biol. 21, 575–581 [DOI] [PubMed] [Google Scholar]
- 4. Gould S. J., Booth A. M., Hildreth J. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10592–10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trams E. G., Lauter C. J., Salem N., Jr., Heine U. (1981) Biochim. Biophys. Acta 645, 63–70 [DOI] [PubMed] [Google Scholar]
- 6. Pan B. T., Johnstone R. M. (1983) Cell 33, 967–978 [DOI] [PubMed] [Google Scholar]
- 7. Fang Y., Wu N., Gan X., Yan W., Morrell J. C., Gould S. J. (2007) PLoS Biol. 5, 1267–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cocucci E., Racchetti G., Meldolesi J. (2009) Trends Cell Biol. 19, 43–51 [DOI] [PubMed] [Google Scholar]
- 9. Théry C., Zitvogel L., Amigorena S. (2002) Nat. Rev. Immunol. 2, 569–579 [DOI] [PubMed] [Google Scholar]
- 10. Skog J., Würdinger T., van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T., Jr., Carter B. S., Krichevsky A. M., Breakefield X. O. (2008) Nat. Cell Biol. 10, 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 12. Liégeois S., Benedetto A., Garnier J. M., Schwab Y., Labouesse M. (2006) J. Cell Biol. 173, 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kolotuev I., Apaydin A., Labouesse M. (2009) Traffic 10, 803–810 [DOI] [PubMed] [Google Scholar]
- 14. Yu X., Harris S. L., Levine A. J. (2006) Cancer Res. 66, 4795–4801 [DOI] [PubMed] [Google Scholar]
- 15. Yu X., Riley T., Levine A. J. (2009) FEBS J. 276, 2201–2212 [DOI] [PubMed] [Google Scholar]
- 16. Keller S., König A. K., Marmé F., Runz S., Wolterink S., Koensgen D., Mustea A., Sehouli J., Altevogt P. (2009) Cancer Lett. 278, 73–81 [DOI] [PubMed] [Google Scholar]
- 17. Alais S., Simoes S., Baas D., Lehmann S., Raposo G., Darlix J. L., Leblanc P. (2008) Biol. Cell 100, 603–615 [DOI] [PubMed] [Google Scholar]
- 18. Leblanc P., Alais S., Porto-Carreiro I., Lehmann S., Grassi J., Raposo G., Darlix J. L. (2006) EMBO J. 25, 2674–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fevrier B., Vilette D., Archer F., Loew D., Faigle W., Vidal M., Laude H., Raposo G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9683–9688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dukers D. F., Meij P., Vervoort M. B., Vos W., Scheper R. J., Meijer C. J., Bloemena E., Middeldorp J. M. (2000) J. Immunol. 165, 663–670 [DOI] [PubMed] [Google Scholar]
- 21. Logozzi M., De Milito A., Lugini L., Borghi M., Calabrò L., Spada M., Perdicchio M., Marino M. L., Federici C., Iessi E., Brambilla D., Venturi G., Lozupone F., Santinami M., Huber V., Maio M., Rivoltini L., Fais S. (2009) PloS One 4, e5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nazarenko I., Rana S., Baumann A., McAlear J., Hellwig A., Trendelenburg M., Lochnit G., Preissner K. T., Zöller M. (2010) Cancer Res. 70, 1668–1678 [DOI] [PubMed] [Google Scholar]
- 23. Booth A. M., Fang Y., Fallon J. K., Yang J. M., Hildreth J. E., Gould S. J. (2006) J. Cell Biol. 172, 923–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krishnamoorthy L., Bess J. W., Jr., Preston A. B., Nagashima K., Mahal L. K. (2009) Nat. Chem. Biol. 5, 244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marsh M., Theusner K., Pelchen-Matthews A. (2009) Biochemical Society transactions 37, 185–189 [DOI] [PubMed] [Google Scholar]
- 26. Hurley J. H. (2008) Curr. Opin. Cell Biol. 20, 4–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. (2009) Nature 458, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saksena S., Sun J., Chu T., Emr S. D. (2007) Trends Biochem. Sci. 32, 561–573 [DOI] [PubMed] [Google Scholar]
