Abstract
Prohead RNA (pRNA) is an essential component of the self-assembling φ29 bacteriophage DNA packaging motor. Different related species of bacteriophage share only 12% similarity in pRNA sequences. The secondary structure for pRNA is conserved, however. In this study, we present evidence for self-assembly in different pRNA sequences and new measurements of the energetics for the quaternary interactions in pRNA dimers and trimers. The energetics for self-assembly in different pRNA sequences are similar despite very different sequences in the loop-loop interactions. The architecture surrounding the interlocking loops contributes to the stability of the pRNA quaternary interactions, and sequence variation outside the interlocking loops may counterbalance the changes in the loop sequences. Thus, the evolutionary divergence of pRNA sequences maintains not only conservation of function and secondary structure but also stabilities of quaternary interactions. The self-assembly of pRNA can be fine-tuned with variations in magnesium chloride, sodium chloride, temperature, and concentration. The ability to control pRNA self-assembly holds promise for the development of nanoparticle therapeutic applications for this biological molecule. The pRNA system is well suited for future studies to further understand the energetics of RNA tertiary and quaternary interactions, which can provide insight into larger biological assemblies such as viruses and biomolecular motors.
Keywords: Bacteriophage, Bioenergetics, Biophysics, Molecular Motors, RNA Folding, RNA Energetics, RNA Nanoparticle, RNA Self-assembly, Prohead RNA
Introduction
Prohead RNA (pRNA)2 is an essential component of the biomolecular motor that packages DNA into φ29 bacteriophage viral capsids (1, 2). At least four very different RNA sequences with conserved secondary structures (Fig. 1) (3) are capable of fulfilling this unique function in bacteriophage. The φ29 RNA interacts with the gp16 ATPase that drives the biological motor (4). Mutations in the pRNA can inactivate the biological motor and packaging activity (4–10). The role of pRNA and the mechanism of this biological motor are a controversial area of active research (11–15). This biological nanomotor can package the 19,300 bases of the φ29 DNA genome into the viral capsid against a load as high as 57 piconewtons with a maximum force exceeding 100 piconewtons, thus filling the capsid to >50% capacity and reaching internal pressures as high as 60 atm (16–19). Cryo-electron microscopy of in vitro assembled packaging motors showed a ring of five pRNAs interacting with a ring of five gp16 ATPases and a ring of 12 gp10 connector proteins to form the core of the nanomotor (12). Biophysical studies of the components of this motor can contribute to understanding how the motor assembles and works to package DNA.
FIGURE 1.
Secondary structures of φ29, M2, SF5, and GA1 pRNAs. Conserved nucleotides are green. Nucleotides that participate in potential Watson-Crick intermolecular tertiary and quaternary interactions between the CE bulge loop and the D loop hairpin are in red. The secondary structures shown were determined experimentally by enzymatic and chemical modification data (3). Three additional pRNA sequences have also been proposed from phylogenetic alignments (9).
pRNA is a natural sequence with the unique property of forming ring nanostructures. pRNA self-assembles in dimers, trimers, pentamers, and hexamers through loop-loop interactions (Fig. 2) (5, 9, 14, 15, 20). HNN-COSY NMR experiments have shown the formation of intermolecular hydrogen bonding in Watson-Crick pairs between two RNA hairpin models of the loop-loop interaction (21). All of the natural sequences for pRNA contain the potential for Watson-Crick pairing within the interlocking loops, i.e. the CE and D loops (Figs. 1 and 2). The novel self-assembly properties of pRNA are the basis for using pRNA as a nanoparticle building block and therapeutic delivery vector (22–29). RNA has a tremendous capacity for forming diverse nanostructures, including tRNA squares (30, 31), cubes (32), polyhedrons (33), nanorings (34), filaments (35), paranemic motifs (36), and nanocrowns (37). Understanding the sequence variation of pRNA self-assembly will enable future design of nanoparticles and nanomotors. This work provides the biophysical foundations for further studies on the structures and applications of this naturally self-assembling RNA nanoparticle.
FIGURE 2.
Ball-and-stick model of pRNA monomer, dimer, and pentamer. Loops are represented as balls, and helices are represented as sticks. The angles between helices are shown as right angles for simplicity, but coaxial stacking and helix orientations have not been experimentally determined yet. The helices are labeled in the monomer diagram. Helix B extends from the 3′-end of helix A in the full-length 174-mer pRNA but is not necessary for self-assembly or motor function and is not shown. The CE bulge loop and the D hairpin loop are shown as orange balls. Base pairing interactions between the CE and D loops is proposed to occur in pRNA dimers and multimers. The arrangement of pRNA in a pentamer is based on cryo-electron microscopy of in vitro assembled packaging motors (12).
Although previous studies have thoroughly explored the self-assembly of the φ29 RNA sequence (5, 9, 14, 15, 20, 38–40), biophysical studies on other pRNA sequences have not been pursued. The potential to form loop-loop interactions occurs in a very different surrounding architecture in the different natural pRNA sequences. The sequences of the multibranch and bulge loops vary, and the lengths of the helices that position the nucleotides to pair in the interlocking loops are very different in different pRNA sequences. This study reports the self-assembly of the M2, SF5, and GA1 pRNAs; the free energies for dimer formation; the effects of swapping interlocking loop sequences; and the differential effects of metal ions and temperature on pRNA multimerization. The self-assembly in the native pRNA sequences shows similar stabilities of dimer formation, although swapping the interlocking loop sequences results in different dimer stabilities. Evidently, sequence variation outside the interlocking loops may compensate for the different stabilities of loop-loop interactions. Thus, the divergent pRNA sequences maintain not only secondary structure and function but also the energetics of quaternary interactions.
