Abstract
The SUMO ligase activity of Mms21/Nse2, a conserved member of the Smc5/6 complex, is required for resisting extrinsically induced genotoxic stress. We report that the Mms21 SUMO ligase activity is also required during the unchallenged mitotic cell cycle in Saccharomyces cerevisiae. SUMO ligase-defective cells were slow growing and spontaneously incurred DNA damage. These cells required caffeine-sensitive Mec1 kinase-dependent checkpoint signaling for survival even in the absence of extrinsically induced genotoxic stress. SUMO ligase-defective cells were sensitive to replication stress and displayed synthetic growth defects with DNA damage checkpoint-defective mutants such as mec1, rad9, and rad24. MMS21 SUMO ligase and mediator of replication checkpoint 1 gene (MRC1) were epistatic with respect to hydroxyurea-induced replication stress or methyl methanesulfonate-induced DNA damage sensitivity. Subjecting Mms21 SUMO ligase-deficient cells to transient replication stress resulted in enhancement of cell cycle progression defects such as mitotic delay and accumulation of hyperploid cells. Consistent with the spontaneous activation of the DNA damage checkpoint pathway observed in the Mms21-mediated sumoylation-deficient cells, enhanced frequency of chromosome breakage and loss was detected in these mutant cells. A mutation in the conserved cysteine 221 that is engaged in coordination of the zinc ion in Loop 2 of the Mms21 SPL-RING E3 ligase catalytic domain resulted in strong replication stress sensitivity and also conferred slow growth and Mec1 dependence to unchallenged mitotically dividing cells. Our findings establish Mms21-mediated sumoylation as a determinant of cell cycle progression and maintenance of chromosome integrity during the unperturbed mitotic cell division cycle in budding yeast.
Keywords: Cell Cycle, Checkpoint Control, Chromosomes, Mitosis, Sumoylation
Introduction
Accurate transmission of genetic information without alteration or loss is important during every cell division cycle. Monitoring systems that survey, alert, and aid in repair of harmful DNA damage help prevent the acquisition of genetic changes (1). Failure of these systems can result in genomic instability often observed in some genetic disorders and in cancer cells (2). Studying mechanisms that suppress genomic instability is therefore important to gain insight into the causes of genetic disorders associated with genomic instability such as cancer.
During the normal unchallenged mitotic cell cycle, spontaneous DNA damage can occur during DNA replication. During replication, DNA is unwound and duplicated and is particularly vulnerable to breakage, especially if any of the protective repair and rescue mechanisms fail. Replication forks can stall or collapse at DNA lesions, resulting in activation of the Mec1/Ataxia telangiectasia and Rad3 related kinase-dependent replication checkpoint response (3, 4). The replication checkpoint stabilizes stalled forks, prevents their breakdown, and coordinates fork restart processes (5). In addition, it delays cell cycle progression to facilitate the repair and completion of DNA replication (3, 4).
Structural alterations in chromosome organization facilitate chromosome segregation and aid in maintenance of chromosome stability. Such alterations in chromosome architecture include sister chromatid cohesion, kinetochore assembly, and chromosome condensation and are brought about by a number of chromatin-associated proteins, e.g. the structural maintenance of chromosomes (Smc)4 protein complexes. SMC proteins are a conserved, essential family of proteins required for chromosome organization and are involved in coordinating a number of chromosomal processes (6–9). The budding yeast Saccharomyces cerevisiae has three Smc protein complexes: Cohesin (Smc1/3 complex), Condensin (Smc2/4 complex), and the Smc5/6 complex required for sister chromatid cohesion, chromosome condensation, and DNA repair (10–12). Each Smc complex consists of a heterodimer of two Smc proteins associated with additional essential non-Smc subunits. For example, the chromatin-associated Smc5/6 complex consists of Smc5, Smc6, and six non-smc elements (Nse1–Nse6) (12–14).
The smc6 gene (also called rad18 in fission yeast) was first discovered in a genetic screen for γ radiation-sensitive mutants in Schizosaccharomyces pombe (15). Smc6 complex mutants are defective in repair of DNA damage induced by exogenous agents. Genetic interaction studies indicate a crucial role for the Smc5/6 complex in recombination-mediated DNA damage repair that is epistatic to the Rad51-dependent homologous recombination repair pathway (16–18). For example, measurement of the survival of rad18-X, rhp51Δ, and rad18-X rhp51Δ after UV irradiation revealed that, although the rad18-X mutant is more sensitive than rhp51Δ to UV irradiation, deletion of rhp51 in the rad18-X mutant background rescues the UV irradiation sensitivity of rad18-X to an extent equivalent to that seen in the rhp51Δ single mutant (15). Fission yeast rhp51Δ mutant cells are more sensitive to ionizing radiation than rad18-X mutant cells as are double mutant rhp51Δ rad18-X cells (15). Furthermore, in budding yeast, deletion of RAD52 (also required for homologous recombination-mediated processes) partially suppresses the temperature-sensitive phenotype of smc6–9 mutants (19), indicating that recombination processes may be toxic in the absence of Smc6 function and suggesting a distinct role for Smc6 in this process.
In budding yeast, Smc5 and Smc6 are required for stability of repetitive chromosomal regions and sister chromatid recombination-mediated repair of induced DNA double strand breaks (19, 20). In particular, the role of the Smc5/6 complex in rDNA maintenance has been well characterized. Smc6 is enriched at ribosomal DNA and telomeres and is required for their segregation (13, 19). Segregation defects in rDNA may arise from non-disjunction of the rDNA region prior to metaphase execution (21) in smc5/6 mutants. In these mutants, replication of rDNA is incomplete, resulting in instability of the rDNA during segregation, which can be alleviated by conditions that may facilitate replication through this locus (21).
Among the Smc protein complexes, the Smc5/6 complex is unique in that one of its essential subunits, Mms21/Nse2, is a small ubiquitin-related modifier (SUMO) E3 ligase. Mms21 of S. cerevisiae was discovered in a screen for methyl methanesulfonate (MMS)-sensitive mutants (22) and has a really interesting new gene (RING) domain at its C-terminal end having SUMO E3 ligase activity. SUMO (23, 24), also known as Smt3 in S. cerevisiae (25), is covalently conjugated to lysine residues of a variety of proteins by SUMO E3 ligases and is deconjugated by SUMO-specific proteases (26, 27). Mms21/Nse2 sumoylates Smc5 and Yku70, a protein involved in non-homologous end joining-mediated repair (28). In S. pombe, Nse2 sumoylates Smc6, whereas in human cells, it is known to sumoylate Smc6 and several telomere-associated proteins such as translin-associated factor X, TRF1 and TRF2, TIN2, and RAP1 (28–30). Although Mms21/Nse2 has been reported to be essential for viability in all organisms analyzed, SUMO ligase domain-defective mutants are hypersensitive to DNA-damaging agents but are viable, indicating that the SUMO ligase activity of Mms21 is dispensable under normal growth conditions. The C-terminal region of Mms21 including the SUMO ligase domain is required for resisting extrinsically induced genotoxic stress, maintenance of nucleolar integrity, and telomere clustering (28). X-shaped DNA intermediates (detected by two-dimensional gel electrophoresis) that are generated at damaged replication forks encountering MMS-induced DNA lesions accumulate in Mms21 SUMO ligase-defective mutants, suggesting a role for Mms21-dependent sumoylation in counteracting accumulation of X-structures at MMS-damaged forks (31).
Although the requirement for the SUMO ligase activity of Mms21/Nse2 in countering genotoxic stress has been established, its requirement under non-stress conditions has not been explored extensively. Previous studies in S. pombe indicated that the SUMO ligase activity of Nse2 was dispensable for its essential function; nse2.SA mutant cells having two mutations in the SPL-RING domain (C195S/H197A) that inactivate the SUMO ligase activity are viable, whereas a deletion of Nse2 is lethal (29). The nse2.SA cells are sensitive to MMS and hydroxyurea, but no defects in survival or optimal growth of SUMO ligase-defective mutants in the absence of extrinsic stressors have been reported. An equivalent mutant in S. cerevisiae having substitutions in analogous residues Cys-200 and His-202 of Mms21 (mms21-CH allele) shows accumulation of X-molecules at MMS-damaged forks (31), but no other defects under normal unperturbed mitotic cell division were reported. In this study, we investigated the role of the SUMO ligase activity of Mms21 during the unchallenged mitotic cell division cycle in S. cerevisiae and found evidence for its requirement in the absence of extrinsically induced genotoxic stress for maintenance of normal growth and chromosome integrity and prevention of DNA damage.
EXPERIMENTAL PROCEDURES
Media and Reagents
Cells were grown in YPD medium or supplemented minimal medium as indicated. We purchased MMS, hydroxyurea (HU), and G418 from Sigma; 12CA5 anti-HA antibody from Roche Applied Science, and HRP-conjugated secondary antibodies from Bangalore Genei.
