Abstract
RET is a tyrosine kinase receptor involved in numerous cellular mechanisms including proliferation, neuronal navigation, migration, and differentiation upon binding with glial cell derived neurotrophic factor family ligands. RET is an atypical tyrosine kinase receptor containing four cadherin domains in its extracellular part. Furthermore, it has been shown to act as a dependence receptor. Such a receptor is active in the absence of ligand, triggering apoptosis through a mechanism that requires receptor intracellular caspase cleavage. However, different data suggest that RET is not always associated with the cell death/survival balance but rather provides positional information. We demonstrate here that caspase cleavage of RET is involved in the regulation of adhesion in sympathetic neurons. The cleavage of RET generates an N-terminal truncated fragment that functions as a cadherin accessory protein, modifying cadherin environment and potentiating cadherin-mediated cell aggregation. Thus, the caspase cleavage of RET generates two RET fragments: one intracellular domain that can trigger cell death in apoptotic permissive settings, and one membrane-anchored ectodomain with cadherin accessory activity. We propose that this latter function may notably be important for the adequate development of the superior cervical ganglion.
Keywords: Apoptosis, Caspase, Cell Adhesion, Cell Surface Receptor, Neuron, Dependence Receptor, RET
Introduction
A limited number of growth factors and their cognate receptors are required to convey the development of extraordinarily complex biological structures. This suggests that cells maximize their molecular resources. It is then tempting to assume that selection of multitasked receptors has been favored during evolution. The RET tyrosine kinase receptor is a good example for the integration of multiple functions in a single protein. RET mediates many different activities during development but also through adulthood. It has been described that RET induces survival, differentiation, proliferation, migration, or axonal guidance via activation of canonical signaling pathways like PI3K and MAPK (for review, see Ref. 1). This ability to promote different biological effects is partially due to an exquisitely coordinated and regulated relationship between RET and its ligands. RET can be activated by the GDNF5 family ligands (GFLs), a family composed of four members: GDNF, neurturin, artemin, and persephin. However, RET cannot directly bind any of these ligands but requires the presence of the accessory proteins GDNF family receptor α1–4 (GFRα1–4) for its tyrosine kinase catalytic activation (2). The general view is that GFL binding to GFRα/RET triggers RET dimerization, autophosphorylation, and tyrosine kinase activation of RET under physiological conditions, although RET tyrosine kinase activity can be activated in a ligand-independent manner under some pathological circumstances (3).
Although many tyrosine kinase receptors can activate multiple pathways to promote different biological effects, RET is yet an atypical tyrosine kinase receptor. RET belongs to the cadherin superfamily, and it has been suggested that the ret gene is the result of a recombination between a gene encoding a tyrosine kinase receptor and a gene encoding an ancestral cadherin at an early stage of evolution (4). The extracellular domain of RET comprises, as observed for other tyrosine kinase receptors, a cysteine-rich domain, but also four cadherin-like domains (5). Bioinformatic alignments of the second and third cadherin-like domains of RET indicate a close relationship to Drosophila melanogaster Fat cadherin (4). However, the recently published structures of the first and second RET cadherin domains resemble the first and second cadherin domains of the glycosylphosphatidylinositol-anchored nonadhesive T-cadherin (6). Furthermore, it has been recently described that RET interacts and phosphorylates protocadherins (7), suggesting that the relationship of RET with cadherin proteins is closer than initially thought.
Another atypical aspect of RET is its ability to trigger apoptosis when expressed in the absence of GFLs. As such, RET belongs to the functional family of dependence receptors (8, 9). These receptors, which include DCC, UNC5H, Neogenin, Ptc, or TrkC among others, share the intrinsic property of mediating apoptosis in the absence of their respective ligand. This activity participates in the dual role of these dependence receptors in the regulation of neuronal development (10–12) and the control of tumor progression (for review, see Ref. 13). It has been shown that a common feature of these dependence receptors is the cleavage of their intracellular domains by proteases, generally caspase, in the absence of their ligand (for review, see Ref. 14). The general view is that the caspase cleavage allows the exposure or the release of an intracellular protein fragment that mediates apoptosis by interacting with proapoptotic proteins like initiator caspase or proapoptotic kinase (15).
During the course of our research, we have found that RET can take advantage of its atypical duality, as a dependence receptor and as a cadherin protein, to generate a caspase-derived fragment that could impact positional information by regulating adhesion. Here we show that RET is cleaved by caspase in cultured superior cervical ganglion (SCG) neurons during trophic starvation. This cleavage allows the stabilization of an N-terminal truncated form of RET. In transfected cells, caspase-truncated RET behaves as a cadherin accessory protein interacting with the cadherin stabilizer P120. This promotes the interaction of P120 with N-cadherin that results in a reduction of N-cadherin degradation with a corresponding increase of its adhesive properties. With these data, we propose a model for caspase-truncated RET involvement during SCG formation.