EXPERIMENTAL PROCEDURES
RNA Preparation
The sequences of M2 and GA1 pRNAs were cloned from genomic DNA provided by Dr. Shelley Grimes (University of Minnesota). A synthesized DNA sequence (Integrated DNA Technologies) was used as template for SF5 sequence amplification. The PCR product was ligated into the pGEM-T vector (Promega Corp.). A plasmid for the φ29 pRNA sequence was kindly provided by Dr. Nicola Stonehouse (University of Leeds). Loop mutants were prepared by site-specific PCR mutation of plasmid pGEM-T pRNA using a Stratagene QuikChange kit. The primers used in this study are listed in supplemental Tables S1 and S2. The identity of all clones was confirmed by the DNA sequencing facility at the University of California at Davis. The RNAs were produced by T7 in vitro transcription (41) and purified by denaturing PAGE. pRNA was 32P-labeled using T4 polynucleotide kinase, and the purity was >90%. The RNA was dialyzed at least three times using Centricon filters to equilibrate the RNA in defined salt and buffer conditions.
Sedimentation Velocity Analysis
Analytical ultracentrifugation experiments were conducted in the same buffer conditions as used for native gels to determine the molecular weight of gel bands. Sedimentation velocity experiments were run at 4 °C and 35,000 rpm in a Beckman XLA centrifuge with an An-60 TI rotor. The rotor and cells were pre-equilibrated at 4 °C, and the samples were renatured from 90 to 4 °C by snap cooling on ice. The buffer for the sample was 90 mm Tris, 200 mm boric acid, 5 mm NaCl, and 10 mm MgCl2 (pH 7.5). Standard double sector cells were loaded with 500 μl of buffer and 500 μl of the appropriate sample solution. Absorbance scans at a wavelength of 260 nm were acquired at 10-min intervals. Buffer density and viscosity were calculated to be 1.0064 g/ml and 1.0871 poise at 4 °C, respectively. A value of 0.55 ml/g was used for the partial specific volume for the RNAs (14, 15). Data were analyzed with UltraScan-9.9 software and fit using van Holde-Weischet analysis, two-dimensional spectrum analysis, and genetic algorithm analysis (42, 43).
Native Gel Electrophoresis
Detailed renaturing protocols for each gel with specific buffers are included in the figure legends. After at least 30 min of equilibration, the pRNA sample was mixed with 40% (w/v) sucrose loading buffer and run on a 10% polyacrylamide gel buffered at pH 7.5 in 90 mm Tris and 200 mm boric acid with different concentrations of NaCl or MgCl2. The gel and buffer were precooled for 4 °C experiments. Gels were exposed to a PhosphorImager screen and read with a GE Storm scanner. Band intensities were quantified with ImageQuant software. The molecular weights for conformations of φ29, M2, GA1, and SF5 at high and low concentrations were determined by analytical ultracentrifugation to accurately identify monomer and dimer bands. DNA ladders were used only as control markers for consistency in running many native gels.
For measurements of dissociation constants of the native pRNA sequences, the same amount of 32P-labeled pRNA was added to serial dilutions of unlabeled pRNA over an ∼1000-fold concentration range. The concentration of monomer was calculated from the ratios of the band intensities and then plotted as monomer concentration versus fraction multimer (supplemental Fig. S1). The slope of the line indicates the equilibrium constant between monomer and dimer formation, according to the following equations for dimer and trimer formation, respectively: 2 pRNA ↔ pRNA2, [mono] = Kd2([total]/[mono] − 1)/2; and 3 pRNA ↔ pRNA3, [mono]2 = Kd3 ([total]/[mono] − 1)/3, where [mono] is the concentration of pRNA in the monomer state, and [total] is the total pRNA concentration. The reported dissociation constants for the conditions shown in Fig. 3 are the average of at least three measurements with an estimated error of ±15%. The dissociation constants for natural pRNAs in other conditions and mutant pRNA sequences were estimated from the results presented in Fig. 4 and supplemental data.
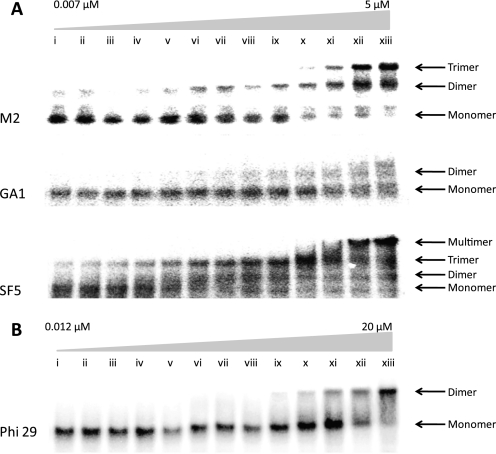
FIGURE 3.
M2, GA1, SF5, and φ29 pRNA self-assembly in serial dilutions. A, the pRNA concentrations in lanes i–xiii are 0.007, 0.009, 0.01, 0.02, 0.03, 0.05, 0.1, 0.2, 0.3, 0.6, 1.3, 2.5, and 5 μm, respectively. The pRNA samples were renatured from 90 to 4 °C by snap cooling on ice in 5 mm NaCl and 10 mm MgCl2 with 90 mm Tris and 200 mm boric acid (pH 7.5). The native polyacrylamide gel was run at 4 °C with the same buffer conditions as the pRNA sample. B, the φ29 pRNA samples were renatured from 90 to 4 °C by snap cooling on ice in 10 mm MgCl2 with 90 mm Tris and 200 mm boric acid (pH 7.5). The pRNA concentrations in lanes i–xiii are 0.012, 0.022, 0.032, 0.051, 0.090, 0.17, 0.33, 0.64, 1.3, 2.5, 5.0, 10, and 20 μm, respectively. The gels were analyzed with ImageQuant software, and background correction values were <10%. Additional bands in the same lanes outside of the region of gel shown were not observed. The positions of monomer and dimer bands were benchmarked using analytical ultracentrifugation experiments. DNA size markers were used only as a reference for consistency in comparing many different gels (supplemental Table S3). Additional gel data at different pRNA concentrations and ionic conditions are included in supplemental Figs. S2–S6.