Yeast Strains and Plasmids
The yeast strains used or generated in this study are listed in Table 1. Oligonucleotides used for construction of strains and plasmids in this study were synthesized by Sigma Genosys; sequences of these oligonucleotides are provided in Table 2.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source/Ref. |
|---|---|---|
| YPH499 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | 59 |
| SLY792 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 mms21Δsl::KanMX6 | This study |
| SLY861 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 MRC1-3HA-HIS5 | This study |
| SLY1095 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 RAD9-3HA-klTRP | This study |
| SLY1100 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 RAD53-3HA-klTRP1 | This study |
| SLY872 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 MRC1-3HA-HIS5 mms21Δsl::KanMX6 | This study |
| SLY1098 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 RAD9-3HA-klTRP mms21Δsl::KanMX6 | This study |
| SLY1104 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 RAD53-3HA-klTRP1 mms21Δsl::KanMX6 | This study |
| VPS106 | MATa ade2 can1r leu2-3,112 Δtrp1 ade3 Δura3 lys2-801 | 60 |
| SLY1351 | MATa ade2 canl r leu2-3,112 Δtrp1 ade3 Δura3 lys2-801 MRC1-3HA-klTRP1 | This study |
| SLY1415 | MATa ade2 canl r leu2-3,112 Δtrp1 ade3 Δura3 lys2-801 MRC1-3HA-klTRP1 mms21Δsl::KanMX6 | This study |
| tel1 | MATa ade2 can1r leu2-3,112 Δtrp1 ade3 Δura3 lys2-801 tel1::MET17 | 60 |
| mec1 | MATa ade2 can1r leu2-3,112 Δtrp1 ade3 Δura3 lys2-801 sml1 mec1::leu2::hisG | 60 |
| SLY1353 | MATa ade2 can1r leu2-3,112 Δtrp1 ade3 Δura3 lys2-801 tel1::MET17 MRC1-3HA-klTRP1 | This study |
| SLY1412 | MATa ade2 can1r leu2-3,112 Δtrp1 ade3 Δura3 lys2-801 sml1 mec1::leu2::hisG mms21Δsl::KanMX6 | This study |
| SLY1416 | MATa ade2 can1r leu2-3,112 Δtrp1 ade3 Δura3 lys2-801 tel1::MET17 MRC1-3HA-klTRP1mms21Δsl::KanMX6 | This study |
| DH1003/102 | MATa leu2 trp1 ura3-52 can1 ade2::KanMX/yWSS1572-1 | 35 |
| DH2004/101 | MATa leu2 trp1 ura3-52 can1 ade2::KanMX/PA3-1 | 35 |
| SLY1348 | MATa leu2 trp1 ura3-52 can1 ade2::KanMX mms21Δsl::LEU2/yWSS1572-1 | This study |
| SLY1344 | MATa leu2 trp1 ura3-52 can1 ade2::KanMX mms21Δsl::LEU2/PA3-1 | This study |
| Y1127 | MATa ade2-1 his3-11, 15 leu2-3,112 can1-100 trp1-1 ura3-1 mrc1Δ-3::his5+ | 40 |
| Y1134 | MATa ade2-1 his3-11, 15 leu2-3,112 can1-100 trp1-1 ura3-1 HIS::MRC1-MYC13 | 40 |
| Y2297 | MATa ade2-1 his3-11, 15 leu2-3,112 can1-100 trp1-1 ura3-1 rad9Δ::HIS3 mrc1Δ-2::HIS3/pMRC1 | 39 |
| SLY1516 | MATa ade2-1 his3-11, 15 leu2-3,112 can1-100 trp1-1 ura3-1 mrc1Δ-3::his5+mms21Δsl::LEU2 | This study |
| SLY1523 | MATa ade2-1 his3-11, 15 leu2-3,112 can1-100 trp1-1 ura3-1 HIS::MRC1-MYC13 mms21Δsl::LEU2 | This study |
| SLY1526 | MATa ade2-1 his3-11, 15 leu2-3,112 can1-100 trp1-1 ura3-1 rad9Δ::HIS3 mrc1Δ-2::HIS3 mms21Δsl::LEU2/pMRC1 | This study |
| SLY1534 | MATa ade2-1 his3-11, 15 leu2-3,112 can1-100 trp1-1 ura3-1 HIS::MRC1-MYC13 rad24::hphNT1 | This study |
| SLY1543 | MATa ade2-1 his3-11, 15 leu2-3,112 can1-100 trp1-1 ura3-1 HIS::MRC1-MYC13 rad24::hphNT1 mms21Δsl::LEU2 | This study |
| JK9-3da | MATa leu2-3,112 ura3-52 trp1 his4 rme1 HMLa | 61 |
| JK350-21a | MATa leu2-3,112 ura3-52 trp1 his4 rme1 HMLa tor1::LEU2-4 tor2::ADE2-3/pJK5 | 61 |
| JK350-18a | MATa leu2-3,112 ura3-52 trp1 his4 rme1 HMLa tor2::ADE2-3/pJK5 | 61 |
| SLY1546 | MATa leu2-3,112 ura3-52 trp1 his4 rme1 HMLa tor2::ADE2-3 mms21Δsl::KanMX6/pJK5 | This study |
| SLY1549 | MATa leu2-3,112 ura3-52 trp1 his4 rme1 HMLa tor1::LEU2-4 tor2::ADE2-3 mms21Δsl::KanMX6/pJK5 | This study |
| SLY1552 | MATa leu2-3,112 ura3-52 trp1 his4 rme1 HMLa mms21Δsl::KanMX6 | This study |
| SLY782 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 mms21::KanMX6 (pRR50) | This study |
| SLY1620 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 mms21::KanMX6 (pRR51) | This study |
| SLY1621 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 mms21::KanMX6 (pRR55) | This study |
| SLY1622 | MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 mms21::KanMX6 (pRR56) | This study |
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| SLO38 | 5′-GGGGATCCACATTCTCTTCAACAAGTTT-3′ |
| SLO60 | 5′-TAGCCAATTTCCAGTCGTCC-3′ |
| SLO61 | 5′-CAACCATCGTATCCGCTAGT-3′ |
| SLO62 | 5′-ATTAATCTAGACCTAAAACATCGATGGCTTGAC-3′ |
| SLO69 | 5′-CAATCGATAAAAAGAAAAGGAGAG-3′ |
| SLO124 | 5′-CGCTATACTGCTGTCGATTC-3′ |
| SLO131 | 5′-AAAACAAAATCGAATAAACTTTTTGAAAGCGGACAAGATAGCTTTGATAATTACCCATACGATGTTCCTGAC-3′ |
| SLO132 | 5′-CAAGACAGCTTCTGGAGTTCAATCAACTTCTTCGGAAAAGATAAAAAACCACGATAAGCTTCGTACGCTGCA-3′ |
| SLO295 | 5′-CACGATGATATTACGGACAATGATATATACAACACTATTTCTGAGGTTAGACGTACGCTGCAGGTCGAC-3′ |
| SLO296 | 5′-TTTAATCGTCCCTTTCTATCAATTATGAGTTTATATATTTTTATAATTTCAATCGATGAATTCGAGCTCG-3′ |
| SLO297 | 5′-CTTCTCTCTTAAAAAGGGGCAGCATTTTCTATGGGTATTTGTCCTTGGTTAATCGATGAATTCGAGCTCG-3′ |
| SLO298 | 5′-AGGGCAAAAATTGGACCAAACCTCAAAAGGCCCCGAGAATTTGCAATTTTCGCGTACGCTGCAGGTCGAC-3′ |
| SLO348 | 5′-CTGACAAGAAGAACATTGCTGATG-3′ |
| SLO355 | 5′-AAAACAAAATCGAATAAACTTTTTGAAAGCGGACAAGATAGCTTTGATAATCGTACGCTGCAGGTCGAC-3′ |
| SLO356 | 5′-ACAGCTTCTGGAGTTCAATCAACTTCTTCGGAAAAGATAAAAAACCACTAATCGATGAATTCGAGCTCG-3′ |
| SLO366 | 5′-ATGGATTCTAGAACAGTTGG-3′ |
| SLO367 | 5′-CCTTAAGTTGAACGGAGTCC-3′ |
| SLO368 | 5′-GCTACATATAAGGAACGTGC-3′ |
| SLO369 | 5′-CTTCCCTTTGCAAATAGTCC-3′ |
| SLLEU2-R1B | 5′-TCGCATTATCCTCGGGTTCAG-3′ |
| SLLEU2-R1F | 5′-GGGCACCACACAAAAAGTTAGG-3′ |
| SLO135 | 5′-TCAAGAAAATGTAATGCTGTCTTCGATAGAGAC-3′ |
| SLO136 | 5′-GTCTCTATCGAAGACAGCATTACATTTTCTTGA-3′ |
| SLO137 | 5′-TACACGACAAGAGATGCTCCGCAAGCAGCGTGT-3′ |
| SLO138 | 5′-ACACGCTGCTTGCGGAGCATCTCTTGTCGTGTA-3′ |
| SLO283 | 5′-ATCGATGAATTCGAGCTCG-3′ |
| SLO408 | 5′-CACGCATTGATATCTGAGAG-3′ |
| SLO409 | 5′-GTTAATAGTACTTGATTCAACACCACTAATTATCAAGTTTGTTCCTGTCTGAATGATATGCGTACGCTGCAGGTCGAC-3′ |
| SLO410 | 5′-ATAGATTTGTGTGGAATATTTCCTGGGGTTTTCTCGTCAAATTTAAAGAGTAAAAAGTTAATCGATGAATTCGAGCTCG-3′ |
Generation of mms21Δsl Strains
The mms21Δsl mutant allele was created by replacing the C-terminal SPL-RING domain of Mms21 with KanMX6 by one-step gene disruption to produce a mutation genetically equivalent to mms21-11, a previously described mutant containing a transposon insertion at the SPL-RING domain (28). A 1.8-kb fragment containing MMS21 was PCR-amplified from S288C genomic DNA with Pfu polymerase using the primers SLO60 and SLO62 and cloned into pCR-Blunt II-TOPO (Invitrogen) to generate pRR52. A 1.1-kb HincII-EcoRI fragment excised from pRR52 was subcloned into HincII-EcoRI sites of pUC19, resulting in pRR53. A 1.4-kb KanMX6 fragment was excised from pFA6a-KanMX6 vector (32) by PvuII and EcoRV digestion and subcloned in the EcoRV site of pRR53, resulting in pRR71. The mms21::KanMX6 disruption cassette was excised from pRR71 by PvuII and XbaI and used for integrative yeast transformation. Transformants were selected on YAPD (YPD supplemented with adenine to a final concentration of 0.01 mg/ml)-G418 (200 μg/ml) plates and screened by colony PCR to identify insertions at the correct genomic locus that would yield a product of 525 bp using the primers KanMX6FP (SLO124) and MMS21R (SLO61).
An mms21Δsl::LEU2 disruption cassette was generated by subcloning LEU2 (PCR-amplified from pRS425 by Pfu Turbo DNA polymerase using the primers SLO348 and SLLEU2-R1B) in the EcoRV site of pRR53, resulting in pRR72. The mms21Δsl::LEU2 disruption cassette was excised from pRR72 by PvuII and Xba1 and used for integrative yeast transformation. The transformants were selected on SC-Leu plates, and the desired mms21Δsl mutants were identified by colony PCR using the primers SLLEU2-R1F and MMS21R (SLO61), resulting in amplification of an 871-bp fragment corresponding to the insertion of the disruption cassette at the correct genomic locus. The results reported in this study were verified with three independent isolates of mms21Δsl mutant strains derived as detailed above.
Generation of rad24 Deletion Strain
RAD24 was disrupted with hygromycin resistance marker (hphNT1) by PCR-mediated one-step gene disruption. Primers (SLO409 and SLO410) that have homology to upstream and downstream sequences of RAD24 were used to amplify ∼2-kb hphNT1 from pYM16 (33). The PCR amplicon was used for integrative yeast transformation, and transformants were selected for hygromycin resistance on YAPD-hygromycin (300 μg/ml) plates. Integration of hphNT1 at the RAD24 locus was confirmed by genomic DNA PCR using the primers SLO283 and SLO408, which produce a ∼2-kb fragment in the case of desired integrants.