EXPERIMENTAL PROCEDURES
Constructs
All RET constructs used derived from pCR3.1 human RET9 kindly given by Dr. U. Arumae. The kinase-dead mutation was obtained by PCR using acg gcc ttc cat ctg aac ggc aga gca ggg tac oligonucleotide and reverse. The D707N mutation was introduced by using cag gtc tcc gtg aat gcc ttc aag atc ctg oligonucleotide and reverse. Stop codons were introduced by using cag gtc tcc gtg tag gcc ttc aag atc ctg and reverse oligonucleotide for RET Stop708 and gcc ttc tgc atc cac tga tac cac aag ttt gcc and reverse oligonucleotide for RET Stop 659. The Myc epitope was introduced at the junction between cadherin domains and the cysteine-rich domain (between amino acids 513 and 514 of human sequence) of the different RET constructs; we performed PCR using ggg tca tat gtg gcc gag gag gcg gaa cag aaa ctg atc tct gaa gaa gac ctg ggc tgc ccc ctg tcc tgt gca gtc and reverse oligonucleotide. We confirmed that all Myc constructs were correctly expressed and folded either by Western blotting and by immunohistochemistry (data not shown). The N-cadherin construct was kindly given by Dr. D. Choquet and Dr. O. Thoumine.
Culture of SCG Neurons
Mouse sympathetic neurons from SCG were cultured as described by others (16). In brief, ganglia were dissected from newborn mice (postnatal day 0–1), digested in collagenase and trypsin (Worthington, Lakewood, NJ), dissociated by trituration, plated on dishes previously coated with rat tail collagen I (BD Biosciences) in DMEM containing 50 ng/ml NGF (Alomone Labs, Jerusalem, Israel), 10% fetal bovine serum, and 5 ng/ml aphidicolin (A. G. Scientific, San Diego, CA) for 5–7 days. Importantly, for optimal detection of caspase-truncated RET sympathetic cell bodies have to be seeded at confluence.
Cell Lines, Transfections, and Cell Viability
SH-SY5Y and COS-7 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and were transfected with Lipofectamine 2000 (Invitrogen). L cells were cultured in DMEM supplemented with 10% fetal bovine serum and were transfected using Jet Pei (Polyplus Transfection, New York, NY). Viability of transfected SH-SY5Y cells was measured by a ToxiLightTM kit used according to the manufacturer's instructions.
Treatments
After being rinsed twice, SCG neurons were exposed to 50 ng/ml NGF, 100 ng/ml GDNF (Abcys, Paris, France) combined with 100 ng/ml Fc-GFRa1 (R&D Systems, Minneapolis, MN) or exposed to 5 μg of anti-NGF (R&D Systems) or anti-NGF combined with 50 μm caspase inhibitor BAF (Sigma). Transfected cells were exposed to 20 nm BAF, 100 ng/ml cycloheximide, and 2 nm DAPT (Sigma).
Western Blotting, Immunoprecipitation, and Immunostaining
The C-19 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used for detection of the RET intracellular epitope. The RET extracellular epitope was detected using MAB482 antibody (R&D Systems) when SCG neurons were analyzed by Western blotting and H-300 antibody (Santa Cruz Biotechnology) when SCG neurons were analyzed by immunofluorescence and when transfected cells were analyzed. The antibodies against ERK2 and actin were from Santa Cruz Biotechnology and Millipore, respectively. Antibodies for p120 and N-cadherin detection were from BD Biosciences. Anti-active caspase 3 antibody was from Cell Signaling (Danvers, MA). Phalloidin Alexa Fluor 488 was from Invitrogen. Anti p-ERK, anti-Myc antibodies, phalloidin-TRITC, and secondary antibodies used for immunostaining were from Sigma. Confocal analysis was performed with LSM 510 confocal laser scanning microscope or confocal Micro Radiance Axioskop 2 from Carl Zeiss (Germany). Analysis and treatment of images were performed using LSM Image Browser, Fiji and Adobe Photoshop.
Native Gel Protein Analysis
Transfected SH-SY5Y cells were lysed using a Native PAGE sample preparation kit (Invitrogen) according to the user's manual. To analyze samples, 4% non-SDS acrylamide gel was prepared using a Native PAGE running buffer kit (Invitrogen). HiMark prestained protein marker (Invitrogen) was used to estimate the size of complexes.
Aggregation Assays
Aggregation assays were performed with little variations of the protocol previously described by Takeichi and Nakagawa (17). In brief, semiconfluent L cells were transfected for 48 h. After transfection or co-transfection, cells were rinsed with ice-cold HCMF buffer (10 mm HEPES, pH 7.4, 137 mm NaCl, 5.4 mm KCl, 0.3 mm Na2HPO4·7H2O, 5.5 mm glucose). Cells were detached using 5 mm calcium and 0.002% trypsin (Worthington), rinsed, and seeded in 1 mm calcium buffer onto BSA-coated dishes. Dishes were placed on a shaker at 80 rpm and incubated during 60 min at 37 °C. Finally, cells were fixed and counted.
Neuronal Nucleofection
SCG neurons were nucleofected using mouse stem cell program (A-033) of the Amaxa device and Basic Neuron SCN Nucleofector solution from Lonza (Basel, Switzerland). Nucleofection was performed according to the manufacturer's instructions.
RESULTS
We showed that human RET is cleaved by caspase in vitro after aspartic acid residues 707 and 1017 (Fig. 1A) and that the fragment lying between these two caspase cleavage sites is able to induce cell death (8). To investigate whether this cleavage occurs in more physiological settings, we used cultured neurons from the SCG, a well established model for caspase-induced cell death during trophic starvation, with high levels of expression of RET and GFRα1 proteins (16). In primary neurons treated for 24 h with NGF, full-length RET protein (170 kDa) was clearly detected by Western blotting using an antibody raised against C-terminal RET intracellular domain (Fig. 1B). Stimulation with GDNF induced proteasomal degradation of full-length RET as described (18), which was delayed using GDNF plus soluble GFRα1 (Fig. 1B). Under trophic starvation, full-length RET protein completely disappeared, except when the same trophic starvation was performed in the presence of BAF, a broad-spectrum caspase inhibitor.
