Abstract
Human α1-acid glycoprotein (hAGP) in serum functions as a carrier of basic drugs. In most individuals, hAGP exists as a mixture of two genetic variants, the F1*S and A variants, which bind drugs with different selectivities. We prepared a mutant of the A variant, C149R, and showed that its drug-binding properties were indistinguishable from those of the wild type. In this study, we determined the crystal structures of this mutant hAGP alone and complexed with disopyramide (DSP), amitriptyline (AMT), and the nonspecific drug chlorpromazine (CPZ). The crystal structures revealed that the drug-binding pocket on the A variant is located within an eight-stranded β-barrel, similar to that found in the F1*S variant and other lipocalin family proteins. However, the binding region of the A variant is narrower than that of the F1*S variant. In the crystal structures of complexes with DSP and AMT, the two aromatic rings of each drug interact with Phe-49 and Phe-112 at the bottom of the binding pocket. Although the structure of CPZ is similar to those of DSP and AMT, its fused aromatic ring system, which is extended in length by the addition of a chlorine atom, appears to dictate an alternative mode of binding, which explains its nonselective binding to the F1*S and A variant hAGPs. Modeling experiments based on the co-crystal structures suggest that, in complexes of DSP, AMT, or CPZ with the F1*S variant, Phe-114 sterically hinders interactions with DSP and AMT, but not CPZ.
Keywords: Drug Transport, Protein-Drug Interactions, Protein Structure, Xenobiotics, X-ray Crystallography, Crystal Structure, Drug-binding Selectivity, Orosomucoid, Variant
Introduction
Human α1-acid glycoprotein (hAGP),2 a serum glycoprotein composed of 183 amino acid residues, contains five N-linked glycans that account for about half of the total 40-kDa mass of the protein (1, 2). hAGP is an acute-phase protein; its blood concentration is therefore significantly increased in inflammatory states (e.g. tumor growth) (3). The protein is a member of the lipocalin family of proteins and is known to be a carrier of hydrophobic ligands (4). hAGP also binds to a variety of drugs that are predominantly apolar but with some basic characteristics (4) and thereby regulates their tissue distribution. For example, the anticancer drug UCN-01 has a long half-life and a small distribution volume in patients because of its high binding affinity for hAGP (5). Moreover, the serum concentration of imatinib, a tyrosine kinase inhibitor used in the treatment of leukemia, was reported to be positively correlated with that of hAGP (6). Treatment regimes in cancer patients can be complicated by the fact that hAGP levels are increased as the result of their inflammatory state, thereby increasing the impact of the protein on the pharmacokinetics of drugs that bind to it (5, 6). Consistent with this, transgenic mice expressing excess rat AGP exhibited a lower clearance and a smaller distribution volume (compared with control mice) for the HIV protease inhibitor saquinavir, which binds to the protein (7).
In most individuals, hAGP exits as a mixture of F1*S and A variants. The molar ratio of the F1*S and A variants in blood typically ranges from 3:1 to 2:1 (8, 9). However, Vékey and co-workers (10) reported that, in plasma of some cancer patients, AGP levels were increased by 4-fold and that the molar ratio of the F1*S and A variants was in the vicinity of 8:1. Therefore, the pharmacokinetics and pharmacodynamics of drugs that bind to hAGP are affected during inflammation not only as the result of increased hAGP concentrations but also by the change in the ratio of the hAGP variants.
Hervé et al. (11) reported that F1*S and A variants showed different drug-binding selectivities. For example, the F1*S variant of hAGP had a higher selectivity for dipyridamole (17-fold) than the A variant; conversely, compared with F1*S, the A variant bound propafenone, amitriptyline, desipramine, imipramine, and nortriptyline with 15-, 46-, 12-, 12-, and 27-fold higher selectivities, respectively. These A variant-specific drugs have similar structures, probably reflecting unique structural features of the drug-binding site on this variant of hAGP. However, the mechanism underlying the drug selectivity of the A variant has yet to be established.
Attempts to map the amino acids involved in drug binding of hAGP have been made using a range of different experimental approaches, including fluorescent probe displacement (12), chemical modification (13, 14), photoaffinity labeling (15, 16), and other spectroscopic methods (17). However, most of these investigations used mixtures of hAGP variants, resulting in some contradictory results. A significant step toward clarifying the structural basis of the drug-binding specificity of hAGP was made recently when Skerra and co-workers (18) reported the first high-resolution x-ray structural analysis of hAGP. They determined the crystal structure of the F1*S variant, produced by overexpression in Escherichia coli, and showed that F1*S hAGP has a drug-binding pocket located in the center of an eight-stranded β-barrel structure, which is characteristic of the lipocalin family of proteins. The binding pocket consists of three distinct “lobes,” I–III. Lobe I is the largest and deepest cavity in the protein and appears to serve as the main hydrophobic drug-binding chamber. Lobes II and III project from the central lobe I and are smaller and negatively charged. Although Skerra and co-workers were unable to obtain co-crystals of hAGP-drug complexes, they used computational docking protocols to model the likely interactions of diazepam and progesterone for their binding to F1*S hAGP. Their model predicted that diazepam binds to lobe I, whereas progesterone binds to lobe II, and they proposed that the unique geometry of the binding pocket of the F1*S variant explains the diverse ligand spectrum of F1*S. This result is in good agreement with our findings indicating that AGP has a wide binding site consisting of three different subsites (19).