Generation of Epitope-tagged Strains
Mediator of replication checkpoint 1 (Mrc1), Rad9, and Rad53 were tagged at the C terminus with three copies of the HA epitope (3HA) by one-step PCR-mediated gene tagging (33) using the primer pairs SLO355/SLO356, SLO295/SLO296, and SLO297/SLO298, respectively. The PCR was carried out using pYM22 (33) as the template. For Mrc1-3HA tagging, p473 (kindly provided by Doug Koshland) was used as a template for PCR (using primers SLO131/SLO132). The linear PCR products were transformed into appropriate yeast strains, and transformants were selected on SC-Trp plates (when pYM22 was used as the template) or on SC-His plates (when p473 was used as the template). In each case, the tagging at the correct chromosomal locus was confirmed by immunoblotting.
Generation of mms21 Point Mutants by Site-directed Mutagenesis
The mms21-C221A and mms21-H202A mutants of MMS21 were generated by introducing the required substitution in the primers, resulting in the desired mutations. Primers having the required missense mutations (SLO135/SLO136 and SLO137/SLO138) were synthesized by Sigma-Genosys and used for PCR amplification as detailed below.
Inverse PCRs for introducing missense mutations were performed using Pfu Turbo DNA polymerase and the plasmid pRR51 (pRS314-MMS21-3HA) grown in the Escherichia coli TOP10 strain as the in vivo methylated DNA template. Two complementary mutagenic primers of length ∼30–35 nucleotides containing the desired mutation(s) flanked by ∼15 nucleotides of wild-type sequences were used as primers in a 50-μl inverse PCR. The PCR cycling parameters were −95 °C for 2 min followed by 20 cycles of 95 °C for 1 min, 55 °C for 1 min, and 72 °C for 5 min followed by a final extension step at 72 °C for 15 min. As a control, mock PCRs using the same plasmid template were set up without addition of Pfu Turbo DNA polymerase. The input methylated circular plasmid DNA template was selectively digested by adding 10 units of the restriction endonuclease DpnI to the completed PCR and allowing 6–8 h of digestion at 37 °C. This treatment leaves the PCR-generated non-methylated mutant DNA intact. 10 μl of the DpnI-treated PCR-amplified DNA was transformed into chemically competent E. coli TOP10 cells. In a successful attempt, the expected colony number is ∼10-fold higher when the mutated PCR-amplified DNA is transformed compared with transformation of control mock PCRs that were also DpnI-treated. Plasmids from the colonies thus obtained were isolated, and the presence of the mutations was verified by sequencing the insert containing plasmid DNA from two independent colonies, thus generating the plasmids pRR55 (mms21-C221A) and pRR56 (mms21-H202A).
Growth and Drug Sensitivity Assays
For assaying growth and sensitivity to various drugs, cells were grown in YAPD medium to midlog phase, and cultures were diluted serially (by 10-fold) and spotted adenine supplemented (10 μg/ml) on YPD plates or on YAPD plates containing the indicated drug. The plates were incubated at 23 °C for 3–5 days and photographed. For analysis of sensitivity to UV radiation, cells were serially diluted and spotted on YPD plates and irradiated with the indicated doses of UV radiation using a UVC-500 cross-linker (Amersham Biosciences). The plates were immediately covered with aluminum foil, incubated at 23 °C for 3–5 days, and photographed.
To assay the growth in liquid medium, cells were grown overnight in YAPD. The cultures were diluted in duplicate with fresh YAPD to A600 of 0.1, and an aliquot of cells was removed at time 0 and every 2 h thereafter to measure A600.
Preparation of Yeast Extracts and Western Blotting
Total protein extracts were prepared from S. cerevisiae as described previously (34). Cells were grown at 25 °C to midlog phase, and 10 A600 units of the culture were harvested and used per extraction. The clear crude preparation was nearly 150 μl of which 25 μl was resolved per lane by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE). The resolved proteins were transferred to a PVDF membrane using a wet electroblotting apparatus in transfer buffer (39 mm glycine, 48 mm Tris-HCl, 20% methanol). The membrane was blocked overnight in 5% nonfat milk (HiMedia) made in 1× TBS (25 mm Tris-HCl, 125 mm NaCl, pH 8.0). The HA epitope was detected using 12CA5 monoclonal anti-HA antibody (Roche Applied Science) at a 1:5000 dilution in 5% nonfat milk prepared in 1× TBS with peroxidase-conjugated anti-mouse IgG secondary antibody (purchased from Bangalore Genei). Mad1 protein was detected using anti-Mad1 antibodies (1:2000 dilution prepared in 5% nonfat milk prepared in 1× phosphate-buffered saline (PBS) with Tween 20), a kind gift from Kevin Hardwick, and peroxidase-conjugated anti-rabbit IgG secondary antibody (1:5000 dilution; purchased from Bangalore Genei). The blots were developed with PerkinElmer Life Sciences Chemiluminescence Plus reagents according to the manufacturer's instructions.
Quantitative Assay for Telomere Marker Loss and Total YAC Loss
The YAC assay for measuring gross chromosomal rearrangements was performed as described previously (35). In brief, single colonies from SC-Ura plates that select for the YAC telomere marker (URA3) were picked and resuspended in 1 ml of sterile water. These cell suspensions were diluted 103-fold and plated onto YPD plates such that ∼150–200 colonies formed on each plate. Colonies on the YPD plates were replica-plated to SC-Trp and SC-Ura, and growth was scored after incubation at 23 °C for 3–4 days. The percentage of cells with telomere marker loss (Trp+ura−) or entire YAC loss (trp−ura−) in medium selective for the full-length YAC is a direct measure of the rate of these events.
PCR Analysis for Assaying YAC Markers to Confirm Chromosome Breakage
For PCR analysis, yeast genomic DNA was isolated and quantified. PCRs were carried out using Taq DNA polymerase (Promega) and standard conditions for amplification. The PCR products were resolved on a 1.5% agarose gel. Gene-specific primers were used to amplify TRP1 (SLO38/SLO69), URA3 (SLO368/SLO369), and ADE2 (SLO366/SLO367), respectively.
Multiple Sequence Alignment
The multiple sequence alignment of Mms21-related proteins was generated using ClustalW2 using default parameters. The sequences of Mms21-related proteins were obtained by using Blastp to search the non-redundant protein sequence database using the Mms21 protein sequence (NCBI reference sequence NP_010896.1) as a query and default search parameters. The sequences of the Mms21 orthologs included in the alignment shown in Fig. 6 are from Homo sapiens (gi|16552560|dbj|BAB71338.1|), Equus caballus (gi|194215096|ref|XP_001497689.2|), Salmo salar (gi| 221221686|gb|ACM09504.1|), Arabidopsis thaliana (gi| 11994504|dbj|BAB02569.1|), Oryza sativa Indica group (gi|218197269|gb|EEC79696.1|), Anopheles gambiae (gi| 158285718|ref|XP_001687938.1|), S. pombe, gi|162312472| ref|XP_001713077.1|), S. cerevisiae (gi|6320817|ref|NP_010896.1|), Caenorhabditis elegans (gi|17538091|ref|NP_495157.1|), and Plasmodium vivax (gi|156100297|ref|XP_001615876.1|).
FIGURE 6.
Genetic interactions of mms21Δsl with mutants defective in replication stress and DNA damage checkpoint pathways. A, the mms21Δsl mutant cells are HU-sensitive. WT (YPH499) and mms21Δsl (SLY792) strains were grown to midlog phase, serially diluted, spotted onto a YAPD plate containing 150 mm HU, and incubated at 23 °C for 5 days. B, genetic interaction of mms21Δsl with checkpoint-defective mutants mrc1 and rad9. WT (Y1134), mrc1 (Y1127), mms21Δsl (SLY1523), mrc1 mms21Δsl (SLY1516), rad9 (Y2297), and rad9 mms21Δsl (SLY1526) strains were grown to midlog phase, serially diluted, spotted on adenine supplemented YPD plates or YAPD plates having the indicated concentrations of MMS and HU, and incubated for 3 days. C, genetic interaction of mms21Δsl with DNA damage checkpoint-defective mutants rad9 and rad24. WT (Y1134), mms21Δsl (SLY1523), rad9 (Y2297), rad9 mms21Δsl (SLY1526), rad24 (SLY1534), and rad24 mms21Δsl (SLY1543) strains were grown to midlog phase and analyzed for MMS and HU sensitivity.
Fluorescence-activated Cell Sorting (FACS) Analysis
For flow cytometric DNA analysis, yeast cells were fixed in 70% ethanol. Cells were then washed in 1× PBS, mildly sonicated (Branson Sonifier; duty cycle, 40%; six to seven pulses; 1 s each), and pelleted briefly. Cells were then resuspended in 1× PBS with 0.25 mg/ml RNase A and incubated overnight at 37 °C. Propidium iodide was added to the sample to a final concentration of 16 μg/ml, and DNA analysis was performed on a BD Biosciences FACScan. The histograms were plotted and overlaid with Win MDI 2.8 software using standard parameters.
RESULTS
Mms21/Nse2 C-terminal SUMO Ligase Domain Is Required during Unchallenged Mitotic Cell Cycle
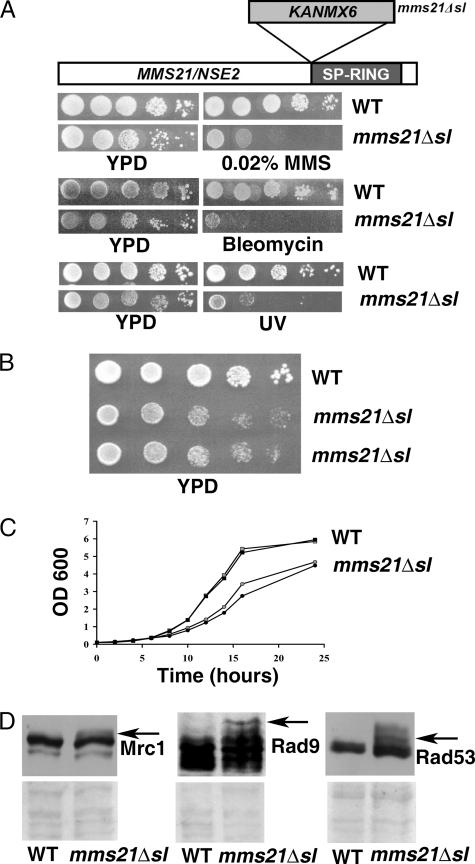
The C-terminal SUMO E3 ligase domain of Mms21/Nse2 is required for resisting extrinsically induced genotoxic stress but has been reported to be dispensable for the essential function of Nse2 (28, 29). To study the function of the SUMO ligase activity of Mms21, we created a mutant mms21Δsl, which results in truncation of the Mms21 protein prior to the E3 ligase domain, by insertion of the KanMX6 selectable marker prior to the start of the E3 ligase coding sequence. The mutant cells were sensitive to various genotoxic agents such as MMS, bleomycin, and UV irradiation as expected (28) (Fig. 1A). Although viable, we observed that the mms21Δsl cells were slow growing when compared with wild-type cells on solid and liquid media (Fig. 1, B and C).