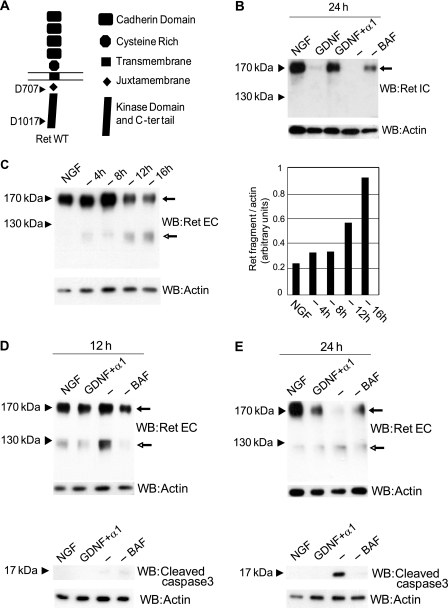
FIGURE 1.
RET is cleaved by caspase in SCG neurons during trophic starvation, and a caspase-truncated fragment of RET is stabilized. A, schematic representation of the RET structure with its different domains. Arrows indicate the Asp-707 and Asp-1017 (human sequence) caspase cleavage site. B, Western blot (WB) analysis of extracts from SCG neurons treated for 24 h with NGF, GDNF, GDNF plus soluble GFRα1, anti-NGF (−), or anti-NGF with the caspase inhibitor BAF and immunostained with intracellular RET-derived antibody. Arrow indicates full-length RET. C, left panel, time course of RET cleavage in extracts of SCG neurons analyzed by Western blotting using extracellular RET-derived antibody. Solid arrow indicates full-length RET, and open-headed arrow indicates the presence of caspase-truncated RET. C, right panel, quantification of levels of caspase-truncated RET compared with actin. D, upper panel, Western blot of SCG neurons extracts treated for 12 h with NGF, GDNF and soluble GFRα1, anti-NGF, or anti-NGF plus caspase inhibitor BAF and stained with extracellular RET-derived antibody. Solid arrow indicates full-length RET, and open-headed arrow indicates the presence of caspase-truncated RET. D, lower panel, same extracts were used for active caspase-3 Western blotting. E, upper panel, Western blot of SCG neuron extracts treated for 24 h with NGF, GDNF and soluble GFRα1, anti-NGF, or anti-NGF plus caspase inhibitor BAF and stained with extracellular RET-derived antibody. Solid arrow indicates full-length RET, and open-headed arrow indicates the presence of caspase-truncated RET. E, lower panel, same extracts were used for an active caspase-3 immunoblotting.
Using an antibody directed against an extracellular epitope of RET, a fragment migrating at approximately 120 kDa was detected increasingly with the duration of trophic starvation, mirroring the decreased detection of full-length RET (Fig. 1C). By size estimation, this fragment could correspond to caspase-truncated RET comprising amino acids 29–708 (mouse sequence, corresponding in human sequence to 29–707). Neurons were then exposed to a 12-h or a 24-h trophic starvation in the presence or absence of BAF. The 120 kDa band was observed in these neurons unless the caspase inhibitor BAF was present (Fig. 1, D, upper panel, and E, upper panel). Interestingly, the appearance of this caspase-dependent 120-kDa fragment was observed before cell death commitment monitored by massive caspase-3 activation (Fig. 1, D, lower panel, and E, lower panel). The detection of this 120-kDa fragment before apoptosis commitment suggests that RET cleavage is achieved by a first round of caspase activated before general caspase-3 activation, e.g. initiator caspases such as caspase-9 or local effector caspase activation. Similar results were obtained with another RET antibody (supplemental Fig. 1A). Altogether, these experiments suggest that caspase cleavage of RET produces a truncated fragment that contains, from its N terminus, the four cadherin domains, the cysteine-rich domain, the transmembrane domain, and nearly all of the RET juxtamembrane region.
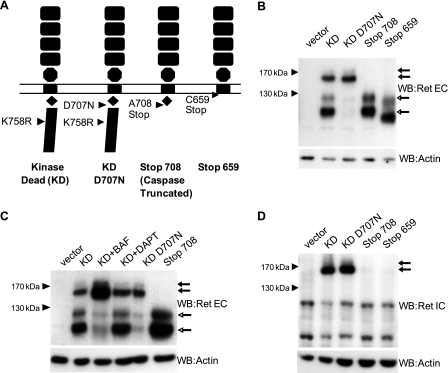
To characterize this 120-kDa fragment further, different RET mutant constructs were prepared (see schematic representation in Fig. 2A), including RET K758R kinase-dead (KD), RET KD mutated at the first caspase cleavage site D707N (RET KD D707N), putative caspase-truncated RET cleaved after aspartic acid 707 (RET 708Stop), and as a control, a construct RET truncated just after the transmembrane region (RET 659Stop). The kinase-dead mutation was introduced into the full-length RET constructs to avoid the constitutive trans-phosphorylation observed with overexpressed tyrosine kinase receptors. RET mutant constructs were then expressed in human neuroblastoma SH-SY5Y cells. Four specific fragments were detected in cells transfected with RET KD using an antibody targeting the RET extracellular domain (Fig. 2B). Two fragments migrating, respectively, at 170–150 kDa correspond to the mature high glycosylated and immature low glycosylated full-length RET proteins, respectively, as described elsewhere (19). However, two specific fragments at 120–100 kDa were also detected. These fragments could correspond to caspase cleavage of mature and immature RET, respectively, as they were not detected in RET KD D707N transfected cells, and their size matched the RET 708Stop fragments. In addition, BAF but not DAPT, a γ-secretase inhibitor, prevented the formation of these fragments (Fig. 2C). The possibility that these fragments were aberrant degradation products was also excluded because they were not recognized by an antibody directed against RET intracellular domain epitope (Fig. 2D). Taken together, these results indicate that mature and immature unphosphorylated RET are cleaved at aspartic 707 by caspase, and the caspase cleavage is associated with the presence of an N-terminal truncated RET form.