Despite these advances, obtaining more direct structural information on the drug-binding properties of hAGP continues to be an important issue. To this end, we previously constructed expression systems in E. coli that allow both F1*S and A variants to be produced (20). We found that the introduction of a C149R mutation into the A variant of hAGP, in which a surface-exposed Cys residue was replaced by an Arg residue (as found in F1*S), yields a much more homogeneous protein sample without any detectable alterations in the drug-binding specificity of the protein. We report here on the crystal structure of this C149R mutant of the A variant (hereafter referred to as the A variant). In addition, we also determined the crystal structures of complexes of this A variant hAGP with disopyramide (DSP) and amitriptyline (AMT), which bind to the A variant with a high degree of selectivity, and chlorpromazine (CPZ), which binds with less selectivity. Comparison of our results with the structure of the F1*S variant provides important new insights into the mechanism of the drug-binding selectivity of hAGP variants.
EXPERIMENTAL PROCEDURES
Materials
The pT-Trx vector was a gift from Dr. Shunsuke Ishii (Laboratory of Molecular Genetics, RIKEN Tsukuba Institute, Tsukuba, Japan). The pET Expression System 3 and Origami B(DE3) were purchased from Novagen. Restriction enzymes, the DNA ligation kit, and the DNA polymerase Premix Taq® (Ex Taq version) were obtained from Takara Biotechnology (Kyoto, Japan). The DNA sequencing kit was obtained from PerkinElmer Life Sciences. Isopropyl β-d-thiogalactopyranoside was purchased from Promega (Madison, WI). AMT, DSP, and CPZ were purchased from Sigma. HiTrap chelating HP and Q-XL columns and Sephadex G-100 were purchased from Amersham Biosciences. All other chemicals and solvents were of analytical grade.
Construction of Recombinant F1*S and A Variant Expression Vectors
For expression of the recombinant F1*S and A variants, the pET-3c vector, which is frequently used as an expression system in E. coli, was utilized (see Fig. 1). The cDNA of the F1*S variant was a gift from Kyowa Hakko Co., Ltd. (Tokyo, Japan). The cDNA of the A variant was prepared according to our previous method (19). A DNA fragment encoding the F1*S or A variant containing a His6 tag was amplified by PCR using each template and the following oligonucleotide primers: 5′-GGTCATATGCAGATCCCATTGTGTGCCAACCTAGTACCGG-3 (5′-NdeI) and 5′-GGTGGATCCTCACTAGTGGTGGTGGTGGTGGTGGGATTCCCCCTCCTCCTGTTTCC-3 (3′-BamHI) and inserted into the restriction enzyme sites NdeI and BamHI in a multicloning site on this vector.
FIGURE 1.
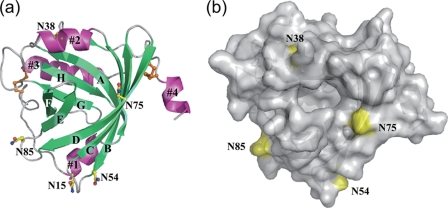
a, crystal structure of the A variant (C149R) of hAGP at a resolution of 2.10 Å shown as a ribbon diagram. Secondary structures are colored lime green (β-strands) and light magenta (α-helices). Cys residues are depicted as orange sticks and balls. Asn residues of five N-linked glycosylation sites are depicted as yellow sticks and balls. Nitrogen and oxygen are colored blue and red, respectively. b, top view into the β-barrel with surface representation. Asn residues corresponding to glycosylation sites are colored yellow.
Introduction of Mutations into the A Variant Coding Region
Site-directed mutagenesis was performed to replace cysteine with arginine at position 149 in A variant hAGP using the QuikChange XL site-directed mutagenesis Kit (Qiagen). Complementary mutagenic primers were synthesized according to the forward sequence 5′-CTCGACTGCTTGCGCATTCCCAGGTCA-3′ (the underlined letter indicates a mismatch). The mutation in the mutated gene was confirmed by DNA sequencing of the entire A variant coding region using the dideoxy chain termination method with a PerkinElmer ABI Prism 310 genetic analyzer.