FIGURE 1.
Characterization of growth properties of SUMO ligase-defective mms21Δsl mutant. A, sensitivity of mms21Δsl cells to DNA damage induction. A schematic showing the strategy for creation of mms21Δsl::KanMX6 mutant is shown at the top. To confirm the sensitivity of the nse2Δsl mutant cells to extrinsic genotoxic stress, WT (YPH499) and mms21Δsl (SLY792) strains were grown to midlog phase, serially diluted (10-fold), spotted onto YPD containing MMS (0.02%) or bleomycin (5 μg/ml) or subjected to UV irradiation (4000 μJ/cm2), and incubated for 3–4 days at 23 °C. B, slow growth of unchallenged mms21Δsl cells. WT (YPH499) and mms21Δsl (SLY792) strains were grown to midlog phase, serially diluted, spotted on a YPD plate, and incubated for 3 days. C, the graph depicts the slow growth of mms21Δsl mutant cells (circles) relative to wild-type cells (squares) in liquid medium. D, accumulation of checkpoint signals in unchallenged mms21Δsl mutant cells. Whole cell extracts from midlog phase asynchronous cultures of MRC1-3HA (SLY861), Mrc1-3HA/mms21Δsl (SLY872), Rad53-3HA (SLY1100), Rad53-3HA/mms21Δsl (SLY1104), Rad9-3HA (SLY1095), and Rad9-3HA/mms21Δsl (SLY1098) strains were resolved by SDS-PAGE and analyzed by immunoblotting using anti-HA antibody. The arrows indicate the locations of modified forms of the activated checkpoint proteins Mrc1, Rad9, and Rad53. The lower panels are corresponding regions from the Ponceau S-stained Western blots.
To investigate whether the slow growth may be the consequence of activation of a checkpoint pathway resulting from enhanced occurrence of spontaneous DNA damage in the mutant cells, we assessed the modification status of various checkpoint proteins by Western blot analysis of cell lysates derived from wild-type or mms21Δsl mutant cells expressing HA epitope-tagged variants of the checkpoint genes. DNA damage induces Mec1 kinase-dependent phosphorylation of Rad9 and Rad53 (36–38). In addition, upon induction of replication stress, Mrc1 is phosphorylated by Mec1 kinase, also ultimately resulting in Rad53 phosphorylation (39, 40). Bands corresponding to phosphorylated forms of Rad9 and Rad53 were clearly visible in lysates from mutant cells but not wild-type cells; subtle modification of Mrc1 was also observed in the corresponding mutant cell lysates (Fig. 1D). This observation suggests that mms21Δsl mutant cells spontaneously incur DNA damage, resulting in activation of the DNA damage checkpoint pathway to a limited extent even in unchallenged mitotically dividing cells.
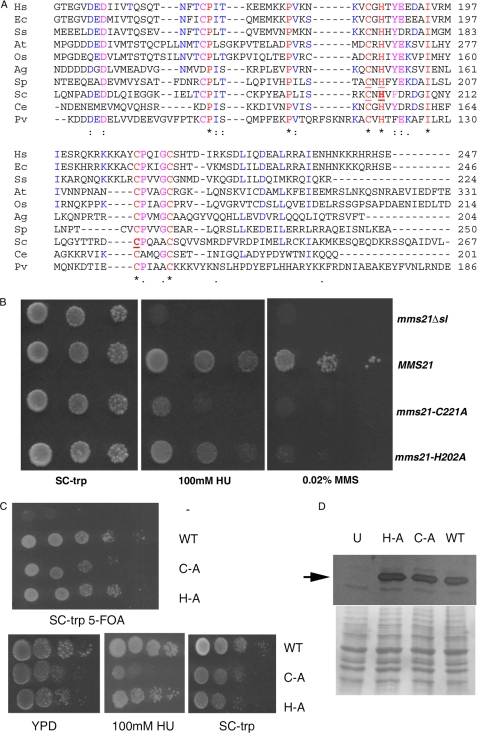
Cysteine 221 of Mms21 SUMO Ligase Domain is Essential for Genotoxic Stress Tolerance and Normal Growth of Unchallenged Mitotically Dividing Cells
The mms21Δsl mutant lacks the SUMO E3 ligase domain and a short C-terminal region of ∼40 amino acids following it. To directly test whether the phenotypes reported above are a direct consequence of a defect in the function of the SUMO ligase domain of Mms21, we attempted to make mutations in conserved residues in the SUMO E3 ligase catalytic domain based on homology-based sequence alignment. Multiple sequence alignment of the sequence of Mms21 with other SUMO ligases revealed a limited number of conserved residues in the SPL-RING domain including four cysteine residues (Cys-184, Cys-200, Cys-221, and Cys-226) and one histidine (His-202) that are highly conserved (Fig. 2A) in agreement with an earlier study (41). A recent structural analysis of Mms21 associated with Smc5 arm revealed that Cys-200, His-202, Cys-221, and Cys-226 are engaged in coordinating a zinc ion in Loop 2 of the SPL-RING domain (41). Inclusion of two worm and insect Mms21 orthologs from C. elegans and A. gambiae revealed that substitutions at position Cys-184 are tolerated; i.e. this conserved residue located in Loop 1 of the SPL-RING domain of Mms21 is not conserved in all orthologs (Fig. 2A). Earlier studies in S. pombe revealed that mutation of Cys-195 and His-197 in the Nse2 RING finger-like motif abolishes Nse2-dependent sumoylation, and nse2.SA cells, which have the nse2.C195S/H197A mutant allele, are sensitive to DNA-damaging agents and to exposure to hydroxyurea (29). Furthermore, mutations in analogous residues Cys-200 and His-202 in S. cerevisiae (mms21-CH allele) result in accumulation of X-molecules at MMS-damaged forks (31). We created new single point mutations in His-202 and Cys-221 (which has not been mutated earlier in any of the Mms21 orthologs) in which these conserved histidine or cysteine residues were substituted by alanine (Fig. 2A). The mms21-C221A and mms21-H202A mutant cells were sensitive to MMS and HU (Fig. 2B) like the mms21Δsl cells. Upon prolonged incubation up to 7 days (HU panel, middle) or longer (9 days; MMS panel, right) we observed that mms21-H202A mutant cells were less sensitive to MMS- or HU-induced genotoxic stress relative to mms21-C221A cells, which are strongly sensitive. Our findings reveal that cysteine 221 is a key functionally important residue of the SPL-RING ligase domain required for the role of Mms21 in resisting genotoxic stress.
FIGURE 2.
Genotoxic stress sensitivity of strains having point mutations in conserved residues of SPL-RING domain of Mms21. A, multiple sequence alignment showing conserved residues (depicted in bold) of S. cerevisiae Mms21 (His-202 and Cys-221) that were mutated to alanine in this study. Other known functionally important residues of S. cerevisiae and S. pombe Mms21/Nse2 proteins are underlined. “*” denotes identical residues (100% identity), “:” denotes conserved substitutions, and “.” denotes semiconserved substitutions. The color denotes regions having 100% identity (red), 80–90% identity (magenta), and 50–70% identity (blue) among the Mms21-related proteins shown. The sequences included are H. sapiens (Hs), E. caballus (Ec), S. salar (Ss), A. thaliana (At), O. sativa Indica group (Os), A. gambiae (Ag), S. pombe (Sp), S. cerevisiae (Sc), C. elegans (Ce), and P. vivax (Pv). B, HU and MMS sensitivity of mutants having C221A and H202A substitutions. Haploid strains having the mms21Δsl mutation (SLY792) and yeast centromeric plasmid pRS314 (no insert), pRR51 (expressing MMS21), pRR55 (mms21-C221A), or pRR56 (mms21-H202A) were grown to midlog phase, serially diluted, spotted onto an SC-Trp (Trp omission) plate and SC-Trp plates containing 100 mm HU or 0.02% MMS, and incubated at 23 °C. Images were recorded after 7 days in the case of SC-Trp and 100 mm HU plates and after 9 days in the case of 0.02% MMS-containing plates. C, slow growth of unchallenged strains having the mms21-C221A and mms21-H202A point mutations. A haploid strain (SLY782) having the mms21::KanMX6 null allele complemented by pRR50 having MMS21 on a centromeric URA3-bearing plasmid was transformed with yeast centromeric plasmid pRS314 (no insert; indicated by −), pRR51 (expressing MMS21; indicated by WT), pRR55 (mms21-C221A; indicated by C-A), or pRR56 (mms21-H202A; indicated by H-A) and subjected to counterselection on SC-Trp 5-FOA plates. Top panel, growth on counterselective medium (3 days) following one round of incubation on 5-FOA. Bottom panel, growth of strains (which had lost pRR50 after plasmid shuffling) having WT (SLY1620), mms21-C221A (C-A) (SLY1621) point mutation, or mms21-H202A (H-A) (SLY1622) point mutation on YPD (2.5 days), 100 mm HU (9 days), or SC-Trp (3 days) plates. D, Western blot analysis of lysates from cells expressing HA epitope-tagged mms21-H202A, mms21-C221A, and MMS21 proteins using anti-HA antibody. U, untagged lysate. The arrow indicates the position of 3HA-tagged Mms21 proteins. The bottom panel is a Ponceau S-stained image of the same blot showing equivalent loading of samples.
We further examined whether the slow growth observed in the mms21Δsl cells resulted from a defect solely in the SPL-RING domain using the mutants having substitutions in the conserved residues of the SPL-RING domain described above. Mutant cells having the mms21::KanMX6 null allele complemented by a centromeric plasmid having MMS21 and URA3 marker were transformed with centromeric plasmids bearing either wild-type MMS21, mms21-H202A, mms21-C221A, or no insert. Strains that had lost the URA3 plasmid bearing MMS21 were obtained by plasmid shuffling on plates having 5-FOA. The mms21-C221A mutant cells showed slow growth on SC-Trp 5-FOA, YPD, and SC-Trp media unlike wild-type MMS21 (Fig. 2C), indicating that Cys-221 of the SPL-RING domain is required for optimal growth of unchallenged mitotically dividing cells. Furthermore, these phenotypic differences are unlikely to arise from differential stability or expression of the mutant proteins as indicated by detection of equivalent levels of wild-type and mutant Mms21 proteins in cell extracts (Fig. 2D, top panel) having comparable protein concentration (Fig. 2D, bottom panel).
