Abstract
Tomosyn is a 130-kDa cytosolic R-SNARE protein that associates with Q-SNAREs and reduces exocytotic activity. Two paralogous genes, tomosyn-1 and -2, occur in mammals and produce seven different isoforms via alternative splicing. Here, we map the structural differences between the yeast homologue of m-tomosyn-1, Sro7, and tomosyn genes/isoforms to identify domains critical to the regulation of exocytotic activity to tomosyn that are outside the soluble N-ethylmaleimide-sensitive attachment receptor motif. Homology modeling of m-tomosyn-1 based on the known structure of yeast Sro7 revealed a highly conserved functional conformation but with tomosyn containing three additional loop domains that emanate from a β-propeller core. Notably, deletion of loops 1 and 3 eliminates tomosyn inhibitory activity on secretion without altering its soluble N-ethylmaleimide-sensitive attachment receptor pairing with syntaxin1A. By comparison, deletion of loop 2, which contains the hypervariable splice region, did not reduce the ability of tomosyn to inhibit regulated secretion. However, exon variation within the hypervariable splice region resulted in significant differences in protein accumulation of tomosyn-2 isoforms. Functional analysis of s-tomosyn-1, m-tomosyn-1, m-tomosyn-2, and xb-tomosyn-2 demonstrated that they exert similar inhibitory effects on elevated K+-induced secretion in PC12 cells, although m-tomosyn-2 was novel in strongly augmenting basal secretion. Finally, we report that m-tomosyn-1 is a target substrate for SUMO 2/3 conjugation and that mutation of this small ubiquitin-related modifier target site (Lys-730) enhances m-tomosyn-1 inhibition of secretion without altering interaction with syntaxin1A. Together these results suggest that multiple domains outside the R-SNARE of tomosyn are critical to the efficacy of inhibition by tomosyn on exocytotic secretion.
Keywords: Exocytosis, Membrane Fusion, Membrane Trafficking, Neurobiology, Secretion
Introduction
Synaptic vesicle fusion and the subsequent release of neurotransmitter require the formation of heterotrimeric SNARE2 complexes formed from plasma membrane proteins syntaxin1A and SNAP-25 (Q-SNAREs) with the synaptic vesicle membrane protein VAMP/synaptobrevin (R-SNARE) (1–3). Present on opposing membranes, these SNAREs combine and engage in thermodynamically stable coiled-coil interactions that bridge the two membranes and catalyze their fusion (4). The formation of SNARE complexes is spatially and temporally controlled by accessory components that lend additional specificity to SNARE pairing, arrest SNARE complex intermediates, and/or lower the energy required for fusion (4–6). Ultimately, it is the functional activity of these regulators on SNARE complex assembly that determines the dynamics of the exocytotic event.
Tomosyn is an important regulator of SNARE complex formation whose mechanism of action remains unclear. Initially identified in neurons (7–8), tomosyn, a soluble R-SNARE protein, was considered to be a negative effector of fusogenic SNARE complex assembly through interactions with syntaxin1A and SNAP-25 that preclude the binding of VAMP2, thereby resulting in “dead-end” nonfusogenic SNARE complexes (7–10). Cumulative biochemical evidence lends much support to this mechanism. Tomosyn overexpression inhibits exocytosis in Caenorhabditis elegans (11–14), in neurons (15–16), and a number of neuroendocrine secretory cell models (17–20). Moreover, gene mutation studies in C. elegans along with a tomosyn knock-out mouse models have demonstrated enhanced synaptic transmission in the absence of functional tomosyn (12, 13, 21). Although there is clear evidence for a negative role for tomosyn in the regulation of neurotransmitter release, additional tomosyn functions have also been reported.
Recent studies have provided evidence for a second permissive role for tomosyn regulation of neurotransmission. Tomosyn depletion by siRNA inhibits acetylcholine release from superior cervical ganglion neurons (22) and insulin secretion from insulin-secreting INS-1E cells (23). Subsequent to strong stimulation, tomosyn-overexpressing cells exhibited enhanced late phase secretion in chromaffin cells (16, 24) and asynchronous release in neurons (22) relative to controls. Moreover, in adipocytes, tomosyn was displaced by VAMP2 in vitro and bound simultaneously to Munc18c and syntaxin4 (19) in a complex that has been suggested to prime syntaxin on the plasma membrane for fusion (25).
In addition, although negative regulation by tomosyn on neurotransmitter release has been attributed to its C-terminal SNARE domain, this SNARE motif is absent from tomosyn homologues in yeast (Sro7p and Sro77p), the Drosophila tumor suppressor lethal giant larvae family, and the mammalian Mg/l family (26–30) whose functions are likewise attributed to interactions with cognate Q-SNAREs. Indeed, the R-SNARE motif of tomosyn occupies less than 10% of its sequence, yet it is the primary domain assigned a functional significance. Notably, one of the first reports on mammalian tomosyn demonstrated that both the N- and C-terminal regions were required for its inhibitory effect on secretion (8). More recently, an N-terminal deletion mutant of tomosyn was reported to still bind to syntaxin1A, yet it lacked the ability to inhibit secretion (24). Moreover, a tomosyn truncation mutant that lacked the R-SNARE domain and could not interact with syntaxin1A still demonstrated partial inhibition of secretion (24). Taken together, these studies indicated additional tomosyn regulatory domains.
Recently, an autoregulatory function has been assigned to a tail domain of tomosyn in a manner likened to the autoregulation of Sro7 (31, 32). Although the N-terminal β-propeller domains of tomosyn compose a substantial proportion of the protein and are required for the full inhibitory effect of tomosyn on secretion (24), only a few specific protein interactions have been identified with this region. For example, synaptotagmin-1 binds in a Ca2+-dependent manner directly to the N-terminal WD40 repeats to negatively regulate synaptotagmin-1-mediated neurotransmitter release (33). However, the specific structural motifs of tomosyn involved in this interaction have not been identified. The N-terminal portion of tomosyn has also been reported to enhance oligomerization of SNARE complexes facilitating inhibition of synaptic transmission (21).
To date, structural and functional analysis of tomosyn has been relegated to one specific mammalian (rat) isoform, m-tomosyn-1. Yet, two tomosyn genes, tomosyn-1 and tomosyn-2, and seven distinct isoforms have been identified in mice that arise from specific differential splicing within a domain termed the hypervariable region (HVR). Splicing of tomosyn-1 generates three distinct isoforms (s, m, and b) (34), whereas splicing tomosyn-2 results in four isoforms (s, m, b, and xb) (35). All structural differences within the isoforms of each gene occur strictly within the HVR. Recent studies indicate the functional importance of the HVR. For example, expression of the HVR with the N-terminal portion of tomosyn was sufficient to elicit an inhibitory effect on vesicle priming (24). In addition, protein kinase A (PKA) phosphorylation within the HVR acts to negatively regulate tomosyn interaction with syntaxin1A and up-regulate the readily releasable vesicle pool in superior cervical ganglion neurons (22). However, this phosphorylation site is not present in all tomosyn isoforms and therefore is not a general regulatory site of tomosyn function.
In this study, we capitalize on differences between tomosyn-1 and tomosyn-2 and their isoforms to identify structural motifs of tomosyn that underlie its functional activity. Based on sequence analysis, we find that rat m-tomosyn-1 shares a nearly identical structural conformation with the yeast homologue Sro7 but, importantly, contains an additional three loops that emanate free from the main β-propeller structure. Remarkably, deletion of loops 1 or 3, but not loop 2, resulted in a complete loss in negative regulation of tomosyn on secretion, although a strong interaction with syntaxin1A remained. We also identify a site within the HVR of m-tomosyn-1 that is subject to SUMOylation and that regulates the extent of tomosyn inhibition on secretion. Taken together, these data identify multiple novel structural motifs of tomosyn that are critical to its negative regulation on secretory activity independent of SNARE-SNARE interactions.
