Abstract
Mammalian mucin-type O-glycosylation is initiated by a large family of ∼20 UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferases (ppGalNAc Ts) that transfer α-GalNAc from UDP-GalNAc to Ser and Thr residues of polypeptide acceptors. Characterizing the peptide substrate specificity of each isoform is critical to understanding their properties, biological roles, and significance. Presently, only the specificities of ppGalNAc T1, T2, and T10 and the fly orthologues of T1 and T2 have been systematically characterized utilizing random peptide substrates. We now extend these studies to ppGalNAc T3, T5, and T12, transferases variously associated with human disease. Our results reveal several common features; the most striking is the similar pattern of enhancements for the three residues C-terminal to the site of glycosylation for those transferases that contain a common conserved Trp. In contrast, residues N-terminal to the site of glycosylation show a wide range of isoform-specific enhancements, with elevated preferences for Pro, Val, and Tyr being the most common at the −1 position. Further analysis reveals that the ratio of positive (Arg, Lys, and His) to negative (Asp and Glu) charged residue enhancements varied among transferases, thus further modulating substrate preference in an isoform-specific manner. By utilizing the obtained transferase-specific preferences, the glycosylation patterns of the ppGalNAc Ts against a series of peptide substrates could roughly be reproduced, demonstrating the potential for predicting isoform-specific glycosylation. We conclude that each ppGalNAc T isoform may be uniquely sensitive to peptide sequence and overall charge, which together dictates the substrate sites that will be glycosylated.
Keywords: Amino Acid, Carbohydrate Biosynthesis, Enzyme Structure, Glycoprotein Biosynthesis, Mathematical Modeling, Edman Sequencing, Amino Acid Preferences, Glycosyltransferase, Mucin-type O-Glycosylation, Random Peptide
Introduction
Mucin-type O-glycosylation is one of the most common post-translational modifications of secreted and membrane-associated proteins. Glycoproteins containing O-glycosylated mucin domains serve many important biological roles chiefly because of their unique biophysical and structural properties that include an extended peptide conformation and robust resistance to proteases. Consequently, glycoproteins containing O-glycosylated mucin domains function in the protection of the cell surface, the modulation of cell-cell interactions, in the inflammatory and immune response, in metastasis and tumorigenesis, and in protein sorting, targeting, and turnover (for examples see Refs. 1–5). It is also likely that such O-glycosylated domains may further present a molecular code for the specific recognition of additional binding partners, enzymes, or even other glycosyltransferases. In some instances, specific mucin-type O-glycosylation may modulate receptor activity (6, 7) and protein hormone processing (8); hence, mucin-type O-glycosylation may be sufficiently regulated to actively serve as a modulator of complex biological processes and even signaling. Recent studies clearly indicate the critical role of mucin-type O-glycosylation in vertebrate and nonvertebrate development (2, 4, 9–13).
Mucin-type protein O-glycosylation is initiated in the Golgi compartment by the transfer of α-GalNAc, from UDP-GalNAc,5 to Ser and Thr residues of polypeptide acceptors by the large family (∼20) of UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferases (ppGalNAc Ts). By subsequent action of a series of specific glycosyltransferases, the O-linked glycan can be further elongated to produce a vast array of glycan structures (14). Unlike the N-glycosylation of Asn residues and the O-xylosylation of Ser residues of proteoglycans, there are no highly specific sequence motifs (i.e. Asn-Xaa (not Pro)-(Ser/Thr) and acidic-acidic-Xaa-Ser-Gly-Xaa-Gly, respectively (15, 16)) that enable the facile prediction or recognition of sites of mucin-type O-glycosylation based on peptide sequence. Nevertheless, data base analysis of known mucin-type O-glycosylation sites have resulted in a number of algorithms (17–19)6 for the approximate prediction of mucin-type O-glycosylation. None of these approaches, however, readily account for the wide range and remarkable reproducibility of the O-glycan site-to-site occupancy observed in the mucins that have been characterized to date (20, 21). Importantly, the predictive approaches do not take into account the different peptide substrate specificities of the various ppGalNAc T isoforms.
Structurally the ppGalNAc Ts consist of an N-terminal catalytic domain tethered by a short linker to a C-terminal ricin-like lectin domain containing three recognizable carbohydrate-binding sites (22). Some members of the ppGalNAc T family prefer substrates that have been previously modified with O-linked GalNAc on nearby Ser/Thr residues, hence having so-called glycopeptide or filling-in activities, i.e. ppGalNAc T7 and T10 (23–25). Others simply possess altered preferences against glycopeptide substrates, i.e. ppGalNAc T2 and T4 (26–29), or may be inhibited by neighboring glycosylation, i.e. ppGalNAc T1 and T2 (20, 21, 25). These latter transferases have been called early or initiating transferases, preferring nonglycosylated over glycosylated substrates. The roles of the catalytic and lectin domains on modulating ppGalNAc T peptide and glycopeptide specificity are not fully understood. It has been shown that the presence of the lectin domain of ppGalNAc T2 significantly shifts the preferred sites of glycosylation on glycopeptide substrates (23, 24, 28, 29), although other studies have demonstrated that the catalytic domain of ppGalNAc T10 is responsible for its near absolute glycopeptide specificity (29, 30). Clearly, detailed studies of the catalytic and lectin domain specificities of these transferases are necessary to fully understand their properties.
Several ppGalNAc T isoforms have been shown to be necessary for, or associated with, normal development, cellular processes, or specific disease states, presumably by possessing specific protein targets that other coexpressed ppGalNAc T isoforms fail to recognize.7 For example, inactive mutations in the fly PGANT35A (the ppGalNAc T11 orthologue in mammals) are lethal (31–33), although mutations in PGANT3 (with no close mammalian homologue) result in wing blistering (34). Both of these transferases play significant roles modulating specific cell-cell interactions in the developing fly (12, 35). In humans, mutations in ppGalNAc T3 cause a form of familial tumoral calcinosis, due to abnormal cleavage and secretion of the phosphaturic factor FGF23 (8, 36, 37). Human ppGalNAc T14 may modulate apoptotic signaling in tumor cells by glycosylating the proapoptotic receptors DLR4 and DLR5 (7), although the specific O-glycosylation of the TGFB-II receptor (ActR-II) by GALNTL1 (ppGalNAc T16) modulates its signaling in Xenopus and mammalian development (6). Specific ppGalNAc Ts have also been linked to Williams-Beuren syndrome (WBSCR17, pt-GalNAc-T, or GALNTL3) (38, 39) and hereditary multiple exostoses (ppGalNAc T5) (40). Genome-wide sequencing studies have also revealed biochemically inactivating germ lines and somatic mutations in GALNT5 and GALNT12 (ppGalNAc T5 and T12) in individuals with breast and colon cancers (41, 42) consistent with previous studies (43). Other genome-wide association scans suggest that GALNT2 (ppGalNAc T2) variants may be associated with levels of HDL cholesterol and coronary artery disease (44–46). Obviously, there is a need for characterizing the peptide substrate specificity of each isoform to further elucidate their specific targets and mechanism of action. This information is critical for our understanding of the biological roles and significance of the ppGalNAc T family of transferases and mucin-type O-glycosylation in general.
Our laboratory has recently reported the use of a series of oriented random peptide and glycopeptide substrate libraries for quantitatively determining the amino acid residue preferences of the catalytic domains of ppGalNAc T1, T2, and T10 and the fly orthologues of T1 and T2 (30, 47, 48). In this study, we extend our studies to three additional members of the family, ppGalNAc T3, T5, and T12, with potential roles in human disease, utilizing two previously reported random peptide substrates and an additional new substrate capable of obtaining preferences for neighboring nonglycosylated Ser residues (Table 1). With these substrates, unique substrate preference data for all amino acid residues except Thr, Trp, and Cys have now been obtained for the following six mammalian ppGalNAc Ts: T1, T2, T3, T5, T10, and T12. Our findings have revealed both common and unique features among the transferases characterized to date. The most striking was the very similar pattern of enhancements for those residues C-terminal to the site of glycosylation, particularly enhancements at +1 and +3 for Pro, found in all the ppGalNAc Ts characterized except the glycopeptide preferring ppGalNAc T10 (24, 25). A structural analysis suggests these enhancements arise from interactions with a common conserved Trp residue found in these transferases. In contrast, residues N-terminal to the site of glycosylation show a range of enhancements that are isoform-specific, with elevated preferences for Pro, Val, Ile, and Tyr being the most common. Further analysis revealed that the ratio of positive (Arg, Lys, and His) to negative (Asp and Glu) charged residue preferences varied among transferases with ppGalNAc T1 and T2 preferring the most acidic substrates, with ppGalNAc T5 and T3 preferring the most basic. We also show that these observations are consistent with the homology modeled structures of the ppGalNAc Ts. Thus, the peptide sequence and overall charge serve to modulate the peptide substrate specificity of each ppGalNAc T. Coupled with the variable sensitivity of each isoform to prior substrate glycosylation (20, 21, 25–29), a wide range of unique and specific substrate preferences is achieved across the ppGalNAc T family of transferases. We further demonstrate that for several of the transferases, these preferences can be used to predict isoform-specific glycosylation patterns consistent with previously reported experimental data.
TABLE 1.
ppGalNAc transferase random substrates utilized in this work
P-VI, P-VII, and P-VIII are random peptide substrates.