Activation of Mec1-dependent Checkpoint Pathway Is Important for Survival of mms21Δsl Cells
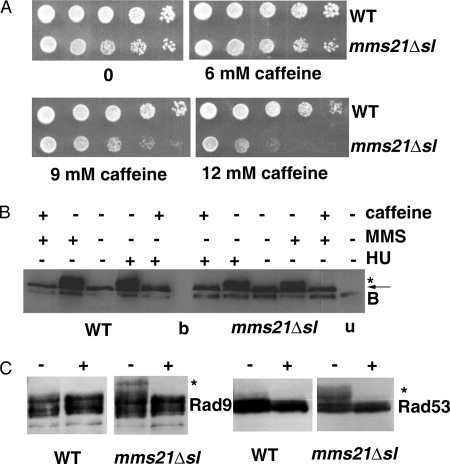
The DNA damage checkpoint pathway is activated in response to DNA damage and delays cell cycle progression in the presence of damaged DNA, thereby facilitating repair of the damage prior to segregation (1, 42). We observed modest activation of the DNA damage checkpoint pathway in the mms21Δsl mutant cells as described earlier. To test whether the activation of this checkpoint pathway was important for survival of the mutant cells, we used a chemical-genetic approach. Caffeine is an inhibitor of the human phosphatidylinositol 3-kinases Ataxia telangiectasia mutated and Ataxia telangiectasia and Rad3 related, which are similar to the Mec1/Tel1 kinases in S. cerevisiae, and it overrides the Ataxia telangiectasia mutated/Ataxia telangiectasia and Rad3 related-dependent checkpoint pathways in irradiated human cells (43–46). We compared the caffeine sensitivity of mms21Δsl mutant cells with wild-type cells and found that unchallenged mutant cells were caffeine-sensitive (Fig. 3A).
FIGURE 3.
Caffeine-sensitive checkpoint pathway signaling is important for survival of unchallenged mms21Δsl cells. A, caffeine-sensitive growth of mms21Δsl mutant cells. Wild type (SLY1351) and mms21Δsl (SLY1415) were grown to midlog phase, serially diluted, and spotted onto a YPD agar plate (top left) and YPD agar plates having the indicated concentrations of caffeine. B, caffeine overrides activation of Mrc1 in HU- and MMS-treated cells. HU (150 mm) or MMS (0.03%) was added to asynchronous cultures of strains Mrc1-3HA (SLY1351) and Mrc1-3HA/mms21Δsl (SLY1415) growing at 25 °C in YPD medium. After 2 h of incubation, caffeine (10 mg/ml) was added to one-half of the culture and further incubated for 1 h. Whole cell extracts were resolved by SDS-PAGE and analyzed by immunoblotting using anti-HA antibody. The arrow indicates the location of Mrc1, * indicates location of the replication stress-induced modified Mrc1, and the label B indicates an anti-HA cross-reactive background band present in these cell lysates. The label b indicates a blank lane, and u denotes a lane having lysate from the parental untagged strain. C, caffeine abrogates activation of Rad9 and Rad53 in unchallenged mms21Δsl cells. Rad53-3HA (SLY1100), Rad53-3HA/mms21Δsl (SLY1104), Rad9-3HA (SLY1095), and Rad9-3HA/mms21Δsl (SLY1098) strains were grown to midlog phase, caffeine (10 mg/ml) was added to one-half of the culture, and strains were incubated further for 1 h. Whole cell extracts were resolved by SDS-PAGE and analyzed by Western blotting using the anti-HA antibody. + indicates samples in which caffeine was added, − indicates that no caffeine was added, and * indicates the location of caffeine-sensitive modified bands of Rad9 (left) or Mrc1 (right).
To confirm whether caffeine indeed inhibits Mec1 kinase in S. cerevisiae, we examined the effect of caffeine on HU- and MMS-induced Mec1-dependent modification of Mrc1 (Fig. 3B). Both HU and MMS induced the modification of Mrc1 upon treatment with MMS and HU in WT and mms21Δsl mutant cells as expected. The modified forms of Mrc1, which are Mec1-dependent (40), were not produced in the presence of caffeine (Fig. 3B), demonstrating that caffeine indeed abrogates the activity of the Mec1 kinase in budding yeast cells. Addition of caffeine also abolished the modified forms of Rad53 and Rad9 that were detected in extracts from unchallenged mms21Δsl mutant cells (Fig. 3C).
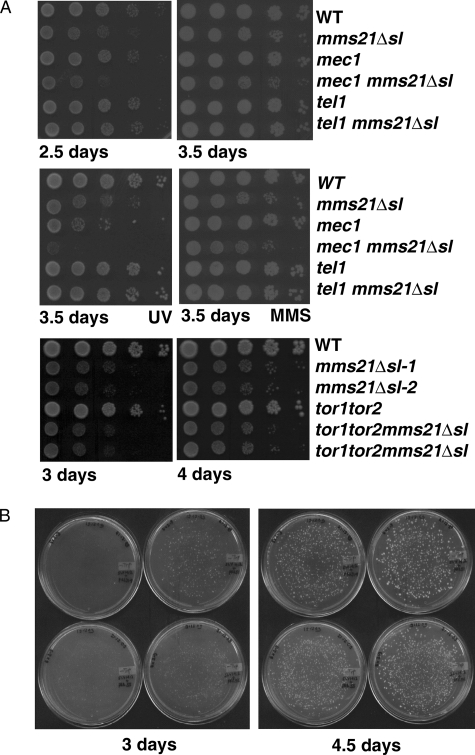
In addition to Mec1, other potential cellular targets of caffeine include the Ataxia telangiectasia mutated-related kinases Tel1 and Tor1 (47). To determine the individual contribution of these kinases to the survival of mms21Δsl mutant cells, we constructed double mutants of mms21Δsl in combination with mec1, tel1, tor1, and tor2. When the SUMO ligase of Mms21 was inactivated in a mec1 mutant strain (also having a deletion of sml1 that bypasses the essential function of MEC1 (48)), we found that the mms21Δsl mec1 double mutant selectively exhibited a clear synthetic sick phenotype (Fig. 4A, top left panel). However, mms21Δsl combined with other putative caffeine targets such as tel1 did not exhibit synthetic lethality, and when the mms21Δsl mutation was created in mutants defective in tor1 and tor2 kinases, only a very subtle growth defect was seen in the tor1 tor2 mms21Δsl triple mutant (Fig. 4A, bottom panels). We also found that the mms21Δsl mec1 double mutant was highly sensitive to UV radiation compared with either single mutant or similarly treated wild-type cells (Fig. 4A, middle left panel). Furthermore, the synthetic sick phenotype of the mec1 mms21Δsl double mutant cells could be complemented efficiently by MMS21 but not by mms21-C221A (Fig. 4B, left panel, top right and bottom left plates), whereas the complementation by mms21-H202A was to an intermediate level (Fig. 4B, left panel, bottom right plate), although the transformation efficiency in all cases was equivalent (Fig. 4B, bottom right panel showing plates incubated for 4.5 days). Collectively, these results suggest that activation of a Mec1 kinase-dependent checkpoint pathway is required for viability of mitotically dividing SUMO ligase SPL-RING domain-defective mms21 mutant cells.
FIGURE 4.
A, genetic interactions of mms21Δsl with mutants defective in known caffeine targets. WT (VP106), mec1 (also having the sml1 mutation), tel1 (SLY1353), mec1 mms21Δsl (also having the sml1 mutation; SLY1412), and tel1 mms21Δsl (SLY1416) strains were grown to midlog phase, serially diluted, and spotted on YPD plates (top panels) and further analyzed for UV radiation (500 μJ/cm2) or MMS (0.0025%) sensitivity (middle panels). The bottom panels show WT (Jka-3da), tor2 (JK350-18a), tor1 tor2 (JK350-21a), mms21Δsl (SLY1552), and tor1 tor2 mms21Δsl (SLY1549) strains that were grown to midlog phase, serially diluted, spotted on YPD plates, and incubated for 3 or 4 days at 23 °C. B, genetic interaction of mms21-C221A and mms21-H202A point mutants with mec1. A haploid strain having the mms21Δsl and mec1 mutations (SLY1412) was transformed with the centromeric plasmid pRS314 (no insert; top left), pRR51(expressing MMS21; top right), pRR55(mms21-C221A; bottom left), or pRR56 (mms21-H202A; bottom right); plated on SC-Trp plates; and incubated at 23 °C. Images were recorded 3 and 4.5 days after plating. Representative data from three independent transformation experiments are shown.
Chromosome Integrity in Wild-type and mms21Δsl Cells
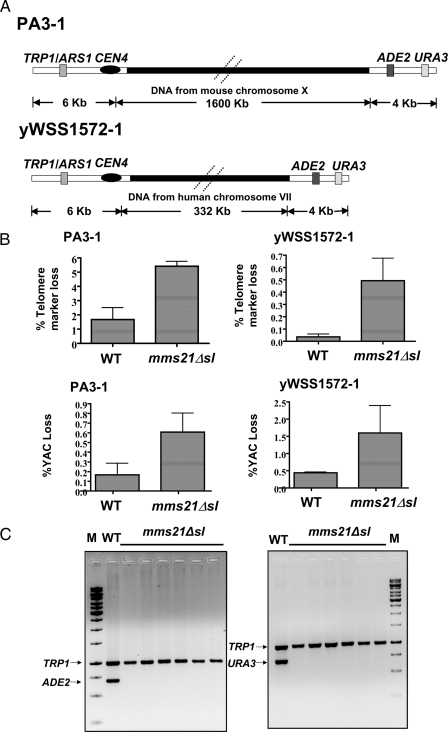
The requirement for Mec1 kinase-dependent signaling for survival of mms21Δsl cells suggests the presence or persistence of damaged DNA in these cells. To investigate whether damaged or broken chromosomes could be produced at a higher frequency in mms21Δsl mutant cells, we tested the maintenance of genomic integrity and stability using a previously described YAC-based genetic assay for scoring gross chromosomal rearrangements (35) in these cells. This assay uses two extremely telocentric YACs (PA3-1 and yWSS1572-1) having a short arm of 6 kb and a long arm of 330 kb (yWSS1572-1) or 1600 kb (PA3-1) (Fig. 5A). The short arm has a selectable marker, TRP1, whereas the telomere proximal end of the long arm has two markers, ADE2 and URA3. Because these YACs are dispensable for growth, DSBs that lead to very large gross chromosomal rearrangements are tolerated. Loss of integrity as well as stability can be measured using this assay by scoring for telomere marker loss (indicating chromosome breakage) or loss of telomere as well as centromere-proximal markers (indicating a chromosome loss event). Telomere marker loss and total YAC loss were quantified for wild-type and mms21Δsl strains harboring the YACs PA3-1 and yWSS1572-1 grown at 23 °C. The telomere marker loss was higher in mms21Δsl cells relative to wild-type cells for the YACs PA3-1 and yWSS1572-1 (Fig. 5B, top panels). A small but reproducible enhancement in the loss of both genetic markers on the YACs (indicating destabilization or loss of the YAC) was also observed in the mutant relative to wild-type cells (Fig. 5B, bottom panels). To confirm that the loss of the selectable phenotype corresponding to the telomere-proximal marker (resulting in uracil auxotrophy) observed in this genetic assay was indeed a consequence of actual loss of the marker and was not due to enhancement of telomeric silencing as was reported in the case of the mms21Δsl mutant earlier (28), we carried out PCR to amplify the YAC-specific markers. Genomic DNA was isolated from random Trp+ura− colonies that were scored as having lost the telomere-proximal marker in the mutant, and PCR was performed to amplify the sequences corresponding to the telomere-proximal markers ADE2 and URA3 and the centromere-proximal marker TRP1. The presence of the TRP1-specific band but absence of ADE2- and URA3-specific bands confirmed the occurrence of a chromosome break in these isolates (Fig. 5C).
