Abstract
Curacin A is a polyketide synthase (PKS)-non-ribosomal peptide synthetase-derived natural product with potent anticancer properties generated by the marine cyanobacterium Lyngbya majuscula. Type I modular PKS assembly lines typically employ a thioesterase (TE) domain to off-load carboxylic acid or macrolactone products from an adjacent acyl carrier protein (ACP) domain. In a striking departure from this scheme the curacin A PKS employs tandem sulfotransferase and TE domains to form a terminal alkene moiety. Sulfotransferase sulfonation of β-hydroxy-acyl-ACP is followed by TE hydrolysis, decarboxylation, and sulfate elimination (Gu, L., Wang, B., Kulkarni, A., Gehret, J. J., Lloyd, K. R., Gerwick, L., Gerwick, W. H., Wipf, P., Håkansson, K., Smith, J. L., and Sherman, D. H. (2009) J. Am. Chem. Soc. 131, 16033–16035). With low sequence identity to other PKS TEs (<15%), the curacin TE represents a new thioesterase subfamily. The 1.7-Å curacin TE crystal structure reveals how the familiar α/β-hydrolase architecture is adapted to specificity for β-sulfated substrates. A Ser-His-Glu catalytic triad is centered in an open active site cleft between the core domain and a lid subdomain. Unlike TEs from other PKSs, the lid is fixed in an open conformation on one side by dimer contacts of a protruding helix and on the other side by an arginine anchor from the lid into the core. Adjacent to the catalytic triad, another arginine residue is positioned to recognize the substrate β-sulfate group. The essential features of the curacin TE are conserved in sequences of five other putative bacterial ACP-ST-TE tridomains. Formation of a sulfate leaving group as a biosynthetic strategy to facilitate acyl chain decarboxylation is of potential value as a route to hydrocarbon biofuels.
Keywords: Crystal Structure, Decarboxylase, Enzyme Structure, Protein Structure, X-ray Crystallography, Natural Products Biosynthesis, Polyketide Synthase, Thioesterase
Introduction
Natural products display a remarkable chemical diversity, providing advantages for the producing plants and microbes to survive and thrive in particular ecological niches. These secondary metabolites and their derivatives have important applications as pharmaceuticals (1), and some have the potential to be developed as biofuels (2). Gene clusters encoding assembly line biosynthetic pathways for polyketide and polypeptide natural products are ubiquitous in bacterial and fungal genomes. Polyketide synthase (PKS)3 and non-ribosomal peptide synthetase (NRPS) pathways have a common modular organization in which intermediates tethered to carrier domains by a thioester linkage pass sequentially through modules of the assembly line. The final step in the assembly line is typically a thioesterase-catalyzed off-loading from the final carrier domain to produce a carboxylate, macrolactone, or cyclic peptide.
In a notable exception to this off-loading paradigm, the curacin A final product contains a terminal alkene moiety. Curacin A, from the marine cyanobacterium Lyngbya majuscula, is a mixed polyketide/non-ribosomal peptide with antimitotic properties (3). The hybrid PKS/NRPS assembly line pathway for curacin A (4) generates several unusual chemical groups in addition to the terminal alkene, including a cyclopropyl ring, a thiazoline ring, and a cis double bond. We have investigated the biosynthetic steps leading to several of these segments (5–9). Herein we investigate the structural basis for the unique off-loading strategy leading to the terminal alkene in the curacin A molecule.
The curacin PKS has an unusual terminal module, the CurM protein, with a C-terminal tridomain comprised of an acyl carrier protein (ACP), a sulfotransferase (ST), and a thioesterase (TE) (Fig. 1A). Annotations of both ST and TE were based on weak sequence similarity to characterized enzymes. The prediction of an ST within a PKS was unprecedented (4). STs are widely distributed and are known to have detoxification, hormone regulation, or signaling functions (10). They catalyze transfer of a sulfonate group from the donor PAPS to a hydroxyl or amine of an acceptor small molecule or protein. CurM TE, although identifiable as a thioesterase, does not resemble any of the previously established PKS or NRPS TE subfamilies (11).
FIGURE 1.
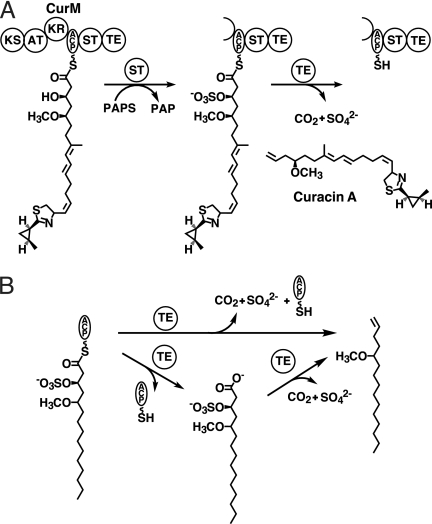
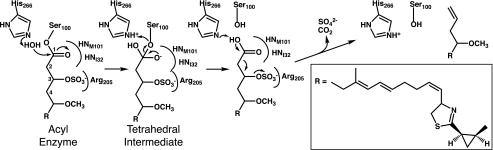
Chain termination in curacin A biosynthesis. A, off-loading reactions in the final module, CurM. Following extension by the CurM ketosynthase (KS) and acyltransferase (AT) and reduction by the ketoreductase (KR), ST catalyzes transfer of sulfonate to the β-hydroxyl followed by TE hydrolysis of the thioester with concomitant decarboxylation and sulfate elimination. The sulfonate group donor is the PAPS cofactor, which is converted to PAP. B, experimental scheme for the assay of CurM TE. Recombinant CurM ACP loaded with a substrate analog and sulfonated by ST was reacted with excised CurM TE. Activity was monitored by HPLC of the reaction mixture and detection of the holo-ACP product.