FIGURE 2.
Characterization of caspase-truncated RET in transfected SH-SY5Y. A, schematic representation of the different RET constructs used to characterize caspase-truncated RET Stop708. B, SH-SY5Y cells transfected with different RET constructs and starved for 24 h. The presence of caspase-truncated RET was analyzed by Western blotting (WB) using a RET extracellular antibody. In all blots the solid arrow indicates mature and nonmature full-length RET, and the open-headed arrow indicates the presence of mature and nonmature caspase-truncated RET. C, SH-SY5Y cells transfected with different RET constructs and starved for 24 h in the presence or absence of BAF and DAPT. D, SH-SY5Y cells transfected with different RET constructs and starved for 24 h. The presence of caspase-truncated RET was analyzed by Western blotting using a RET intracellular antibody (lower panel). Arrows indicate mature and nonmature caspase-truncated RET.
We next investigated the biological activity of this fragment. Because RET cleavage by caspase is a prerequisite for RET-induced apoptosis (8), we first analyzed whether the RET 708stop truncated fragment could mediate apoptosis. SH-SY5Y cells were then transiently transfected. In this setting, RET 708Stop similarly to the caspase cleavage mutant RET D707N failed to induce cell death (data not shown). We next investigated whether this caspase-truncated fragment could act as a dominant negative mutant of full-length RET that could titrate out GDNF. We first tested this hypothesis by measuring GDNF-induced ERK phosphorylation in SH-SY5Y cells expressing endogenous RET and GFRα1. When cells were stimulated with 100 ng/ml GDNF, ERK phosphorylation was not affected by RET 708Stop (or as a control RET 659Stop) expression compared with mock-transfected cells (Fig. 3A and supplemental Fig. 2). Noteworthy, uncleavable RET KD D707N and RET KD appeared as efficient blockers for GDNF-induced ERK activation (Fig. 3A and supplemental Fig. 2).
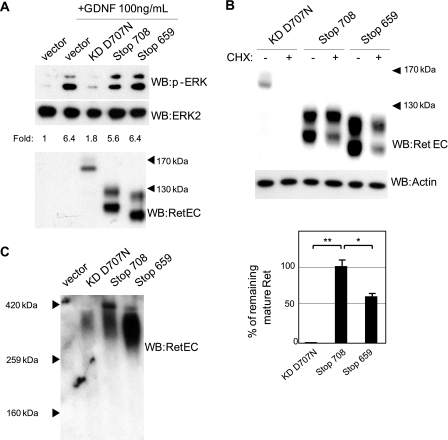
FIGURE 3.
Different behavior of caspase-truncated RET compared with full-length RET in transfected SH-SY5Y. A, SH-SY5Y cells were transfected with different RET constructs, starved for 4 h, and stimulated with 100 ng/ml GDNF. Activation of RET downstream pathways was analyzed by Western blotting (WB) using p-ERK (upper panel) and ERK2 antibody (middle panel). Quantification is given as -fold induction of the p-ERK/ERK2 ratio and corresponds to the representative experiment shown. B, SH-SY5Y cells were transfected with different RET constructs. After 24 h, transfected cells were starved and exposed to cycloheximide (CHX) for 24 h. RET stability was quantified after Western blotting using RET extracellular antibody. Quantification corresponds to the average of three experiments. **, p < 0.01; *, p < 0.05 analyzed by Student's test. C, SH-SY5Y cells were transfected with different RET constructs and starved for 24 h. The formation of RET complexes was analyzed using native gel eletrophoresis and Western blotting using RET extracellular antibody.
One possibility for the lack of dominant negative effect of both RET 708Stop and RET 659Stop could be that the deletion of the kinase domain generates a short lived fragment. Therefore, the stability of different RET constructs was studied in transfected SH-SY5Y by analyzing their expression in the presence or absence of a 24-h cycloheximide treatment (Fig. 3B). Unexpectedly, both constructs lacking the kinase domain of RET were less degraded than RET KD D707N. The mature caspase-truncated RET displayed a remarkable stability (Fig. 3B) with very little degradation detected after 24-h exposure to cycloheximide. Then, rather than being a short lived fragment, RET 708Stop increased stability, supporting the view that the deletion of the kinase domain of RET forces a structural rearrangement of its ectodomain, promoting interactions with different proteins rather than the known formation of RET dimers. Native PAGE of transfected SH-SY5Y cells extracts was then performed (Fig. 3C). Fig. 3C shows that RET KD D707N migrated at approximately 340 kDa, the predicted size of the RET dimer (2 × 170 kDa), as described in cells with overexpressed RET (20). However, both RET 708Stop and RET 659Stop mainly migrated above, respectively, 320 kDa and 280 kDa, sizes higher than expected in predicted dimer forms (2 × 120 kDa and 2 × 100 kDa; Fig. 3C), suggesting alternative conformations like multimers of RET 708Stop and RET 659Stop or the formation of stable complexes with proteins other than RET. Altogether, these results demonstrate that caspase-truncated RET is not a dominant negative for full-length RET and suggest that the highly stable caspase-truncated RET displays a conformation allowing either multimeric conformation or the interaction with unknown proteins.