FIGURE 5.
Measurement of chromosome integrity in wild-type and mms21Δsl mutant cells. A, quantitative genetic assay for telomere marker loss and total YAC loss. The schematic shows the organization of the YACs used for the gross chromosomal rearrangement assay (adapted from Ref. 35). B, chromosome instability in mms21Δsl cells. Shown is the quantification of telomere marker loss (YAC integrity; top panels) and total YAC loss (YAC stability; bottom panels) plotted as percent loss on the y axis in WT (DH2004-1/101 and DH1003/102) and mms21Δsl (SLY1344 and SLY1348) cells that harbored the YACs PA3-I (left panels) and yWSS1572-1 (right panels). C, confirmation of telomere marker loss or chromosome breakage. Genomic DNA was prepared from randomly isolated Trp+ura− mms21Δsl cells identified in B and subjected to PCR analysis (to amplify telomere-proximal markers ADE2 and URA3 or the centromere-proximal marker TRP1) to confirm chromosome breakage in samples scored as telomere marker loss events. M, molecular weight marker; WT, PCR analysis of DNA isolated from wild-type cells bearing the intact YAC.
Replication Stress Sensitivity of SUMO Ligase-deficient mms21 Mutant Cells
Because the function of the Smc5/6 complex has been ascribed to S phase, we tested the sensitivity of mms21Δsl cells to the replication inhibitor HU, which causes replication delay due to reduction of production of deoxyribonucleotides by selective inhibition of ribonucleoside-diphosphate reductase, an enzyme required to convert ribonucleoside diphosphates into deoxyribonucleoside diphosphates (49). HU induces replication stress resulting in the activation of the DNA replication stress checkpoint pathway, which delays S phase progression until DNA replication is completed (5). We found that mms21Δsl cells were strongly sensitive to HU (Fig. 6A).
Genetic interactions of mms21Δsl mutant with various caffeine targets revealed a requirement for Mec1 kinase-dependent signaling for survival of mms21Δsl cells (Fig. 4). Mec1 initiates signal transduction in response to both DNA damage and replication stress (for a review, see Ref. 50). Therefore, to test the contribution of these two overlapping but independent pathways for optimal growth of mms21Δsl cells, we extended our genetic interaction studies to include additional checkpoint pathway genes such as MRC1, RAD9, and RAD24.
Mrc1 is a replication fork component that is required to activate Rad53 in response to replication stress (39). We found that unchallenged mrc1 mms21Δsl cells did not exhibit any additional growth defect when compared with either single mutant alone (Fig. 6B). Because both mrc1 and mms21Δsl cells are exquisitely sensitive to HU, we used low doses of HU to investigate whether there was any genetic interaction between these mutants. Slight growth retardation of each single mutant relative to wild-type cells was observed at doses of 10 and 25 mm HU (Fig. 6B). However, the double mutant did not exhibit significant enhancement in sensitivity to HU relative to the single mutants under these conditions (Fig. 6B, top panel). Likewise, the phenotype of mrc1 mms21Δsl cells was indistinguishable from that of mms21Δsl single mutant cells when subjected to low doses of MMS (Fig. 6B, bottom panels). These observations suggest that the effect of the mms21Δsl mutation is epistatic to Mrc1.
Genetic Interactions of mms21Δsl with DNA Damage Checkpoint Pathway Genes
Rad9 and Rad24 participate in two additive branches of the DNA damage checkpoint pathway that converge on Mec1 and Rad53 and are required to modify and activate Rad53 in response to DNA damage (51). Mutants defective in Mms21 SUMO ligase and DNA damage checkpoint genes encoding Rad9 or Rad24 also do not exhibit any additional growth defect compared with the single mutants under normal growth conditions (Fig. 6C) perhaps due to functional overlap between the two respective branches of the damage response pathway that may be sufficient to cope with low levels of DNA damage occurring in unchallenged cells. However, upon treatment with the replication inhibitor HU, both rad9 mms21Δsl and rad24 mms21Δsl double mutants exhibited a synthetic sick phenotype compared with the respective single mutants. Likewise, MMS treatment revealed a clear synthetic sick growth phenotype in the rad9 mms21Δsl and rad24 mms21Δsl double mutants compared with the single mutants (Fig. 6C). Thus, replication-stressed mms21Δsl cells require both Rad9- and Rad24-dependent signaling to cope with resultant DNA damage.
Cell Cycle Progression Defects in Transiently Replication-stressed SUMO Ligase-deficient Cells
The activation of the DNA damage checkpoint generally results in a delay in cell cycle progression to enable recovery from the DNA damage. To test whether replication-stressed Mms21 SUMO ligase-deficient cells show a delay in cell cycle progression, we subjected wild-type and mms21 mutant cells to HU-induced replication stress and subsequently released the S phase-arrested cells from replication stress by transfer to medium lacking HU. We analyzed the distribution of cells having varying DNA content by determining the FACS profile of wild-type, mms21Δsl, and mms21-C221A cells at varying intervals after release from arrest. Following release from replication stress, all three types of cell populations were able to progress and double their DNA content as revealed by the appearance of peaks corresponding to 2C DNA content (Fig. 7A). As expected, wild-type cells were able to progress further and partition DNA to give rise to haploid cells as suggested by the appearance of a prominent peak having 1C DNA content in these samples. However, only a very small peak corresponding to 1C DNA content could be detected in the case of mms21Δsl and mms21-C221A cells even after several hours of release from HU-induced replication stress (Fig. 7A), suggesting that these cells are defective in progression through mitosis. Similar results were obtained in the case of cells having MMS21 (wild type) or mms21-C221A expressed from centromeric plasmids in a strain background having a null allele, mms21::KanMX6, as the sole chromosomal copy (Fig. 7B). In addition, mms21Δsl cells consistently showed a small peak corresponding to cells having greater than 2C DNA content, indicating formation of hyperploid cells (Fig. 7A, middle panel).
FIGURE 7.
Cell cycle arrest in SUMO ligase-deficient cells released from transient HU-induced replication stress. A, FACS profiles showing the distribution of cells having varying DNA content were determined for cells having the mms21Δsl mutation (SLY792) transformed with yeast centromeric plasmid pRR51 (expressing MMS21; top), pRS314 (no insert; middle; labeled as mms21Δsl), or pRR55 (mms21-C221A; bottom). In each case, the series of histograms represent (from bottom to top) untreated cycling cells having an A600 of ∼0.2, cells treated with 150 mm hydroxyurea for 3 h, and cells that were released from HU-induced arrest for varying intervals (10 min, 30 min, 40 min, 60 min, 1 h 20 min, 1 h 40 min, 2 h, 2 h 20 min, 2 h 40 min, 3 h, 3.5 h, 4 h, 4.5 h, and 5 h). The arrows indicate the position of peaks corresponding to 1C and 2C DNA content. B, FACS analysis of strains having mms21::KanMX6 null allele complemented by centromeric plasmid expressing wild-type (MMS21, SLY1620; top panel) or SUMO ligase-defective mutant (mms21-C221A, SLY1621; bottom panel) Mms21 proteins. Samples were treated with HU as in A and released into YPD for varying intervals (20 min, 40 min, 1 h, 1 h 20 min, 1 h 40 min, 2 h, 3 h, 4 h, 5 h, 6 h, 8 h, and 10 h).
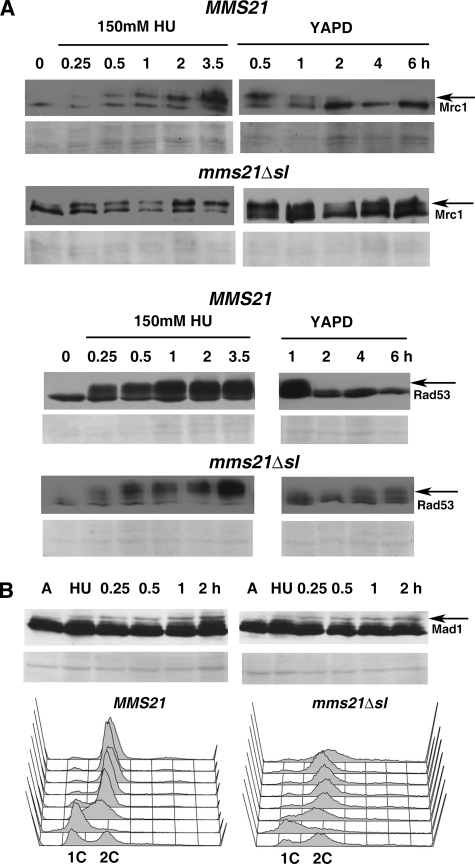
Because transiently HU-stressed mutant cells showed a mitotic progression defect, we tested whether this could result from a defect in the activity of one or more mitotic checkpoints. We compared the activation of the replication stress checkpoint between wild-type and mms21Δsl mutant cells by analyzing HU-induced Mec1-dependent modification of Mrc1, the mediator of the replication checkpoint, and Rad53, which is downstream in this pathway (Fig. 8). Modified forms of Mrc1 and Rad53 could be readily detected after addition of HU in both wild-type and mms21Δsl mutant cells (Fig. 8A). However, upon release from HU-induced replication stress, we found that modified forms of Mrc1 and Rad53 persisted in mutant cells up to 6 h after release, whereas they could no longer be detected in wild-type cells after 2 h of release. The persistence of replication stress checkpoint signals indicates that mutant cells are unable to recover from replication stress fully perhaps due to defects downstream of Rad53 signaling. Upon progression, these cells may accumulate DNA damage resulting in mitotic arrest as observed (Fig. 7). Phosphorylation of Rad53, as observed in mutant cells, is known to contribute to the G2M DNA damage checkpoint (50) and may explain the observed mitotic arrest in mutant cells. Because mms21Δsl cells accumulate tetraploid cells when they are subjected to transient replication stress, we tested the activity of the spindle assembly checkpoint in these cells in the presence of nocodazole, a drug that inhibits proper spindle assembly. We observed that when cells that had been treated with HU for 3.5 h (as in our earlier experiments) were released from HU-induced arrest into medium having nocodazole both wild-type and mms21Δsl mutant cells were able to activate the spindle assembly checkpoint as evidenced by the appearance of a modified form of Mad1, a spindle assembly checkpoint protein that is known to be phosphorylated in response to nocodazole (Fig. 8B). Furthermore, both wild-type and mms21Δsl mutant cells were arrested with a 2C DNA content up to 4 h after nocodazole treatment (Fig. 8B) as would be expected if the spindle assembly checkpoint were functional.