FIGURE 4.
Native gel analysis of pRNA mutants. The pRNA samples were renatured from 90 to 4 °C by snap cooling on ice in 5 mm NaCl and 10 mm MgCl2 with 90 mm Tris and 200 mm boric acid (pH 7.5), but there was no MgCl2 in the RNA samples or gels in the right panel in B. A, 20 μm mutant 1 pRNA formed only a monomer. B, 0.1 μm mutants 2–5 formed dimers in the presence of MgCl2 but formed only monomers in the absence of MgCl2. C, concentration dependence of dimer formation for mutants 4 and 7. The pRNA concentrations in lanes i–iv are 0.1, 0.3, 1.1, and 5.1 μm, respectively. D, concentration dependence of dimer formation for mutants 8–10. The pRNA concentrations in lanes i–iv are 0.1, 2.6, 7.6, and 20.1 μm, respectively. E, concentration dependence of dimer formation for mutants 11–13. The pRNA concentrations in lanes i–iv are 0.1, 0.3, 1.1, and 5.1 μm, respectively. F, concentration dependence of dimer formation for mutants 14 and 15. The pRNA concentrations for mutant 14 in lanes i–iv are 0.1, 0.2, 0.3, and 0.7 μm, respectively, and those for mutant 15 are 0.1, 0.3, 1.1, and 5.1 μm, respectively. The same amount of labeled 32P was added to all samples, and the amount of monomer and dimer was calculated as a ratio of the measured intensities in monomer and dimer bands.
UV Optical Melting
UV absorbance was measured with a Beckman DU800 spectrometer equipped with a temperature controller. Absorbance versus temperature curves of the pRNA were measured at 260 nm in 100 mm NaCl and 10 mm sodium cacodylate (pH 7.0) containing various concentrations of MgCl2. The heating rate was 1 °C/min. Before melting, the pRNA sample was renatured from 90 to 4 °C by snap cooling on ice.
Circular Dichroism Spectroscopy
The CD spectra of pRNA sequences at 4 °C were measured for 3 μm pRNA using a J-820 spectropolarimeter (JASCO Co., Ltd., Hachioji, Japan) with a 0.1-cm path length quartz cell. The CD spectrum was obtained by taking the average of three scans made at 0.5-nm intervals from 200 to 400 nm. Before measurement, the pRNA sample was renatured from 90 to 4 °C by snap cooling on ice. The temperature of the cell holder was regulated by a PTC-348 temperature controller (JASCO Co., Ltd.), and the cuvette-holding chamber was flushed with a constant stream of dry N2 gas to avoid water condensation on the cuvette.
RESULTS AND DISCUSSION
All Natural pRNA Sequences Form Monomers, Dimers, and Higher Order Multimers
Table 1 shows the molecular weights of pRNA conformations measured at low and high concentrations of pRNA. Analytical ultracentrifugation separates different conformations on the basis of mass, size, and shape. For φ29, M2, GA1, and SF5, the first species had a molecular weight consistent with a monomer pRNA, and the second species had a molecular weight consistent with dimer formation. The percentage of the population in the dimer conformation increased at higher pRNA concentrations. Accurate analysis became more difficult for more than two species and at higher molecular weights; however, the next species appeared to be consistent with trimer formation. The SF5 pRNA sequence showed many higher order species. The pRNA sequence with only U nucleotides in the interlocking loops, mutant 1, formed only one conformation with a molecular weight consistent with monomer formation. The analytical ultracentrifugation experiments identified monomer and dimer conformations of φ29, M2, GA1, and SF5, providing benchmarks for native gel experiments.
TABLE 1.
Molecular weight distribution of φ29, M2, SF5, GA1, and mutant 1 pRNAs in analytical ultracentrifugation
The pRNA samples are renatured from 90 to 4 °C by snap cooling on ice in 5 mm NaCl and 10 mm MgCl2 with 90 mm Tris and 200 mm boric acid (pH 7.5). The reported molecular weights in grams/mol are from the genetic algorithm analysis with an estimated error of 10% from all sources of experimental error. The molecular weights of species 1 are consistent with monomer molecular weights. The molecular weights of species 2 are consistent with a dimer conformation. —, no conformation was observed with this molecular weight.
| pRNA | Concentrationa | Species 1 Mr | Population | Species 2 Mr | Population | Higher order species | Population |
|---|---|---|---|---|---|---|---|
| g/mol × 104 | % | g/mol × 104 | % | g/mol × 104 | % | ||
| φf29 | High | — | — | 8.62 ± 0.86 | 76.3 | 11.28 ± 1.13 | 23.7 |
| φ29 | Low | 3.31 ± 0.33 | 11.6 | 7.34 ± 0.73 | 70.1 | 11.95 ± 1.20 | 18.3 |
| M2 | High | 4.25 ± 0.43 | 12.2 | 8.13 ± 0.81 | 43.9 | 11.21 ± 1.12 | 44.0 |
| M2 | Low | 3.94 ± 0.39 | 22.7 | 8.34 ± 0.83 | 77.3 | — | — |
| GA1 | High | 3.70 ± 0.37 | 82.8 | 7.95 ± 0.80 | 17.2 | — | — |
| GA1 | Low | 4.21 ± 0.42 | 100 | — | — | — | — |
| SF5 | High | 3.54 ± 0.35 | 2.2 | 6.25 ± 0.63 | 24.5 | 13.63 ± 1.36 | 73.2 |
| SF5 | Low | 3.90 ± 0.39 | 28.8 | 6.95 ± 0.70 | 19.2 | 13.72 ± 1.37 | 52.0 |
| Mutant 1b | High | 4.42 ± 0.44 | 100 | — | — | — | — |
| Mutant 1b | Low | 4.09 ± 0.41 | 100 | — | — | — | — |
a The low and high concentrations are 0.3 A260 and 0.8 A260. The extinction coefficients of φ29, M2, GA1, SF5, and mutant 1 pRNAs are 1,168,000, 1,203,400, 1,192,900, 1,178,000, and 1,152,900 (51).
b Mutant 1 is a pRNA sequence with only U nucleotides in the interlocking loops.