Expression of Recombinant F1*S and A Variants and C149R in E. coli (Origami B(DE3) Strains)
pET-3c vectors containing the inserted cDNA for the A variant (pET-3c/A) were transformed in the Origami B(DE3) strain that had been pretransformed with pT-Trx. After culturing this transformant in LB medium containing 2% glucose, 50 μg/ml ampicillin, and 25 μg/ml chloramphenicol at 30 °C for 18 h, a 20-ml aliquot of the medium was cultured for 18 h in 2× yeast extract/tryptone medium (1 liter) containing 2% glucose and 50 μg/ml ampicillin. After reaching an appropriate A value at 600 nm (A600 = 0.5), recombinant AGP expression was induced for 3 h by the addition of 0.1 mm isopropyl β-d-thiogalactopyranoside. After centrifugation (3000 × g, 15 min, 4 °C), the pellet was suspended in 20 mm Tris-HCl (pH 8.0), 50 mm NaCl, 20 mm imidazole, and 0.1% CHAPS and sonicated. After centrifugation (15,000 × g, 20 min, 4 °C), the soluble and insoluble proteins were recovered in the supernatant and pellet, respectively. The supernatant was loaded on a HiTrap chelating HP column. The target protein was eluted with 20 mm Tris-HCl (pH 8.0), 50 mm NaCl, 100 mm imidazole, and 0.1% CHAPS. The eluate was then loaded on a HiTrap Q-XL column. The protein was eluted with 20 mm Tris-HCl (pH 8.0) and 0.1% CHAPS using a linear gradient of 0–1.0 m NaCl. Finally, the eluate was loaded onto a Sephadex G-100 column and eluted with 20 mm Tris-HCl (pH 7.4) containing 50 mm NaCl.
Crystallization of C149R and Complexes with DSP, AMT, and CPZ
In a previous study (20), we reported that the recombinant wild-type A variant expressed from E. coli underwent oligomerization due to the formation of intermolecular disulfide bonds but that a mutation of Cys-149 to Arg yielded a monomeric protein that could be purified to homogeneity and had a drug-binding capacity comparable with that of the A variant isolated from serum. Therefore, crystallization trials of the recombinant A variant were carried out using the mutant (C149R) (see Table 1). Crystallization was performed by hanging-drop vapor diffusion at 20 °C. Crystals of the A variant were grown from a droplet (3 μl) consisting of equal volumes of the protein solution (18 mg/ml) and the reservoir solution containing 0.2 m ammonium acetate, 0.1 m sodium acetate (pH 4.6), and 26% (w/v) polyethylene glycol 4000. For co-crystallization experiments with drugs, the protein solutions were incubated with 5 mm AMT, 5 mm DSP, and 5 mm CPZ at 20 °C for >12 h, respectively. All complex crystals (A variant-AMT complex, A variant-DSP, and A variant-CPZ complex) were obtained under conditions similar to those for the drug-free A variant. (The concentration of polyethylene glycol 4000 was 21–25%.) The crystals were transferred into a cryoprotectant solution composed of reservoir solution containing 10% glycerol and flash-frozen in a stream of nitrogen at 100 K. Diffraction data were collected on beamline BL44XU at SPring-8 (Harima, Japan) and on beamlines NW12A, BL-5A, and BL-17A at the Photon Factory (Tsukuba, Japan). The data sets for A variant, A variant-AMT complex, and A variant-CPZ complex were indexed, integrated, and scaled with HKL2000 (21). The data set for A variant-DSP complex was indexed, integrated, and scaled with iMOSFLM (22) and SCALA (23) in the CCP4 program suite. Crystals of the A variant, A variant-AMT complex, and A variant-DSP complex belong to space group C2 with the unit cell dimensions listed in Table 1. The crystal of A variant-CPZ complex belongs to space group P212121.
TABLE 1.
Data collection and refinement statistics
Values in parentheses correspond to the highest resolution shell. PF, Photon Factory; r.m.s.d., root mean square deviation.
| Apo form (PEG complex) | AMT complex | DSP complex | CPZ complex | |
|---|---|---|---|---|
| Data collection | ||||
| Beamline | SPring-8 BL44XU | PF BL-5A | PF BL-17A | PF BL-5A |
| Space group | C2 | C2 | C2 | P212121 |
| Unit cell constants | a = 67.1, b = 44.9, c = 120.8 Å; β = 91.9° | a = 66.7, b = 45.9, c = 120.7 Å; β = 92.1° | a = 66.7, b = 45.5, c = 120.6 Å; β = 91.7° | a = 42.1, b = 63.1, c = 65.0 Å |
| Resolution range (Å) | 50.0–2.10 (2.18–2.10) | 50.0–2.15 (2.23–2.15) | 40.19–2.20 (2.32–2.20) | 50.0–2.20 (2.28–2.20) |
| No. of observed reflections | 152,696 | 133,660 | 139,452 | 47,798 |
| No. of unique reflections | 21,258 | 20,082 | 18,846 | 9216 |
| Completeness (%) | 99.9 (99.3) | 98.8 (93.8) | 99.6 (100.0) | 99.2 (95.2) |
| Rmerge (%)a | 4.4 (32.6) | 5.9 (17.7) | 5.8 (10.5) | 7.9 (16.1) |
| 〈I/σI〉 | 56.8 (6.7) | 52.4 (8.1) | 26.2 (15.2) | 27.6 (8.7) |
| Refinement statistics | ||||
| Resolution range (Å) | 31.99–2.10 | 36.31–2.15 | 40.19–2.20 | 30.83–2.20 |
| No. of reflections used | 20,189 | 18,955 | 17,931 | 8671 |
| Completeness (%) | 99.9 | 98.9 | 99.5 | 99.3 |
| Rcryst/Rfree (%)b | 20.0/23.2 | 21.9/26.7 | 22.8/26.3 | 20.2/26.8 |
| No. of atoms | ||||
| Protein | 2882 | 2871 | 2945 | 1503 |
| Water | 83 | 96 | 74 | 138 |
| Ligand | 26 | 46 | 50 | 25 |
| Ramachandran plot (%) | ||||
| Most favored | 94.0 | 92.0 | 93.1 | 95.7 |
| Additionally allowed | 6.0 | 8.0 | 6.9 | 4.3 |
| Generously allowed | 0 | 0 | 0 | 0 |
| Disallowed | 0 | 0 | 0 | 0 |
| r.m.s.d. | ||||
| Bonds (Å) | 0.012 | 0.013 | 0.012 | 0.008 |
| Angles | 1.3° | 1.4° | 1.3° | 1.1° |
a Rmerge = 100 × Σ|Ihkl − 〈Ihkl〉|/ΣIhkl, where 〈Ihkl〉 is the mean value of Ihkl.