Using a simplified analog of the penultimate pathway intermediate, we recently demonstrated that off-loading and terminal alkene formation require ST-mediated sulfonation of the β-hydroxyl group from the PAPS cofactor (9) (Fig. 1A). This was the first observation of biological substrate activation by formation of a sulfate leaving group. CurM TE acts upon the β-sulfate intermediate to yield a decarboxylated product with a terminal double bond resulting from sulfate elimination (Fig. 1B). CurM TE catalyzes thioester hydrolysis 800-fold more slowly on the corresponding substrate bearing a β-hydroxyl group. The unprecedented requirement of a β-sulfate for thioester hydrolysis as well as the decarboxylation and sulfate elimination suggests a unique catalytic strategy and active site structure for the TE. Moreover, this curacin pathway decarboxylation strategy provides an opportunity to investigate a new biological route to hydrocarbon production from fatty acids. Thus, in addition to interest in curacin biosynthesis as a route to a potent anti-cancer compound (3), the pathway also has relevance to biofuel production.
Off-loading TEs from many PKSs have been studied including the TEs of the pikromycin (Pik TE) (12–14), erythromycin (14), and tautomycetin (15) PKS. PKS off-loading TEs typically catalyze either hydrolysis to produce a linear carboxylic acid or the attack of an intramolecular hydroxyl to produce a large-ring macrolactone. PKS TEs are members of the α/β-hydrolase superfamily with a catalytic triad active site located at the top of an α/β core and covered by an α-helical lid subdomain. All PKS off-loading TEs of known structure are dimers in which two N-terminal α-helices in the lid form a lid-to-lid dimer interface (12–15). A classic Ser-His-Asp catalytic triad is positioned at the center of a narrow tunnel formed by the lid. The tunnel architecture with open ends is fixed by the dimer interface. In contrast to these dimeric off-loading TEs, many PKS and NRPS pathways also have a second monomeric thioesterase called a TE II, which performs an editing function within the pathway. TE IIs, as well as NRPS off-loading TEs, are monomers with a flexible lid domain that appears to control access to the active site (16–20). The curacin TE sequence has low similarity to sequences in all parts of the TE phylogenic tree (11), lacks an N-terminal extension for dimerization, and has a longer internal lid than other PKS off-loading TEs.
To gain further insights and enable mechanistic studies of the novel decarboxylation and sulfate elimination, we report here the crystal structure of CurM TE. The structure of the TE lid and an unusual dimer interface appear to fix the active site in a perpetually open state. A model for β-sulfate recognition was tested by site-directed mutagenesis. The similarity of CurM to other conserved ACP-ST-TE tridomain sequences strongly suggests that CurM TE is part of a new subfamily of thioesterases.
EXPERIMENTAL PROCEDURES
Cloning and Site-directed Mutagenesis
A construct encoding the TE (CurM residues 1929–2211) was amplified from the cosmid pLM14 (9) and was inserted into pMoCR, a vector encoding the fusion protein His6-Mocr for enhanced solubility (21). Site-directed mutagenesis was performed using the QuikChange protocol (Stratagene). All constructs were verified by sequencing. The CurM ACP and ST expression plasmids were previously described (9).
Protein Expression and Purification
Escherichia coli strain BL21(DE3) was transformed with an expression plasmid, grown at 37 °C in 500 ml of TB with 4% glycerol to an A600 of 1.0, cooled to 18 °C, induced with isopropyl β-d-thiogalactopyranoside (final concentration, 0.2 mm), and grown for an additional 18 h. Selenomethionyl (SeMet) protein was produced in the same E. coli strain in SelenoMet medium (AthenaES) containing 100 μg/ml of seleno-dl-methionine.
All steps were performed at 4 °C. The cell pellet from 500 ml of cell culture was resuspended in 40 ml of Buffer A (20 mm Tris, pH 7.9, 500 mm NaCl, 20 mm imidazole, 10% glycerol), incubated 30 min with DNase (2 mg), lysozyme (5 mg), and MgCl2 (4 mm), lysed by sonication, and the soluble fraction loaded onto a 5-ml HisTrap Ni-NTA column (GE Healthcare). CurM TE was eluted with a linear gradient from 20 to 650 mm imidazole (Buffer B). The His6-Mocr fusion partner was removed by a 2-h incubation with 1 mm DTT and tobacco etch virus protease (1 mg of protease/50 mg of TE) at room temperature. After overnight dialysis at 4 °C in Buffer C (20 mm Tris, pH 7.9, 500 mm NaCl, 10% glycerol) with 1 mm DTT, the remaining His-tagged proteins were removed by nickel-affinity chromatography, followed by size exclusion chromatography with a HiLoad 16/60 Superdex 200 column (GE Healthcare) pre-equilibrated with Buffer C. CurM TE was concentrated to 5 mg/ml, flash frozen in liquid N2, and stored at −80 °C. Of 14 TE variants purified as the wild type, only 6 yielded enough soluble protein for assay. SeMet TE was purified as the wild type with addition of 2 mm DTT to all buffers. Yields per 500 ml of culture were 5 mg of TE and 2 mg of SeMet TE.
Crystallization
Crystals grew at 4 °C within 24–48 h by vapor diffusion from a 1:1 mixture of protein stock (2 mg/ml of TE, 20 mm Tris, pH 7.9, 200 mm NaCl, 2.5% glycerol) and well solution (27–32% PEG3350, 100 mm Tris, pH 8.3–8.5). Micro-seeding was required for crystal growth of the SeMet protein in similar conditions with 1 mm DTT in the protein solution. Crystals were cryoprotected in a well solution with 15% glycerol, harvested in loops, and flash cooled in liquid N2.