Dissociation Constants for Different pRNA Sequences Are Similar Despite Different Interlocking Loop Sequences
Fig. 3 shows the native gel analysis of natural pRNA sequences forming dimers and trimers. At low concentrations, M2 and GA1 pRNA sequences formed only monomers and dimers (Fig. 3 and supplemental Fig. S2). These lanes were thus used to calculate the equilibrium constant for dimer formation. At very low concentrations, SF5 formed a monomer and trimer. Then as the concentration increased, more SF5 monomer formed dimer and trimer and higher order multimers. Thus, a simple dissociation constant for SF5 dimerization could not be calculated. Instead, the low SF5 concentration lanes were used to calculate the equilibrium constant for trimer formation. An estimate of the SF5 dimerization dissociation constant could be calculated with the assumption that dimer formation is not an intermediate in trimer formation.
The φ29 sequence showed a sharper transition from monomer to dimer in the absence of sodium chloride (Fig. 3B), which is the best ionic condition for the calculation of φ29 dimerization dissociation constants. The measured dissociation constant for φ29 dimerization is 6.45 × 10−7 m in the presence of 10 mm MgCl2, which agrees within experimental error with previous measurements made by equilibrium sedimentation (44). M2 and GA1 also formed dimers in 10 mm MgCl2 (supplemental Figs. S3 and S4). Each natural sequence responded slightly differently to changes in ionic conditions, which can complicate direct comparisons of energetics measurements. However, in 10 mm MgCl2 with or without 5 mm NaCl, the dissociation constants are within experimental error for the calculation of free energies.
The dissociation constants and free energies for pRNA self-assembly are summarized in Table 2. All four natural pRNA interlocking loop sequences contain possible Watson-Crick pairing. The predicted free energies for the Watson-Crick pairing between loops were calculated using the INN-HB model and Turner rules (45). If Watson-Crick pairing were the only contribution to the stability of pRNA self-assembly, then a range of 5 orders of magnitude in dissociation constants would be predicted for pRNA self-assembly. In contrast, the stabilities of pRNA self-assembly in M2, GA1, and φ29 sequences are within experimental error. The trimer dissociation constant for SF5 (1.88 × 10−15 m2) is similar to the trimer dissociation constant for φ29 (8.58 × 10−14 m2) previously measured by equilibrium sedimentation (44). Thus, similar stabilities of tertiary and quaternary interactions occur in pRNA assembly of different natural sequences.
TABLE 2.
Dissociation constants and free energy of pRNA multimer formation at 4 °C
| pRNA | Ion condition |
Kd | −ΔG40 | Interacting bp | −ΔG40a | Kdb | |
|---|---|---|---|---|---|---|---|
| [Mg2+] | [Na+] | ||||||
| mm | m | kcal/mol | kcal/mol | m | |||
| φ29 | 10 | 0 | 6.45 × 10−7c | 7.85 | AACC/GGUU | 9.11 | 6.54 × 10−8 |
| AACC/GGUU | 4.33f | 3.85 × 10−4f | |||||
| M2 | 10 | 0 | 9.30 × 10−7 | 7.65 | AUC/GAU | 5.41 | 5.41 × 10−5 |
| 10 | 5 | 1.11 × 10−6c | 7.55 | ||||
| GA1 | 10 | 0 | 1.52 × 10−6 | 7.37 | CA/UG | 2.98 | 4.47 × 10−3 |
| 10 | 5 | 2.73 × 10−6c | 7.06 | ||||
| SF5 | 10 | 5 | 5.67 × 10−7d | 7.92d | AAA/UUU | 3.11 | 3.53 × 10−3 |
| 1.88 × 10−1c,e | 18.67 e | ||||||
a Free energy of Watson-Crick base pairing was calculated with nearest neighbor parameters (45).
b The predicted dissociation constant of dimer formation was calculated from the predicted ΔG40 for the Watson-Crick interacting base pairs.
c The reported dissociation constants are the average of at least three measurements with an estimated error of ±15%.
d The Kd for dimer formation was calculated assuming that dimer and trimer formation in SF5 pRNA follows two independent pathways.
e The value is the Kd for trimer formation.
f The free energy for the φ29 base pairing considers the two GC base pairs, consistent with the NMR data of this interaction (21).
Any Combination of Consecutive Watson-Crick or GU Pairs Facilitates pRNA Dimer Formation
To test the potential of the interlocking loops to direct pRNA self-assembly, several mutations swapping the interlocking loop sequences were studied (Fig. 4 and Table 3). Surprisingly, all interlocking loop sequences with at least two consecutive Watson-Crick or GU pairs formed dimers. In the absence of magnesium, no dimer was formed, which is consistent with previous studies of the magnesium dependence of φ29 dimerization (38, 39). The overall architecture in pRNA may be conducive to forming dimers and place the loops in the correct position to interact, and then the pairing interactions between the loops may simply stabilize the dimer.