b Rcryst = 100 × Σ‖Fo| − |Fc‖/Σ|Fo|. Rfree was calculated from the test set (5% of the total data).
The structure of the A variant was determined by molecular replacement with MOLREP (24) using the structure of the F1*S variant as a search model (Protein Data Bank code 3BX6) (18). The model was refined with CNS (25) and rebuilt with COOT (26). The structures of A variant-AMT complex and A variant-DSP complex were refined with the coordinates of the A variant, respectively. The structure of A variant-CPZ complex was determined by molecular replacement with MOLREP using the structure of A variant as a search model. At the final stages, the refinement of each structure was carried out with REFMAC using the TLS parameters (27). The stereochemical qualities of all structures were checked by PROCHECK (28). Data collection and refinement statistics are given in Table 1. Superpositions of the hAGP structures were carried out with LSQKAB (29). All figures of protein structures were generated with PyMOL (30) and CCP4mg (31). The electrostatic surface potential was calculated and drawn with CCP4mg.
RESULTS
Tertiary Structure of the A Variant
The crystal structure of drug-free A variant hAGP was determined at a resolution of 2.1 Å. As shown in Fig. 1, the A variant is composed of four α-helices, 1–4, corresponding to Asn-15–Ile-21, Glu-35–Gln-42, Lys-135–Cys-147, and Glu-166–Gln-171, respectively, and eight β-strands, A–H, corresponding to Gly-23–Phe-32, Ile-44–Asn-54, Thr-59–Arg-68, Gln-71–Gln-82, Thr-87–Glu-92, Arg-95–Leu-102, Thr-109–Ser-114, and Gly-123–Ala-128, respectively. These eight β-strands form a β-barrel, which is a common structural feature in other proteins of the lipocalin family. The entrance to the ligand-binding pocket is located at the open end of the β-barrel. The five glycans of AGP are attached to Asn-15, Asn-38, Asn-54, Asn-75, and Asn-85, respectively; among these residues, only Asn-75 is in close proximity to the entrance of the binding pocket (Fig. 1b). It was observed that a PEG molecule derived from the reservoir solution is bound in the binding pocket (supplemental Fig. S1).
Modes of Binding of DSP, AMT, and CPZ to the A Variant
We successfully obtained the crystal structures of complexes of A variant hAGP with DSP, AMT, and CPZ at resolutions of 2.20, 2.15, and 2.20 Å, respectively. The complexes with DSP and AMT crystallized in the same space group (C2) as drug-free hAGP and with essentially the same unit cell dimensions (Table 1), and each crystal contains two protein-drug complexes in the asymmetric unit (molecules A and B). In both cases, the A and B structures were very similar to one another, so, for the sake of simplicity, the discussion below refers to molecule A. The hAGP-CPZ complex crystallized in a different space group (Table 1) with only one complex in the asymmetric unit.
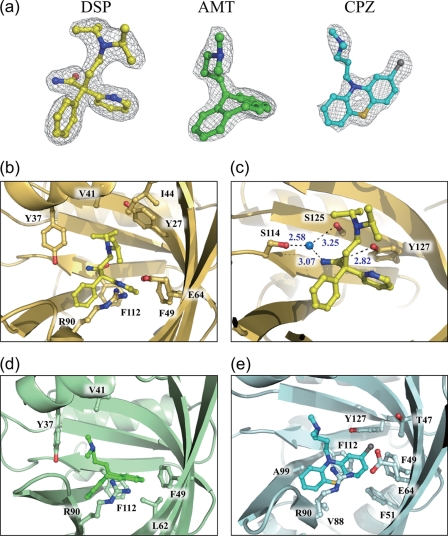
We were able to clearly observe the electron density for DSP in the binding pocket of A variant hAGP (Fig. 2a). Although there are both S- and R-forms of DSP, it is not easy to determine from the electron density whether the S- or R-form is bound because of the similarity in electron densities between the pyridinyl and phenyl rings. Because hAGP is known to bind preferentially to the S-form of DSP (32), the structure of the hAGP-DSP complex was refined using the S-form of DSP (Fig. 2b). DSP makes multiple contacts with residues lining the binding pocket. The two aromatic rings of DSP are in direct contact with Phe-49 and Phe-112, resulting in CH-π interactions (edge to face); in addition, van der Waals contacts with Glu-64 and Arg-90 are observed. In the S-form of DSP, the nitrogen atom of the pyridinyl ring can be located near the guanidium group of Arg-90, which might contribute to preference for the S-form by the A variant. The amide group of DSP forms a hydrogen bond with the hydroxyl group of Tyr-127. Further specific hydrogen bonds with the amide group of the drug in molecule A are made via a water molecule (Fig. 2c), which is held in position by hydrogen bonding with Ser-114 and Ser-125. Finally, the alkyl chain of DSP makes van der Waals interactions with Tyr-27, Tyr-37, Val-41, and Ile-44 (Fig. 2b).