EXPERIMENTAL PROCEDURES
Antibodies and Chemicals
The following antibodies were used: α-syntaxin1A clone HPC-1 (Sigma); α-SNAP25 clone 71.1 (Synaptic Systems, Göttingen, Germany); α-tomosyn-1 clone 15 (BD Transduction Laboratories); α-V5 epitope (Invitrogen); α-FLAG antibody (Sigma); α-SUMO 1 and α-SUMO 2/3 (Abcam); and IRDye 800CW goat α-mouse IgG (H + L) and IRDye 680 Goat α-Rabbit IgG (H + L) (LiCoR Biosciences, Lincoln, NE). MG132, a cell-permeable, proteasome inhibitor was obtained from Sigma.
Expression Constructs
Tomosyn loop deletion mutants were initially created in plasmid pSFV. The segment N-terminal to each deletion region was PCR-amplified while adding restriction sites BamHI to the 5′ end and AscI and BssHII to the 3′ end. DNA sequence corresponding to glycine residues was added upstream of the AscI sites to replace the missing loop regions and prevent disruptions of the three-dimensional structure of the protein. Then the segment was ligated to the vector (pSFV-GFP) using the BamHI and BssHII restriction sites. The segment C-terminal to the deletion area was PCR-amplified in a similar manner, adding the restriction enzyme sites AscI and BssHII. This segment also included sequence corresponding to glycine residues to replace the deleted region. This plasmid construction was repeated for all loop deletion mutant constructs. As a final step, these mutated cDNA sequences were subcloned pEGFP for imaging in PC12 cells and pFLAG for growth hormone secretion assay.
Plasmids containing full-length cDNA inserts for mouse (Mus musculus) m-tomosyn-1, in plasmid pcDNA3.1, and tomosyn-2 isoforms (s-, m-, b-, and xb-), in plasmid pCR-ScriptSKII+, were generously provided by Dr. Alexander J. A. Groffen (Vrije Universiteit, Amsterdam, The Netherlands) and subcloned into vector pDNR-CMV (Clontech). Monomeric mutants (A206K) of pECER-C1 (cerulean) and pEcYFP-C1 (citrine) vectors containing the LoxP sequence were used as recipient vectors for subcloning mouse m-tomosyn-1 and mouse (s-, m-, b-, and xb-) tomosyn-2 to the C terminus of cerulean and citrine fusion tags using the Cre-recombinase-mediated Creator system (Clontech). Mouse s-tomosyn-1 was obtained by PCR-based deletion of the region corresponding to exon 22 of mouse m-tomosyn-1 in vector pDNR-CMV using the QuickChange® site-directed mutagenesis kit (Stratagene). The resulting mutant was subsequently transferred by recombination to vectors pLoxP-cerulean and pLoxP-citrine.
SDM was also used to generate mutants of rat m-tomosyn-1. Mutants include a point mutation that introduces a premature stop codon at residue 1068 resulting in a tomosyn that lacks the C-terminal SNARE domain (ΔSNARE) and a Lys to Arg or Ala mutation at lysine 730. The sequence fidelity of all constructs was confirmed by DNA sequencing (University of Michigan DNA Sequencing Core).
Protein Expression of GST, GST-Syntaxin1A, and Biotinylated Tomosyn
BL21 DE3 cells were transformed with a pGex-KG vector encoding GST or a soluble form of GST-syntaxin1A (residues 4–263). Cells were grown to an A600 of 0.4–0.6, induced with 0.1 mm isopropyl β-d-1-thiogalactopyranoside, and grown overnight with shaking at 23 °C. Cells were harvested by centrifugation for 20 min at 6000 × g (Beckman, Fullerton, CA, JA-14 rotor). The resulting pellet was resuspended in French press buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.05% Tween 20, 1 mm DTT) supplemented with a protease inhibitor mixture (Roche Applied Science) at 20 ml/liter of culture, passed twice through the press (10,000 p.s.i.), and centrifuged at 20,000 × g for 20 min at 4 °C. The supernatant (S1) was recovered, and GST or GST-syntaxin1A was purified from the S1 using glutathione-Sepharose 4B beads (Amersham Biosciences), according to the manufacturer's protocol. A small sample of the bound beads was fractionated by SDS-PAGE alongside standard amounts of BSA followed by staining with Coomassie. The staining intensities of the Coomassie-stained gels were captured and analyzed by Odyssey Infrared Imaging Systems (LiCoR).
Mouse tomosyn-1 isoforms (s- and m-) and tomosyn-2 isoforms (m- and xb-) were PCR-amplified and TOPO-cloned into Gateway® entry vector pENTR/D-TOPO (Invitrogen) according to the manufacturer's specifications. N-terminally tagged mammalian expression constructs were generated by performing LR recombination into the Gateway® destination vector pcDNATM3.2/capTEVTM/V5-DEST. Mouse s- and b-tomosyn-2 mammalian expression constructs were generated by PCR-based deletion of the regions corresponding to exon 20 in mouse m-tomosyn-2 and xb-tomosyn-2 in Gateway® destination vector pcDNATM3.2/capTEVTM/V5-DEST using site-directed mutagenesis. Similar mutagenesis methods were used to generate additional tomosyn-2 deletion constructs by deleting the regions corresponding to exon 21 from s-tomosyn-2, m-tomosyn-2, and b-tomosyn-2 in mammalian expression vector pcDNATM3.2/capTEVTM/V5-DEST to create null (ΔHVR), exon 20, and exon 22 tomosyn-2 mutants, respectively. The sequence fidelity of all constructs was confirmed by DNA sequencing (University of Michigan DNA Sequencing Core).
Cell Culture and Transfection
Rat adrenal pheochromocytoma (PC12) cells were plated and cultured in Kaighn's media supplemented with 15% horse serum, 2.5% fetal bovine serum, and 1% penicillin/streptomycin (Invitrogen) at 37 °C and 5% CO2. Human embryonic kidney (HEK) 293A cells were cultured at 37 °C and 5% CO2 in DMEM supplemented with 10% fetal bovine serum, 1% nonessential amino acids, 1% Glutamax (Invitrogen), and 1% penicillin/streptomycin (Invitrogen). All transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. One hour before transfection, cells were placed into OptiMEM (Invitrogen) lacking antibiotics and serum. 4–6 h after the transfection, cells were returned to normal growth medium.
Real Time Quantitative PCR
Quantitative analysis of mRNA levels was performed in duplicate by real time monitoring of SYBR Green I fluorescence on a Mastercycler® ep realplex2 real time PCR system (Eppendorf). Prior to cDNA synthesis, RNA was extracted from cells transfected with a mammalian expression plasmid coding for biotinylated tomosyn-1 (s- or m-), tomosyn-2 (s-, m-, b-, or xb-), or their mutants (ΔHVR, s-tomosyn-2 (20) or s-tomosyn-2 (22)) using the RNeasy mini kit (Qiagen). Following treatment with RNase-free DNase I (Qiagen) to degrade trace amounts of genomic DNA, 3 μl of total RNA was used for oligo(dT)15-primed cDNA synthesis by avian myeloblastosis virus reverse transcriptase (Promega) for 15 min at 42 °C. Each qPCR was composed of 2 μl of template cDNA, 70 nm primers, 12.5 μl of 2× FastStart SYBR Green qPCR master mix (Roche Applied Science), and PCR-grade water in a total reaction volume of 25 μl. The thermal cycler program included an initial polymerase activation step (10 min at 95 °C), followed by 40 cycles of denaturation (15 s at 95 °C), annealing (30 s at 50 °C), and polymerization (20 s at 68 °C). Finally, a dissociation curve was determined for each sample (15 s at 95 °C and 15 s at 50 °C, followed by a slow ramp to 95 °C under continuous monitoring of fluorescence intensity). In all cases, the dissociation curves excluded a significant contribution of relatively short by-products to the measured fluorescence intensities. All measurements were performed in duplicate. To allow for normalization with respect to the amount of template cDNA in different samples, we also measured the mRNA levels for the small subunit 18 S rRNA housekeeping gene measured in duplicate. For each reaction, the cycle threshold value was calculated as the fractional cycle number at which the fluorescence intensity exceeds a threshold value of 10 standard deviations above background fluorescence. Relative gene expression was calculated using the comparative CT, or 2−ΔΔCT, method (36, 37).