TABLE 3.
pRNA CE and D loop mutants
| Mutant | Surrounding sequence | Loop |
Loop sequence |
Estimated Kda | Estimated −ΔG40b | ||
|---|---|---|---|---|---|---|---|
| CE bulge loop | D hairpin loop | CE bulge loop | D hairpin loop | ||||
| m | kcal/mol | ||||||
| 1 | φ29 | GUUUUUUU | UUUUU | c | |||
| 2 | φ29 | SF5 | φ29 | GUUGUUUU | UGGUU | 1.10 × 10−6 | 7.56 |
| 3 | φ29 | φ29 | SF5 | GAUUAAACC | AAA | 6.50 × 10−7 | 7.85 |
| 4 | φ29 | SF5 | SF5 | GUUGUUUU | AAA | 2.76 × 10−5 | 5.78 |
| 5 | φ29 | M2 | φ29 | GUAUAUC | UGGUU | 6.21 × 10−7 | 7.87 |
| 6 | φ29 | φ29 | M2 | GAUUAAACC | GAU | 6.85 × 10−7 | 7.82 |
| 7 | φ29 | M2 | M2 | GUAUAUC | GAU | 1.26 × 10−5 | 6.21 |
| 8 | φ29 | GA1 | φ29 | GCAUCC | UGGUU | d | |
| 9 | φ29 | φ29 | GA1 | GAUUAAACC | GUG | 2.10 × 10−4 | 4.66 |
| 10 | φ29 | GA1 | GA1 | GCAUCC | GUG | 1.97 × 10−4 | 4.70 |
| 11 | SF5 | φ29 | φ29 | GAUUAAACC | UGGUU | d | |
| 12 | M2 | φ29 | φ29 | GAUUAAACC | UGGUU | 9.73 × 10−6 | 6.36 |
| 13 | GA1 | φ29 | φ29 | GAUUAAACC | UGGUU | 3.12 × 10−5 | 5.71 |
| 14e | φ29 | φ29 | GAUUAGCGA | UGGUU | 9.73 × 10−5 | 5.09 | |
| 15e | φ29 | φ29 | GAUUAAACC | UUCGC | 3.18 × 10−5 | 5.70 | |
a Dissociation constants were estimated at the concentrations of mutant pRNA in gels shown in Fig. 4 in the ionic condition of 10 mm Mg2+ and 5 mm Na+.
b The estimated free energy was calculated with the dissociation constants.
c No dimer was formed.
d The pRNA formed intermediates between monomer and dimer.
e The four consecutive bases (AACC) in CE loop for intermolecular interaction were mutated to GCGA, and those in D loop (GGUU) were mutated to UCGC. Previous experiments studying φ29 pRNA dimerization with UV absorbance did not detect any dimerization with these mutants (7). More sensitive 32P labeling detected dimerization of these mutants in the conditions reported under “Experimental Procedures” and for Fig. 4F.
All of the mutants except mutant 1 could form Watson-Crick or GU pairs. For example, mutant 3 has 5′-UU-3′ and 5′-AA-3′ in the CE and D loops, respectively; and mutant 6 has 5′-AU-3′ and 3′-UA-5′ in the CE and D loops, respectively. Mutant 2 could form only GU pairs between the loops. However, consecutive terminal GU pairs can stabilize RNA helices, and helices with only GU pairs can form (46). Mutant 5 could form a mixture of Watson-Crick and GU pairs, 5′-AUC-3′ and 3′-UGG-5′ in the CE and D loops, respectively. Some mutated pRNA sequences had two lower bands, suggesting that the monomer may fold into more than one conformation. However, at least one of these conformations was able to form dimers. Secondary structure predictions from RNAstructure 4.6 (39) for all of the φ29 mutants suggested that all of the sequences can fold into similar monomer secondary structures as wild-type φ29. The combinations of mutants 2 and 3 and mutants 5 and 6 have the potential to form heterodimers, although the heterodimer and homodimers could not be distinguished on a gel (supplemental Fig. S5).
The only sequence that did not form dimers under any conditions is mutant 1, with only U nucleotides in the loop. The molecular weight of mutant 1 was confirmed by analytical ultracentrifugation (Table 1). The inability of mutant 1 to form a dimer even at 20 μm suggests that the renaturing process does not cause the monomers to completely unfold all helices and then refold as one long duplex with internal loops but rather supports the interpretation that dimers are forming between two folded monomers. The wide variation in dissociation constants in the mutants is also inconsistent with the possibility of forming one long duplex. The 3 orders of magnitude range of dissociation constants for the mutants is much greater than would be expected for internal loop sequence variation in a long duplex (47, 48). Chemical modification experiments show many changes upon dimerization but do not conclusively prove monomer or dimer formation (49). The snap cooling procedure and the formation of trimers and higher order multimers also support the interpretation of self-assembly among folded monomers.
Mutants 14 and 15 form a small amount of dimer with increasing concentration (Fig. 4F). Previous studies of these pRNA mutants did not detect dimer formation using less sensitive UV absorption detection (5, 7, 9). The increased sensitivity of 32P-labeled RNA enables detection of dimer formation at 2 orders of magnitude lower concentration than the wild-type sequence. These mutants have the potential to form AU and GU pairs in the interlocking loops. The formation of base pairs in the interlocking loops in mutants 14 and 15 is consistent with the low levels of packaging activity retained by these mutants (5).
Sequence Variation outside the Interlocking Loops Contributes to the Stability of pRNA Self-assembly
The predicted free energies of Watson-Crick pairing between the interlocking loops does not fully account for the stabilities of pRNA self-assembly in natural sequences (Table 2). The assembly of the mutant pRNA sequences suggests that the pairing in the loop-loop interaction stabilizes the intrinsic propensity of the pRNA molecule's overall shape and architecture to dimerize. Within the context of the φ29 pRNA architecture or surrounding sequence, the CE and D loop sequences for SF5, M2, and GA1 were studied as single and double mutants. For example, mutants 2–4 contain the SF5 sequence in the CE loop, D loop, or both loops, respectively, in the φ29 pRNA architecture (Fig. 4 and Table 3). Similarly, mutants 5–7 and mutants 8–10 contain the M2 and GA1 loop sequences in the context of the φ29 pRNA architecture. In addition, the φ29 CE and D loop sequences were studied in the context of the SF5, M2, and GA1 surrounding sequences (mutants 11–13).