FIGURE 2.
Interactions of DSP, AMT, and CPZ with the A variant. a, 2Fo − Fc electron density maps of DSP, AMT, and CPZ in the complex structures, contoured at 1.2σ. b, structure of the binding pocket in the A variant-DSP complex. c, structure of the binding pocket in the A variant-DSP complex. A water molecule is depicted as a blue sphere, and hydrogen bonds are indicated as black dotted lines. d, structure of the binding pocket in the A variant-AMT complex. e, structure of the binding pocket in the A variant-CPZ complex. DSP, AMT, and CPZ molecules are depicted as yellow, green, and cyan, respectively.
In the case of AMT (Fig. 2d), again the electron density clearly indicated the binding of a single drug molecule. Similar to DSP, the aromatic rings of AMT are involved in CH-π interactions (edge to face) with Phe-49 and Phe-112 and van der Waals interactions with Leu-62 and Arg-90. The tip of the alkyl chain of AMT makes van der Waals contacts with Tyr-37 and Val-41. It is noteworthy that the side chain of Tyr-37 is rotated compared with its position in the complex with DSP. (The difference in the side chain torsion angle (χ2) is ∼39°.) This difference in the rotation of the Tyr-37 side chain may contribute to the ability of this protein to bind drugs with different lengths of alkyl chains.
Interestingly, CPZ shows a different interaction mode in the binding pocket compared with DSP and AMT (Fig. 2e). In this case, the fused aromatic ring system of CPZ interacts with Phe-112 by π-π stacking interactions; in addition, it makes CH-π interactions with Phe-49 and Ala-99. Further van der Waals contacts are made with Phe-51, Val-88, and Arg-90. The chlorine atom attached to one end of the aromatic ring system in CPZ is accommodated at the end of the binding pocket formed by the side chains of Thr-47, Phe-49, Glu-64, and Tyr-127.
Comparison with the F1*S Variant of hAGP
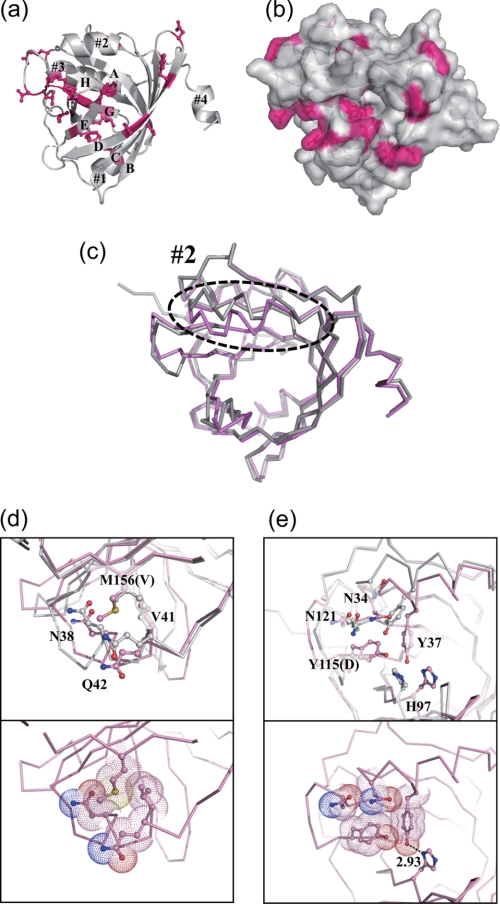
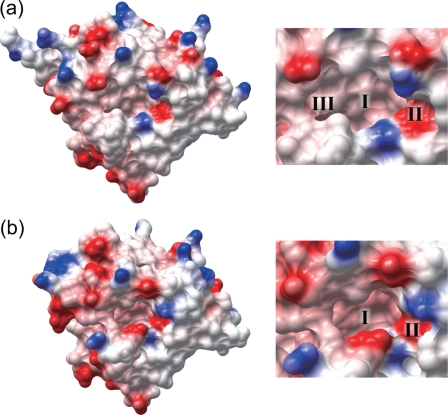
In Fig. 3a, the 21 amino acid residues of the A variant that are different from the F1*S variant are shown in red. These are distributed throughout the molecule, but many are found lining the interior of the pocket (there is a near-continuous stretch of substitutions in strand G and along the GH loop) and near its entrance (Fig. 3, a and b). Superposition of the tertiary structures of the A and F1*S variants of hAGP reveals that their overall folds are similar (root mean square deviation of 1.37 Å for 168 Cα atoms) (Fig. 3c). However, there are some notable differences in both structures: the angle of α-helix 2 relative to strands A and B is substantially altered, being rotated by ∼16° compared with its position in the F1*S variant (Fig. 3c). This rotation appears to be due primarily to the presence of the bulkier side chain of Met-156 in the A variant (compared with Val-156 in the F1*S variant), which changes the nature of the interactions with Asn-38, Val-41, and Gln-42, moving α-helix 2 toward the pocket (Fig. 3d). In the loop between strands G and H (positions 112–117), Tyr-115 in the A variant is also observed to project inside the binding pocket (Fig. 3e), whereas Asp-115 in the F1*S variant enters into hydrogen bond formation with the side chain of His-100 at the side of the pocket. These collective differences account for the formation of the van der Waals interactions among Asn-34, Tyr-37, Tyr-115, and Asn-121 and a hydrogen bond between Tyr-37 and His-97 in the structure of the A variant (Fig. 3e), which reduce the width of its binding pocket. Indeed, these changes effectively eliminate the lobe III subcompartment of the drug-binding pockets that were identified in F1*S hAGP by Schönfeld and co-workers (18) (Fig. 4, a and b).