Data Collection and Structure Determination
Data were collected at GM/CA-CAT beamline 23ID-D at the Advanced Photon Source (APS) at the Argonne National Lab (Argonne, IL). Among 25 SeMet TE crystals, only one diffracted beyond 4 Å, but was visibly two crystals and had multiple lattices in the diffraction pattern. A region visually identified as a single crystal was probed in three 10-μm steps using a 20-μm mini-beam (22). The crystal was centered at the position with the strongest diffraction and the least interference from the second lattice, and Friedel data were collected in inverse-beam geometry (φ = 0–90° and 180–270° as wedges of 45° with 1° images) at the wavelength of peak absorption at the selenium edge. The diffraction images showed signs of decay at the end of the collection. The crystal was translated to expose a fresh region to the beam, and rotated 90° from the start of data collection. Each of two regions was probed with the 10-μm mini-beam in a 3 × 3 raster of 10-μm steps. Data were collected at the strongest diffracting position with a single lattice, again in inverse beam geometry (φ = 90–150° and 270–330° as 30° wedges with 0.5° images). The two partial datasets were integrated separately and scaled together using the HKL2000 suite (23). Using Phaser (24) in the PHENIX (25) software suite, selenium sites were found for all Met residues of the four polypeptides in the asymmetric unit. Nine Met side chains had two partially occupied selenium sites and another Met had three sites for 39 total selenium sites (average figure of merit = 0.401). After density modification and 4-fold noncrystallographic symmetry averaging in RESOLVE (26) (figure of merit = 0.81), an 86% complete initial model was built by AUTOBUILD (27) and completed manually in COOT (28). A 1.7-Å native dataset was used for refinement. REFMAC5 (29), from the CCP4 suite (30), was used for refinement with 5 translation/libration/screw groups per monomer (29–31). Non-crystallographic symmetry was not used during any stage of the refinement. Electron density was complete throughout the polypeptide chain except for two loop regions, which had different disordered residues in the four polypeptide chains (monomer A, 132–134; B, 205–206; C, 133–136 and 205–216; D, 206–207). Although no single monomer is a complete view of these loops, superposition of the monomers provides a complete model.
Sequence Alignment, Structure Alignment, and Substrate Modeling
The search for ACP-ST-TE homologs was done with BLAST (32), MUSCLE (33) was used for multiple sequence alignment, and PyMOL was used to align structures and prepare figures (34). CurM TE was aligned with affinity-labeled Pik TE (Protein Data Bank code 2H7X, root mean square deviation = 3.309) by superposition of the core domains (residues 55–176 and 232–292 in Pik TE to residues 1–126 and 217–282 in CurM TE). The PRODRG2 server (35) was used to generate coordinates and a topology file for modeling the acyl-enzyme intermediate, which was modeled manually in COOT using the affinity label in the active site of PikTE (12, 13) as a guide.
Enzyme Assay
CurM TE activity was assayed using a modification of our previous protocol (9). Apo-ACP was loaded with a substrate analog by a 2-h incubation of 50 μm apo-ACP, 100 μm (3R)-hydroxy-5-methoxytetradecanoyl-CoA (9), 10 μm S. verticillus Svp (36), 10 mm MgCl2, 100 mm Tris, pH 7.9, at 30 °C. Complete loading was confirmed by reverse phase HPLC using a Jupiter C4 column (250 × 2.0 mm, 5 μm, 300 Å, Phenomenex) and a linear elution gradient from 30 to 90% CH3CN (0.1% CF3CO2H)/H2O (0.1% CF3CO2H) over 45 min. After exchange into Buffer C and concentration (Amicon Ultra 10-kDa concentrators Millipore), substrate-loaded ACP was flash frozen and stored at −80 °C. To generate the sulfated substrate for the TE assay, 225 μm loaded ACP was incubated with 5 μm ST, 1.75 mm PAPS (Sigma), 100 mm Tris, pH 7.9, at room temperature for 10 min. Complete sulfonation was confirmed by HPLC. The TE reaction was initiated by addition of TE (4 μm). After 1 min the reaction was quenched with 10% formic acid. Conversion of loaded to holo-ACP was quantified by HPLC as described above. Assays with the non-sulfated substrate were performed by incubating 1 mm (3R)-hydroxy-5-methoxytetradecanoyl-CoA, 50 mm Tris, pH 7.9, with 40 μm TE for 16 h, including 6% glycerol and 300 mm NaCl for protein stability. The reaction was quenched with equal volume of 1 m CH3CO2H and neutralized with 1 m NaOH, and crotonyl-CoA was added as an internal standard. Hydrolysis was analyzed using a Luna C18 column (250 × 4.60 mm, 5 μm, 100 Å, Phenomenex) with a linear gradient from 10 to 90% CH3OH/H2O (10 mm CH3CO2NH4) over 20 min.
RESULTS
Overall Structure
The curacin TE is the C-terminal domain of the 2211-residue CurM polypeptide. Because low sequence identity to other TEs prevented accurate definition of the domain N terminus, expression plasmids were constructed with three different start sites corresponding to amino acids 1917, 1929, and 1934. For all three domain variants, an N-terminal fusion of the protein Mocr (21) was necessary to obtain sufficient soluble protein for purification and crystallization. The construct encoding amino acids 1929–2211 of CurM (here denoted 1–283) yielded crystals after removal of the Mocr fusion partner. In the CurM TE crystal structure, solved by single anomalous diffraction phasing using selenomethionyl CurM TE (Table 1), the polypeptide chain was ordered to the ends of the construct.
TABLE 1.
Crystallographic summary
| CurM TE (SeMet) | CurM TE (native) | |
|---|---|---|
| Diffraction data | ||
| Space group | P21 | P21 |
| X-ray source | APS 23-ID-D | APS 23-ID-D |
| a, b, c (Å) | 74.1, 86.9, 87.1 | 74.5, 86.9, 87.6 |
| α, β, γ (o) | 90, 90.4, 90 | 90, 90.8, 90 |
| Wavelength (Å) | 0.97948 | 1.0332 |
| dmin (Å) | 2.14 (2.22-2.14)a | 1.68 (1.74-1.68) |
| Avgerage I/σI | 16.3 (5.5) | 18.1 (2.1) |
| Rsymb | 0.113 (0.362) | 0.059 (0.384) |
| Completeness | 99.5 (99.9) | 90.2 (49.8) |
| Average redundancy | 6.4 (6.3) | 3.4 (2.0) |
| Unique reflections | 60,147 | 114,584 |
| Refinement | ||
| Data range (Å) | 39.10–1.68 | |
| No. reflections | 108,718 | |
| Rwork/Rfreec | 0.164/0.209 | |
| Root mean square deviations | ||
| Bonds (Å) | 0.011 | |
| Angles (o) | 1.268 | |
| Avg B-factors (Å2) | ||
| Protein | 29.5 | |
| Water | 39.9 | |
| Ramachandran | ||
| Allowed | 99.9% | |
| Outliers | 0.1% | |
| Protein atoms | 8817 | |
| Water molecules | 1203 | |
a Outermost shell in parentheses.
b Including anomalous differences.
c The Rfree data set included a random 5% of reflections.