For the SF5 loop sequences in the φ29 surrounding sequences, changing either the CE or D loop (mutant 2 or 3) did not significantly decrease the stability of dimerization compared with wild-type φ29. Changing both the CE and D loops to the SF5 sequences (mutant 4) decreased the dimerization stability by 2 orders of magnitude, however. Similarly, the single M2 loop mutation 5 and 6 had little effect, whereas the double M2 loop mutation 7 decreased dimerization stability by over an order of magnitude. In the case of the GA1 loop sequence mutants in the φ29 architecture (mutants 8–10), the measureable single mutant 9 and the double mutant 10 decreased the stability of dimerization by 2 orders of magnitude relative to wild-type φ29. Thus, changing both the CE and D loops together does not recover stability by providing the matching interlocking loop sequence but rather further decreases stability in a context-dependent manner.
All of the double CE-D loop mutants (mutants 4, 7, and 10) were not only less stable dimers than wild-type φ29 but also less stable than any of the natural wild-type pRNA sequences (Tables 2 and 3). For example, mutant 10 with the GA1 interlocking loop sequences in the context of the φ29 architecture has a Kd of 1.97 × 10−4 m; the dimerization dissociation constant for mutant 10 is 2 orders of magnitude less favorable than the natural GA1 pRNA sequence (Kd = 2.73 × 10−6 m). Thus, additional tertiary and quaternary interactions outside of the loop-loop interaction are likely responsible for the 2 orders of magnitude difference in dimerization between these double mutants and the natural pRNA sequences.
The contributions to dimer stability of the loop sequences and the surrounding sequences are interdependent and not simply additive. For example, the GA1 surrounding architecture may provide very stabilizing interactions for the GA1 CE and D interlocking loops because φ29 and GA1 pRNAs have similar stabilities in dimer formation, but mutant 10 dimerization (GA1 loop sequences and the φ29 surrounding sequence) is much less favorable. One prediction from these observations could be that the combination of φ29 loop sequences in the GA1 surrounding architecture (mutant 13) would be exceptionally stable. However, both mutant 10 (GA1 loop sequences in the φ29 surrounding sequence) and mutant 13 (φ29 loop sequences in GA1 surrounding sequences) formed less stable dimers than either natural φ29 or GA1 pRNA. Thus, the contributions of the loop sequences and the surrounding architecture are synergistic in pRNA dimer formation.
When the φ29 CE and D loop sequences were inserted into the surrounding sequences of SF5, M2, and GA1 (mutants 11–13), the stability of dimer formation was reduced by 1–2 orders of magnitude in a dissociation constant. Thus, the decrease in dimer formation stability when mutating the loop sequences is not unique to the φ29 architecture. Interestingly, mutant 4 with the SF5 CE and D loops in the context of the φ29 architecture and mutant 11 with the φ29 CE and D loops in the SF5 architecture did not show any propensity for trimer formation or high order multimers.
Thus, the sequence of the interlocking loops and the surrounding sequence together create stable dimers. The surrounding sequences may correctly position the nucleotides to form Watson-Crick or GU pairs. The surrounding sequences may have complementary shapes that facilitate dimer formation but still require stable pairing in the interlocking loops. Forming pairs in the interlocking loops may change the conformation and stability of the surrounding sequence, for example, by stabilizing a particular coaxial stacking arrangement in the multibranch loop that is dynamic in the monomer but more rigid in the dimer. The bulged nucleotides in the surrounding architecture may then further stabilize dimer formation but only when the right combination of pairing interactions and conformational changes occurs during dimerization.
All Natural pRNA Sequences Require Magnesium for Self-assembly
Magnesium often stabilizes RNA tertiary interactions (50) and also stabilizes the quaternary interactions in pRNA assembly. For example, reducing the magnesium concentration increases the φ29 pRNA concentration necessary for trimer formation. In the presence of 2 mm MgCl2 and 5 mm NaCl, the φ29 pRNA did not form a trimer until 10.1 μm pRNA, whereas trimer formation occurred at 0.64 μm in 10 mm MgCl2 and 5 mm NaCl (supplemental Fig. S6). None of the natural pRNA sequences formed dimers without magnesium in up to 100 mm NaCl, which suggests that higher monovalent ionic conditions do not compensate for magnesium.
Fig. 5 shows the effects of adding magnesium on the thermal stability of φ29 and GA1 pRNAs. The thermal unfolding of pRNA is highly cooperative as indicated by the sharp transition in UV absorbance. The temperature at which this transition occurred did not change with increasing concentration of RNA from 3 to 10 μm, which is consistent with unimolecular unfolding. No transition was observed below 20 °C. In φ29 and GA1 sequences, both the monomer and dimer formed at 4 °C in the optical melting buffer conditions of 100 mm NaCl and 10 mm MgCl2 (supplemental Fig. S7). If there is any hyperchromicity associated with the loss of intermolecular base pairs, then this transition occurs together with the unfolding of the pRNA molecule. In the absence of magnesium, the φ29 and GA1 pRNAs showed similar melting temperatures. The addition of 5 mm MgCl2 significantly stabilized φ29 and GA1 pRNAs and increased the melting temperatures from 49.6 to 63.5 °C and from 50.2 to 63.9 °C, respectively. Further increases in MgCl2 to 10 mm provided no additional stability (supplemental Fig. S7), which indicates a specific ion effect rather than a general increase in stability from increased salt concentration.
FIGURE 5.
Optical melting curves of φ29 (A) and GA1 (B) pRNAs at 3 μm RNA and 100 mm NaCl (yellow); 10 μm RNA and 100 mm NaCl (pink); 3 μm RNA, 100 mm NaCl, and 5 mm MgCl2 (blue); and 10 μm RNA, 100 mm NaCl, and 5 mm MgCl2 (purple). Abs, absorbance.