FIGURE 3.
Comparison of the amino acid residues and the structure between the hAGP variant. Shown are the overall structure of the A variant represented in schematic form (a) and the top view into the β-barrel with surface representation (b). A superposed view of the F1*S and A variants is presented (c). The dashed oval represents the difference in the position of α-helix 2 between the hAGP variants. Local structural differences in the GH loop (d) and near the binding pocket (e) between hAGP variants in the region indicated by the dashed oval in c are shown. Amino acid residues in the F1*S variant (Protein Data Bank code 3BX6) are colored hot pink (a and b), and the F1*S and A variants are colored gray and pink (c and d).
FIGURE 4.
Comparison of the drug-binding pockets in the hAGP variants. a and b, electrostatic surface representation of the F1*S and A variants, respectively. Negatively and positively charged areas are colored red (−20 kBT/e) and blue (20 kBT/e).
DISCUSSION
Tertiary Structure of the A Variant
The structural and functional properties of hAGP have been a subject of considerable interest since the protein was first purified from serum in the 1950s. In these prior investigations, hAGP was found to have a high content of glycan structure, which makes up about half of the total mass of this glycoprotein (1, 2). These glycans, which are thought to contribute to the aqueous solubility of hAGP, undoubtedly add to its structural heterogeneity and have made it difficult to obtain the single crystals needed for structure determination by x-ray crystallography. However, Skerra and co-workers (18) recently succeeded in determining the crystal structure of the recombinant F1*S variant of hAGP expressed from E. coli in the form of an unglycosylated protein. They found that the F1*S variant has a typical lipocalin fold consisting of an eight-stranded β-barrel, which forms a deep, wide drug-binding pocket. In this study, we employed a similar approach to determine the crystal structure of an unglycosylated form of the A variant of hAGP. Our results confirm that this protein has the same overall folding as the F1*S variant. The carbohydrate groups on hAGP are thought to have little effect on drug-binding properties and secondary structure (33, 34). Our previous findings also demonstrated that the drug-binding capacity of a mutant, C149R, was equivalent to that of the A form purified from serum (35). Although it should be noted that Asn-75, which is glycosylated in vivo in both variants, was found to be located near the entrance of the drug-binding pocket, it seems that glycosylation at this position has no impact on the binding of drugs by hAGP.
Interactions between hAGP and Drugs
For the first time, we were able to obtain co-crystal structures of hAGP bound to drug molecules. Our results provide new and detailed insights into the structural basis of the different drug-binding specificities of the two major variants of the protein.
DSP and AMT are well known as ligands that bind to A variant hAGP with a high degree of selectivity. These two drugs, which are structurally quite similar, were observed to bind in essentially the same manner to the central cavity (lobe I) in hAGP, making contact with many of the same amino acids lining the binding pocket (Fig. 5, b and c). In particular, both complexes reveal conserved edge-face contacts between the two aromatic rings on the drugs and the aromatic side chains of Phe-112 and Phe-49. This suggests that Phe-112 and Phe-49 are critical residues for the binding of these drugs to the A variant. Notably, the residue at position 112 in the F1*S variant is leucine; therefore, this difference may contribute to the reduced binding affinity of DSP and AMT for the F1*S variant. There are additional differences that explain why DSP and AMT preferentially bind to the A variant. Whereas Ser-114 in the A variant is involved in a water-mediated hydrogen bond with the amide group of DSP, this residue is replaced by Phe-114 in the F1*S variant. Thus, not only would the hydrogen bond to DSP be lost, but the larger side chain in F1*S would be expected to impede the binding of DSP (and AMT) due to steric hindrance in the binding pocket (Fig. 5a). Therefore, the differences in the amino acid residues between the two major hAGP variants at positions 112 and 114 appear to be crucial for the high selectivity of the A variant for DSP, AMT, and other A variant-specific drugs that contain two aromatic rings with similar configurations.