Homologous Proteins for Structural Modeling
Homologues were collected to build a multiple sequence alignment, from which the pairwise alignment between target and template would be extracted. The homologues taken here for each family, Sro7, LGl, and tomosyn, are the same as in Ref. 26.
Multiple Sequence Alignment
A multiple sequence alignment (MSA) of all the homologues was created for each protein family (Sro7, tomosyn, and LGL) separately, using the alignment software MUSCLE (38). Each MSA was edited according to secondary structures as indicated in Ref. 26 and cross-referenced with secondary structure prediction by PSIPRED (39). This was followed by a profile-to-profile alignment between the tomosyn and LGL profiles, with editing of the MSA (see above), and finally running a profile-to-profile alignment between the tomosyn-LGL profile and the Sro7 profile. This last MSA was modified again, according to secondary structure elements. Ultimately, the pairwise alignment of interest, between the m-tomosyn-1 and Sro7 of the S. cerevisiae, was extracted and used for structural modeling.
Modeling
The model structure of the tomosyn protein was built with the NEST modeling software (40). The modeling was implemented with optimization refinement in insertion and deletion regions (usually loop regions).
Model Validation
The compatibility of the model structure with general properties of proteins was examined by calculating Ramachandran plots. Another indicator of structural quality is the correlation between the degree of evolutionary conservation of amino acid positions and their location; buried residues are usually highly conserved, whereas peripheral residues are variable. The evolutionary conservation was calculated using MSA of homologues (Sro7, Lgl, and tomosyn) and the ConSurf server (41).
Human Growth Hormone (hGH) Secretion Assay
PC12 cells were plated onto poly-d-lysine-coated 24-well plates and cotransfected with a plasmid coding for hGH, in addition to full-length tomosyn-1 (s or m isoform), tomosyn-2 (s, m, b, or xb isoform), or an empty vector control. Each of the mouse tomosyn genes was expressed as N-terminal biotinylated V5 epitope-tagged fusions in pcDNA3.2/capTEV/V5-DEST vectors (Invitrogen). The total concentration of transfected DNA was held equal across all treatments. Secretion assays were performed 48 h following transfection. To measure secretion, cells were rinsed for 5 min in a physiological saline solution (145 mm NaCl, 5.6 mm KCl, 15 mm NaHEPES, 0.5 mm MgCl2, 2.2 mm CaCl2, pH 7.3, containing 5.6 mm glucose, 2 mg/ml sodium ascorbate, and 2 mg/ml fatty acid-free bovine serum albumin). Cells were subsequently incubated in either an additional application of saline or stimulated to secrete by treatment with saline supplemented with 100 mm K+ (equimolar replacement for Na+), each for 10 min. The saline solution containing the secreted hGH was collected, and cells were lysed (2% Triton X-100) to determine total hGH content. Secreted and total hGH was measured from sample aliquots using an hGH enzyme-linked immunosorbent assay kit (Roche Applied Science). Each experiment was performed with duplicate replicates for each treatment with experiments repeated a minimum of three times.
Syntaxin1A Q-SNARE-Tomosyn V-SNARE Binding Analysis
GST or soluble GST-syntaxin1A was prepared as described above. GST or GST-syntaxin1A bound to glutathione-Sepharose 4B beads was then incubated for 16 h at 4 °C with lysates prepared from HEK293A cells expressing mouse tomosyn isoforms that were N-terminally biotinylated, V5, or of rat m-tomosyn-1 variants that were FLAG-tagged. The lysis buffer contained 100 mm Tris-HCl, pH 8.0, 100 mm KCl, 200 μm EDTA, 1.5 mm MgCl2, 0.1% Nonidet P-40, and protease inhibitors (Roche Applied Science). The beads were then collected by centrifugation and washed three times with lysis buffer. Bound proteins were analyzed by SDS-PAGE followed by immunoblot analysis with α-V5 antibody (Invitrogen).
Analysis of Tomosyn SUMOylation
N-terminally fused V5 epitope-tagged constructs of m-tomosyn-1, s-tomosyn-1, or an empty vector control were transfected into HEK293 cells. At 48 h post-transfection, the cells were lysed, and tomosyn was immunoprecipitated using Dynabeads® protein G (Invitrogen) beads prebound with α-V5 antibody. After extensive washing, immunoprecipitates were resuspended in 2× SDS reducing sample buffer and fractionated by SDS-PAGE followed by immunoblot analysis with α-SUMO 1 or α-SUMO 2/3 antibodies.
Fluorescence Microscopy of Cerulean and Citrine Fusion Proteins in Live Cells
PC12 cells were cultured for 48 h post-transfection on poly-d-lysine-coated glass coverslips (No. 1, Fisher) attached to the bottom of 35-mm culture dishes. Prior to fluorescence imaging, culture media bathing the cells were removed and replaced with physiological saline solution. Confocal imaging of the subcellular localization of expressed fluorophore-tagged proteins was performed on an Olympus FluoView 500 laser scanning confocal microscope (Olympus, Melville, NY) using an LD405 laser, 60× objective (1.2 numerical aperture), and a pinhole aperture of 260 μm.
Sensitized emission FRET was detected using an Olympus IX71 microscope equipped with a 60× 1.49 NA oil immersion objective. Two lasers, a 45-milliwatt, 442-nm diode-pumped solid state laser (Toptica Photonics, Grafelfing, Germany) and a 225-milliwatt argon-ion laser (National Laser Co., Salt Lake City, UT), were combined into an accousto-optic tunable filter whose output was coupled via a unimodal fiber optic into the microscopes back port. The filter cube of the microscope contains a triple band laser clean-up filter (z442/514/594×) and laser polychroic mirror (z442/514/594rpc). Fluorescence emission from cerulean and citrine were separated with a Quad-View beam splitter (Mag Biosystems, Pleasanton, CA) containing two dichroic mirrors and emission filters for Cerulean (HQ480/40m) and citrine (HQ540/30m), as well as mCherry (HQ630/50m) and infrared (D700/10m). All filters were from Chroma Technology (Rockingham, VT). Images were acquired with a Photometrics QuantEM 512SC EM-CCD camera (Roper Scientific, Tucson, AZ). A final pixel size of 67 nm (object coordinates) was achieved by insertion of a 1.6× slide magnifier and a 2.5× beam expander in the emission light path. Raw FRET image sets (donor excitation, donor emission; donor excitation, acceptor emission; acceptor excitation, acceptor emission) were aligned, background-subtracted, and analyzed off line using MATLAB scripts (The MathWorks, Natick, MA). Sensitized emission FRET analysis used the FRET stoichiometry method (42). Calibration parameters specific to the optical system and fluorophores that are required for FRET analysis were determined as described previously (43). The apparent FRET efficiency of the acceptor in complex with the donor (EA), apparent FRET efficiency of the donorin complex with the acceptor (ED), and molar ratio of total acceptor/donor (Ratio) were determined by pixel-by-pixel analysis of the three required images.
Data Analysis and Statistics
Statistical analysis was performed using IGOR PRO (Wavemetrics Inc.) and Excel (Microsoft Office 2007) software. Population data were expressed as means ± S.E., and statistical significance was determined using Student's unpaired t tests or for multiple comparisons using analysis of variance with Dunnett's post hoc test or a Mann-Whitney U test for nonparametric data. Significant differences are defined in the corresponding figure legends and indicated by asterisks.