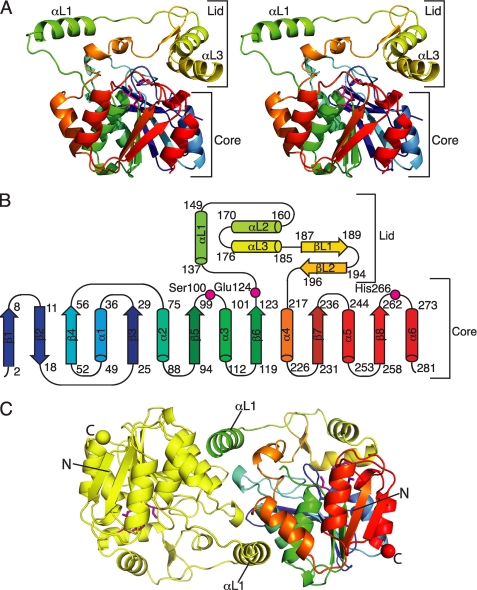
CurM TE possesses the α/β-hydrolase fold, as expected, with residues 1–132 and 215–283 comprising the structurally conserved core domain (Fig. 2, A and B). Residues of the catalytic triad (Ser100, Glu124, and His266) are located at the top of the core domain as in other TEs, but with a Glu in place of the more common Asp (Fig. 2B). The catalytic triad faces into a cleft between the core and lid. The lid subdomain (residues 133–214) is ∼20 residues longer than the analogous region in other TEs, and includes three helices (αL1, αL2, and αL3) and a small β-hairpin (βL1 and βL2) (Fig. 2, A and B). The first lid helix (αL1) is designated the “protruding helix” because it has few contacts with the lid and none with the core of the polypeptide.
FIGURE 2.
Structure of curacin A thioesterase. A, CurM TE polypeptide. The stereo ribbon diagram is colored as a rainbow from blue at the N terminus to red at the C terminus with the catalytic triad residues in stick form with a magenta C. B, topology diagram. CurM TE has an α/β-hydrolase fold in the core domain and a novel lid topology. Residues of the catalytic triad (Ser100, Glu124, and His266) are labeled. C, backbone trace of the CurM TE dimer viewed along the molecular dyad. Monomers are colored as a rainbow (right) and in yellow (left), with the catalytic triad as in A, and N and C termini shown as spheres of the same color as the terminal residue.
The orientation of the lid with respect to the core is identical in the four independent copies of the CurM TE polypeptide in the asymmetric unit of crystals (root mean square deviation = 0.29 Å for 216 Cα atoms, supplemental Fig. S1A), demonstrating that the active site cleft is identically open in all four polypeptides. The position of the lid is maintained by complementary surface contacts of lid helix αL3 (residues 176–186) with several loops in the core. Most of the contacts are hydrophobic. Specificity for the fixed lid-core orientation is provided by an “arginine anchor” in which the side chain of Arg185 in lid helix αL3 extends into the core domain where it forms a full set of five hydrogen bonds with core residues, including a buried salt bridge with Asp57 and hydrogen bonds with Gln35 and Glu3 (supplemental Fig. S2). Additionally, the Gln35 side chain amide is hydrogen bonded with the backbone carbonyl of Leu182 in lid helix αL3. In contrast to the remarkable lid-core surface complementarity on one side of the active site cleft, the linker peptides (residues 129–136 and 205–215) on the opposite side of the cleft are dynamic with some residues disordered in some subunits (supplemental Fig. S1A).
Novel Dimer Interface
CurM TE is dimeric in solution (supplemental Fig. S3) and also in the crystal structure (Fig. 2C). The mode of dimer formation is radically different from other PKS off-loading TEs even though the core structures and catalytic triads are similar (Fig. 3A). The primary dimer contact is between the protruding helix (αL1) in the lid of one subunit and three helices (α2, α3, and α4) in the core of the partner subunit (Fig. 2C), with additional core-to-core contacts of the β4-α2 loops of the two subunits. The subunit interface is predominantly hydrophobic and large (buried surface area of 1220 Å2 per monomer). The N termini are distant from the dimer interface (Fig. 2C), allowing for fusion to the monomeric ST domain as well as dimerization of the TE domain within the dimeric CurM module. This is consistent with the observation that the natural CurM ST-TE di-domain is dimeric in solution (data not shown).
FIGURE 3.
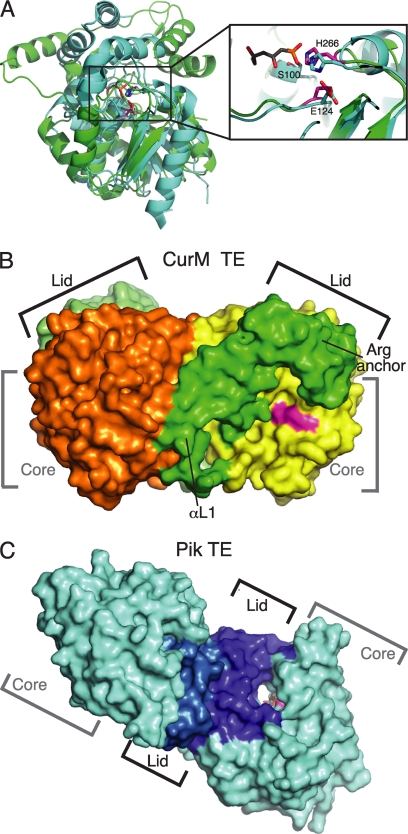
Comparison of curacin and pikromycin TEs. A, structure alignment of the core of CurM TE (green) and Pik TE (cyan, Protein Data Bank code 2H7X, (12)) (root mean square deviation = 1.5 Å for 95 Cα atoms). Both structures have the conserved α/β-hydrolase core, but the lids differ. The zoom view shows the active site conservation in the catalytic triad of CurM TE (magenta) and Pik TE (cyan) with a triketide affinity label (gray). The view is similar to Fig. 2A. B, surface representation of the CurM TE dimer. The primary dimer contact is between the lid (subunits in two shades of green) and core (subunits in yellow and orange). The active site (magenta) is in an open cleft between the core and lid. C, surface representation of the Pik TE dimer. The dimer contact is exclusively between the lid subdomains (subunits in two shades of blue) with no contacts of core domains (cyan). The active site (magenta) with an affinity label (gray sticks) is at the center of an open-ended tunnel (12). The views in B and C highlight the differences in active site access, which are distinct for the CurM TE and Pik TE enzymes.