EXPERIMENTAL PROCEDURES
Transferases
Soluble recombinant bovine ppGalNAc T1 was a gift of Ake Elhammer (Kalamazoo, MI). The soluble bovine ppGalNAc T1, human ppGalNAc T2, and human ppGalNAc T10 used in this work have been characterized previously (30, 47, 48). The expression and use of the N-terminal affinity tag immobilized ppGalNAc T5 and T12 have also been described previously (42). The cloning, expression, and stem sequence modification of h-ppGalNAc T3 from Pichia pastoris is described in the supplemental material. h-ppGalNAc T3 was utilized either as a media supernatant or after affinity purification.
Random Peptide Substrates, Additional Reagents, and Procedures
Random peptide substrates P-VI P-VII and P-VIII (Table 1) were custom-synthesized by QCB Inc. (Hopkinton, MA) or AnaSpec Inc. (San Jose CA). All peptide substrates were pH-adjusted to ∼7.5 with dilute NaOH and/or HCl and lyophilized multiple times from water prior to use. Fully N-acetylated UDP-[3H]GalNAc was purchased from ARC Inc. (St. Louis). GalNAc binding-immobilized lectins SJA (Sophoro japonica), SBA (Glycine max), and HPA (Helix pomatia) were obtained from EY Laboratories (San Mateo, CA); immobilized VVA (Vicia villosa) was acquired from Vector Laboratories (Burlingame, CA). Sephadex G-10 and Dowex (1-X8) ion exchange resin (100–200 mesh) were purchased from Amersham Biosciences and Acros Organics, respectively. Liquid scintillation counting was performed on a Beckman model LS5801 or LS6500. Edman amino acid sequencing was performed on an Applied Biosystems Procise 494 peptide sequencer as described previously (30, 47, 48).
Glycosylation of Random Peptide Substrates
The glycosylation of random peptide P-VIII with soluble transferases T1, T2, and T10 was performed as described previously for the glycosylation of random peptides P-VI and P-VII (10 mg/ml peptide, 1–2 mm UDP-GalNAc (3H-labeled, ∼0.01 mCi/ml), 100 mm HEPES, pH 7.5, 10 mm MnCl2, 1 mm EDTA, protease inhibitor mixtures (Sigma) P8340 and P8849 (1:100 dilution), and 16–240 μg/ml transferase, incubated overnight at 37 °C) (30, 47, 48).
ppGalNAc T3 reactions, performed at 32 °C, consisted of 10 mg/ml random peptide (P-VI, P-VII, or P-VIII), 50 mm sodium cacodylate, pH 6.5, 0.1% Triton X-100, 0.3–1.5 mm 2-mercaptoethanol, 10 mm MnCl2, 0.5–2 mm UDP-GalNAc (3H-labeled, ∼0.01 mCi/ml), protease inhibitor mixtures P8340 and P8849 (1:100 dilution), 0.3% sodium azide, and 50 μl of media supernatant from Pichia expressing WT-h-ppGalNAc T3 or ∼1–10 μg/ml affinity-purified furin cleavage site-eliminated ppGalNAc T3, in a final volume of 250 μl. Residue enhancement values obtained from the WT-h-ppGalNAc T3 (media) were indistinguishable from the affinity-purified furin site-eliminated h-ppGalNAc T3 and were therefore combined.
Incubations with ppGalNAc T5 were performed as described for ppGalNAc T3 except 50–100 μl of immobilized transferase was added to the reaction buffers that were shaken overnight on a thermostated microplate shaker (Taitec Microincubator M-36) at 37 °C. Incubations for ppGalNAc T12 were performed as described for ppGalNAc T5 except 25 mm Tris, pH 7.4, was used as buffer.
For all transferases, overnight reaction incubations were quenched with EDTA and passed through Dowex 1-X8 to remove unreacted UDP-GalNAc. Random peptide glycosylation was typically 3% or less based on radiolabel incorporation. Isolation of the glycosylated peptide was performed via mixed bed lectin column chromatography as described previously by Gerken et al. (47, 48). GalNAc-eluted fractions were pooled based on 3H content, lyophilized, and rechromatographed on Sephadex G-10. After lyophilization several times from distilled water, random glycopeptides were Edman-sequenced and the integrated peak areas processed as described previously (47). Amino acid residue enhancement factors were obtained at each randomized residue position by comparing the glycopeptide mole fractions to the peptide mole fractions obtained prior to the lectin column as performed previously (47, 48). At least two determinations were obtained for each peptide resulting in a minimum of six determinations obtained for each transferase.
Enhancement Product Values
For predictive purposes, transferase-specific enhancement values flanking (positions −3 to +3) a potential site of Thr or Ser glycosylations were multiplied together to obtain a so-called enhancement factor product. Calculations were performed manually utilizing an Excel spreadsheet containing the full matrix of random peptide-derived enhancement values. For Cys, Thr, Trp, and the missing N- or C-terminal residues, a value of 1 was utilized as the enhancement value.8 Enhancement values for T-synthase were obtained previously (49).
Site-specific Glycosylation Determination
Peptides EA2 (PTTDSTTPAPTTK) and MUC5AC (GTTPSPVPTTATTSA) were glycosylated by ppGalNAc T5 and T12, respectively, utilizing [3H]UDP-GalNAc as described previously (42). Glycopeptide products after isolation on Sephadex G-10 were Edman-sequenced, and the radiolabeled content of each cycle was determined by liquid scintillation counting as described previously (30).
Isoelectric Point Calculations
Isoelectric point calculations were performed using the EMBOSS iep program available on line and written by Alan Bleasby, European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK.
Homology Modeling and Electrostatic Surface Charge Calculations
To identify conserved sequence elements of the known ppGalNAc T sequences, comparative alignments were produced using T-Coffee (50). To date, only ppGalNAc T1 (murine), T2 (human), and T10 (human) x-ray crystal structures have been solved (22, 51, 52). To identify conserved functional regions of the known ppGalNAc Ts whose crystal structures are not know, protein structure homology models were created using an automated comparative protein modeling server, the Swiss-Model Swiss Institute of Bioinformatics Service (53, 54). Target template alignments were created as input for Swiss-Model Alignment Mode in DeepView Swiss-PDB Viewer (version 4.0) (55) using the human ppGalNAc T2 2FFU template crystal structure. Visualization of homology models was performed in Visual Molecular Dynamics version 1.8.7 molecular graphics software (56).
To evaluate the comparative electrostatic potential surfaces within the ppGalNAc T family, electrostatic surface calculations were performed on the homology model structures using the Adaptive Poisson-Boltzmann Solver web service (57). Adaptive Poisson-Boltzmann Solver input files were created using the on-line Protein Data Bank 2PQR service (58, 59). The Poisson-Boltzmann equations were solved at pH 7 within the Adaptive Poisson-Boltzmann Solver, while enabling the PROPKA (60) pKa predictions. Visualization of the electrostatic surface computation results was performed in Visual Molecular Dynamics overlapping the ABPS results onto molecular surface models created using MSMS (61).
RESULTS
Determination of Ser-OH ppGalNAc T Preferences
We have previously used random peptide substrates P-VI and P-VII (Table 1) for the characterization of the specificity of ppGalNAc T1, T2, and T10 yielding preferences for all amino acid residues except Ser, Thr, Trp, and Cys for these transferases (30, 48). In an attempt to obtain preferences for the hydroxyl amino acid residue Ser, which in addition to Thr is common to heavily O-glycosylated domains, we designed random peptide P-VIII, which contains free Ser residues in the randomized X regions (Table 1). We reasoned that because Ser residues are typically at least an order of magnitude less rapidly glycosylated by the ppGalNAc Ts characterized to date (17), a substrate containing randomized Ser residues could be utilized to obtain Ser preferences under the appropriate conditions of limited glycosylation. Studies utilizing P-VIII with ppGalNAc T1, T2, and T10 proved successful yielding random glycopeptides showing GalNAc glycosylation at the central Thr residue (at residue 10) and no detectable Ser-O-GalNAc glycosylation (30, 62) in the random X regions based on the Edman sequencing of the lectin-isolated glycopeptide product (data not shown). The remaining amino acid preferences (i.e. Gly, Ala, Pro, Val, Tyr, Glu, Asn, Arg, and Lys) obtained from P-VIII were also consistent with those previously obtained from P-VI and P-VII for these transferases (data not shown) (30, 48). Together, these findings confirm that random peptide P-VIII can be used to successfully provide enhancement factors for unglycosylated Ser residues.
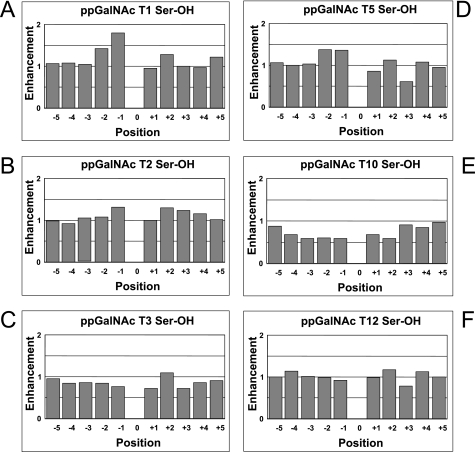
Plots of the Ser enhancement factors for ppGalNAc T1, T2, T3, T5, T10, and T12 obtained from random peptide P-VIII are shown in Fig. 1. Interestingly the enhancement factors for all of the transferases except ppGalNAc T10 range near 1, suggesting that Ser residues are neither greatly favored nor disfavored by these transferases. (Note that enhancement values of less than 1 indicate decreased preference, while values of greater than 1 indicate increased preference for the given residue by the transferase.) Only ppGalNAc T1 displays significantly elevated Ser preference values of 1.4 and 1.8 at the −2 and −1 sites (N-terminal of the site of glycosylation), respectively. Interestingly, at all but the flanking positions, the Ser enhancement values for the glycopeptide preferring ppGalNAc T10 are significantly less than 1. Thus, despite the high prevalence of Ser residues in heavily O-glycosylated mucin-type domains (typically ∼10–30%), the ppGalNAc Ts studied to date have not evolved highly elevated neighboring residue preferences for these residues.