RESULTS
Homology Modeling of Tomosyn
The crystal structure of a close homologue of tomosyn, Saccharomyces cerevisiae Sro7 (gi: 6325289), has been determined at 2.4 Å resolution (Protein Data Bank code 2OAJ) and reveals a structure (residues 61–962) composed of two, seven-bladed WD40 β-propellers, which lead to a sequence “tail” domain bound to the β-propeller loops (Fig. 1A) (26). This structure was used as a template for modeling of the rat m-tomosyn-1 (gi: 3790389) protein. The sequence identity between target and template was low (∼19%), but the core domains of m-tomosyn-1 (Fig. 1B) were generally well modeled based on the Sro7 template (Fig. 1A). This is reflected in the compatibility of the evolutionary conservation profile of the protein with the structure (Fig. 1D, right panel). As anticipated, the structural core is highly conserved, although the periphery is variable, which is the same as in the Sro7 template (Fig. 1D, left panel). However, the primary tomosyn sequence is significantly longer and exhibits three loops that deviate from the Sro7 template. These loops result from insertions in m-tomosyn-1 compared with Sro7, creating regions in the model with no template. The loops emanate from the main β-propeller backbone as shown in Fig. 1B. These loops are conserved among the tomosyn genes and isoforms and encompass amino acid residues of rat b-tomosyn-1 and xb-tomosyn-2 as shown in Fig. 1C. Loop two is the most extensive, occurring in an exposed region of the protein. A separate homologous template for this region was not found. The individual loops are likely to be functionally important, as phosphorylation of a serine residue within loop 2 of rat m-tomosyn-1 has been reported to regulate tomosyn interaction with syntaxin1A and the extent of tomosyn-mediated secretory inhibition (22).
FIGURE 1.
Homology modeling of m-tomosyn-1 sequence to Sro7. A, model representing the crystal structure of Sro7, including the N-terminal (purple) and C-terminal (green) sequence. B, homology model of m-tomosyn-1 based on Sro7 demonstrating preservation of the β-propellers and addition of three undefined loops (L1, L2, and L3). C, schematic of the two longest isoforms of mouse tomosyn genes 1 and 2. Regions corresponding to the left-sided (gray) and right-sided (blue) β-propeller structures, conserved tail domain (green), R-SNARE (black), and residues comprising the three predicted loops (L1, L2, and L3) are shown. D, evolutionary conservation profiles of Sro7 (left) and tomosyn (right). The residues are colored according to their conservation grades, with turquoise-through-maroon indicating variable-through-conserved. The most highly conserved amino acids are presented using space-filled atoms.
To validate the accuracy of the tomosyn model, a Ramachandran plot was created using the Swiss PDB-viewer (44). The plot revealed discrepancies that rendered the model inadequate. However, going over the residues in the disallowed regions, it was clear that they were all part of the three template-less loops that were arbitrarily modeled. Consequently, a second Ramachandran plot was created without the three undefined loops resulting in a highly significant correlation. Residues that remained located in the disallowed regions were restricted to sequence defined above as the loop regions.
Effect of Tomosyn Loop Regions on Inhibition of Secretion in PC12 Cells
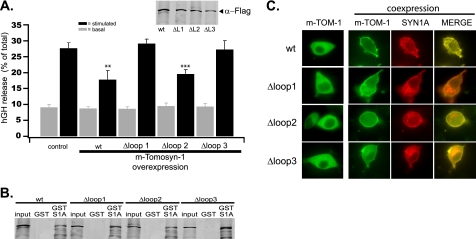
To test for functional effects of each loop region, deletion mutants of rat m-tomosyn-1 were constructed and transfected individually into PC12 cells along with a reporter of the regulated secretory pathway, hGH (45). As shown in Fig. 2A, wild-type m-tomosyn-1 inhibited elevated K+-induced hGH secretion by ∼36% (27.7 ± 1.8% of total hGH, control; 17.8 ± 2.8%, m-tomosyn-1; mean ± S.E., n = 4) relative to control cells transfected with empty vector. Importantly, deletion of loop 1 (Δ-loop 1) or loop 3 (Δ-loop 3) resulted in complete elimination of inhibition by tomosyn of K+-induced hGH secretion. By comparison, deletion of loop 2 (Δ-loop 2) had no significant effect on the ability of tomosyn to inhibit secretion. Notably, these differences did not result from reduced expression of the Δ-loop mutants relative to wild-type m-tomosyn-1 (Fig. 2A, inset). In addition, there were no significant effects of m-tomosyn-1 or the Δ-loop mutants on basal secretion.
FIGURE 2.
Effect of tomosyn loop regions on secretion from PC12 cells. A, comparison of basal (gray bars) or elevated K+-induced (100 mm, 10 min; black bars) secretion of hGH from PC12 cells expressing full-length m-tomosyn 1 (wt) or m-tomosyn-1 deletion constructs (Δloop 1, Δloop 2, or Δloop 3) from which the indicated loop domain regions were deleted. hGH secreted was calculated as a percentage of the total cell content using duplicate samples and then normalized to the control condition. Bar graphs depict the mean values ± S.E. (n = 4 replicate experiments; **, p < 0.05 compared with control values). Tomosyn protein (WT and loop deletion mutants) was expressed at nearly equivalent levels as shown by Western blot of corresponding PC12 cell lysates probed with α-FLAG antibody (inset). Each tomosyn construct was epitope FLAG-tagged on its N terminus. B, representative immunoblot showing in vitro GST-syntaxin1A pulldown of indicated FLAG-tagged tomosyn proteins. Recombinant GST-syntaxin1A or GST (control) was immobilized on glutathione-Sepharose beads and incubated with HEK293A cell lysates expressing tomosyn WT or indicated tomosyn loop deletion construct. Tomosyn immunoreactivity in each fraction was determined using a α-FLAG antibody. C, confocal fluorescence images of EGFP-tomosyn (WT and Δloop deletions) expressed alone (left) or in combination with mRFP-syntaxin1A (right) in live PC12 cells.
Tomosyn inhibition of secretion is believed to depend primarily on its high affinity interaction with syntaxin1A and SNAP25 via its C-terminal VAMP-like R-SNARE domain. Therefore, we next determined if the lack of secretory inhibition by the Δ-loop 1 and Δ-loop 3 tomosyn mutants resulted from reduced syntaxin1A binding relative to wild-type m-tomosyn-1. Experiments used GST-syntaxin1A immobilized on glutathione-Sepharose beads to pull down each Δ-loop mutant from the HEK293 cell lysate that was then compared with pulldown by wild-type tomosyn. The recombinant tomosyn constructs were expressed in HEK293A cells to facilitate maturation and folding of the protein in a eukaryotic environment. The HEK293 cell line does not demonstrate endogenous expression of neuronal SNARE proteins (46) and therefore provides a clean background upon which to test interactions with the various tomosyn deletions. Immunoblots of the pulldown reactions show that each of the Δ-loop mutants bound GST-syntaxin1A equivalent to that of wild-type m-tomosyn-1 (Fig. 2B). N-terminal epitope FLAG tags on each tomosyn construct were used for detection to avoid potential differences in reactivity of the tomosyn deletion mutants to the tomosyn antibody. Membrane targeting of tomosyn occurs via direct interaction with syntaxin1A at the plasma membrane (20, 24) Therefore, as an additional test for interaction between the tomosyn mutants and syntaxin1A, we compared membrane targeting of each Δ-loop tomosyn mutant relative to wild-type tomosyn when coexpressed with mCherry-syntaxin1A in PC12 cells. Fig. 2C shows that each tomosyn mutant targeted to the membrane, indicating the capacity of these mutants to interact with syntaxin1A in vivo. Thus, loop domains one and three are required for the inhibitory activity of m-tomosyn-1 on regulated secretion, independent of the ability of tomosyn to bind syntaxin1A.