The crystal asymmetric unit contains two dimers that differ by a slight flexure (3°) at the dimer interface, accounting for the poorer overall fit of dimers (root mean square deviation = 1.05 Å for 486 Cα atoms) compared with monomers. The protruding lid helix, αL1, moves with the partner subunit as the subunits flex (supplemental Fig. S1B). Thus, the lid-to-core dimer contact helps fix the lid in an open orientation (Fig. 3B). This is surprising and unique compared with off-loading TEs of other PKSs in which lid-to-lid dimer contacts fix the lid in a closed orientation. In these other TEs, the lid-to-lid dimer contact stabilizes an open-ended tunnel with the active site at its center (Fig. 3C). In contrast, the dimer-enforced open lid of CurM TE results in a highly exposed, open cleft active site (Fig. 3B). Thus, the dimer interface creates a very different active site environment in CurM TE and the other off-loading PKS TEs.
CurM TE Is Not Alone
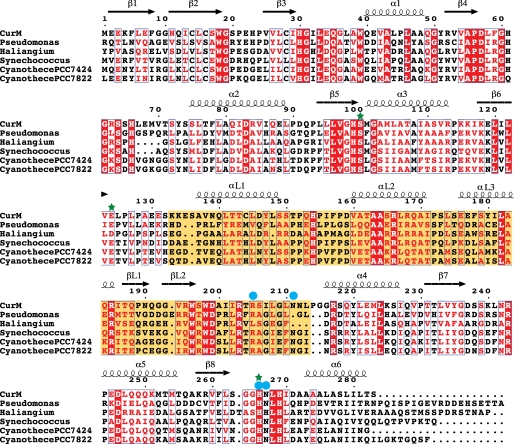
We identified five open reading frames in the sequence data base with substantial identity (33 to 51%) to the CurM ACP-ST-TE tridomain. The sequences are uncharacterized protein products from genomes of cyanobacteria (Synechococcus and Cyanothece) and proteobacteria (Pseudomonas and Haliangium). Based on the conservation of all three domains, including, for example, the TE catalytic triad, we assume that the other gene products also catalyze sequential sulfonation, hydrolysis, decarboxylation, and sulfate-elimination reactions, and that conserved residues within these sequences may illuminate areas of the structure that are important for function. Conserved residues in CurM TE and the five other putative TE sequences (Fig. 4) were mapped onto the CurM TE structure, revealing a dense area of conservation in the active site cleft (Fig. 5A). Sequence conservation indicates that CurM TE and the other gene products have the same lid structure and orientation and the same dimer interface. The essential features of the lid-to-core arginine anchor (Arg185, Asp57, and Gln35) are conserved. The protruding helix (αL1) has a conserved hydrophobic character, as do the surfaces of the core that it contacts in the dimer. The lid-to-core TE dimer interface, the open active site cleft, and the surface of the cleft are likely conserved among these TEs.
FIGURE 4.
Sequence alignment of CurM TE with putative TEs from open reading frames encoding tandem ACP-ST-TE tridomains. Invariant residues are highlighted in red, conserved residues are printed in red. The conserved lid domain is highlighted in yellow. Green stars indicate active site residues and blue circles indicate residues probed by site-directed mutagenesis. Sequence alignment was performed by MUSCLE (33), and the figure was prepared using ESPript (42). GenBank entries are: Pseudomonas entomophila L48 (GenBank YP_610919), H. ochraceum DSM 14365 (YP_003265308), Synechococcus PCC 7002 (YP_001734428), Cyanothece PCC 7424 (YP_002377174), and Cyanothece PCC 7822 (ZP_03153601).
FIGURE 5.
CurM TE active site. A, surface of the active site cleft. In the molecular surface view, the active site (magenta catalytic triad) is in a cleft in which the presumed phosphopantetheine entrance is lined with conserved residues (cyan), with other parts of the protein surface in green and the modeled acyl-enzyme intermediate in gray sticks. B, modeled acyl-enzyme intermediate (gray C). In this stereo view, the intermediate is surrounded by conserved amino acid residues (cyan C) and the catalytic triad (Ser100, Glu124, and His266) (magenta C). The carbonyl oxygen is bound in the oxyanion hole (hydrogen bonds to the NHs of the Ile32 and Met101).