FIGURE 1.
Random peptide-derived ppGalNAc T serine residue enhancement factors. Serine enhancement factors for ppGalNAc T1 (panel A), T2 (panel B), T3 (panel C), T5 (panel D), T10 (panel E), and T12 (panel F) are relative to the site of glycosylation (position 0). Negative positions are N-terminal, although positive positions are C-terminal to the site of glycosylation. Values represent the average of two or more determinations on random peptide P-VIII (Table 1) as described under “Experimental Procedures.” Enhancement values of less than 1 indicate decreased preference, although values greater than 1 indicate increased preference for the given residue by the transferase.
Hydrophobic and Hydrophilic Enhancement Values for ppGalNAc T3, T5, and T12
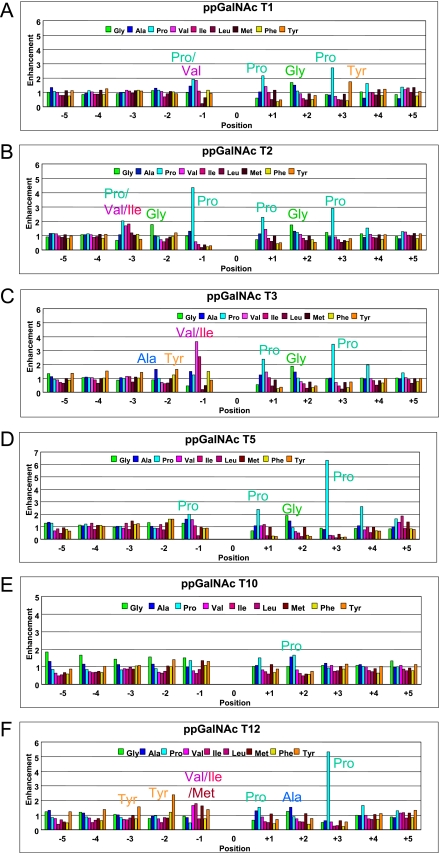
ppGalNAc T3, T5, and T12 were further characterized against random peptides P-VI and P-VII, and the preference data were combined with that from P-VIII as given in Figs. 2 and 3 (panels C, D, and F) for the hydrophobic (plus Gly) and hydrophilic residues, respectively. For comparison, the corresponding enhancement values for ppGalNAc T1, T2, and T10 (incorporating the newly obtained P-VIII data) are also given in each figure. As was observed for ppGalNAc T1 and T2 (but not ppGalNAc T10 (30)), the transferases display both common and unique features and appear to be most sensitive to substrate positions −3 to +3 relative to the site of glycosylation (position 0). The most obvious similarity among transferases is the nearly identical patterns of hydrophobic residue enhancements for residues +1 to +3 observed in all the ppGalNAc Ts except the glycopeptide-preferring ppGalNAc T10 (Fig. 2). The similarity of the hydrophobic residue patterns at the +1 and +2 positions is particularly striking for the five transferases. These transferases all possess enhanced Pro preferences (∼1.5–2.5-fold) at the +1 position and Gly or Ala enhancements (∼1.5-fold) at the +2 position. Large variable Pro preferences are also found at the +3 position, with ppGalNAc T5 and T12 having the largest enhancements (5–6-fold). Interestingly, ppGalNAc T1 is unique in having an additional nearly 2-fold Tyr enhancement at +3. The x-ray crystal structure of ppGalNAc T2 bound the EA2 peptide (22), and our molecular docking studies9 show that the Pro residues in the ∼TPAP∼ substrate sequence clasp a conserved Trp residue (Trp-282 in h-ppGalNAc T2) that is found in the peptide binding cleft of most ppGalNAc Ts, which is absent in ppGalNAc T10 (i.e. Arg-373) (see supplemental Fig. S1). We suggest that this Trp residue along with two additional Phe residues (ppGalNAc T2 Phe-280 and Phe-361), also common to the five ppGalNAc Ts (see supplemental Fig. S1), may represent a structural motif predictive for the preference of substrates with a C-terminal ∼TP(G/A)P∼ sequence.
FIGURE 2.
Random peptide-derived hydrophobic residue enhancement factors. Hydrophobic amino acid residues, including Gly, enhancement factors for ppGalNAc T1 (panel A), T2 (panel B), T3 (panel C), T5 (panel D), T10 (panel E), and T12 (panel F) were obtained from random peptides P-VI, P-VII, and P-VIII (Table 1). Note that factors for ppGalNAc T1, T2 (panel B), and T10 include previously reported random peptide data for P-VI and P-VII (30, 47, 48). Key common and unique amino acid residues are labeled.
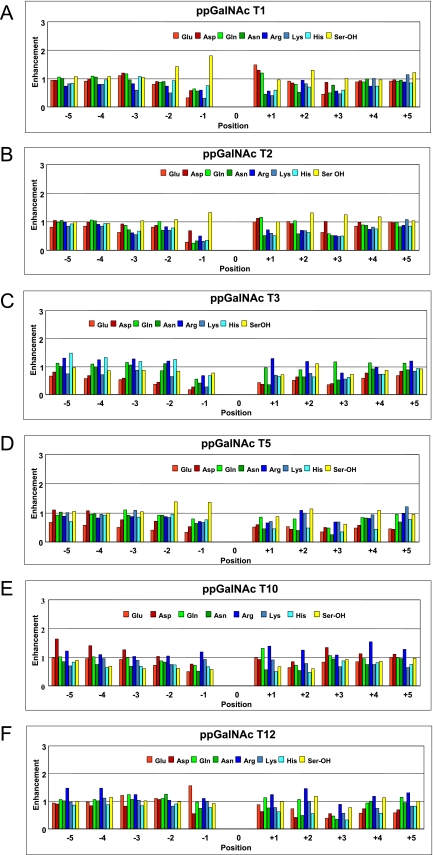
FIGURE 3.
Random peptide-derived hydrophilic residue enhancement factors. Hydrophilic amino acid residue enhancement factors for ppGalNAc T1 (panel A), T2 (panel B), T3 (panel C), T5 (panel D), T10 (panel E), and T12 (panel F) were obtained from random peptides P-VI, P-VII, and P-VIII (Table 1). Note that factors for ppGalNAc T1, T2 (panel B), and T10 include previously reported random peptide data for P-VI and P-VII (30, 47, 48).
The C-terminal hydrophilic residue substrate enhancements (Fig. 3) are relatively unremarkable, showing few obvious common patterns, with values typically clustering around 1. However, on closer examination, it is clear that the charged residue enhancements vary between transferases, with ppGalNAc T1 and T2 having larger acidic residue (Glu and Asp) enhancements and ppGalNAc T3 and T5 having larger basic residue (Arg, Lys, and His) enhancements. The role of charge residues will be discussed below.
In contrast to the C-terminal preferences, the N-terminal hydrophobic residue preferences show a range of isoform-specific enhancements, particularly at the −1 position, typically displaying elevated preferences for Pro, Val, Ile, or Tyr (Fig. 2). Thus, ppGalNac T1 and T5 show very similar N-terminal enhancements, displaying nearly equal Pro and Val enhancements (∼1.5–2-fold) at the −1 position, although ppGalNAc T3 and T12 exhibit elevated Val and Ile enhancements (∼1.5–3.5-fold) and weak to neutral Pro enhancements at this position. In addition, ppGalNAc T12 displays unique Tyr enhancements (∼1.5–2.5-fold) at positions −3, −2, and −1 along with an elevated Met enhancement (∼1.5-fold) at position −1. ppGalNAc T2 is the only transferase thus far characterized to possess a highly elevated (∼4.5-fold) Pro enhancement at the −1 position thus dominating its specificity. It is interesting to note that the least preferred residue at the −1 position, for nearly all of the transferases, is Leu, although Val, Ile, and Met are typically highly favored or neutral. Molecular docking experiments on ppGalNAc T2 tend to confirm this trend.9 At the −2 position, the enhancements are relatively neutral, although several transferases display elevated Tyr (and Phe) preferences, and ppGalNAc T2 has an elevated Gly enhancement. Again, the preferences at the −3 position are relatively neutral, except for ppGalNAc T2 having elevated (∼2-fold) enhancements for Pro, Val, and Ile. As with the C-terminal preferences, the N-terminal uncharged hydrophilic preferences are typically neutral, although the charged residue preferences vary between transferase isoforms (Fig. 3).