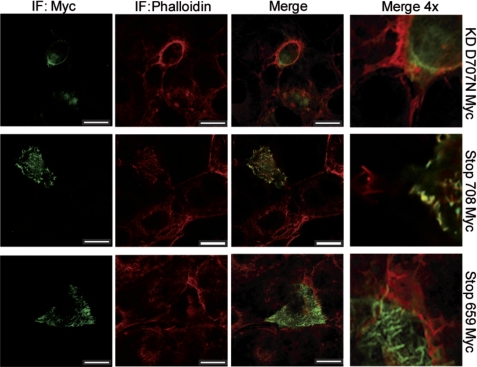
To understand the possible functions of caspase-truncated RET, we studied its cellular localization. We first designed different RET constructs tagged with the Myc epitope in its ectodomain. We thus performed immunostaining in transfected SH-SY5Y and COS-7 cells, and we analyzed Myc localization using confocal microscopy. All three constructs tested, RET KD D707N Myc, RET 708Stop Myc, and RET 659Stop Myc showed a diffuse localization within the cell and along the membrane (Fig. 4). However, the cells expressing the Myc tagged caspase-truncated RET displayed a punctuate distribution on cell membrane with a marked co-localization with phalloidin at the cell borders (Fig. 4 and supplemental Fig. 3).
FIGURE 4.
Transfected caspase-truncated RET co-localizes with actin. COS-7 cells were transfected with different RET-Myc-tagged constructs and starved for 24 h. Localization of RET-Myc-tagged constructs was analyzed by immunocytochemistry using a Myc antibody (green) and phalloidin (red) to stain F-actin. Pictures are representative of confocal sections, and co-localization appears in yellow. Confocal analysis was repeated in two different transfections. Scale bars, 20 μm.
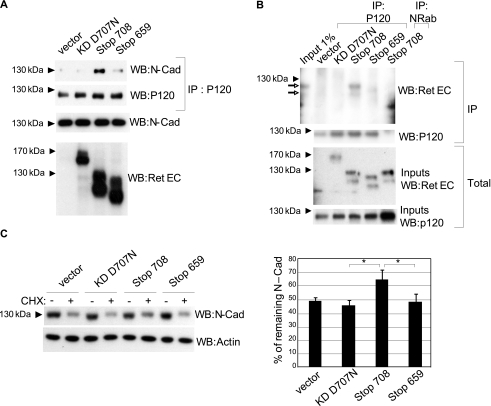
Because caspase-truncated RET comprises the four cadherin-like domains in its ectodomain, we hypothesized that it may act as a cadherin or a cadherin-like protein. Cadherins form a family of proteins that regulate cell adhesion by the establishment of trans-homophilic interactions via their extracellular domains, whereas their intracellular domains bind actin filaments via different catenins (21). We failed to show a reproducible interaction of the caspase-truncated RET with itself, i.e. trans-homophilic interaction, or with cadherin (data not shown). We thus investigated whether the caspase-truncated RET could affect the behavior of endogenous cadherins. The caspase-truncated RET was expressed in SH-SY5Y cells known to express N-cadherin, and we analyzed the activity of the latter via its interaction with the cadherin stabilizer protein P120. As shown in Fig. 5A, in cells transfected with RET KD D707N or the RET 659Stop, only a weak interaction between P120 and N-cadherin was evidenced by co-immunoprecipitation. In contrast, the presence of caspase-truncated RET was associated with a clear recruitment of P120 to N-cadherin although the interaction of N-cadherin with β-catenin was unaffected (supplemental Fig. 4). This suggests a specific effect on P120 mediated by caspase-truncated RET.
FIGURE 5.
Caspase-truncated RET interacts with P120 and increases N-cadherin stability. A, SH-SY5Y cells were transfected with different RET constructs and starved for 24 h. Formation of a P120-N-cadherin complex was analyzed by immunoprecipitation of P120 followed by N-cadherin Western blotting (WB). B, SH-SY5Y cells were transfected with different RET constructs and starved for 24 h. The formation of a complex between P120 and caspase-truncated RET was analyzed by immunoprecipitation of P120 or nonrelated antibody (NRab) followed by RET extracellular Western blotting. Open-headed arrow indicates the presence of mature and nonmature caspase-truncated RET. C, SH-SY5Y cells were transfected with different RET constructs. After 24 h, transfected cells were starved and exposed to cycloheximide (CHX) for 24 h. N-cadherin stability was quantified by Western blotting. Quantification corresponds to the average of three independent experiments. *, p < 0.05 analyzed by Student's test.