Active Site
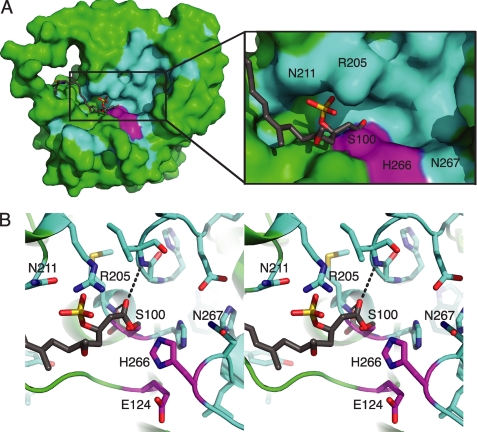
The open cleft active site and intact catalytic triad (Fig. 3) appear poised to hydrolyze a broad array of thioester substrates. To understand how CurM TE selects a sulfated substrate, we modeled the acyl-enzyme intermediate of the hydrolytic reaction (Scheme 1) using our knowledge of catalytic triad catalysis and geometry and the structure of affinity-labeled Pik TE (12) (Fig. 5B). Constraints on the model included acylation of the Ser100 nucleophile, carbonyl-oxygen binding in the oxyanion hole (NHs of Ile32 and Met101), the (R)-β-sulfate isomer, contact of sulfate with the enzyme, and placement of the long acyl chain away from the catalytic His266. Given these constraints, the acyl-enzyme was easily modeled with favorable bond rotations and without change to the protein structure. The acyl chain lies in a narrow cleft between the lid and core domains, and the (R)-β-sulfate occupies a niche adjacent to Arg205 (Fig. 5A). The model is consistent with the strong stereoselectivity of CurM TE for the (R)-β-sulfate over the corresponding (S) isomer (9), as modeling of the (S)-β-sulfate form of the substrate resulted in sulfate contacts with the hydrophobic surface of the protein. The model also suggests that the hydrogen-bonded side chains of Arg205 and Asn211 may recognize the β-sulfate moiety. To explore these issues further, the functions of the presumed sulfate-recognition residues and others in the active site cleft were probed by site-directed mutagenesis. Three substitutions were made at Arg205 (Gln, Glu, and Ala), as well as Ala substitutions at Asn267, which is adjacent to the catalytic His, and at Asn211, which is hydrogen bonded with Arg205 (Fig. 5B).
SCHEME 1.
Predicted reaction sequence for CurM TE hydrolysis, sulfate group elimination, and decarboxylation. Arg205 recognizes the (R)-β-sulfate, facilitating hydrolysis by the catalytic triad. The catalytic His266 is positioned to assist in decarboxylation followed by sulfate elimination.
The TE wild type and variants were tested using a one-pot, multistep assay (9) in which recombinant CurM ACP was loaded with a synthetic substrate mimic (Fig. 1B) and reacted sequentially with recombinant CurM ST and CurM TE. ACP substrates and ACP hydrolysis products were analyzed by HPLC (supplemental Fig. S3). Activities of the CurM TE variants were normalized to the activity of the wild type (Table 2). The three substitutions at Arg205 (R205Q, R205E, and R205A) resulted in significantly reduced activity, comparable with a negative-control substitution in the catalytic triad (H266R). Substitutions at two Asn residues (N211A and N267A) that are conserved in the ACP-ST-TE group of sequences but not in the wider TE family resulted in only 2–3-fold reduced catalytic activity. To test the role of Arg205 in sulfate recognition, the Arg205 variants were assayed with a non-sulfated substrate. Due to the ∼800-fold lower activity of wild type TE on non-sulfated substrates than on sulfated substrates (9), these assays were performed with a CoA-linked β-OH substrate available at a higher concentration than the ACP-linked substrate. In striking contrast to their lack of activity with sulfated substrates, the Arg205 variants had activity comparable with the wild type and non-sulfated substrate (supplemental Table S1). This result supports a vital role for Arg205 in sulfate recognition.
TABLE 2.
Activity of CurM TE variants
| Activitya | |
|---|---|
| % | |
| WT | 100 |
| R205Q | 1.8 ± 0.3 |
| R205E | 2 ± 2 |
| R205A | 6 ± 3 |
| N267A | 34 ± 3 |
| N211A | 55 ± 2 |
| H266R | 1.4 ± 0.2 |
a Raw HPLC chromatogram peak area for the holo-ACP product with background subtracted and normalized with an internal standard to correct for differences in sample injection. The area for each mutant was normalized to the wild type within each replicate. Mean ± S.D. from duplicate experiments are shown.
DISCUSSION
A New Thioesterase Subfamily
The CurM TE structure defines a new thioesterase subfamily with the remarkable catalytic activity of hydrolysis, decarboxylation, and sulfate elimination to form alkene products from β-sulfated acyl-ACP substrates. The structural features that distinguish CurM TE from other thioesterase subfamilies are conserved in sequences of five other putative ACP-ST-TE tridomains, indicating that all of them catalyze terminal alkene formation. The CurM TE lid subdomain, which sits above the active site, is substantially longer than the analogous region in other TE subfamilies. The lid creates an open active site cleft, and is fixed in an open orientation by key interactions unique to this TE subfamily (Figs. 2C and 3B). One side of the lid is secured by the surface complementarity of lid helix αL3 with the core domain and by an arginine anchor in which Arg185 in the lid extends deep into the core domain to form a salt bridge with Asp57. The other side of the lid is secured by dimer contacts of lid helix αL1 with the core domain of the partner subunit.
The characteristics of the CurM TE lid provide a striking contrast to other PKS and NRPS TE subfamilies. Dimeric PKS off-loading TEs have a closed lid, formed by a lid-to-lid dimer contact, which creates an open-ended tunnel with the active site at its center (Fig. 3C). Monomeric editing TE II enzymes and NRPS off-loading TEs have a flexible lid that may control access of the phosphopantetheine arm to the active site by its ability to open and close. The region of substrate entrance in these other TEs (the active site tunnel in other dimeric TEs and the flexible part of the movable lid of monomeric TEs) is analogous to the region in CurM TE secured by the arginine anchor. Hence, the usual entrance route for acyl-ACP substrates is blocked in CurM TE by the anchoring of lid helix αL3 to the core domain. Instead, we presume that the substrate enters the CurM TE active site via the conserved cleft that extends away from the catalytic triad toward the top of the lid (Fig. 5A). In contrast to the firm anchoring on the entrance side of the CurM TE active site cleft, the opposite side of the cleft has flexible lid-to-core linkers and less conservation.