Correlation of Charged Residue Enhancement Values with Transferase Isoelectric Point and Electrostatic Surface Charge
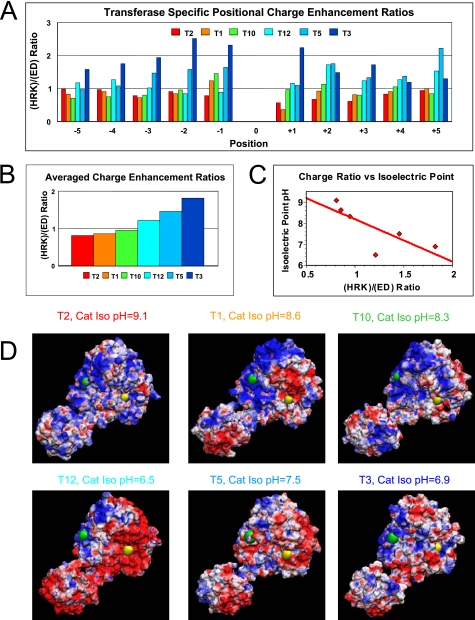
To further assess the role of charged residues in modulating substrate glycosylation, the averages of the basic residue (His, Arg, and Lys) enhancement values were compared with the average of the acidic residue (Asp and Glu) enhancements by obtaining the (HRK)/(ED) ratio. These transferase-specific charge ratios are plotted relative to substrate position in Fig. 4, panel A, and as an overall average in Fig. 4, panel B. The plots clearly show that at most positions ppGalNAc T2 and T1 prefer acidic residues (ratios less than 1), although ppGalNAc T12, T5, and T3 prefer basic residues (ratios greater than 1). This suggests that the overall peptide-binding surface of the transferases may possess a net positive or negative charge, respectively, that would serve to modulate peptide substrate specificity. To confirm that our derived charge preferences relate to the primary sequence of the transferase, we calculated the isoelectric point for the catalytic domain of each transferase and plotted these values versus the average charge enhancement ratio. As shown in Fig. 4, panel C, the plot gave a linear inverse correlation, with only one outlier (ppGalNAc T12) thus confirming that the obtained charge ratios reflect an actual property of the transferase. To further confirm this trend, transferases were homology modeled against the ppGalNAc T2 structure (22), and their surface electrostatic potentials were calculated (see “Experimental Procedures”). The results of these calculations are displayed in Fig. 4, panel D, where the homologous N- and C-terminal residues of the proposed peptide substrate-binding site were labeled as green and yellow spheres, respectively. Based on their charge residue enhancement ratios, ppGalNAc T2 and T1 would be expected to display the most positive or basic surface charge (Fig. 4, panel D, blue), although ppGalNAc T5 and T3 would be expected to display the most negative or acidic (red) surface. Indeed, this general trend is observed in the figures, except again for ppGalNAc T12 whose surface is significantly more acidic than that of ppGalNAc T5 or T3. We conclude that indeed the transferase electrostatic charge plays a role in modulating substrate specificity. The discrepancy in the ranking of ppGalNAc T12 may be partly due to the presence of a basic Lys residue directly above the substrate binding cleft that is not present in ppGalNAc T5 and T3 (see supplemental Fig. S1) (22), in addition to different reaction buffers and pH used for ppGalNAc T12 (25 mm Tris, pH 7.4) compared with ppGalNAc T3 and T5 (50 mm sodium cacodylate, pH 6.5).
FIGURE 4.
Transferase-specific charged residue enhancement ratios ((HRK)/(ED) ratio) roughly correlate with calculated isoelectric point and electrostatic surface charge. Panels A and B, plots of experimental charged residue enhancement ratios for each transferase as a function of position (panel A) or averaged over all positions (panel B). Panel C, plot of charged residue enhancement value versus catalytic domain isoelectric point. Line represents least squares fit to the data. Panel D, homology-modeled structures of the ppGalNAc Ts colored by surface electrostatic charge (red, negative charge; blue, positive charge). The catalytic domain is located at the upper right, and the lectin domain is at the lower left of each panel. The approximate catalytic domain peptide substrate-binding cleft is marked by green and yellow spheres representing an N- to C-terminal orientation based on the substrate peptide-bound x-ray structure of ppGalNAc T2 (22). See under “Experimental Procedures” for details.
Enhancement Factor Products
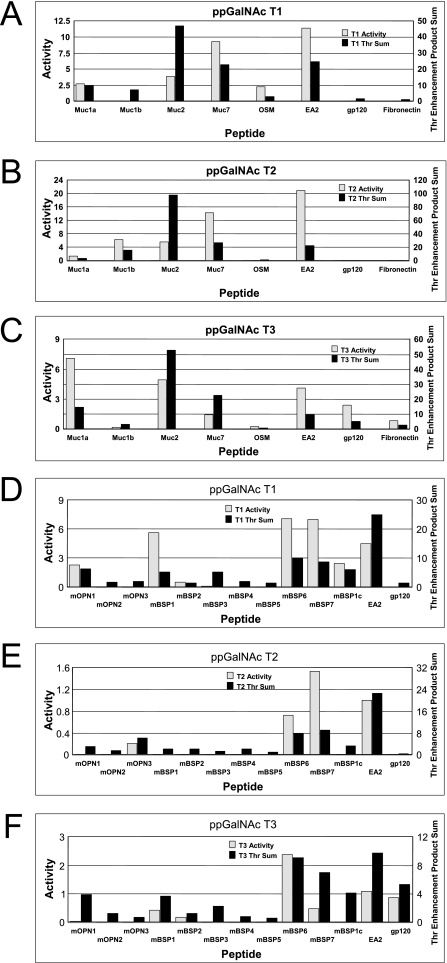
We have previously suggested that the product of the enhancement factors flanking a potential site of glycosylation (positions −3 to +3) can roughly predict glycosylation sites and/or relative rates of glycosylation for ppGalNAc T1 and T2 for a number of selected peptides substrates (47, 63). We would like to further extend this analysis to include ppGalNAc T3, T5, and T1210 while presenting additional examples for ppGalNAc T1 and T2 that further demonstrate the modulating effect of peptide charge. There are several reports comparing the activity of ppGalNAc T3 and other transferases (usually ppGalNAc T1 and T2) against a range of substrates (8, 63–65) but only one report each for ppGalNAc T5 and T12 (66, 67). Unfortunately, in most of the studies the actual site(s) of glycosylation have typically not been determined, and hence, we can only compare the reported relative activities to our enhancement factor products. We will initially focus on the work of Bennett et al. (65) where the relative activities of a large series of peptide substrates for ppGalNAc T1, T2, and T3 have been reported. In Fig. 5, panels A–C, we compare the reported experimental activities (gray bars) for each peptide with the sum of the Thr enhancement product values calculated for each Thr in the peptide (black bars). We chose to utilize only the Thr residue enhancement products in our glycosylation predictions/rankings because Ser residues are typically an order of magnitude less reactive than Thr residues (17) and therefore would be expected to be minor contributors to the experimentally observed rate. The individual preference products from which the data in Fig. 5, panels A–C was generated are given in supplemental Table S1. As shown in supplemental Table S1 and in Fig. 5, panels A–C (compare data for each peptide across panels A–C), the sum of the Thr enhancement factor products (i.e. sum of product for all Thr residues in the peptide) for ppGalNAc T1, T2, and T3 indeed identify the experimentally determined optimal transferase for seven of the eight peptides, although the maximum individual Thr enhancement factor product identifies six of the eight peptides (supplemental Table S1). Furthermore, as shown in Fig. 5, panels A–C, the enhancement products in nearly all cases can distinguish active from inactive peptides for a given transferase, although the enhancement product value rankings may not fully correlate with the experimental activity. It is important to note that comparing the experimental activities of the different transferase isoforms is not straightforward as the activity of each is based on a different substrate. Likewise, the intrinsic activities of different isoforms are likely to differ, and thus the same enhancement product for different transferases will not necessarily translate to identical rates of glycosylation. Regardless, two of the three ppGalNAc T1 inactive, or low activity, peptides can be identified (gp120 and fibronectin), although all of the low activity peptides for ppGalNAc T2 and T3 can be identified (i.e. OSM, gp120, and fibronectin for T2 and Muc1b and OSM for T3) based on their enhancement products. Importantly, Fig. 5, panels A–C, also shows that the transferase-selective peptides, i.e. OSM for ppGalNAc T1 and gp120 and fibronectin for ppGalNAc T3, can be correctly predicted based on the Thr enhancement product sums (or by the maximum Thr residue product).
FIGURE 5.
Transferase-specific threonine enhancement products roughly correlate with experimental transferase activity. Panels A–C, plots of glycosyltransferase activity (gray) and transferase-specific Thr enhancement product sums (black) for the peptides Muc1a, Muc1b, Muc2, Muc7, OSM, EA2, gp120, and fibronectin whose activity against ppGalNAc T1 (panel A), ppGalNAc T2 (panel B), and ppGalNAc T3 (panel C) was taken from Bennett et al. (65). Peptide sequences for panels A–C are defined in supplemental Table 1. Panels D–F, plots of glycosyltransferase activity (gray) and transferase-specific Thr enhancement product sums (black) for the peptides mOPN1 to mOPN3, mBSP1 to mBSP7, EA2, and gp120 whose activity against ppGalNAc T1 (panel D), ppGalNAc T2 (panel E), and ppGalNAc T3 (panel F) was taken from Miwa et al. (63). Peptide sequences for panels D–F are defined in supplemental Table 2. Enhancement value products were obtained as described under “Experimental Procedures.”