- 29. Hanson P. I., Shim S., Merrill S. A. (2009) Curr. Opin. Cell Biol. 21, 568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. (2008) Science 319, 1244–1247 [DOI] [PubMed] [Google Scholar]
- 31. Deneka M., Pelchen-Matthews A., Byland R., Ruiz-Mateos E., Marsh M. (2007) J. Cell Biol. 177, 329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Welsch S., Keppler O. T., Habermann A., Allespach I., Krijnse-Locker J., Kräusslich H. G. (2007) PLoS Pathog. 3, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. (2002) Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- 34. Raposo G., Moore M., Innes D., Leijendekker R., Leigh-Brown A., Benaroch P., Geuze H. (2002) Traffic 3, 718–729 [DOI] [PubMed] [Google Scholar]
- 35. Mager D. L., Freeman J. D. (2000) J. Virol. 74, 7221–7229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roth J. F. (2000) Yeast 16, 785–795 [DOI] [PubMed] [Google Scholar]
- 37. Göttlinger H. G., Sodroski J. G., Haseltine W. A. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5781–5785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. (2006) Science 314, 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zimmermann P., Meerschaert K., Reekmans G., Leenaerts I., Small J. V., Vandekerckhove J., David G., Gettemans J. (2002) Mol. Cell 9, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 40. Parent C. A., Blacklock B. J., Froehlich W. M., Murphy D. B., Devreotes P. N. (1998) Cell 95, 81–91 [DOI] [PubMed] [Google Scholar]
- 41. James S. R., Downes C. P., Gigg R., Grove S. J., Holmes A. B., Alessi D. R. (1996) Biochem. J. 315, 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haugh J. M., Codazzi F., Teruel M., Meyer T. (2000) J. Cell Biol. 151, 1269–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Adamson P., Marshall C. J., Hall A., Tilbrook P. A. (1992) J. Biol. Chem. 267, 20033–20038 [PubMed] [Google Scholar]
- 44. Brodsky J. L., Wojcikiewicz R. J. (2009) Curr. Opin. Cell Biol. 21, 516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gillooly D. J., Raiborg C., Stenmark H. (2003) Histochem. Cell Biol. 120, 445–453 [DOI] [PubMed] [Google Scholar]
- 46. De Matteis M. A., Di Campli A., Godi A. (2005) Biochim. Biophys. Acta 1744, 396–405 [DOI] [PubMed] [Google Scholar]
- 47. Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirchhausen T., Albanesi J. P., Roth M. G., Yin H. L. (2003) Cell 114, 299–310 [DOI] [PubMed] [Google Scholar]
- 48. Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., De Matteis M. A. (2004) Nat. Cell Biol. 6, 393–404 [DOI] [PubMed] [Google Scholar]
- 49. Coffin J. M., Hughes S. H., Varmus H. E. (eds) (1997) Retroviruses, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York: [PubMed] [Google Scholar]
- 50. Charollais J., Van Der Goot F. G. (2009) Mol. Membr. Biol. 26, 55–66 [DOI] [PubMed] [Google Scholar]
- 51. Nguyen D. G., Booth A., Gould S. J., Hildreth J. E. (2003) J. Biol. Chem. 278, 52347–52354 [DOI] [PubMed] [Google Scholar]
- 52. Chen K., Bachtiar I., Piszczek G., Bouamr F., Carter C., Tjandra N. (2008) Biochemistry 47, 1928–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Freed E. O. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11101–11102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hamard-Peron E., Juillard F., Saad J. S., Roy C., Roingeard P., Summers M. F., Darlix J. L., Picart C., Muriaux D. (2010) J. Virol. 84, 503–515 [DOI] [PMC free article] [PubMed] [Google Scholar]