FIGURE 8.
Activation of mitotic checkpoint pathways in SUMO ligase-deficient cells. A, analysis of the activity of the replication stress checkpoint pathway. Wild-type (MMS21) or mms21Δsl cells having HA-tagged Mrc1 were subjected to HU-induced replication stress for varying time periods up to 3.5 h (left panels), and total lysates were analyzed by SDS-PAGE followed by Western blotting using anti-HA antibody (left panels). Modification of Mrc1 was observed in wild-type (top left panel) as well as mutant (bottom left panel) cell extracts. Cells were released from HU-induced arrest into YAPD medium lacking HU for varying time periods and analyzed for persistence of modification of Mrc1 (right panels), which was preferentially observed in the case of mutant cells. Mrc1 indicates the location of unmodified form; the arrow indicates the location of replication stress-induced modified forms. Similarly, wild-type (MMS21) or mms21Δsl cells having HA-tagged Rad53 were analyzed to detect the formation of HU-induced modified forms of Rad53 (position indicated by arrows). A, asynchronous cells prior to addition of HU. The lower panels are the corresponding regions from the Ponceau S-stained Western blots. B, activation of the spindle assembly checkpoint pathway in replication-stressed cells that were released into nocodazole (30 μg/ml)-containing medium for varying time intervals. Total lysates from wild-type (left) or mms21Δsl (right) cells were analyzed by SDS-PAGE followed by Western blotting using anti-Mad1 antibody (top panels). A, asynchronous cells prior to addition of HU; HU, cells treated with 150 mm HU for 3.5 h; the arrow indicates the position of the modified form of Mad1 that was observed after treatment with nocodazole. The lower panels are the corresponding regions from the Ponceau S-stained Western blots. Histograms show the results of FACS analysis, confirming mitotic arrest in nocodazole-treated wild-type (MMS21; left) and mms21Δsl (right) cell populations that were treated with HU for 3.5 h and released into nocodazole-containing medium (from bottom to top: asynchronous, 150 mm HU for 3.5 h, and released into nocodazole (30 μg/ml)-containing medium for 0.5, 1, 1.5, 2, 3, and 4 h).
DISCUSSION
Maintenance of genomic stability is achieved by a number of mechanisms that act in concert to ensure accurate transfer of unaltered genetic information to progeny cells. These include DNA repair systems, cell cycle checkpoint signaling pathways, and elaborate machinery including the mitotic spindle, kinetochores, and the chromosomes themselves, which facilitate high fidelity chromosome segregation. Post-translational modifications of proteins (e.g. phosphorylation, ubiquitination, acetylation, etc.) play an important role in regulating these complex events. Sumoylation is a recently described modification in which a ubiquitin-like protein, SUMO, is attached to various protein targets by an enzymatic mechanism similar to ubiquitination involving E1-activating, E2-conjugating, and E3 ligase enzymes (27, 52). The requirement of sumoylation in resisting genotoxic stress has been documented previously (28). In this study, we report the requirement for the SUMO ligase associated with the Smc5/6 complex in the maintenance of chromosomal stability in unchallenged mitotically dividing cells.
Previous studies have established a role for Mms21/Nse2 SUMO ligase in resisting extrinsically induced genotoxic stress (28). We found that the Mms21 SUMO ligase activity was required even when no external stress was imposed on the cells. Our findings that the DNA damage checkpoint was activated and that Mec1 kinase was required for the survival of cells lacking Mms21 SUMO ligase activity suggest that these cells may incur DNA damage resulting in lesions that activate the DNA damage checkpoint and delay the growth rate. Although the nature of these lesions is not known, it is likely that they occur at a high enough frequency and severity in unchallenged cells such that activation of the Mec1-dependent checkpoint pathway becomes crucial for the survival of these cells. We speculate that Mms21-mediated sumoylation of one or more Mms21 sumoylation targets may be crucial for maintenance of genomic integrity in cells undergoing mitotic cell division in S. cerevisiae. This observation contrasts with the role of Nse2-mediated sumoylation in S. pombe in which the SUMO ligase activity of Nse2 was reported to be dispensable for the viability of unchallenged mitotically dividing cells (29). It is possible that this disparity may arise from a difference in the functions and/or repertoire of Mms21/Nse2 sumoylation targets in these different organisms.
Our chemical and genetic interaction studies with various replication stress inducers and mutants defective in checkpoint pathway proteins revealed that Mms21 SUMO ligase activity was required to counter replication stress, and its inability to do so may result in DNA damage. Mms21 SUMO ligase-defective mutants were sensitive to hydroxyurea, a replication inhibitor. Our finding that the sensitivity of the Rad9/Rad24 and Mms21 SUMO ligase-deficient cells to HU was enhanced suggests that the activation of a (Rad9- or Rad24-dependent) DSB-specific checkpoint is important for survival for Mms21 SUMO ligase-defective cells that are subjected to replication stress. This suggests that replication-stressed mms21Δsl cells may generate DSBs that are lethal in the absence of active DNA damage checkpoint pathway signaling. DSBs could result if there is a defect in the initiation or transduction of the replication stress checkpoint signal or in one or more downstream events (e.g. maintenance of cell cycle arrest or defective recovery of stalled or collapsed replication forks) required to counter replication stress. Likewise, similar Mms21 SUMO ligase-dependent defects in intra-S phase events may also occur when replication forks spontaneously collapse in unchallenged cells (albeit at a lower frequency relative to HU- or MMS-treated cells), resulting in the persistence of DSBs, slow growth, dependence on Mec1 signaling, and chromosome breakage as observed in our study.
Our finding that the sensitivity of the mrc1 mms21Δsl double mutant to HU was not significantly greater than either single mutant suggests that mms21Δsl is epistatic with mrc1 and may function within the intra-S phase signaling or related events. Failure of proper recovery from HU-induced replication stress resulted in a delay in progression through mitosis in transiently replication-stressed Mms21 SUMO ligase-deficient cells. Although the reason for HU-induced replication stress sensitivity of mms21Δsl budding yeast cells is unclear, insight into the resultant molecular defects in mms21Δsl cells has been obtained in the case of MMS-induced DNA damage. Branzei et al. (31) have found that mutants defective in Mms21 accumulate branched X-shaped DNA structures at damaged forks in the presence of MMS but not in the presence of HU. In budding yeast, Smc6 accumulates at replication forks in RAD53-deleted cells when arrested with HU (13). Because arrested forks collapse in the absence of an intact checkpoint, the accumulation of Smc5/6 at these sites indicated that this complex is recruited to collapsed forks. In light of these observations, it is plausible that Mms21-dependent sumoylation may be required for the role of Smc5/6 complex in recovery/rescue events at these sites even when these errors occur spontaneously during unperturbed DNA replication. Failure of recovery from fork collapse may result in DSBs, cell cycle arrest, and broken chromosomes as observed in our study.
Recent findings reveal additional roles for the Smc5/6 complex in chromosome maintenance. For example, in mammalian cells, MMS21 has a role in sister chromatid cohesion and mitotic progression (53). In human cancer cells that have inactive telomerase and show alternative lengthening of telomeres (ALT cells) by telomere homologous recombination, the Smc5/6 complex is required for telomere maintenance and prevention of senescence (54). RNA interference-mediated knockdown of SMC5/6 subunits inhibits telomere homologous recombination, resulting in telomere shortening and senescence in ALT cells. Furthermore, human MMS21 sumoylates two subunits, TRF2 and RAP1, of the telomere-associated shelterin complex. Inhibition of TRF2 sumoylation prevents formation of ALT-associated promyelocytic leukemia bodies, which are ALT-specific promyelocytic leukemia bodies with which telomeres are associated. In another related study (55) using yeast telomerase-deficient tlc1Δ mutant cells that show increased telomere loss and senescence, it was observed that SMC5/6 facilitates resolution of sister chromatid homologous recombination intermediates and slows senescence. Although these observations were made using altered (i.e. telomerase-deficient) cell lines, it is possible that defects in analogous MMS21-mediated chromosomal transactions in normal cells could also result in DNA damage in Mms21 SUMO ligase-deficient mutants.
More compelling evidence for a requirement of Mms21 SUMO ligase in normal growth and development comes from recent studies in plant development. In two independent studies, A. thaliana mutants lacking functional AtMMS21 show defects in root meristem development and severe dwarfism (56, 57). Loss of AtMMS21/HPY2 (high ploidy 2) results in premature transition from the mitotic cycle to the endocycle (56). Thus, MMS21 is a negative regulator of the switch from the mitotic to the endoreduplication cycle during which some cell types double their DNA content without corresponding cell division in plants. Flow cytometry analysis of nuclei from aerial tissues of hpy2-1 mutant plants (having a C-terminally deleted allele of AtMMS21) also show additional small 64C and 128C peaks that are not normally present in wild-type plants, suggesting that hpy2-1 cells have undergone additional rounds of endoreduplication (56). These findings are consistent with our observation of accumulation of a subset of hyperploid cells in the Mms21 SUMO ligase-defective mutants, indicating that Mms21 may be involved in a similar switch that represses endoreduplication in yeast cells. We speculate that repression of endoreduplication may represent a conserved function of Mms21 in different organisms.
Because Mms21 is a SUMO E3 ligase, it is likely that many of the phenotypes reported in ligase-defective mutants may arise from impairment of sumoylation of one or more of its substrates involved in mitotic processes. In budding yeast, there are several known sumoylation targets of Mms21 that are involved in the mitotic cycle, e.g. Smc5, Smc6, Yku70 (28), Smc1, Smc3, and Smc2 (58). These substrates are known to affect many processes required for chromosomal integrity (e.g. DNA double strand break repair, cohesion, condensation, etc.), and it is possible that defects in sumoylation of some of these substrates may result in compromised chromosome integrity and slow growth during the unperturbed mitotic cycle.
In conclusion, in this study, we report a requirement for the SUMO ligase activity of Mms21 for the maintenance of chromosome integrity and stability during the unchallenged mitotic cell cycle in S. cerevisiae. Our findings suggest that impairment of Mms21-dependent sumoylation can result in broken chromosomes causing slow growth and dependence on Mec1-mediated signaling. Mms21 SUMO ligase is required for maintaining proper progression of events in the mitotic cell cycle.