A similar analysis has been performed on a series of murine osteopontin and bone sialoprotein model peptides recently characterized against ppGalNAc T1, T2, and T3 (63). In this work, the initial rates of glycosylation for all three transferases were obtained and the sites of glycosylation determined for ppGalNAc T1 and T2 (63). These data and our enhancement products are given in supplemental Table S2 and are summarized in Fig. 5, panels D–F. As shown in supplemental Table S2, the sites of glycosylation and optimal transferase are typically correctly predicted by the enhancement products. However, a few peptides in Fig. 5, panels D–F, were predicted to be glycosylated that are not (i.e. mBSP3 for T1 and mOPN1, mBSP3, and mBSP1c for T3). Nevertheless, the enhancement value products predicted mOPN3 as ppGalNAc T2-specific, that mBSP1 would be glycosylated by both ppGalNAc T1 and T3, and that mBSP6, mBSP7, and EA2 would be glycosylated by all three transferases. The discrepancy between our predictions and the experimentally determined activities may arise from several sources. One is the end effects present in the short peptides,11 and another is the conformational effects that may alter access of a peptide to the transferase catalytic domain. Additional confounding effects due to multiple glycosylation sites may further alter the glycosylation of these substrates that are not presently taken into account.
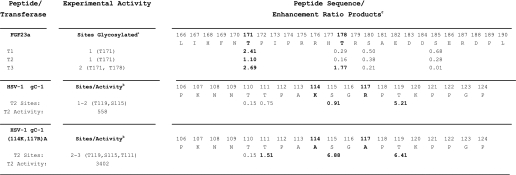
The loss of an active ppGalNAc T3 results in an improperly processed and secreted phosphaturic factor FGF23 due to the absence of O-glycosylation at Thr-178 (8). Kato et al. (8) have demonstrated, utilizing synthetic peptide substrates, that the glycosylation of Thr-178 in FGF23 is performed by ppGalNAc T3 but not by ppGalNAc T1 or T2. Furthermore, all three transferases will glycosylate a neighboring site, Thr-171 (8). As shown in Table 2, the enhancement ratio products for these two sites in FGF23 for ppGalNAc T1, T2, and T3 indeed predict the experimentally observed behavior. All three transferases have product preferences greater than 1 for Thr-171 suggesting all three could glycosylate this site. However, for the Thr-178 site, ppGalNAc T1 and T2 have product preferences significantly less than 1 (0.29 and 0.16, respectively) suggesting these transferases would not readily glycosylate this site, although ppGalNAc T3 gave a preference of ∼1.8 suggesting this transferase would indeed be capable of glycosylating this site. It is interesting to note that the enhanced preference for glycosylation by ppGalNAc T3 is principally due to the elevated basic charges flanking this site and reflecting the charge ratio preferences shown in Fig. 4 for this transferase, compared with ppGalNAc T1 or T2. Further experimental evidence of the significant role that basic residues play in reducing the activity of ppGalNAc T2 can be found in the studies of a wild type and mutant herpes simplex virus type 1 glycoprotein gC peptide (68). As shown in Table 2, the WT HSV-1 gC-1 peptide is glycosylated in vitro by ppGalNAc T2 in the order of Thr-119 and Ser-115, for which preference products of 5.2 and 0.9 are obtained, respectively. In the case of a mutant HSV-1 gC-1(114K, 117R)A peptide, where the basic residues Lys-114 and Arg-117 are replaced by Ala, up to three sites are glycosylated by ppGalNAc T2, Thr-119, Ser-115, and Thr-111, again consistent with the preference products of 6.4, 6.9, and 1.5 respectively. Furthermore, the mutant peptide shows an ∼6-fold increase in activity (incorporation) compared with the WT peptide (Table 2). Neighboring acidic charge residues have previously been shown to alter O-glycosylation in in vivo assays, although the nature of the participating transferase isoforms in these studies had not been fully characterized (69, 70).
TABLE 2.
Random peptide enhancement value products predict experimental transferase-specific sites of glycosylation and confirm the role of charged residues
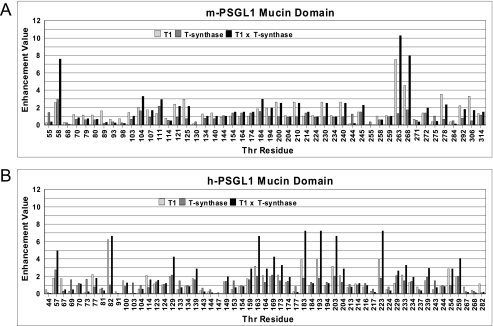
The leukocyte P-selectin glycoprotein ligand-1 (PSGL1) binds P-selectin on endothelial cells via cell surface O-glycosylated mucin domain of PSGL1 (71). In addition to specific tyrosine sulfation, the binding of PSGL1 to its receptor requires the presence of an extended Core 2-O-sLex O-linked glycan5 at a specific N-terminal Thr residue of PSGL1 (Thr-57 in human or Thr-58 in murine PSGL1 (72, 73)). The targeted biosynthesis of this O-glycan structure at this site is not understood; however, in the ppGalNAc T1 knock-out mouse PSGL1 is significantly reduced (74). This suggests that in the mouse ppGalNAc T1 may be required to initiate the O-glycosylation of the PSGL1 mucin domain, perhaps Thr-58 in particular. To evaluate this possibility, we have obtained the ppGalNAc T1 and T-synthase enhancement products for the Thr residues of the mouse and human PSGL1 mucin domains (Fig. 6, panels A and B, light and gray bars, respectively). T-synthase transfers β(1–3)Gal to the peptide-linked GalNAc and represents the second step in the biosynthesis of the Core 2-O-sLex structure, and its random peptide substrate enhancement values have recently been obtained (49). A rough probability that a site would be occupied by the disaccharide and thereby initiate the formation of the Core 2-O-sLex structure can be derived from the product of the ppGalNAc T1 and T-synthase enhancement products as given in Fig. 6 (black bars). The higher this value the more likely that the site could be sLex-substituted. Indeed, the values obtained for the murine PSGL1 show significantly elevated product values for Thr-58, compared with most of the other Thr residues of the mucin domain. This indeed suggests that its local peptide sequence is targeted for optimal elongation enabling the formation of the sLex structure. Only two other sites in murine PSGL1, Thr-263 and Thr-268, show comparable T1-T-synthase product values, although these sites are located relatively close to the cell surface (Fig. 6, panel A). In the human PSGL1 mucin domain (Fig. 6, panel B) Thr-57 is also clearly indicated as a target for elongation, but there are several other Thr residues that could also be targets based on their enhancement value products. Importantly, Thr-58 in mouse and Thr-57 in human PSGL1 each have the highest T-synthase enhancement product values of all of the Thr residues in either PSGL1, further suggesting the peptide sequence at these Thr residues was designed for optimal/rapid elongation. An examination of the sequences surrounding these Thr residues, DDFEDPDYTYNT*DPPELLK and EYEYLDYDFLPET*EPPEMLR (murine and human sequences, respectively), reveal that these are the most acidic regions of PSGL1 and contain ppGalNAc T1 favoring residues at positions +1 and +3, i.e. Glu/Asp and Pro, respectively (see Figs. 2, panel A, and 3, panel A). T-synthase also prefers acidic peptide residues over basic residues (49). Thus, peptide charge and sequence appears to play a significant role in targeting the specific glycosylation of both human and murine PSGL1. Whether the subsequent transferases involved in the biosynthesis of the Core-2 sLex structure recognize peptide charge or sequence remains to be determined (75).
FIGURE 6.
ppGalNAc T1 and T-synthase random peptide enhancement products for the PSGL1 mucin domain. Plots of the Thr residue enhancement products for ppGalNAc T1 and T-synthase (open bars and gray bars) for the mouse (panel A) and human (panel B) PSGL1 mucin domain. The products of the ppGalNAc T1 and T-synthase enhancement products are also plotted (black bars). Note that the functionally important Thr-57 (mouse) and Thr-58 (human) residues display elevated enhancement values relative to the surrounding Thr residues suggesting these residues would more likely be fully elongated. Enhancement value products were obtained as described under “Experimental Procedures.”
We have also calculated the Thr enhancement products for ppGalNAc T2, T3, T5, T10, and T12 for the murine and human PSGL1, which are plotted in supplemental Fig. S3 along with those of ppGalNAc T1. From the plots, we conclude that other transferases, such as ppGalNAc T5 and T12, could target Thr-58/57, although these transferases could also glycosylate several additional sites in the PSGL1 mucin domain. Whether these or any other ppGalNAc Ts are involved in specific PSGL1 O-glycosylation remains to be determined. Furthermore, from the plot in supplemental Fig. S3, it is clear that there are common regions of high and low glycosylation probabilities for nearly all of the transferases characterized. This may suggest some regions of the PSGL1 mucin domain may be specifically designed to be highly O-glycosylated compared with others. Again, this has not been experimentally determined to date.