Effect of the HVR Tomosyn Loop 2 on Tomosyn Expression
All structural differences between the isoforms of each tomosyn gene occur strictly within the HVR that is contained within the loop 2 region. Differential splicing of tomosyn-1 generates three distinct isoforms (s, m, and b), whereas splicing of tomosyn-2 results in four isoforms (s, m, b, and xb) (Fig. 3A). As noted above, deletion of loop 2 did not reduce the level of inhibition exerted on secretion by rat m-tomosyn-1. However, allosteric regulation within loop 2 via PKA-mediated phosphorylation (Ser-724) has been reported to reduce inhibition of secretion for tomosyn (22). This suggests that variation in loop 2 between the tomosyn isoforms may be a critical component of tomosyn regulation. Indeed, the PKA phosphorylation site is absent in the s- and m-tomosyn-2 isoforms, as well as s-tomosyn-1. To compare the physiological effects of the different loop 2 region splice variants, mouse tomosyn-1 (except b-tomosyn-1) and tomosyn-2 isoforms were transfected into HEK293 cells. Surprisingly, although m- and s-tomosyn-1 were well expressed, only the m-tomosyn-2 isoform and, to a lesser extent, xb-tomosyn-2 demonstrated protein overexpression detectable on Western blots (Fig. 3B). These expression differences resulted from differences in the HVR, as each tomosyn isoform was subcloned similarly into the pcDNATM3.2 vector, and DNA sequencing confirmed that structural differences were limited to the HVR for each tomosyn gene. Moreover, RT-PCR of RNA extracted from HEK293A cells transfected with each tomosyn isoform demonstrated the presence of each construct of appropriate size in all cases (data not shown). To determine whether differences in protein expression resulted from differences in levels of mRNA expression, we employed real time qPCR to quantify the mRNA of each splice variant in transfected cells relative to that of m-tomosyn-1. As a control for differences in transfection efficiency, an additional set of primers was included in the qPCRs that corresponded to the neomycin resistance gene found in the pcDNA3.2 plasmids. No significant differences were found in the levels of mRNA between the endogenous tomosyn isoforms relative to that of m-tomosyn-1 (Fig. 3C).
FIGURE 3.
Effect of loop 2/HVR region on tomosyn protein expression. A, schematic of the HVR of tomosyn genes/isoforms and recombinant mutant deletion constructs. B, representative immunoblots resulting from lysate from HEK293 cells transfected with the tomosyn gene/isoform or mutant construct indicated. Tomosyn was detected by probing Western blots with antibodies against the N-terminal V5 (tomosyn) or FLAG (tomosyn Δloop2) epitopes or β-actin (loading control). C, averaged qPCR results comparing relative RNA transcript expression levels in transfected HEK293 cells for corresponding conditions shown in B. Relative tomosyn (top, black bars; normalized to m-tomosyn-1 expression) and control gene (neomycin; bottom, gray bars) transcript levels were calculated using the comparative CT method. D, effects of MG132 treatment on tomosyn protein expression levels. Immunoblot analysis was performed on lysates from transfected HEK293A cells. Cells were treated ± MG132 (100 μm) for 6 h prior to lysis. Region of tomosyn immunoreactivity is indicated (▶). Actin is shown as a loading control (α-β actin). A degradation product detected in the s-tomosyn-2 protein sample is indicated by an asterisk. All experiments shown were performed in triplicate.
To identify specific exons within the HVR that may be responsible for the lowered protein levels of most of the tomosyn-2 variants relative to m-tomosyn-2, we generated additional mutants where the sequence corresponding to exon 21 in the s-, m-, and b-tomosyn-2 isoforms was deleted to create tomosyn-2 cDNA with no HVR (null), exon 20 only, or exon 22 only, respectively (Fig. 3A). Each construct was then transfected into HEK293 cells and protein and mRNA expression examined. As shown in the Western blot of Fig. 3B, none of these mutants generated appreciable tomosyn protein relative to that exhibited by m-tomosyn-2. Importantly, mRNA levels quantified by real time qPCR demonstrated no significant difference in mRNA expression between an exon null and exon 20 only tomosyn 2 construct with that of m-tomosyn-2 (Fig. 3C). RT-PCR confirmed mRNA products expressed were of appropriate size (data not shown). The combination of exons for tomosyn-2 that leads to the highest protein levels is the combination of exon 20 and 21 (m-tomosyn-2), with the addition of exon 22 (xb-tomosyn-2) demonstrating limited expression. By comparison, each tomosyn-1 HVR splice variant, as well as the null mutant, expressed mRNA and protein similar to that of m-tomosyn-1, indicating effects of HVR splicing on protein levels were specific to tomosyn-2. Considered together, the differences in protein levels between tomosyn genes/isoforms indicate that exon 19 in tomosyn-1 may stabilize the protein and that the combination of exons 20 and 21 in tomosyn-2 acts in a partial manner to reduce degradation/turnover of tomosyn-2 protein.
To evaluate if ubiquitin proteosome-mediated tomosyn-2 degradation may be responsible for limited accumulation of tomosyn-2 protein, the effect of a proteosome inhibitor, MG132, was studied. Fig. 3D illustrates that the levels of m-tomosyn-1, m-tomosyn-2, and to a lesser extent xb-tomosyn-2, but not s-tomosyn-2, were substantially increased following MG132 (100 μm) treatment in transfected PC12 cells.
Functional Differences between Tomosyn-1 and Tomosyn-2 on Secretion from PC12 Cells
To date, functional evaluation of tomosyn has relied exclusively on the m-tomosyn-1 isoform. To extend analysis to the tomosyn-2 gene, we compared the effects of overexpression of mouse m-tomosyn-1, s-tomosyn-1, m-tomosyn-2, and xb-tomosyn-2 on secretion using PC12 cells cotransfected with hGH. Notably, each of the tomosyn genes/isoforms inhibited elevated K+-induced hGH secretion to a similar extent (Fig. 4A). By comparison, a significant increase in basal secretion occurred with m-tomosyn-2 overexpression when expressed relative to the empty plasmid control (Fig. 4A). Real time qPCR and Western blots performed on samples of the cells used to assess secretory responsiveness demonstrated only slightly lower expression of m-tomosyn-2 relative to that of the tomosyn-1 isoforms (Fig. 4C). Differences in resting [Ca2+]i cannot account for the m-tomosyn-2 enhancement of basal secretion, as no significant difference in resting free [Ca2+]i was found between the empty vector transfected controls and PC12 cells overexpressing m-tomosyn-2 or m-tomosyn-1 (Fig. 4D). Together, these data demonstrate a lack of functional distinction between the tomosyn genes/isoforms relative to inhibition of evoked secretion, but they pinpoint a novel function for m-tomosyn-2 as a positive regulator of basal secretion.
FIGURE 4.
Effect of overexpression of tomosyn gene/isoforms on regulated secretion. PC12 cells expressing tomosyn gene/isoforms indicated were incubated in physiological saline (A) or stimulated with 100 mm K+-containing saline (B) for 10 min at 30 °C. Collection of aliquots of saline and cell lysate for each condition was used to determine the percent of total hGH content secreted. (n >4 replicate experiments with duplicate sample analysis; **, p < 0.05; ***, p < 0.005 compared with control.) C, relative tomosyn gene expression determined by qPCR analysis of transfected PC12 cells. Inset shows representative immunoblot confirming V5 epitope-tagged tomosyn genes/isoform protein expression corresponding to each transfection condition. D, comparison of averaged measurements of resting intracellular calcium for m-tomosyn-1 and m-tomosyn-2 transfected PC12 cells (control, citrine alone expressing). Relative [calcium] expressed as F340/F380 ratio based on imaging of Fura-2 (Molecular Probes).