P120 is an “armadillo” protein that binds different cadherins at their juxtamembrane region, stabilizing these proteins at the membrane and increasing their adhesive properties (22, 23). As the caspase-truncated RET differs from the RET Stop659 by the conservation of its juxtamembrane region (Fig. 5A), we hypothesized that caspase-truncated RET could interact with P120 via its exposed juxtamembrane region. We analyzed co-immunoprecipitation of P120 with various RET constructs in transfected SH-SY5Y cells exposed to trophic starvation for 24 h. As shown in Fig. 5B, the caspase-truncated RET co-immunoprecipitated with P120 whereas other RET constructs did not.
Because the caspase-truncated RET interacts with P120 and facilitates the N-cadherin/P120 interaction, we investigated whether the caspase-truncated fragment affects N-cadherin stability. The expression of N-cadherin was analyzed in SH-SY5Y cells transfected with different RET-expressing constructs and cultured for 24 h in the presence or absence of cycloheximide. Although cells transfected with RET KD D707N, RET Stop659, or the empty vector showed an ∼50% reduction of their total amount of N-cadherin in presence of cycloheximide (Fig. 5C), the reduction was only around 25% in cells expressing the caspase-truncated RET. Thus, together, these data support the view that the presence of caspase-truncated RET promotes N-cadherin stability via P120.
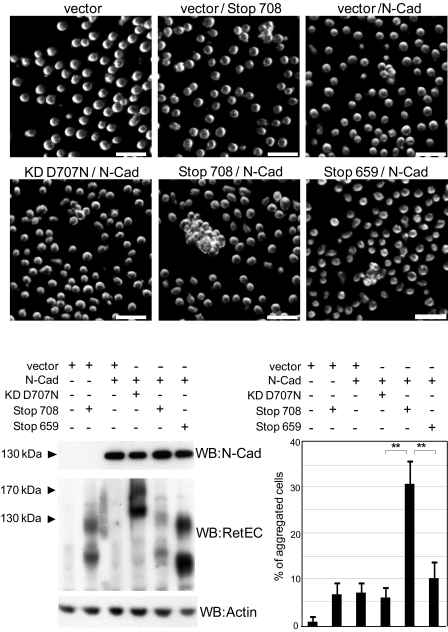
To investigate the cellular relevance of the truncated RET-mediated increase of cadherin stability, we performed aggregation assays on L cells, a mouse fibroblast cell line that does not show spontaneous adhesive properties. We co-transfected L cells with a low amount of N-cadherin together with the different RET constructs and performed aggregation assays. Fig. 6 shows that either ectopic expression of N-cadherin or RET Stop708 alone induced a weak L cell aggregation. However, when N-cadherin was co-transfected with caspase-truncated RET, a synergistic effect was observed, resulting in a striking increased cell aggregation (Fig. 6, left lower panel, expression of the different proteins; right lower panel, quantification). These results suggest that caspase-truncated RET, by stabilizing N-cadherin, improves N-cadherin adhesion capabilities.
FIGURE 6.
Caspase-truncated RET increases N-cadherin aggregative properties. L cells were co-transfected with N-cadherin plasmid (1/5 of total cDNA) and vector or different RET constructs (4/5 of total cDNA). After 48 h of transfection, expression of N-cadherin and RET constructs was analyzed by Western blotting (WB; lower left panels), and in parallel, aggregation assays were performed. Upper panels are representative dark field microscopy images. Scale bars, 50 μm. Lower right panel, quantification corresponds to the average of three independent experiments. **, p < 0.01 analyzed by Student's test.
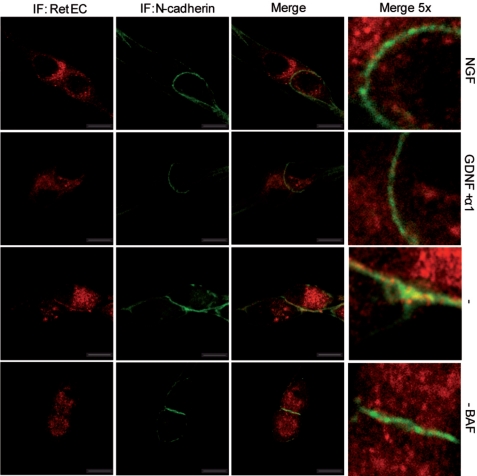
Altogether, these results suggested the formation of a hypothetical ternary complex formed by caspase-truncated RET-P120-N-cadherin. To ensure that this complex is formed in starved SCG neurons, we studied the immunolocalizations of RET and N-cadherin in sympathetic neurons (Fig. 7). Starvation was applied for 16 h, a time period apparently well tolerated by the SCG neurons and a time frame long enough to detect caspase-truncated RET (Fig. 1C). In the presence of NGF or GDNF, no co-localization of RET and N-cadherin was detected (Fig. 7, top two rows). RET extracellular domain staining was diffused within the neuronal soma whereas N-cadherin staining was observed at the membrane. However, in trophic-deprived neurons, a fraction of RET clearly co-localized with N-cadherin especially in the regions of contacts between somas (Fig. 7, third row from top; quantified in supplemental Fig. 5). As expected, this co-localization was not observed when the caspase inhibitor BAF was added to the medium (Fig. 7, bottom row). These results suggest that the complex formed by caspase-truncated RET- P120-N-cadherin occurred in neurons with reduced trophic status. Of interest, this complex is preferentially formed at the contact sites between somas, which could explain why the detection of caspase-truncated RET was facilitated when SCG somas were seeded at confluence (data not shown; see “Experimental Procedures”).