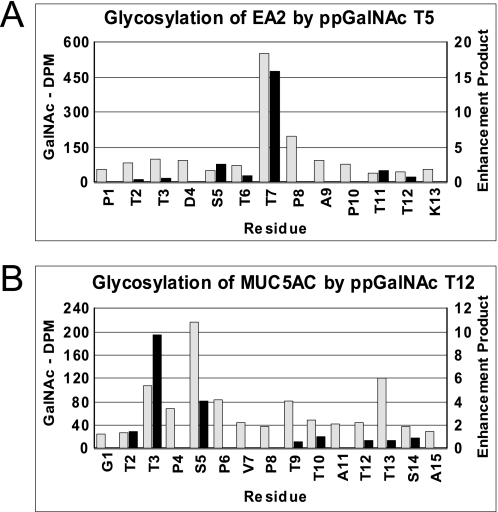
In contrast to what is observed for ppGalNAc T1, T2, and T3, the activities reported for ppGalNAc T5 and T12 do not correlate as well with the random peptide enhancement products obtained for the peptide substrates utilized for these transferases (see supplemental Tables S3 and S4 and Fig. S2). We presently do not understand the origins of these differences, because nearly all of the substrates utilized in these two studies contain between 2 and 10 Ser and Thr residues, and commonly in clusters, and we cannot discount the fact that additional factors such as multiple glycosylation events may be further modulating these transferases by interactions with either the catalytic or lectin domains as has been observed by ppGalNAc T2 (29). Importantly, the actual sites of glycosylation of these substrates has not been determined for either transferase. Therefore, we have obtained the glycosylation patterns of the EA2 and MUC5AC peptide substrates previously used to characterize wild type and mutant ppGalNAc T5 and T12, respectively (Fig. 7, A and B, gray and black bars) (42). As shown in Fig. 7, panel A, glycosylation Thr-7 of EA2 by ppGalNAc T5 is correctly predicted from the random peptide enhancement products, thus validating the use of the enhancement values for this transferase. However, as shown in Fig. 7, panel B, the enhancement products do not fully predict the ppGalNAc T12 sites of glycosylation rather than maximally glycosylating Thr-3 as the enhancement products suggest; experimentally Ser-5 is maximally glycosylated followed by T13 and T3. Again these differences may be due to end effects,10 with Thr-3 being three residues from the N terminus, or possibly due to the absence of preferences for neighboring Thr residues.
FIGURE 7.
ppGalNAc T5 and T12 enhancement products versus experimentally determined sites of glycosylation. ppGalNAc T5 against EA2 (panel A) and ppGalNAc T12 against MUC5AC (panel B). Gray bars represent experimentally determined [3H]GalNAc incorporation obtained by Edman sequencing (uncorrected for background or sequence lag), and black bars represent random peptide-derived enhancement value products.
DISCUSSION
In this study, we have extended our earlier studies of ppGalNAc T1, T2, and T10 to three additional transferases, ppGalNAc T3, T5, and T12, which have been variously implicated in human disease (37, 40–43). In addition, we have introduced a unique random peptide substrate, not previously described, capable of providing preferences for free Ser residues flanking the site of glycosylation for all six transferases (Fig. 1). Thus, by utilizing the oriented random peptide substrates given in Table 1 (P-VI, P-VII, and P-VIII), we have systematically characterized the peptide substrate specificities of 6 of the 20 ppGalNAc Ts for all nonglycosylated amino acid residues except Cys, Trp, and Thr (Figs. 2 and 3). Remarkably, we observe that the C-terminal preferences for the majority of the isoforms are very similar, i.e. having a nearly identical TP(G/A)P-like motif. We concluded that this motif is principally governed by the presence of conserved Trp and Phe residues in the peptide-binding site of the catalytic domain (Trp-282, Phe-280, and Phe-361 in ppGalNAc T2) (supplemental Fig. S1) and that for ppGalNAc T10, which lacks these residues, this preference is lost. On this basis, we suggest that the remaining relatively uncharacterized transferases containing the analogous residues would likely possess similar C-terminal preferences, although those transferases, i.e. ppGalNAc T7, T8, T9, T18, and T20 and WBSCR17 (GALNTL3), lacking these specific residues would not. Indeed, similar to ppGalNAc T10, ppGalNAc T7, and T20 are glycopeptide-preferring transferases that have significant activity only against GalNAc-containing glycopeptide substrates (23, 76). The (S/T)PXP motif is furthermore observed in the data base analysis of O-glycosylation sites (17–19)6 further confirming the commonality of the C-terminal preferences of the majority of the ppGalNAc T isoforms.
As shown in Figs. 2 and 3, transferase specificity against nonglycosylated substrates principally arises from differential recognition at the N-terminal positions of the site of glycosylation with the −1 position showing the most selectivity. Thus, ppGalNAc T2 is distinguished by its elevated −1 Pro enhancement and ppGalNAc T3 and T1 by their Val preferences, although ppGalNAc T12 is distinguished by the presence of Tyr enhancements from the −1 to −3 sites. In addition, we observed that specificity is further modulated by the overall charge of the peptide substrate; ppGalNAc T3 and T5 prefer more basic substrates and ppGalNAc T1 and T2 more acidic substrates. As shown in Fig. 4, these trends were roughly correlated to the isoelectric point and surface charge of the catalytic domain of each transferase.
We have further demonstrated that the preferences can be used to roughly predict rates and sites of glycosylation for a number of common substrates for ppGalNAc T1, T2, and T3 (Fig. 5). More importantly, the preferences predict the isoform-specific glycosylation patterns of the FGF23 peptide and of a HSV-1 gC1 peptide (Table 2). We have further demonstrated the possibility that the ppGalNAc T1 and T-synthase preferences together can predict the site-specific glycosylation of a critical Thr residue in mouse and human PSGL1 (Fig. 6). Although the initial published work on ppGalNAc T5 and T12 do not fully agree with our enhancement value predictions (supplemental Tables S3 and S4), we have shown that for ppGalNAc T5 the enhancements indeed approximate the experimental glycosylation patterns (Fig. 7). We suggest that many of the discrepancies, particularly with the short peptide substrates, arise from end effects,10 the presence of multiple glycosylation sites, and potential conformational effects. It should be further noted that by the nature of the random peptide approach it is possible that highly cooperative specific sequences with strong preferences could go undetected. It is anticipated that these, if present, will be revealed in future studies. Nevertheless, the general success of the transferase-specific random peptide preference values to roughly predict sites of glycosylation will significantly advance our ability to understand site-specific mucin-type O-glycosylation. This is likely to lead to our ability to identify and/or confirm isoform-specific glycosylation sites.
Our work further demonstrates the uniqueness of the different ppGalNAc T isoforms. Although several isoforms appear to have similar enhancement value patterns (Figs. 2 and 3), we find that when the enhancement products are obtained, clear differences between isoforms emerge. An example of this is shown for the Thr residues of the mucin domain of the mouse and human PSGL1 plotted in supplemental Fig. S3. It is clear from the plots that the patterns for the different isoforms can vary significantly from site to site. Of further interest, we find regions of very high and very low probabilities for all of the transferases examined thus far, suggesting that some regions of the peptide core may be designed to be more heavily O-glycosylated than others.
Our understanding of the biological significance of ppGalNAc T substrate preferences remains incomplete. However, several human diseases and conditions are now known to be the result of aberrant O-glycosylation, and several are related to specific ppGalNAc T mutations (reviewed in Tabak (2)).7 With the explosion of data from genome-wide association studies, additional ppGalNAc T associations and biological processes will be uncovered (e.g. GALNT2 (ppGalNAc T2) is associated with the level of blood lipids (44–46)). The ability to predict sites of O-glycosylation in a more precise manner will no doubt speed analysis of associations and help with the functional evaluation.
Acknowledgments
We acknowledge the helpful assistance of Joseph Thome, Jayalakshmi Raman, and Hazuki Miwa.
This work was supported, in whole or in part, by National Institutes of Health Grants CA78834 (to T. A. G.), P30-CA43703, and CA130901 (to S. D. M.) from the NCI and NIDDK intramural program grant (to L. A. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Tables S1–S4, Figs. S1–S3, and additional references.
See also OGEPT version 1.0, a program for predicting mucin-type O-glycosylation sites (Torres, Jr., R., Almeida, I. C., Dayal, Y., and Leung, M.-Y., O-Glycosylation Prediction Electronic Tool, University of Texas, El Paso).
Note that there is also a possibility that the observation of the loss of a glycosylation site in vivo may simply be that no other ppGalNAc Ts are expressed and not necessarily because the mutant transferase is necessarily substrate-specific. The possibility of mis-trafficking/localization at the Golgi membrane could also result in the apparent loss of transferase activity.
This will be available soon on OGEPT version 1.0.
J. C. Collette and T. A. Gerkin, unpublished data.
Note that ppGalNAc T10 is not included here as its peptide substrate preference values are nearly unity and that its major preference is a Ser/Thr-O-GalNAc at the +1 position (30).
As shown in Table 2, the predictions seem to be more concordant with experiments at sites of glycosylation located within the central portions of relatively long peptide substrates five or more residues from the N or C terminus.
- UDP-GalNAc
- uridine diphosphate N-α-acetylgalactosamine
- ppGalNAc T, UDP-α-GalNAc
- polypeptide N-α-acetylgalactosaminyltransferase
- T-synthase, UDP-Gal
- glycoprotein-α-GalNAc β3 galactosyltransferase
- Core 2-O-sLex
- NeuNAcα2–3Galβ1–4 (Fucα1-3)GlcNAcβ1–6 (Galβ1–3)GalNAcα1-O-Thr/Ser
- h
- human.