The CD spectra are consistent with the addition of magnesium stabilizing a transition from one folded conformation to another folded conformation rather than inducing a transition from unfolded to folded conformations. The CD spectra of φ29 and GA1 pRNAs in different ionic conditions showed a maximum peak near 260 nm, a minimum peak near 210 nm, and a small negative peak near 290 nm (supplemental Fig. S8), which are characteristic of A-form RNA helices. The addition of 10 mm MgCl2 reduced slightly the maximum peak at 260 nm, which also suggests a specific ion effect.
Sodium Ion Concentrations and Temperature Affect pRNA Assembly Differently for Different Natural Sequences
Sodium ions and renaturing temperature synergistically affected pRNA self-assembly. For example, at room temperature in the presence of 10 mm MgCl2 (supplemental Fig. S9, lane 3 of each pRNA), φ29 pRNA formed monomers and dimers; M2 pRNA formed dimers and a multimer; GA1 pRNA formed almost only monomer; and SF5 pRNA formed many higher order multimers. SF5 pRNA showed extraordinary competence of multimerization at room temperature but no self-assembly with snap cooling to 4 °C in 10 mm MgCl2 and 100 mm NaCl (supplemental Fig. S10). In contrast, SF5 formed a monomer and trimer with snap cooling to 4 °C in lower sodium chloride concentrations, i.e. 10 mm MgCl2 and 5 mm NaCl (Fig. 3). The complex sequence-dependent effects of sodium ion concentration and renaturation temperature may result from the differences in the architectures of different pRNA natural sequences. The different multibranch loops and bulge loops in different pRNA sequences may have different interactions with metal ions or different probabilities for orientations of the helices that could affect pRNA self-assembly.
In conclusion, the four natural pRNA sequences contain very few conserved nucleotides (Fig. 1). The single nucleotide bulges vary in sequence and position in the helix, and the multibranch loops between the A, C, and D helices also vary in size and sequence. Thus, each pRNA sequence offers different opportunities for additional stabilizing tertiary and quaternary interactions. This sequence variation may explain the difference in stabilities between the natural pRNA sequences and the double mutants with the interlocking loop sequences transplanted into different surrounding sequences, differing propensities for multimer formation, and different magnesium ion dependences in different pRNA sequences. Interestingly, the free energy of pRNA dimerization is approximately the same for all pRNA sequences despite different predicted stabilities for the Watson-Crick base pairing interactions in the interlocking loops. Thus, perhaps the pRNA sequences evolved to maintain a minimum stability in multimerization, and as the CE and D loop sequences evolved, the single nucleotide bulges and multibranch loop sequences also evolved to make energetically compensating changes. Thus, pRNA self-assembly may be stabilized by many noncovalent tertiary and quaternary interactions and magnesium ion interactions in addition to the base pairing in the interlocking loops.
Acknowledgments
We thank Dr. Shelley Grimes for the M2 and GA1 genomic DNAs, Dr. Nicola Stonehouse for the plasmid containing the φ29 sequence, Dr. Marc Dreyfus (CNRS) for the T7 polymerase mutant, Dr. Haiqing Yu (University of Arizona) for CD data collection, and Becky Myers for T7 preparation in the Schroeder laboratory.
This work was supported in part by Grant HR09-160 from the Oklahoma Center for the Advancement of Science and Technology and by Institutional Research Grant 090039 from the American Chemical Society (to the University of Oklahoma Cancer Center).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S10 and Tables S1–S3.
- pRNA
- prohead RNA.
REFERENCES
- 1. Guo P. X., Erickson S., Anderson D. (1987) Science 236, 690–694 [DOI] [PubMed] [Google Scholar]
- 2. Grimes S., Anderson D. L. (1990) J. Mol. Biol. 251, 559–566 [DOI] [PubMed] [Google Scholar]
- 3. Bailey S., Wichitwechkarn J., Johnson D., Reilly B. E., Anderson D. L., Bodley J. W. (1990) J. Biol. Chem. 265, 22365–22370 [PubMed] [Google Scholar]
- 4. Zhao W., Morais M. C., Anderson D. L., Jardine P. J., Grimes S. (2008) J. Mol. Biol. 383, 520–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reid R., Zhang F., Benson S., Anderson D. (1994) J. Biol. Chem. 269, 18656–18661 [PubMed] [Google Scholar]
- 6. Reid R. J., Bodley J. W., Anderson D. (1994) J. Biol. Chem. 269, 9084–9089 [PubMed] [Google Scholar]
- 7. Reid R. J., Bodley J. W., Anderson D. (1994) J. Biol. Chem. 269, 5157–5162 [PubMed] [Google Scholar]
- 8. Zhang C., Tellinghuisen T., Guo P. (1997) RNA 3, 315–323 [PMC free article] [PubMed] [Google Scholar]
- 9. Chen C., Zhang C., Guo P. (1999) RNA 5, 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garver K., Guo P. (1997) RNA 3, 1068–1079 [PMC free article] [PubMed] [Google Scholar]
- 11. Yu J., Moffitt J., Hetherington C. L., Bustamante C., Oster G. (2010) J. Mol. Biol. 400, 186–203 [DOI] [PubMed] [Google Scholar]
- 12. Morais M. C., Koti J. S., Bowman V. D., Reyes-Aldrete E., Anderson D. L., Rossmann M. G. (2008) Structure 16, 1267–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiao F., Zhang H., Guo P. (2008) Nucleic Acids Res. 36, 6620–6632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo P., Zhang C., Chen C., Garver K., Trottier M. (1998) Mol. Cell 2, 149–155 [DOI] [PubMed] [Google Scholar]