FIGURE 7.
Extracellular RET co-localizes with N-cadherin in starved SCG neurons. SCG neurons were treated for 16 h with NGF, GDNF, anti-NGF, or anti-NGF plus caspase inhibitor BAF. Localization of RET and N-cadherin was analyzed by immunocytochemistry using an extracellular RET-derived antibody (red) and an N-cadherin antibody (green). Images are representative confocal sections, and co-localization appears in yellow. Confocal analysis was repeated in two different SCG cultures. Scale bars, 10 μm.
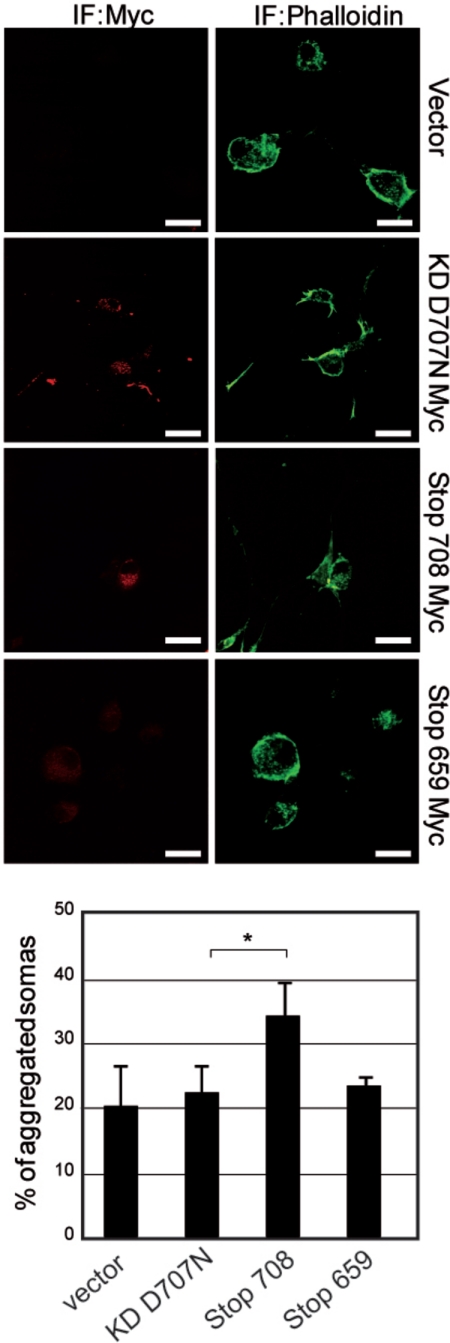
Regulation of N-cadherin adhesion is crucial for sympathetic ganglia formation (24). To challenge a function of caspase-truncated RET as a cadherin accessory protein improving cadherin-mediated adhesion in sympathetic neurons, we nucleofected different Myc-tagged RET constructs in dissociated SCG neurons, and we analyzed neurons by immunofluorescence after 48 h of culture at low density. As it usually occurs, somas of dissociated sympathetic neurons seeded for culture tend to reaggregate with time (25). However, nucleofection of caspase-truncated RET accelerated this tendency, and at 48 h we observed a higher number of aggregated neuronal somas (Fig. 8) than in controls. Interestingly, this effect not only occurred between Myc-positive neurons, but we also observed pairs of Myc-positive/Myc-negative neurons with cell contacts, suggesting that the impact of caspase-truncated RET on neuronal aggregation could occur in a cell-autonomous and non-cell-autonomous manner.
FIGURE 8.
Nucleofection of caspase-truncated RET accelerates soma reaggregation in SCG neurons. SCG neurons were nucleofected with different RET-Myc tagged constructs. 48 h after nucleofection SCG neurons were immunostained to detect Myc-positive cells (red). The total number of neurons was determined using phalloidin (green). Images are representative fluorescence microscopy. Quantification of aggregated somas corresponds to the average of two independent experiments. *, p < 0.05 analyzed by Student's test. Scale bars, 20 μm.
DISCUSSION
We and others have shown that RET and caspase can regulate the cell survival/death balance when RET acts as a dependence receptor (8, 9). RET is a target for caspase in the absence of GFLs, and its intracellular domain is cleaved at aspartic acid 707 and aspartic acid 1017 (human sequence). We proposed that the fragment comprising residues 707–1017 has a proapoptotic function (8). However, several data suggest that the impact of RET on the cell survival/death balance depends on the cellular context: for example, fusimotor motoneurons exhibit a crucial dependence of RET for survival (26), whereas RET, during development of SCG, provides an appropriate positional information for ganglion migration and proper target innervation rather than promoting the activation of survival (27). On the other hand, caspases have been recently involved in nonapoptotic functions such as proliferation, differentiation and migration (for review, see Ref. 28). In this report, we show that RET is cleaved by caspase in trophic-starved SCG neurons. Caspase cleavage of RET generates several fragments (8). Our interest focuses on the N-terminal caspase-truncated RET fragment. This fragment is stabilized in primary culture of SCG neurons during the early phase of trophic starvation, before neurons irreversibly commit to cell death. It is yet fair to say that at this point we cannot discard that this caspase-truncated fragment only appears in setting of apoptosis induction because even though cell death commitment was not achieved, trophic withdrawal may be associated with local caspase activation. However, in transfected cells, we show that caspase-truncated RET is not a cell death inducer, as the 707–1017 fragment, but functions as a cadherin accessory protein. This clearly indicates that besides a fine regulation of the caspase cleavage of RET, its signaling relies on the stabilization of one or the other fragment which is likely contingent on the cellular context and the specific state of cells.