To determine whether m-tomosyn-2 differed in its interaction with syntaxin1A relative to other tomosyn isoforms, we next compared the subcellular distribution patterns of citrine-tomosyn isoforms when expressed individually and when coexpressed with cerulean-syntaxin1A in PC12 cells. Alone, citrine-tomosyn isoforms showed a diffuse cytosolic distribution (Fig. 5A) consistent with the previously described localization of rat m-tomosyn-1 (20). By comparison, coexpression with cerulean-syntaxin1A dramatically altered the citrine-tomosyn distribution resulting in strong targeting of each tomosyn isoform to the plasma membrane region. To confirm that the redistribution reflects a direct interaction between the citrine-tomosyn isoforms and cerulean-syntaxin1A, we performed sensitized emission FRET imaging. The FRET results demonstrate that each of the tomosyn isoforms directly interact with syntaxin1A in vivo (Fig. 5B). Indeed, m-tomosyn-1 and s-tomosyn-1 show equivalent apparent FRET efficiency with syntaxin1A (ED, 10.92 ± 0.783%, m-tomosyn-1; 10.94 ± 0.817% s-tomosyn-1), although FRET efficiencies for m-tomosyn-2 and xb-tomosyn-2 were found to be significantly greater. Taken together, these data indicate that facilitation of basal secretion by m-tomosyn-2 overexpression relative to other tomosyn isoforms does not result from a reduced interaction with syntaxin1A.
FIGURE 5.
Syntaxin1A-dependent plasma membrane targeting of tomosyn gene/isoforms. A, confocal fluorescence images of indicated tomosyn isoforms, expressed alone or in combination with syntaxin1A in live PC12 cells. Monomeric mutants of citrine and cerulean were fused to the N terminus of tomosyn and syntaxin1A proteins, respectively. B, averaged apparent FRET efficiency between citrine-tomosyn and cerulean-syntaxin1A proteins measured by sensitized emission FRET in live transfected PC12 cells.
Tomosyn HVR Is a Substrate for SUMOylation
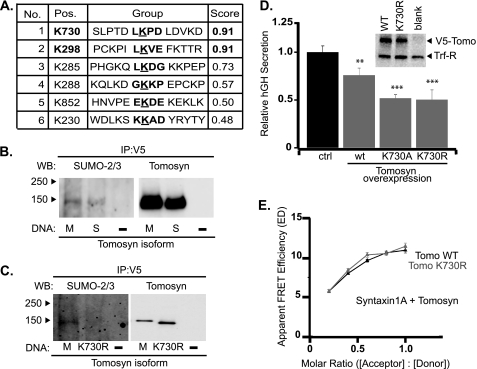
SUMO modification of proteins can alter their localization, activity, stability, or transcriptional regulation as a result of alteration of inter- or intramolecular interactions (47, 48). SUMO acceptor sites map to a consensus sequence ΨKX(D/E), where Ψ is a hydrophobic residue. SUMO modification results in an isopeptide bond between the C-terminal glycine residue of SUMO and the lysine side chain. Examination of the rat m-tomosyn-1 sequence using SUMOplotTM Prediction (Abgent, San Diego) revealed two highly predicted SUMOylation sites, Lys-730 and Lys-298 (Fig. 6A). The Lys-730 site (LKPD) resides in exon 22 of m-tomosyn-1. As a result of alternative splicing of exons within the HVR, the Lys-730 sumoylation site is absent in s-tomosyn-1, although all tomosyn-1 isoforms carry a Lys-298 site. Initial experiments examined if m-tomosyn-1 and s-tomosyn-1 were substrate targets for SUMOylation. For these experiments, N-terminal fused V5 epitope-tagged constructs of m-tomosyn-1 and s-tomosyn-1 and empty vector were transfected into HEK293 cells. Following a 48-h expression period, the cells were lysed and overexpressed tomosyn immunoprecipitated using α-V5 antibody. Immunoprecipitates were then fractionated by SDS-PAGE, and the resulting immunoblots were probed with α-SUMO antibody recognizing SUMO 2/3 isoforms. Both m-tomosyn-1 and s-tomosyn-1 demonstrated SUMO 2/3 labeling, with specificity confirmed by a lack of signal for the empty vector control (Fig. 6B). Importantly, comparison of anti-SUMO immunoreactivity demonstrated no significant difference between s- and m-tomosyn-1 samples when corrected for sample input, suggesting either SUMOylation at an alternative site than Lys-730 or that multiple SUMOylation sites are present. To further evaluate whether Lys-730 is sumoylated in m-tomosyn-1, we repeated the experiment but compared the level of SUMO immunoreactivity between wild-type and an m-tomosyn-1 mutant K730R. The data demonstrate a selective loss of SUMOylation in the mutant (Fig. 6C), thereby demonstrating Lys-730 as a substrate site for m-tomosyn-1 SUMOylation.
FIGURE 6.
SUMOylation of m-tomosyn-1 within HVR of loop 2. A, table of predicted SUMOylation sites in rat m-tomosyn-1 based on sequence analysis. Sites were predicted and scored from high to low probability using SUMOplotTM analysis program (Abgent). B, immunoblots probed with α-SUMO 2/3 (left) antibody to detect SUMOylation of V5 epitope-tagged s- or m-tomosyn-1 immunoprecipitated and detected using α-V5 antibody (right). HEK293 cells were transfected with the tomosyn constructs, or empty vector control, 48 h prior to cell lysis and immunoprecipitation. C, SUMOylation is strongly reduced in a K730R point mutation of m-tomosyn-1 expressed in HEK293 cells. D, effect of mutation of tomosyn SUMOylation site (Lys-730) on tomosyn inhibition of hGH secretion from transfected PC12 cells (mean ± S.E., n = 3). Inset, immunoblot probed with α-V5 and α-transferrin receptor antibodies to demonstrate equivalent expression upon transfection of HEK293 cells of wild-type and a K730R mutant of tomosyn. E, averaged apparent FRET efficiency between citrine-tomosyn (wild type or K730R) and cerulean-syntaxin1A proteins measured by sensitized-emission FRET over a range of acceptor:donor fluoroprotein expression levels in live transfected PC12 cells.
To evaluate the functional consequence of SUMOylation at Lys-730 on regulated secretion, we compared the effects of overexpressed wild-type m-tomosyn-1 to SUMOylation site mutants of m-tomosyn-1. As shown in Fig. 6D, mutation of Lys-730 to either alanine or arginine significantly enhanced the inhibitory effect of tomosyn (normalized hGH secretion, tomosyn (WT) 0.76 ± 0.07; tomosyn K730A 0.52 ± 0.04; and tomosyn K730R 0.50 ± 0.1). The enhanced inhibition of secretion by the K730R mutant was not attributable to enhanced expression compared with wild-type tomosyn, as demonstrated in Western blots of HEK cell lysates overexpressing V5-tagged tomosyn constructs (Fig. 6D, inset). Moreover, the similarity between the K730R and K730A mutants suggests that the increased inhibitory function was unlikely to be related to mutation of the positive charge of the lysine residue.
Typically, only a small fraction of a given protein is SUMOylated, and this modification has been shown to result in altered protein localization and binding partners. We hypothesized that if the inhibitory function of tomosyn was related to its ability to form SNARE complexes, then mutation of the Lys-730 residue should be associated with an increase in FRET with syntaxin1A compared with wild-type tomosyn. However, we found that FRET between tomosyn K730R and syntaxin1A was no different from that with wild-type tomosyn, when measured across a range of molar expression ratios (Fig. 6E).
DISCUSSION
Homology modeling of m-tomosyn-1 onto its yeast counterpart, Sro7, was used in this study to identify novel structural motifs in tomosyn that may be important for its activity on the regulated secretory pathway. Our findings reveal an evolutionarily conserved N-terminal β- propeller structure in which tomosyn differs by three unique regions of loops. Remarkably, deletion of loop 1 and loop 3, but not loop 2, of m-tomosyn-1 resulted in the complete elimination of its ability to inhibit secretion. By comparison, loop 2 was found important in determining the extent of tomosyn protein accumulation, although this effect was restricted to tomosyn-2 isoforms. However, the HVR of loop 2 in those tomosyn-1 isoforms containing exon 22 (b- and m-tomosyn-1) was demonstrated to be a target substrate for SUMOylation. Finally, although functional analysis of tomosyn-1 and tomosyn-2 activity had previously been restricted to the m-tomosyn-1 isoform, our data indicate that tomosyn-1 and tomosyn-2 isoforms exert similar inhibitory activity on exocytotic secretion.