REFERENCES
- 1. Tian E., Ten Hagen K. G. (2009) Glycoconj. J. 26, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tabak L. A. (2010) Semin. Cell Dev. Biol. 21, 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kato K., Takeuchi H., Kanoh A., Miyahara N., Nemoto-Sasaki Y., Morimoto-Tomita M., Matsubara A., Ohashi Y., Waki M., Usami K., Mandel U., Clausen H., Higashi N., Irimura T. (2010) Glycoconj. J. 27, 267–276 [DOI] [PubMed] [Google Scholar]
- 4. Zhang L., Ten Hagen K. G. (2010) J. Biol. Chem. 285, 34477–34484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee S. H., Yu S. Y., Nakayama J., Khoo K. H., Stone E. L., Fukuda M. N., Marth J. D., Fukuda M. (2010) J. Biol. Chem. 285, 37683–37692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herr P., Korniychuk G., Yamamoto Y., Grubisic K., Oelgeschläger M. (2008) Development 135, 1813–1822 [DOI] [PubMed] [Google Scholar]
- 7. Wagner K. W., Punnoose E. A., Januario T., Lawrence D. A., Pitti R. M., Lancaster K., Lee D., von Goetz M., Yee S. F., Totpal K., Huw L., Katta V., Cavet G., Hymowitz S. G., Amler L., Ashkenazi A. (2007) Nat. Med. 13, 1070–1077 [DOI] [PubMed] [Google Scholar]
- 8. Kato K., Jeanneau C., Tarp M. A., Benet-Pagès A., Lorenz-Depiereux B., Bennett E. P., Mandel U., Strom T. M., Clausen H. (2006) J. Biol. Chem. 281, 18370–18377 [DOI] [PubMed] [Google Scholar]
- 9. Xia L., Ju T., Westmuckett A., An G., Ivanciu L., McDaniel J. M., Lupu F., Cummings R. D., McEver R. P. (2004) J. Cell Biol. 164, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fu J., Gerhardt H., McDaniel J. M., Xia B., Liu X., Ivanciu L., Ny A., Hermans K., Silasi-Mansat R., McGee S., Nye E., Ju T., Ramirez M. I., Carmeliet P., Cummings R. D., Lupu F., Xia L. (2008) J. Clin. Invest. 118, 3725–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. ten Hagen K. G., Zhang L., Tian E., Zhang Y. (2009) Glycobiology 19, 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang L., Tran D. T., Ten Hagen K. G. (2010) J. Biol. Chem. 285, 19491–19501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin Y. R., Reddy B. V., Irvine K. D. (2008) Dev. Dyn. 237, 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brockhausen I. (2007) in Comprehensive Glycoscience: From Chemistry to System Biology (Johannis P. K. ed) Vol. 3, pp. 33–59, Elsevier, Oxford [Google Scholar]
- 15. Walmsley A. R., Hooper N. M. (2003) Glycobiology 13, 641–646 [DOI] [PubMed] [Google Scholar]
- 16. Kearns A. E., Campbell S. C., Westley J., Schwartz N. B. (1991) Biochemistry 30, 7477–7483 [DOI] [PubMed] [Google Scholar]
- 17. Elhammer A. P., Poorman R. A., Brown E., Maggiora L. L., Hoogerheide J. G., Kézdy F. J. (1993) J. Biol. Chem. 268, 10029–10038 [PubMed] [Google Scholar]
- 18. Gupta R., Birch H., Rapacki K., Brunak S., Hansen J. E. (1999) Nucleic Acids Res. 27, 370–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Y. Z., Tang Y. R., Sheng Z. Y., Zhang Z. (2008) BMC Bioinformatics 9, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gerken T. A., Gilmore M., Zhang J. (2002) J. Biol. Chem. 277, 7736–7751 [DOI] [PubMed] [Google Scholar]
- 21. Gerken T. A., Tep C., Rarick J. (2004) Biochemistry 43, 9888–9900 [DOI] [PubMed] [Google Scholar]
- 22. Fritz T. A., Raman J., Tabak L. A. (2006) J. Biol. Chem. 281, 8613–8619 [DOI] [PubMed] [Google Scholar]
- 23. Bennett E. P., Hassan H., Hollingsworth M. A., Clausen H. (1999) FEBS Lett. 460, 226–230 [DOI] [PubMed] [Google Scholar]
- 24. Cheng L., Tachibana K., Zhang Y., Guo J., Kahori Tachibana K., Kameyama A., Wang H., Hiruma T., Iwasaki H., Togayachi A., Kudo T., Narimatsu H. (2002) FEBS Lett. 531, 115–121 [DOI] [PubMed] [Google Scholar]
- 25. Pratt M. R., Hang H. C., Ten Hagen K. G., Rarick J., Gerken T. A., Tabak L. A., Bertozzi C. R. (2004) Chem. Biol. 11, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 26. Hassan H., Reis C. A., Bennett E. P., Mirgorodskaya E., Roepstorff P., Hollingsworth M. A., Burchell J., Taylor-Papadimitriou J., Clausen H. (2000) J. Biol. Chem. 275, 38197–38205 [DOI] [PubMed] [Google Scholar]
- 27. Hanisch F. G., Reis C. A., Clausen H., Paulsen H. (2001) Glycobiology 11, 731–740 [DOI] [PubMed] [Google Scholar]
- 28. Wandall H. H., Irazoqui F., Tarp M. A., Bennett E. P., Mandel U., Takeuchi H., Kato K., Irimura T., Suryanarayanan G., Hollingsworth M. A., Clausen H. (2007) Glycobiology 17, 374–387 [DOI] [PubMed] [Google Scholar]
- 29. Raman J., Fritz T. A., Gerken T. A., Jamison O., Live D., Liu M., Tabak L. A. (2008) J. Biol. Chem. 283, 22942–22951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perrine C. L., Ganguli A., Wu P., Bertozzi C. R., Fritz T. A., Raman J., Tabak L. A., Gerken T. A. (2009) J. Biol. Chem. 284, 20387–20397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ten Hagen K. G., Tran D. T. (2002) J. Biol. Chem. 277, 22616–22622 [DOI] [PubMed] [Google Scholar]
- 32. Schwientek T., Bennett E. P., Flores C., Thacker J., Hollmann M., Reis C. A., Behrens J., Mandel U., Keck B., Schäfer M. A., Haselmann K., Zubarev R., Roepstorff P., Burchell J. M., Taylor-Papadimitriou J., Hollingsworth M. A., Clausen H. (2002) J. Biol. Chem. 277, 22623–22638 [DOI] [PubMed] [Google Scholar]
- 33. Tian E., Ten Hagen K. G. (2007) Glycobiology 17, 820–827 [DOI] [PubMed] [Google Scholar]
- 34. Zhang L., Zhang Y., Hagen K. G. (2008) J. Biol. Chem. 283, 34076–43086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tian E., Ten Hagen K. G. (2007) J. Biol. Chem. 282, 606–614 [DOI] [PubMed] [Google Scholar]
- 36. Topaz O., Shurman D. L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., Sprecher E. (2004) Nat. Genet. 36, 579–581 [DOI] [PubMed] [Google Scholar]
- 37. Bennett E. P., Chen Y. W., Schwientek T., Mandel U., Schjoldager K. B., Cohen S. M., Clausen H. (2010) Glycoconj. J. 27, 435–444 [DOI] [PubMed] [Google Scholar]
- 38. Nakamura N., Toba S., Hirai M., Morishita S., Mikami T., Konishi M., Itoh N., Kurosaka A. (2005) Biol. Pharm. Bull. 28, 429–433 [DOI] [PubMed] [Google Scholar]
- 39. Merla G., Ucla C., Guipponi M., Reymond A. (2002) Hum. Genet. 110, 429–438 [DOI] [PubMed] [Google Scholar]
- 40. Simmons A. D., Musy M. M., Lopes C. S., Hwang L. Y., Yang Y. P., Lovett M. (1999) Hum. Mol. Genet. 8, 2155–2164 [DOI] [PubMed] [Google Scholar]
- 41. Wood L. D., Parsons D. W., Jones S., Lin J., Sjöblom T., Leary R. J., Shen D., Boca S. M., Barber T., Ptak J., Silliman N., Szabo S., Dezso Z., Ustyanksky V., Nikolskaya T., Nikolsky Y., Karchin R., Wilson P. A., Kaminker J. S., Zhang Z., Croshaw R., Willis J., Dawson D., Shipitsin M., Willson J. K., Sukumar S., Polyak K., Park B. H., Pethiyagoda C. L., Pant P. V., Ballinger D. G., Sparks A. B., Hartigan J., Smith D. R., Suh E., Papadopoulos N., Buckhaults P., Markowitz S. D., Parmigiani G., Kinzler K. W., Velculescu V. E., Vogelstein B. (2007) Science 318, 1108–1113 [DOI] [PubMed] [Google Scholar]
- 42. Guda K., Moinova H., He J., Jamison O., Ravi L., Natale L., Lutterbaugh J., Lawrence E., Lewis S., Willson J. K., Lowe J. B., Wiesner G. L., Parmigiani G., Barnholtz-Sloan J., Dawson D. W., Velculescu V. E., Kinzler K. W., Papadopoulos N., Vogelstein B., Willis J., Gerken T. A., Markowitz S. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 109, 12921–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guo J. M., Chen H. L., Wang G. M., Zhang Y. K., Narimatsu H. (2004) Oncology 67, 271–276 [DOI] [PubMed] [Google Scholar]