Substrate Specificity
Despite the departure from precedent in the lid architecture and substrate entrance, the catalytic triad of CurM TE is fully formed and appears to be poised for hydrolysis (Fig. 3A). The question remains why CurM TE does not hydrolyze any thioester substrate given the positions of the Ser100 nucleophile, His266 base, and Ile32/Met101 oxyanion hole. An answer is found in the wide open architecture of the active site, which provides few obvious substrate recognition elements. CurM TE does not need exquisite recognition of a substrate that is present at a highly effective concentration via tethering to CurM ACP, which itself is fused to the TE. However, CurM TE must select the (R)-β-sulfated substrate and not other acylated forms of CurM ACP (malonyl, β-keto, and (R)-β-hydroxy), which are also present at a highly effective concentration before they are transformed by the other catalytic domains of CurM (ketosynthase, ketoreductase, and sulfotransferase, respectively). Our model of the TE acyl-enzyme intermediate suggested that Arg205 recognizes the (R)-β-sulfate moiety (Fig. 5), and this was consistent with the site-directed mutagenesis data (Table 2, supplemental Table S1). The Arg205 variants lacked activity with a sulfated substrate but had activity comparable with the wild type in the much slower reaction with a non-sulfated substrate. Thus Arg205 may be the only positive substrate-recognition element, the active site may have little or no affinity for the rest of the acyl chain, and non-sulfated substrates may be only rarely positioned properly for catalysis. In this manner, the β-sulfate acts as a “handle” for Arg205 to assist in effective thioester binding to the catalytic machinery in the large open active site cleft, whereas non-sulfated substrates have no such guidance. The geometry of Arg205 with relation to the active site also ensures that (R)-β-sulfated substrates can be properly positioned for catalysis, whereas steric clashes would occur for the corresponding (S)-β-sulfated isomer. Intriguingly, Arg205 is among the flexible residues in the linker connecting the CurM TE core and lid, and thus may provide an early interaction with incoming sulfated substrates. The flexibility of this region is consistent with the lack of conservation of Arg205 in the homolog from Haliangium ochraceum, in which nearby arginine side chains may serve the same function.
Catalytic Mechanism
The CurM TE structure reveals an intact hydrolytic active site into which an acyl-enzyme intermediate was modeled easily without the need to move interacting groups (Fig. 5). This strongly suggests that acyl-enzyme formation precedes other catalytic events. Therefore, we propose that hydrolysis of the acyl-enzyme precedes decarboxylation and sulfate group elimination, based on our detection of both the terminal alkene product and the substrate analog bearing intact β-sulfate and carboxylic acid groups (i.e. the initial TE serine ester hydrolysis product) (9). The CurM TE assures decarboxylation and sulfate group elimination in vivo, as the corresponding carboxylated and sulfated curacin A intermediate was not detected in cultures of L. majuscula (Scheme 1) (37). Through interactions with the oxyanion hole and Arg205, the carboxylic acid hydrolysis product remains bound to CurM TE long enough for the enzyme to promote loss of CO2 and SO42- in one concerted step. In the reaction scheme, CurM TE assists decarboxylation and sulfate elimination in at least three ways. First, the enzyme binds the carboxylic acid hydrolysis product in a conformation that prevents resonance of COOH with the C1–C2 bond to be broken upon decarboxylation. Arg205, through its interaction with the sulfate at C3, may assist in achieving the optimal conformation. Second, the catalytic His266 likely promotes deprotonation of the carboxylic acid hydrolysis product to facilitate loss of CO2. Stabilization of the additional negative charge on the departing SO42- offered by Arg205 may further reduce energy barriers for initiating this remarkable decarboxylative elimination process.
Potential for Hydrocarbon Production
CurM TE is the first member of a branch of the TE family optimized to work in concert with a sulfotransferase to eliminate a carboxyl functional group and create a terminal double bond. The ST-TE system is the first example of a biological sulfonation employed for chemical activation (9). The ACP-ST-TE system exists as an uncharacterized open reading frame in genome sequences of five other bacteria, with a high level of conservation of residues within and surrounding the TE active site and in key contacts between domains and subunits. In CurM and in some ST-TE homologs, the sulfonation-decarboxylation system seems designed to eliminate the terminal carboxylate that would result from canonical TE hydrolysis of an acyl-ACP. Elimination of the terminal carboxyl group is a challenge for efforts to develop liquid biofuels. Thus the ST-TE system is of potential benefit in the development of a direct biosynthetic route to medium and long chain hydrocarbons from fatty acids. This could be achieved through engineering a β-hydroxy fatty acid biosynthetic pathway for off-loading using the ST-TE mechanism. The ST-TE route to hydrocarbons is biochemically unique compared with other recent approaches, including a cyanobacterial system using acyl-ACP reductase and aldehyde decarbonylase with fatty-acyl-ACP substrates (38) and an E. coli system with fatty acid biosynthesis engineered for hydrocarbon production (39). Other organisms found to produce hydrocarbons with terminal alkenes, such as Botryococcus braunii (40), probably employ a similar ST-TE off-loading strategy. Indeed, B. braunii is known to use very long chain fatty acids activated at the β position to produce terminal olefin dienes and trienes through a decarboxylative process (41).
Conclusion
The curacin TE illustrates the remarkable adaptability of the thioesterase structural framework, here adapted for only those acyl-ACP substrates bearing a sulfate substituent at the β position. The CurM TE is a key component of an unprecedented system for thioester hydrolysis and decarboxylation, working in concert with a fused sulfotransferase to generate a highly favorable leaving group. The CurM TE crystal structure reveals a fully developed hydrolytic active site, an arginine residue for (R)-β-sulfate recognition, and an active site cleft that may accommodate a wide variety of acyl substrates.
Acknowledgments
We thank Drs. Amol Kulkarni and Sarang Kulkarni for the synthesis of substrate analogs. Beamline 23ID-D is supported by the National Institutes of Health, National Institute of General Medical Sciences Grant Y1-GM-1104 and National Cancer Institute Grant Y1-CO-1020 through the GM/CA Collaborative Access Team at the APS, which is supported by the United States Department of Energy.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK42303 (to J. L. S.), R01 CA108874 (to D. H. S., W. H. G., and P. W.), and U01 TW007404, and a Hans W. Vahlteich Professorship (to D. H. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1.
The atomic coordinates and structure factors (code 3QIT) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PKS
- polyketide synthase
- NRPS
- non-ribosomal peptide synthetase
- TE
- thioesterase
- ACP
- acyl carrier protein
- ST
- sulfotransferase
- PAPS
- adenosine 3′-phosphate 5′-phosphosulfate
- PAP
- adenosine 3′-phosphate 5′-phosphate
- SeMet
- selenomethione.