Tomosyn is largely believed to inhibit secretion via direct competition with VAMP for reactive Q-SNARE proteins (49). This interpretation is consistent with studies demonstrating that tomosyn overexpression substantially reduces exocytosis in neurosecretory cells (8, 10, 15–20) and neurons (21–22) by inhibiting vesicle priming reactions that generate fusogenic SNARE core complexes. Moreover, mutations of the orthologue to vertebrate tomosyn in C. elegans, Tom-1, demonstrated that Tom-1 normally negatively regulates vesicle priming at neuromuscular synapses in vivo (11–14, 50). These functional studies are complemented by an extensive analysis of tomosyn interactions attained through in vitro binding studies and coimmunoprecipitation, which demonstrate that R-SNARE domain of tomosyn is required for high affinity interaction with syntaxin1A (10, 21, 34), and it also mediates interaction with SNAP25, SNAP23, and syntaxin4 (19). The affinity of the interaction is sufficient to compete with and dissociate Munc18-1 from a conformational closed form of syntaxin1A (8, 20). Although tomosyn exhibits heterodimer interaction with these Q-SNAREs, it also readily forms into stable heterotrimeric SNARE core complexes in vitro (10, 19, 21). Structurally, these tomosyn-containing SNARE core complexes are very similar to fusion competent syntaxin1A-SNAP25-VAMP SNARE complexes, although tomosyn complexes failed to exhibit tight binding of complexin (9).
Despite extensive biochemical evidence demonstrating the capacity of tomosyn to form tomosyn-SNARE core complexes, it remains uncertain if this association alone is necessary and sufficient to limit availability of reactive Q-SNAREs in vivo for interaction with VAMP and inhibit vesicle priming. Indeed, in this study deletion of loops 1 or 3 (44 and 22 residues in size, respectively) in the N-terminal region of tomosyn ablate the inhibitory action of tomosyn on exocytosis. We believe that the loop deletion is unlikely to have induced loss of the integrity of the large β-propeller structure, as Sro7, which lacks these loops, is a β-propeller structure. In addition, these tomosyn mutants retained membrane targeting and syntaxin1A interaction, suggesting that the mutants were still capable of forming tomosyn-containing heterotrimeric SNARE complexes. Overall, these results suggest that tomosyn R-SNARE/plasma membrane Q-SNARE interactions do not, per se, limit Q-SNARE availability for vesicle priming but instead they dictate targeting of tomosyn to sites where Q-SNAREs are actively transitioning into SNARE core complexes. These data complement previous reports demonstrating that the R-SNARE domain of tomosyn is essential in live cells for targeting of tomosyn to the plasma membrane via direct interaction with syntaxin1A (20) and for forming a SNARE complex containing SNAP25 (10). Of specific importance, deletion data of loops 1 and 3 demonstrate that domains of tomosyn outside the R-SNARE mediate control over formation of VAMP-containing SNARE complexes in vesicle priming and consequent exocytotic activity.
In support of an inhibitory function to regions N-terminal to the R-SNARE of tomosyn are reports demonstrating that tomosyn continued to exert secretory inhibition in transfected PC12 cells (18, 20) and chromaffin cells (24) when key R-SNARE residues required for efficient binding of syntaxin were mutated (18) or when the R-SNARE domain was deleted (20, 24). In addition, the tomosyn gene family includes Drosophila tumor suppressor lethal giant larvae with homologues in yeast (Sro7p and Sro77p) (29, 30) and mammals (Lgl) (28, 51), which all lack a defined R-SNARE motif yet retain the ability to interact with Q-SNARE proteins and perhaps regulate vesicle targeting and SNARE complex assembly.
Recently, three separate but not mutually exclusive mechanisms have been proposed by which the N-terminal region of tomosyn may control calcium-regulated exocytotic activity. These include the following: 1) an N-terminally mediated oligomerization of SNARE core complexes that reduces the probability of a vesicle SNARE to form reactive monomeric SNARE complexes at presumptive sites of exocytosis (21); 2) a structural rearrangement of the 60-residue tail motif found immediately proximal to the SNARE domain that regulates the efficacy and stability R-SNARE interaction of tomosyn with Q-SNAREs (31, 32); and 3) direct binding of the calcium sensor synaptotagmin-1 to the N-terminal region of tomosyn, which results in negative regulation of synaptotagmin-1 function (33).
It is notable that loops 1 and 3 of tomosyn, identified here as essential for the inhibition of secretion of tomosyn, are located on the bottom surface of the N-terminal β-propeller. By comparison, the bottom surface of the β-propeller region of Sro7p forms the binding interface upon which the Sro7p tail interacts, with many of the residues conserved in tomosyn. The additional sequence forming the loop regions in tomosyn raises the possibility that these loops may exert regulation on conformational rearrangements of the tail domain of tomosyn. Indeed, although the low affinity Q-SNARE-binding site within the N-terminal region of tomosyn has not yet been mapped, allosterically regulated conformational shifts in the tail region of Sro7p, which lacks an R-SNARE motif, enables direct binding of the Q-SNARE Sec9 (26). An orthologous tail region in tomosyn may therefore regulate low affinity Q-SNARE interactions. Alternatively, loops 1 and 3 may present scaffolding platforms for effectors that allosterically regulate Q-SNARE binding and assembly of SNARE complex oligomers or that stabilize synaptotagmin-1 interaction.
The level of tomosyn interaction with Q-SNAREs has been reported to be regulated by protein kinase A-mediated phosphorylation of Ser-724 of m-tomosyn-1 within the HVR (22) and of Rho/ROCK phosphorylation of serine 14 of syntaxin1A (20, 52). In this study, we have extended potential for allosteric regulation of tomosyn-1 by identifying a SUMOylation site (Lys-730) within the loop 2 HVR region of b- and m-tomosyn-1. This site was sumoylated by SUMO 2/3 proteins, and SUMO modification was eliminated in a (K730R) site directed mutant of m-tomosyn-1. In addition, functional analysis of tomosyn carrying mutations of the SUMOylation site resulted in an increase in tomosyn inhibition of secretion without alteration in the extent of tomosyn interaction with Q-SNAREs. The signaling pathway utilized to regulate SUMOylation at this site as well as the mechanism by which SUMOylation reduces the inhibitory activity of tomosyn remain to be elucidated. However, SUMOylation frequently allows novel interactions of the modified target protein (53). Furthermore, our data imply that the HVR within loop 2 is a regulatory unit for adjusting the extent of accumulation of tomosyn-2 but not tomosyn-1 protein. This regulation may involve susceptibility of specific tomosyn 2 isoforms to rapid turnover via proteosomal degradation.
In summary, our findings are important as they identify and characterize new structural motifs of tomosyn N-terminal to the SNARE region that substantially impact the ability of tomosyn to inhibit secretion independent of alterations in the strength of the interaction of tomosyn with syntaxin1A. Furthermore, the results establish that regulation of secretion by tomosyn involves cooperative action between the newly identified N-terminal domains of tomosyn that regulate assembly of tomosyn-containing SNARE complexes and the R-SNARE of tomosyn that targets tomosyn to the plasma membrane.
Acknowledgments
We greatly appreciate the gift of mouse tomosyn cDNAs from Alexander J. A. Groffen (Vrije Universiteit Amsterdam, The Netherlands) and for the critical commentary on the work. We also thank Natasha Snider for helpful advice on SUMOylation experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant NS053978. D. M. was supported by the Edmond J. Safra Bioinformatics Program, Tel Aviv University, Tel Aviv, Israel.
- SNARE
- soluble N-ethylmaleimide-sensitive attachment receptors
- VAMP
- vesicle-associated membrane protein
- HVR
- hypervariable region
- hGH
- human growth hormone
- qPCR
- quantitative PCR
- MSA
- multiple sequence alignment
- SUMO
- small ubiquitin-related modifier.