- 44. Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., Strait J., Duren W. L., Maschio A., Busonero F., Mulas A., Albai G., Swift A. J., Morken M. A., Narisu N., Bennett D., Parish S., Shen H., Galan P., Meneton P., Hercberg S., Zelenika D., Chen W. M., Li Y., Scott L. J., Scheet P. A., Sundvall J., Watanabe R. M., Nagaraja R., Ebrahim S., Lawlor D. A., Ben-Shlomo Y., Davey-Smith G., Shuldiner A. R., Collins R., Bergman R. N., Uda M., Tuomilehto J., Cao A., Collins F. S., Lakatta E., Lathrop G. M., Boehnke M., Schlessinger D., Mohlke K. L., Abecasis G. R. (2008) Nat. Genet. 40, 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., Wahlstrand B., Hedner T., Corella D., Tai E. S., Ordovas J. M., Berglund G., Vartiainen E., Jousilahti P., Hedblad B., Taskinen M. R., Newton-Cheh C., Salomaa V., Peltonen L., Groop L., Altshuler D. M., Orho-Melander M. (2008) Nat. Genet. 40, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., Johansen C. T., Fouchier S. W., Isaacs A., Peloso G. M., Barbalic M., Ricketts S. L., Bis J. C., Aulchenko Y. S., Thorleifsson G., Feitosa M. F., Chambers J., Orho-Melander M., Melander O., Johnson T., Li X., Guo X., Li M., Shin Cho Y., Jin Go M., Jin Kim Y., Lee J. Y., Park T., Kim K., Sim X., Twee-Hee Ong R., Croteau-Chonka D. C., Lange L. A., Smith J. D., Song K., Hua Zhao J., Yuan X., Luan J., Lamina C., Ziegler A., Zhang W., Zee R. Y., Wright A. F., Witteman J. C., Wilson J. F., Willemsen G., Wichmann H. E., Whitfield J. B., Waterworth D. M., Wareham N. J., Waeber G., Vollenweider P., Voight B. F., Vitart V., Uitterlinden A. G., Uda M., Tuomilehto J., Thompson J. R., Tanaka T., Surakka I., Stringham H. M., Spector T. D., Soranzo N., Smit J. H., Sinisalo J., Silander K., Sijbrands E. J., Scuteri A., Scott J., Schlessinger D., Sanna S., Salomaa V., Saharinen J., Sabatti C., Ruokonen A., Rudan I., Rose L. M., Roberts R., Rieder M., Psaty B. M., Pramstaller P. P., Pichler I., Perola M., Penninx B. W., Pedersen N. L., Pattaro C., Parker A. N., Pare G., Oostra B. A., O'Donnell C. J., Nieminen M. S., Nickerson D. A., Montgomery G. W., Meitinger T., McPherson R., McCarthy M. I., McArdle W., Masson D., Martin N. G., Marroni F., Mangino M., Magnusson P. K., Lucas G., Luben R., Loos R. J., Lokki M. L., Lettre G., Langenberg C., Launer L. J., Lakatta E. G., Laaksonen R., Kyvik K. O., Kronenberg F., König I. R., Khaw K. T., Kaprio J., Kaplan L. M., Johansson A., Jarvelin M. R., Janssens A. C., Ingelsson E., Igl W., Kees Hovingh G., Hottenga J. J., Hofman A., Hicks A. A., Hengstenberg C., Heid I. M., Hayward C., Havulinna A. S., Hastie N. D., Harris T. B., Haritunians T., Hall A. S., Gyllensten U., Guiducci C., Groop L. C., Gonzalez E., Gieger C., Freimer N. B., Ferrucci L., Erdmann J., Elliott P., Ejebe K. G., Döring A., Dominiczak A. F., Demissie S., Deloukas P., de Geus E. J., de Faire U., Crawford G., Collins F. S., Chen Y. D., Caulfield M. J., Campbell H., Burtt N. P., Bonnycastle L. L., Boomsma D. I., Boekholdt S. M., Bergman R. N., Barroso I., Bandinelli S., Ballantyne C. M., Assimes T. L., Quertermous T., Altshuler D., Seielstad M., Wong T. Y., Tai E. S., Feranil A. B., Kuzawa C. W., Adair L. S., Taylor H. A., Jr., Borecki I. B., Gabriel S. B., Wilson J. G., Holm H., Thorsteinsdottir U., Gudnason V., Krauss R. M., Mohlke K. L., Ordovas J. M., Munroe P. B., Kooner J. S., Tall A. R., Hegele R. A., Kastelein J. J., Schadt E. E., Rotter J. I., Boerwinkle E., Strachan D. P., Mooser V., Stefansson K., Reilly M. P., Samani N. J., Schunkert H., Cupples L. A., Sandhu M. S., Ridker P. M., Rader D. J., van Duijn C. M., Peltonen L., Abecasis G. R., Boehnke M., Kathiresan S. (2010) Nature 466, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gerken T. A., Raman J., Fritz T. A., Jamison O. (2006) J. Biol. Chem. 281, 32403–32416 [DOI] [PubMed] [Google Scholar]
- 48. Gerken T. A., Ten Hagen K. G., Jamison O. (2008) Glycobiology 18, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Perrine C., Ju T., Cummings R. D., Gerken T. A. (2009) Glycobiology 19, 321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 51. Fritz T. A., Hurley J. H., Trinh L. B., Shiloach J., Tabak L. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15307–15312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kubota T., Shiba T., Sugioka S., Furukawa S., Sawaki H., Kato R., Wakatsuki S., Narimatsu H. (2006) J. Mol. Biol. 359, 708–727 [DOI] [PubMed] [Google Scholar]
- 53. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 54. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) Nucleic Acids Res. 37, D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 56. Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 57. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) Nucleic Acids Res. 35, W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li H., Robertson A. D., Jensen J. H. (2005) Proteins 61, 704–721 [DOI] [PubMed] [Google Scholar]
- 61. Sanner M. F., Olson A. J., Spehner J. C. (1996) Biopolymers 38, 305–320 [DOI] [PubMed] [Google Scholar]
- 62. Gerken T. A., Owens C. L., Pasumarthy M. (1997) J. Biol. Chem. 272, 9709–9719 [DOI] [PubMed] [Google Scholar]
- 63. Miwa H. E., Gerken T. A., Jamison O., Tabak L. A. (2010) J. Biol. Chem. 285, 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bennett E. P., Hassan H., Clausen H. (1996) J. Biol. Chem. 271, 17006–17012 [DOI] [PubMed] [Google Scholar]
- 65. Bennett E. P., Hassan H., Mandel U., Hollingsworth M. A., Akisawa N., Ikematsu Y., Merkx G., van Kessel A. G., Olofsson S., Clausen H. (1999) J. Biol. Chem. 274, 25362–25370 [DOI] [PubMed] [Google Scholar]
- 66. Ten Hagen K. G., Hagen F. K., Balys M. M., Beres T. M., Van Wuyckhuyse B., Tabak L. A. (1998) J. Biol. Chem. 273, 27749–27754 [DOI] [PubMed] [Google Scholar]
- 67. Guo J. M., Zhang Y., Cheng L., Iwasaki H., Wang H., Kubota T., Tachibana K., Narimatsu H. (2002) FEBS Lett. 524, 211–218 [DOI] [PubMed] [Google Scholar]
- 68. Mårdberg K., Nyström K., Tarp M. A., Trybala E., Clausen H., Bergström T., Olofsson S. (2004) Glycobiology 14, 571–581 [DOI] [PubMed] [Google Scholar]
- 69. Nehrke K., Hagen F. K., Tabak L. A. (1996) J. Biol. Chem. 271, 7061–7065 [DOI] [PubMed] [Google Scholar]
- 70. Hagen F. K., Gregoire C. A., Tabak L. A. (1995) Glycoconj. J. 12, 901–909 [DOI] [PubMed] [Google Scholar]
- 71. Li F., Wilkins P. P., Crawley S., Weinstein J., Cummings R. D., McEver R. P. (1996) J. Biol. Chem. 271, 3255–3264 [PubMed] [Google Scholar]
- 72. Liu W., Ramachandran V., Kang J., Kishimoto T. K., Cummings R. D., McEver R. P. (1998) J. Biol. Chem. 273, 7078–7087 [DOI] [PubMed] [Google Scholar]
- 73. Xia L., Ramachandran V., McDaniel J. M., Nguyen K. N., Cummings R. D., McEver R. P. (2003) Blood 101, 552–559 [DOI] [PubMed] [Google Scholar]
- 74. Tenno M., Ohtsubo K., Hagen F. K., Ditto D., Zarbock A., Schaerli P., von Andrian U. H., Ley K., Le D., Tabak L. A., Marth J. D. (2007) Mol. Cell. Biol. 27, 8783–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Brockhausen I., Dowler T., Paulsen H. (2009) Biochim. Biophys. Acta 1790, 1244–1257 [DOI] [PubMed] [Google Scholar]
- 76. Peng C., Togayachi A., Kwon Y. D., Xie C., Wu G., Zou X., Sato T., Ito H., Tachibana K., Kubota T., Noce T., Narimatsu H., Zhang Y. (2010) Biochem. Biophys. Res. Commun. 402, 680–686 [DOI] [PubMed] [Google Scholar]