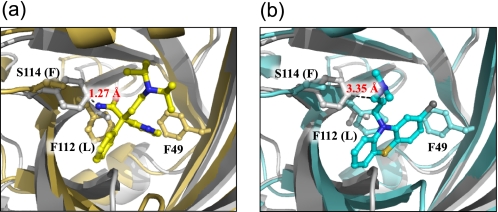
FIGURE 5.
Simulated mode of binding of DSP (a) and CPZ (b) to the F1*S variant. The DSP and CPZ molecules are depicted as yellow and cyan sticks and balls, respectively. The F1*S and A variants are colored gray and yellow orange (a) or light cyan (b), respectively.
CPZ was also observed to bind to the central cavity (lobe I). CPZ binds to the F1*S and A variants with equal affinity (11). Although some structural similarities exist between CPZ and DSP and AMT, its aromatic ring system is more planar and has a chlorine atom attached to one end. Collectively, these features appear to result in a reorientation of the ring system in the CPZ complex compared with the conformations of bound DSP and AMT. In particular, CPZ is involved in parallel stacking interactions with the Phe-112 side chain of the A variant. Modeling studies suggest that CPZ may bind with the same orientation in the pocket of the F1*S variant, despite the replacement of Phe-112 by Leu in this variant. Moreover, maintenance of the same orientation would not result in any steric hindrance due to the substitution of Ser-114 by Phe in the F1*S variant, in contrast to our predictions for DSP and AMT (Fig. 5b). This provides a plausible explanation for the nonselectivity of hAGP variants for CPZ.
Skerra and co-workers (18) modeled the mode of binding of nonspecific diazepam and progesterone to the crystal structure of F1*S hAGP and found that, among the amino acid residues predicted to contact diazepam, Val-92, Leu-112, and Phe-114 are different from the residues for the A variant. As discussed above, our structures suggest that the residues at positions 112 and 114 are critical for the selectivity of the hAGP variant. Glu-92 in the A variant of hAGP, which is located near the top of the drug-binding cavity, is replaced by Val in the F1*S variant. We recently found that this residue is an important determinant for the binding of propafenone, a representative A variant-specific drug (35). The mutation of Glu-92 to Val in the A variant reduced propafenone binding, whereas introduction of the converse V92E mutation into the F1*S variant increased the binding capacity of this drug. These results suggest that the negatively charged side chain on Glu-92 may be required for the binding of propafenone to hAGP. In this study, Glu-92 is located near Arg-90 (supplemental Fig. S2), which interacts with DSP or AMT. Therefore, Glu-92 in the A variant may be indirectly involved in the binding of propafenone. Alternatively, Glu-92 might contribute directly to binding by forming a hydrogen bond with the amide group in the alkyl tail structure of propafenone.
Skerra and co-workers (18) found that the binding pocket of the F1*S variant is wide and consists of three lobes (I–III). Our study indicates that the A variant maintains lobes I and II, but not lobe III. This result shows that the binding region of the A variant of hAGP is narrower than that of the F1*S variant, a difference that may be a factor in its distinctive ligand selectivity. In fact, the selectively of ligand binding to the A variant displays some common structural features, but this is not seen for the case of the F1*S variant, which can recognizes ligands with diverse structures such as warfarin, prazosin, dipyridamole, and imatinib, etc. (11). To further explore this possibility, additional crystallographic and ligand binding studies of F1*S variant-ligand complexes will be needed in future studies.
Acknowledgments
We thank the staff at the Photon Factory and SPring-8 for assistance with the x-ray diffraction experiments.
This work was supported in part by Grant-in-aid for Scientific Research KAKENHI 21390161 from the Japan Society for the Promotion of Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3APU, 3APV, 3APW, and 3APX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- hAGP
- human α1-acid glycoprotein
- DSP
- disopyramide
- AMT
- amitriptyline
- CPZ
- chlorpromazine.