REFERENCES
- 1. Kweon D. H., Kim C. S., Shin Y. K. (2003) Nat. Struct. Biol. 10, 440–447 [DOI] [PubMed] [Google Scholar]
- 2. Gerst J. E. (2003) Biochim. Biophys. Acta 1641, 99–110 [DOI] [PubMed] [Google Scholar]
- 3. Lang T., Margittai M., Hölzler H., Jahn R. (2002) J. Cell Biol. 158, 751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Südhof T. C., Rothman J. E. (2009) Science 323, 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brose N. (2008) Traffic 9, 1403–1413 [DOI] [PubMed] [Google Scholar]
- 6. Südhof T. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13541–13542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Masuda E. S., Huang B. C., Fisher J. M., Luo Y., Scheller R. H. (1998) Neuron 21, 479–480 [DOI] [PubMed] [Google Scholar]
- 8. Fujita Y., Shirataki H., Sakisaka T., Asakura T., Ohya T., Kotani H., Yokoyama S., Nishioka H., Matsuura Y., Mizoguchi A., Scheller R. H., Takai Y. (1998) Neuron 20, 905–915 [DOI] [PubMed] [Google Scholar]
- 9. Pobbati A. V., Razeto A., Böddener M., Becker S., Fasshauer D. (2004) J. Biol. Chem. 279, 47192–47200 [DOI] [PubMed] [Google Scholar]
- 10. Hatsuzawa K., Lang T., Fasshauer D., Bruns D., Jahn R. (2003) J. Biol. Chem. 278, 31159–31166 [DOI] [PubMed] [Google Scholar]
- 11. McEwen J. M., Madison J. M., Dybbs M., Kaplan J. M. (2006) Neuron 51, 303–315 [DOI] [PubMed] [Google Scholar]
- 12. Gracheva E. O., Burdina A. O., Touroutine D., Berthelot-Grosjean M., Parekh H., Richmond J. E. (2007) J. Physiol. 585, 705–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gracheva E. O., Burdina A. O., Touroutine D., Berthelot-Grosjean M., Parekh H., Richmond J. E. (2007) J. Neurosci. 27, 10176–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gracheva E. O., Burdina A. O., Holgado A. M., Berthelot-Grosjean M., Ackley B. D., Hadwiger G., Nonet M. L., Weimer R. M., Richmond J. E. (2006) PLoS Biol. 4, e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yizhar O., Ashery U. (2008) PLoS ONE 3, e2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yizhar O., Matti U., Melamed R., Hagalili Y., Bruns D., Rettig J., Ashery U. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2578–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang W., Lilja L., Mandic S. A., Gromada J., Smidt K., Janson J., Takai Y., Bark C., Berggren P. O., Meister B. (2006) Diabetes 55, 574–581 [DOI] [PubMed] [Google Scholar]
- 18. Constable J. R., Graham M. E., Morgan A., Burgoyne R. D. (2005) J. Biol. Chem. 280, 31615–31623 [DOI] [PubMed] [Google Scholar]
- 19. Widberg C. H., Bryant N. J., Girotti M., Rea S., James D. E. (2003) J. Biol. Chem. 278, 35093–35101 [DOI] [PubMed] [Google Scholar]
- 20. Gladycheva S. E., Lam A. D., Liu J., D'Andrea-Merrins M., Yizhar O., Lentz S. I., Ashery U., Ernst S. A., Stuenkel E. L. (2007) J. Biol. Chem. 282, 22887–22899 [DOI] [PubMed] [Google Scholar]
- 21. Sakisaka T., Yamamoto Y., Mochida S., Nakamura M., Nishikawa K., Ishizaki H., Okamoto-Tanaka M., Miyoshi J., Fujiyoshi Y., Manabe T., Takai Y. (2008) J. Cell Biol. 183, 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baba T., Sakisaka T., Mochida S., Takai Y. (2005) J. Cell Biol. 170, 1113–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheviet S., Bezzi P., Ivarsson R., Renström E., Viertl D., Kasas S., Catsicas S., Regazzi R. (2006) J. Cell Sci. 119, 2912–2920 [DOI] [PubMed] [Google Scholar]
- 24. Yizhar O., Lipstein N., Gladycheva S. E., Matti U., Ernst S. A., Rettig J., Stuenkel E. L., Ashery U. (2007) J. Neurochem. 103, 604–616 [DOI] [PubMed] [Google Scholar]
- 25. Jewell J. L., Oh E., Thurmond D. C. (2010) Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R517–R531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hattendorf D. A., Andreeva A., Gangar A., Brennwald P. J., Weis W. I. (2007) Nature 446, 567–571 [DOI] [PubMed] [Google Scholar]
- 27. Gangar A., Rossi G., Andreeva A., Hales R., Brennwald P. (2005) Curr. Biol. 15, 1136–1142 [DOI] [PubMed] [Google Scholar]
- 28. Katoh M., Katoh M. (2004) Int. J. Oncol. 24, 737–742 [PubMed] [Google Scholar]
- 29. Lehman K., Rossi G., Adamo J. E., Brennwald P. (1999) J. Cell Biol. 146, 125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kagami M., Toh-e A., Matsui Y. (1998) Genetics 149, 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamamoto Y., Mochida S., Kurooka T., Sakisaka T. (2009) J. Biol. Chem. 284, 12480–12490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamamoto Y., Fujikura K., Sakaue M., Okimura K., Kobayashi Y., Nakamura T., Sakisaka T. (2010) Biochem. Biophys. Res. Commun. 399, 24–30 [DOI] [PubMed] [Google Scholar]
- 33. Yamamoto Y., Mochida S., Miyazaki N., Kawai K., Fujikura K., Kurooka T., Iwasaki K., Sakisaka T. (2010) J. Biol. Chem. 285, 40943–40955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yokoyama S., Shirataki H., Sakisaka T., Takai Y. (1999) Biochem. Biophys. Res. Commun. 256, 218–222 [DOI] [PubMed] [Google Scholar]
- 35. Groffen A. J., Jacobsen L., Schut D., Verhage M. (2005) J. Neurochem. 92, 554–568 [DOI] [PubMed] [Google Scholar]
- 36. Schmittgen T. D., Livak K. J. (2008) Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 37. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 38. Edgar R. C. (2004) Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jones D. T. (1999) J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 40. Petrey D., Xiang Z., Tang C. L., Xie L., Gimpelev M., Mitros T., Soto C. S., Goldsmith-Fischman S., Kernytsky A., Schlessinger A., Koh I. Y., Alexov E., Honig B. (2003) Proteins 53, Suppl. 6, 430–435 [DOI] [PubMed] [Google Scholar]
- 41. Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. (2005) Nucleic Acids Res. 33, W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hoppe A., Christensen K., Swanson J. A. (2002) Biophys. J. 83, 3652–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu J., Ernst S. A., Gladycheva S. E., Lee Y. Y., Lentz S. I., Ho C. S., Li Q., Stuenkel E. L. (2004) J. Biol. Chem. 279, 55924–55936 [DOI] [PubMed] [Google Scholar]
- 44. Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 45. Wick P. F., Senter R. A., Parsels L. A., Uhler M. D., Holz R. W. (1993) J. Biol. Chem. 268, 10983–10989 [PubMed] [Google Scholar]
- 46. Gladycheva S. E., Ho C. S., Lee Y. Y., Stuenkel E. L. (2004) J. Physiol. 558, 857–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao J. (2007) Cell. Mol. Life Sci. 64, 3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sen N., Snyder S. H. (2010) Trends Neurosci. 33, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ashery U., Bielopolski N., Barak B., Yizhar O. (2009) Trends Neurosci. 32, 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dybbs M., Ngai J., Kaplan J. M. (2005) PLoS Genet. 1, 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Müsch A., Cohen D., Yeaman C., Nelson W. J., Rodriguez-Boulan E., Brennwald P. J. (2002) Mol. Biol. Cell 13, 158–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sakisaka T., Baba T., Tanaka S., Izumi G., Yasumi M., Takai Y. (2004) J. Cell Biol. 166, 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]