Acknowledgments
We are grateful to Steve Elledge, Michael Hall, Doug Koshland, Kevin Hardwick, and Virginia Zakian for providing yeast strains and reagents used in this work. We thank Imran Siddiqi, Jeff Smith, and Doug Koshland for helpful comments on the manuscript; Sumedha Gattani and Monica Agarwal for technical assistance; and members of our laboratory for discussions. Technical support from the Department of Biotechnology-funded FACS facility at the Indian Institute of Science is acknowledged.
This work was supported by a Wellcome Trust International Senior Research Fellowship in Biomedical Science (Grant GR063263MA) and by a grant from the Council for Scientific and Industrial Research, India (to S. L.).
- Smc
- structural maintenance of chromosomes
- MMS
- methyl methanesulfonate
- HU
- hydroxyurea
- Mms21
- MMS-sensitive 21
- SUMO
- small ubiquitin-related modifier
- Nse
- non-smc element
- Mrc1
- mediator of replication checkpoint 1
- DSB
- double strand break
- YAC
- yeast artificial chromosome
- Mec1
- mitosis entry checkpoint 1
- Tor
- target of rapamycin
- Rad
- radiation-sensitive
- SPL-RING
- Siz/PIAS (Protein Inhibitor of Activated STAT)-like-really interesting new gene
- rDNA
- ribosomal DNA
- YPD
- yeast extract-peptone-dextrose
- SC
- synthetic complete medium
- 5-FOA
- 5-fluoroorotic acid
- ALT
- alternative lengthening of telomeres.
REFERENCES
- 1. Hartwell L. H., Weinert T. A. (1989) Science 246, 629–634 [DOI] [PubMed] [Google Scholar]
- 2. Hartwell L. H., Kastan M. B. (1994) Science 266, 1821–1828 [DOI] [PubMed] [Google Scholar]
- 3. Paulsen R. D., Cimprich K. A. (2007) DNA Repair 6, 953–966 [DOI] [PubMed] [Google Scholar]
- 4. Friedel A. M., Pike B. L., Gasser S. M. (2009) Curr. Opin. Cell. Biol. 21, 237–244 [DOI] [PubMed] [Google Scholar]
- 5. Branzei D., Foiani M. (2009) DNA Repair 8, 1038–1046 [DOI] [PubMed] [Google Scholar]
- 6. Losada A., Hirano T. (2005) Genes Dev. 19, 1269–1287 [DOI] [PubMed] [Google Scholar]
- 7. Nasmyth K., Haering C. H. (2005) Annu. Rev. Biochem. 74, 595–648 [DOI] [PubMed] [Google Scholar]
- 8. Huang C. E., Milutinovich M., Koshland D. (2005) Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hirano T. (2006) Nat. Rev. Mol. Cell Biol. 7, 311–322 [DOI] [PubMed] [Google Scholar]
- 10. Peters J. M., Tedeschi A., Schmitz J. (2008) Genes Dev. 22, 3089–3114 [DOI] [PubMed] [Google Scholar]
- 11. Hudson D. F., Marshall K. M., Earnshaw W. C. (2009) Chromosome Res. 17, 131–144 [DOI] [PubMed] [Google Scholar]
- 12. De Piccoli G., Torres-Rosell J., Aragón L. (2009) Chromosome Res. 17, 251–263 [DOI] [PubMed] [Google Scholar]
- 13. Lindroos H. B., Ström L., Itoh T., Katou Y., Shirahige K., Sjögren C. (2006) Mol. Cell 22, 755–767 [DOI] [PubMed] [Google Scholar]
- 14. Sergeant J., Taylor E., Palecek J., Fousteri M., Andrews E. A., Sweeney S., Shinagawa H., Watts F. Z., Lehmann A. R. (2005) Mol. Cell. Biol. 25, 172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lehmann A. R., Walicka M., Griffiths D. J., Murray J. M., Watts F. Z., McCready S., Carr A. M. (1995) Mol. Cell. Biol. 15, 7067–7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McDonald W. H., Pavlova Y., Yates J. R., 3rd, Boddy M. N. (2003) J. Biol. Chem. 278, 45460–45467 [DOI] [PubMed] [Google Scholar]
- 17. Morikawa H., Morishita T., Kawane S., Iwasaki H., Carr A. M., Shinagawa H. (2004) Mol. Cell. Biol. 24, 9401–9413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pebernard S., McDonald W. H., Pavlova Y., Yates J. R., 3rd, Boddy M. N. (2004) Mol. Biol. Cell. 15, 4866–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Torres-Rosell J., Machín F., Farmer S., Jarmuz A., Eydmann T., Dalgaard J. Z., Aragón L. (2005) Nat. Cell. Biol. 7, 412–419 [DOI] [PubMed] [Google Scholar]
- 20. De Piccoli G., Cortes-Ledesma F., Ira G., Torres-Rosell J., Uhle S., Farmer S., Hwang J. Y., Machin F., Ceschia A., McAleenan A., Cordon-Preciado V., Clemente-Blanco A., Vilella-Mitjana F., Ullal P., Jarmuz A., Leitao B., Bressan D., Dotiwala F., Papusha A., Zhao X., Myung K., Haber J. E., Aguilera A., Aragón L. (2006) Nat. Cell. Biol. 8, 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Torres-Rosell J., De Piccoli G., Cordon-Preciado V., Farmer S., Jarmuz A., Machin F., Pasero P., Lisby M., Haber J. E., Aragón L. (2007) Science 315, 1411–1415 [DOI] [PubMed] [Google Scholar]
- 22. Prakash S., Prakash L. (1977) Genetics 87, 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matunis M. J., Coutavas E., Blobel G. (1996) J. Cell. Biol. 135, 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. (1997) Cell 88, 97–107 [DOI] [PubMed] [Google Scholar]
- 25. Johnson E. S., Schwienhorst I., Dohmen R. J., Blobel G. (1997) EMBO. J. 16, 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saitoh H., Pu R. T., Dasso M. (1997) Trends Biochem. Sci. 22, 374–376 [DOI] [PubMed] [Google Scholar]
- 27. Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 28. Zhao X., Blobel G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andrews E. A., Palecek J., Sergeant J., Taylor E., Lehmann A. R., Watts F. Z. (2005) Mol. Cell. Biol. 25, 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Potts P. R., Yu H. (2005) Mol. Cell. Biol. 25, 7021–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., Seki M., Enomoto T., Ohta K., Foiani M. (2006) Cell 127, 509–522 [DOI] [PubMed] [Google Scholar]
- 32. Wach A., Brachat A., Pöhlmann R., Philippsen P. (1994) Yeast 10, 1793–1808 [DOI] [PubMed] [Google Scholar]
- 33. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 34. Foiani M., Marini F., Gamba D., Lucchini G., Plevani P. (1994) Mol. Cell. Biol. 14, 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang D., Koshland D. (2003) Genes Dev. 17, 1741–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emili A. (1998) Mol. Cell 2, 183–189 [DOI] [PubMed] [Google Scholar]
- 37. Pellicioli A., Lucca C., Liberi G., Marini F., Lopes M., Plevani P., Romano A., Di Fiore P. P., Foiani M. (1999) EMBO. J. 18, 6561–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gilbert C. S., Green C. M., Lowndes N. F. (2001) Mol. Cell 8, 129–136 [DOI] [PubMed] [Google Scholar]
- 39. Osborn A. J., Elledge S. J. (2003) Genes Dev. 17, 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alcasabas A. A., Osborn A. J., Bachant J., Hu F., Werler P. J., Bousset K., Furuya K., Diffley J. F., Carr A. M., Elledge S. J. (2001) Nat. Cell. Biol. 3, 958–965 [DOI] [PubMed] [Google Scholar]
- 41. Duan X., Sarangi P., Liu X., Rangi G. K., Zhao X., Ye H. (2009) Mol. Cell 35, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weinert T. A., Hartwell L. H. (1988) Science 241, 317–322 [DOI] [PubMed] [Google Scholar]
- 43. Blasina A., Price B. D., Turenne G. A., McGowan C. H. (1999) Curr. Biol. 9, 1135–1138 [DOI] [PubMed] [Google Scholar]
- 44. Hall-Jackson C. A., Cross D. A., Morrice N., Smythe C. (1999) Oncogene 18, 6707–6713 [DOI] [PubMed] [Google Scholar]
- 45. Sarkaria J. N., Busby E. C., Tibbetts R. S., Roos P., Taya Y., Karnitz L. M., Abraham R. T. (1999) Cancer Res. 59, 4375–4382 [PubMed] [Google Scholar]
- 46. Tolmach L. J., Jones R. W., Busse P. M. (1977) Radiat. Res. 71, 653–665 [PubMed] [Google Scholar]
- 47. Reinke A., Chen J. C., Aronova S., Powers T. (2006) J. Biol. Chem. 281, 31616–31626 [DOI] [PubMed] [Google Scholar]
- 48. Zhao X., Muller E. G., Rothstein R. (1998) Mol. Cell 2, 329–340 [DOI] [PubMed] [Google Scholar]
- 49. Krakoff I. H., Brown N. C., Reichard P. (1968) Cancer Res. 28, 1559–1565 [PubMed] [Google Scholar]
- 50. Weinert T. (1998) Cell 94, 555–558 [DOI] [PubMed] [Google Scholar]
- 51. de la Torre-Ruiz M. A., Green C. M., Lowndes N. F. (1998) EMBO J. 17, 2687–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ulrich H. D. (2009) Methods Mol. Biol. 497, 3–16 [DOI] [PubMed] [Google Scholar]
- 53. Behlke-Steinert S., Touat-Todeschini L., Skoufias D. A., Margolis R. L. (2009) Cell Cycle 8, 2211–2218 [DOI] [PubMed] [Google Scholar]
- 54. Potts P. R., Yu H. (2007) Nat. Struct. Mol. Biol. 14, 581–590 [DOI] [PubMed] [Google Scholar]
- 55. Chavez A., George V., Agrawal V., Johnson F. B. (2010) J. Biol. Chem. 285, 11922–11930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ishida T., Fujiwara S., Miura K., Stacey N., Yoshimura M., Schneider K., Adachi S., Minamisawa K., Umeda M., Sugimoto K. (2009) Plant Cell 21, 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang L., Yang S., Zhang S., Liu M., Lai J., Qi Y., Shi S., Wang J., Wang Y., Xie Q., Yang C. (2009) Plant J. 60, 666–678 [DOI] [PubMed] [Google Scholar]
- 58. Takahashi Y., Dulev S., Liu X., Hiller N. J., Zhao X., Strunnikov A. (2008) PLoS Genet. 4, e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tsukamoto Y., Taggart A. K., Zakian V. A. (2001) Curr. Biol. 11, 1328–1335 [DOI] [PubMed] [Google Scholar]
- 61. Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]