- 15. Zhang F., Lemieux S., Wu X., St-Arnaud D., McMurray C. T., Major F., Anderson D. (1998) Mol. Cell 2, 141–147 [DOI] [PubMed] [Google Scholar]
- 16. Moffitt J. R., Chemla Y. R., Aathavan K., Grimes S., Jardine P. J., Anderson D. L., Bustamante C. (2009) Nature 457, 446–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith D. E., Tans S. J., Smith S. B., Grimes S., Anderson D. L., Bustamante C. (2001) Nature 413, 748–752 [DOI] [PubMed] [Google Scholar]
- 18. Chemla Y. R., Aathavan K., Michaelis J., Grimes S., Jardine P. J., Anderson D. L., Bustamante C. (2005) Cell 122, 683–692 [DOI] [PubMed] [Google Scholar]
- 19. Rickgauer J. P., Fuller D. N., Grimes S., Jardine P. J., Anderson D. L., Smith D. E. (2008) Biophys. J. 94, 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fang Y., Cai Q., Qin P. Z. (2005) Biochemistry 44, 9348–9358 [DOI] [PubMed] [Google Scholar]
- 21. Kitamura A., Jardine P. J., Anderson D. L., Grimes S., Matsuo H. (2008) Nucleic Acids Res. 36, 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee T. J., Schwartz C., Guo P. (2010) Ann. Biomed. Eng. 37, 2064–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo P. (2005) J. Nanosci. Nanotechnol. 5, 1964–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo S., Tschammer N., Mohammed S., Guo P. (2005) Human Gene Ther. 16, 1097–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shu D., Huang L. P., Hoeprich S., Guo P. (2003) J. Nanosci. Nanotechnol. 3, 295–302 [DOI] [PubMed] [Google Scholar]
- 26. Guo S., Huang F., Guo P. (2006) Gene Ther. 13, 814–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ko S. H., Chen Y., Shu D., Guo P., Mao C. (2008) J. Am. Chem. Soc. 130, 17684–17687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu H., Guo S., Roll R., Li J., Diao Z., Shao N., Riley M. R., Cole A. M., Robinson J. P., Snead N. M., Shen G., Guo P. (2007) Cancer Biol. Ther. 6, 697–704 [DOI] [PubMed] [Google Scholar]
- 29. Zhang H. M., Su Y., Guo S., Yuan J., Lim T., Liu J., Guo P., Yang D. (2009) Antiviral Res. 83, 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Severcan I., Geary C., Verzemnieks E., Chworos A., Jaeger L. (2009) Nano Lett. 9, 1270–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chworos A., Severcan I., Koyfman A. Y., Weinkam P., Oroudjev E., Hansma H. G., Jaeger L. (2004) Science 306, 2068–2072 [DOI] [PubMed] [Google Scholar]
- 32. Afonin K. A., Bindewald E., Yaghoubian A. J., Voss N., Jacovetty E., Shapiro B. A., Jaeger L. (2010) Nat. Nanotechnol. 5, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Severcan I., Geary C., Chworos A., Voss N., Jacovetty E., Jaeger L. (2010) Nat. Chem. 2, 772–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grabow W. W., Zakrevsky P., Afonin K. A., Chworos A., Shapiro B. A., Jaeger L. (2011) Nano Lett. 11, 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nasalean L., Baudrey S., Leontis N. B., Jaeger L. (2006) Nucleic Acids Res. 34, 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Afonin K. A., Cieply D. J., Leontis N. B. (2008) J. Am. Chem. Soc. 130, 93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koyfman A. Y., Braun G., Magonov S., Chworos A., Reich N. O., Jaeger L. (2005) J. Am. Chem. Soc. 127, 11886–11887 [DOI] [PubMed] [Google Scholar]
- 38. Chen C., Guo P. (1997) J. Virol. 71, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fuller D. N., Rickgauer J. P., Jardine P. J., Grimes S., Anderson D. L., Smith D. E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11245–11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Atz R., Ma S., Gao J., Anderson D. L., Grimes S. (2007) J. Mol. Biol. 369, 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guillerez J., Lopez P. J., Proux F., Launay H., Dreyfus M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5958–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Demeler B., van Holde K. E. (2004) Anal. Biochem. 335, 279–288 [DOI] [PubMed] [Google Scholar]
- 43. Brookes E., Demeler B. (2006) Prog. Colloid Polymer Sci. 131, 32–38 [Google Scholar]
- 44. Robinson M. A., Wood J. P., Capaldi S. A., Baron A. J., Gell C., Smith D. A., Stonehouse N. J. (2006) Nucleic Acids Res. 34, 2698–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xia T., SantaLucia J., Jr., Burkard M. E., Kierzek R., Schroeder S. J., Jiao X., Cox C., Turner D. H. (1998) Biochemistry 37, 14719–14735 [DOI] [PubMed] [Google Scholar]
- 46. Nguyen M. T., Schroeder S. J. (2010) Biochemistry 49, 10574–10581 [DOI] [PubMed] [Google Scholar]
- 47. Mathews D. H., Disney M. D., Childs J. L., Schroeder S. J., Zuker M., Turner D. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7287–7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Turner D. H., Mathews D. H. (2010) Nucleic Acids Res. 38, D280–D282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trottier M., Mat-Arip Y., Zhang C., Chen C., Sheng S., Shao Z., Guo P. (2000) RNA 6, 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cate J. H., Gooding A. R., Podell E., Zhou K., Golden B. L., Kundrot C. E., Cech T. R., Doudna J. A. (1996) Science 273, 1678–1685 [DOI] [PubMed] [Google Scholar]
- 51. Borer P. N. (1975) in Handbook of Biochemistry and Molecular Biology: Nucleic Acids (Fasman C. D. ed) Vol. 1, 3rd Ed., p. 589, CRC Press, Cleveland, OH [Google Scholar]