REFERENCES
- 1. Schmid K., Nimerg R. B., Kimura A., Yamaguchi H., Binette J. P. (1977) Biochim. Biophys. Acta 492, 291–302 [DOI] [PubMed] [Google Scholar]
- 2. Yoshima H., Matsumoto A., Mizuochi T., Kawasaki T., Kobata A. (1981) J. Biol. Chem. 256, 8476–8484 [PubMed] [Google Scholar]
- 3. Hochepied T., Berger F. G., Baumann H., Libert C. (2003) Cytokine Growth Factor Rev. 14, 25–34 [DOI] [PubMed] [Google Scholar]
- 4. Kremer J. M., Wilting J., Janssen L. H. (1988) Pharmacol. Rev. 40, 1–47 [PubMed] [Google Scholar]
- 5. Fuse E., Tanii H., Kurata N., Kobayashi H., Shimada Y., Tamura T., Sasaki Y., Tanigawara Y., Lush R. D., Headlee D., Figg W. D., Arbuck S. G., Senderowicz A. M., Sausville E. A., Akinaga S., Kuwabara T., Kobayashi S. (1998) Cancer Res. 58, 3248–3253 [PubMed] [Google Scholar]
- 6. Gambacorti-Passerini C., Zucchetti M., Russo D., Frapolli R., Verga M., Bungaro S., Tornaghi L., Rossi F., Pioltelli P., Pogliani E., Alberti D., Corneo G., D'Incalci M. (2003) Clin. Cancer Res. 9, 625–632 [PubMed] [Google Scholar]
- 7. Holladay J. W., Dewey M. J., Michniak B. B., Wiltshire H., Halberg D. L., Weigl P., Liang Z., Halifax K., Lindup W. E., Back D. J. (2001) Drug Metab. Dispos. 29, 299–303 [PubMed] [Google Scholar]
- 8. Eap C. B., Baumann P. (1989) Prog. Clin. Biol. Res. 300, 111–125 [PubMed] [Google Scholar]
- 9. Yuasa I., Weidinger S., Umetsu K., Suenaga K., Ishimoto G., Eap B. C., Duche J. C., Baumann P. (1993) Vox Sang. 64, 47–55 [DOI] [PubMed] [Google Scholar]
- 10. Budai L., Ozohanics O., Ludányi K., Drahos L., Kremmer T., Krenyacz J., Vékey K. (2009) Anal. Bioanal. Chem. 393, 991–998 [DOI] [PubMed] [Google Scholar]
- 11. Hervé F., Caron G., Duché J. C., Gaillard P., Abd Rahman N., Tsantili-Kakoulidou A., Carrupt P. A., d'Athis P., Tillement J. P., Testa B. (1998) Mol. Pharmacol. 54, 129–138 [DOI] [PubMed] [Google Scholar]
- 12. Imamura H., Maruyama T., Otagiri M. (1993) Biol. Pharm. Bull. 16, 926–929 [DOI] [PubMed] [Google Scholar]
- 13. Kute T., Westphal U. (1976) Biochim. Biophys. Acta 420, 195–213 [DOI] [PubMed] [Google Scholar]
- 14. Katsuki M., Chuang V. T., Nishi K., Suenaga A., Otagiri M. (2004) Pharm. Res. 21, 1648–1655 [DOI] [PubMed] [Google Scholar]
- 15. Chuang V. T., Hijioka M., Katsuki M., Nishi K., Hara T., Kaneko K., Ueno M., Kuniyasu A., Nakayama H., Otagiri M. (2005) Biochim. Biophys. Acta 1725, 385–393 [DOI] [PubMed] [Google Scholar]
- 16. Katsuki M., Chuang V. T., Nishi K., Kawahara K., Nakayama H., Yamaotsu N., Hirono S., Otagiri M. (2005) J. Biol. Chem. 280, 1384–1391 [DOI] [PubMed] [Google Scholar]
- 17. Kopecký V., Jr., Ettrich R., Hofbauerová K., Baumruk V. (2003) Biochem. Biophys. Res. Commun. 300, 41–46 [DOI] [PubMed] [Google Scholar]
- 18. Schönfeld D. L., Ravelli R. B., Mueller U., Skerra A. (2008) J. Mol. Biol. 384, 393–405 [DOI] [PubMed] [Google Scholar]
- 19. Maruyama T., Otagiri M., Takadate A. (1990) Chem. Pharm. Bull. 38, 1688–1691 [DOI] [PubMed] [Google Scholar]
- 20. Nishi K., Fukunaga N., Ono T., Akuta T., Yumita N., Watanabe H., Kadowaki D., Suenaga A., Maruyama T., Otagiri M. (2010) Drug Metab. Pharmacokinet. 25, 200–207 [DOI] [PubMed] [Google Scholar]
- 21. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 22. Leslie A. G. W. (1992) Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography, No. 26, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 23. Evans P. R. (1997) Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography, No. 33, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 24. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 25. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 26. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27. Winn M. D., Murshudov G. N., Papiz M. Z. (2003) Methods Enzymol. 374, 300–321 [DOI] [PubMed] [Google Scholar]
- 28. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 29. Kabsch W. (1976) Acta Crystallogr. Sect. A 32, 922–923 [Google Scholar]
- 30. DeLano W. L. (2009) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 31. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 32. Kishino S., Itoh S., Nakagawa T., Miyazaki K. (2001) Eur. J. Clin. Pharmacol. 57, 583–587 [DOI] [PubMed] [Google Scholar]
- 33. Schmid K., Buringame R. W., Paulson J. C., Hill R. L. (1978) Fed. Proc. 37, 1298–1309 [Google Scholar]
- 34. Friedman M. L., Wermeling J. R., Halsall H. B. (1986) Biochem. J. 236, 149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nishi K., Ueno M., Murakami Y., Fukunaga N., Akuta T., Kadowaki D., Watanabe H., Suenaga A., Maruyama T., Otagiri M. (2009) J. Pharm. Sci. 98, 4316–4326 [DOI] [PubMed] [Google Scholar]