REFERENCES
- 1. Newman D. J., Cragg G. M. (2007) J. Nat. Prod. 70, 461–477 [DOI] [PubMed] [Google Scholar]
- 2. Fortman J. L., Chhabra S., Mukhopadhyay A., Chou H., Lee T. S., Steen E., Keasling J. D. (2008) Trends Biotechnol. 26, 375–381 [DOI] [PubMed] [Google Scholar]
- 3. Verdier-Pinard P., Lai J. Y., Yoo H. D., Yu J., Marquez B., Nagle D. G., Nambu M., White J. D., Falck J. R., Gerwick W. H., Day B. W., Hamel E. (1998) Mol. Pharmacol. 53, 62–76 [DOI] [PubMed] [Google Scholar]
- 4. Chang Z., Sitachitta N., Rossi J. V., Roberts M. A., Flatt P. M., Jia J., Sherman D. H., Gerwick W. H. (2004) J. Nat. Prod. 67, 1356–1367 [DOI] [PubMed] [Google Scholar]
- 5. Gu L., Geders T. W., Wang B., Gerwick W. H., Håkansson K., Smith J. L., Sherman D. H. (2007) Science 318, 970–974 [DOI] [PubMed] [Google Scholar]
- 6. Geders T. W., Gu L., Mowers J. C., Liu H., Gerwick W. H., Håkansson K., Sherman D. H., Smith J. L. (2007) J. Biol. Chem. 282, 35954–35963 [DOI] [PubMed] [Google Scholar]
- 7. Akey D. L., Razelun J. R., Tehranisa J., Sherman D. H., Gerwick W. H., Smith J. L. (2010) Structure 18, 94–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gu L., Wang B., Kulkarni A., Geders T. W., Grindberg R. V., Gerwick L., Håkansson K., Wipf P., Smith J. L., Gerwick W. H., Sherman D. H. (2009) Nature 459, 731–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gu L., Wang B., Kulkarni A., Gehret J. J., Lloyd K. R., Gerwick L., Gerwick W. H., Wipf P., Håkansson K., Smith J. L., Sherman D. H. (2009) J. Am. Chem. Soc. 131, 16033–16035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chapman E., Best M. D., Hanson S. R., Wong C. H. (2004) Angew. Chem. Int. Ed. Engl. 43, 3526–3548 [DOI] [PubMed] [Google Scholar]
- 11. Roongsawang N., Washio K., Morikawa M. (2007) Chembiochem 8, 501–512 [DOI] [PubMed] [Google Scholar]
- 12. Giraldes J. W., Akey D. L., Kittendorf J. D., Sherman D. H., Smith J. L., Fecik R. A. (2006) Nat. Chem. Biol. 2, 531–536 [DOI] [PubMed] [Google Scholar]
- 13. Akey D. L., Kittendorf J. D., Giraldes J. W., Fecik R. A., Sherman D. H., Smith J. L. (2006) Nat. Chem. Biol. 2, 537–542 [DOI] [PubMed] [Google Scholar]
- 14. Tsai S. C., Miercke L. J., Krucinski J., Gokhale R., Chen J. C., Foster P. G., Cane D. E., Khosla C., Stroud R. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14808–14813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scaglione J. B., Akey D. L., Sullivan R., Kittendorf J. D., Rath C. M., Kim E. S., Smith J. L., Sherman D. H. (2010) Angew. Chem. Int. Ed. Engl. 49, 5726–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bruner S. D., Weber T., Kohli R. M., Schwarzer D., Marahiel M. A., Walsh C. T., Stubbs M. T. (2002) Structure 10, 301–310 [DOI] [PubMed] [Google Scholar]
- 17. Samel S. A., Wagner B., Marahiel M. A., Essen L. O. (2006) J. Mol. Biol. 359, 876–889 [DOI] [PubMed] [Google Scholar]
- 18. Koglin A., Löhr F., Bernhard F., Rogov V. V., Frueh D. P., Strieter E. R., Mofid M. R., Güntert P., Wagner G., Walsh C. T., Marahiel M. A., Dötsch V. (2008) Nature 454, 907–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frueh D. P., Arthanari H., Koglin A., Vosburg D. A., Bennett A. E., Walsh C. T., Wagner G. (2008) Nature 454, 903–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Claxton H. B., Akey D. L., Silver M. K., Admiraal S. J., Smith J. L. (2009) J. Biol. Chem. 284, 5021–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DelProposto J., Majmudar C. Y., Smith J. L., Brown W. C. (2009) Protein Expr. Purif. 63, 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fischetti R. F., Xu S., Yoder D. W., Becker M., Nagarajan V., Sanishvili R., Hilgart M. C., Stepanov S., Makarov O., Smith J. L. (2009) J. Synchrotron Radiat. 16, 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., Adams P. D. (2008) Methods Mol. Biol. 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 25. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 26. Terwilliger T. C. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Terwilliger T. C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30. Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 31. Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 32. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 33. Edgar R. C. (2004) BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 35. Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 36. Sánchez C., Du L., Edwards D. J., Toney M. D., Shen B. (2001) Chem. Biol. 8, 725–738 [DOI] [PubMed] [Google Scholar]
- 37. Gerwick W. H., Proteau P. J., Nagle D. G., Hamel E., Blokhin A., Slate D. L. (1994) J. Org. Chem. 59, 1243–1245 [Google Scholar]
- 38. Schirmer A., Rude M. A., Li X., Popova E., del Cardayre S. B. (2010) Science 329, 559–562 [DOI] [PubMed] [Google Scholar]
- 39. Steen E. J., Kang Y., Bokinsky G., Hu Z., Schirmer A., McClure A., Del Cardayre S. B., Keasling J. D. (2010) Nature 463, 559–562 [DOI] [PubMed] [Google Scholar]
- 40. Metzger P., Largeau C. (2005) Appl. Microbiol. Biotechnol. 66, 486–496 [DOI] [PubMed] [Google Scholar]
- 41. Chan Young T., Largeau C., Casadevall E. (1986) New J. Chem. 10, 701–707 [Google Scholar]
- 42. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]