Full-length RET is a nonclassical cadherin with no adhesive properties (19) but with high affinity for complexes formed by GFLs-GFRαs. In contrast, caspase-truncated RET does not compete with endogenous RET for binding of GDNF, but functions as a cadherin accessory protein. This suggests that the removal of the RET intracellular tyrosine kinase domain by caspase cleavage promotes conformational changes in the ectodomain, coupled with the acquisition of functional properties different from those of full-length RET protein. Caspase-truncated RET shows a strong stability, enough for a caspase fragment to perform a relevant biological function (29). Recent resolution of the structures of first and second cadherin domains of RET reveals structural similarities with T-cadherin (6), a truncated cadherin anchored to the membrane by a glycosylphosphatidylinositol (30). In both cases, cadherin domains show a closed “head to tail” conformation instead of the classic cadherin open conformation required for the adequate formation of trans-homophilic interactions (6). This observation may explain why we failed to detect any trans-homophilic interaction of caspase-truncated RET and its low ability to induce aggregation. However, caspase-truncated RET is still able to regulate adhesion.
After cleavage by caspase, RET remaining fragment conserves its juxtamembrane region. In contrast to the full-length RET protein, the juxtamembrane region of the caspase-truncated RET is exposed and allows the interaction with the cadherin stabilizer P120. The high stability of caspase-truncated RET could rely on the lack of degradation motifs but also on its interaction with P120 as RET 708Stop showed higher stability than RET 659Stop. Juxtamembrane region of caspase-truncated RET also permits N-cadherin stabilization with the corresponding increase in its adhesive capabilities. Here, we have demonstrated that caspase-truncated RET co-localizes with N-cadherin. However, this co-localization at the border of contact between somas does not match totally. This fact and the evidence that caspase-truncated RET co-immunoprecipitates with P120, a stabilizer of different cadherins, support the view that N-cadherin may not be the only cadherin regulated by caspase-truncated RET. It is also important to note that the membrane-associated P120 is a cadherin stabilizer whereas the cytosolic P120 is a regulator of small GTPases (31). So, we cannot exclude the possibility that caspase-truncated RET may also affect the behavior of RhoA, Rac1, and cdc42, but this will require further studies.
Finally our results recall the data suggesting that GDNF is a master regulator of cellular adhesion. GDNF is not only a ligand for RET, but also a ligand for the adhesion molecule p140 NCAM when bound to GFRα1 (32). A complex formed by GDNF, GFRα1, and p140 NCAM inhibits NCAM cell adhesion and activates ERK and Fyn signaling, inducing migration of neuronal precursors (32). Moreover, it has been proposed recently that GDNF and GFRα1 proteins could act as ligand-induced cell adhesion molecules at synapses (33). It could be thus interesting to speculate on a role of this GDNF-dependent caspase cleavage in the establishment of region of weak/strong adhesive properties. In regions near sources of GDNF, we could hypothesize that it will be “weak adhesive regions” due to NCAM and full-length RET signaling activation with the corresponding reduced NCAM and cadherin adhesion. On the opposite side, while moving out of GNDF sources, the generation of the caspase-truncated RET, acting as a cadherin accessory protein could reinforce cadherin adhesion and could help to define “strong adhesive regions” with reduced motility. It is in this manner, and coordinated with N-cadherin or other cadherins, that we propose that the availability of GDNF provides positional information important for SCG formation.
Acknowledgments
We thank L. Giraud for technical advice, U. Arumae for the RET9 wild-type construct; D. Choquet and O. Thoumine for the N-cadherin construct; J. G. Delcros for text correction; and F. Wandosell, V. Castellani, J. J. Garrido, and M. Encinas for helpful discussions.
This work was supported by an institutional grant from CNRS (to P. M.), the Centre Léon Bérard (to P. M.), the Université de Lyon (to P. M.) and from the Ligue Contre le Cancer (to P. M.), Institut National du Cancer (to P. M.), Agence Nationale de la Recherche blanche (to P. M.), Specific Targeted Research Projects Hermione (to P. M.), and Apoptosis Mechanisms in Cancer and AIDS (to P. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- GDNF
- glial cell derived neurotrophic factor
- GFL
- GDNF family ligand
- GFRα
- GDNF family receptor α
- KD
- kinase-dead
- SCG
- superior cervical ganglion
- BAF
- boc-aspartyl-(OMe)-fluoromethyl-ketone.
REFERENCES
- 1. Airaksinen M. S., Saarma M. (2002) Nat. Rev. Neurosci. 3, 383–394 [DOI] [PubMed] [Google Scholar]
- 2. Treanor J. J., Goodman L., de Sauvage F., Stone D. M., Poulsen K. T., Beck C. D., Gray C., Armanini M. P., Pollock R. A., Hefti F., Phillips H. S., Goddard A., Moore M. W., Buj-Bello A., Davies A. M., Asai N., Takahashi M., Vandlen R., Henderson C. E., Rosenthal A. (1996) Nature 382, 80–83 [DOI] [PubMed] [Google Scholar]
- 3. Plaza-Menacho I., Burzynski G. M., de Groot J. W., Eggen B. J., Hofstra R. M. (2006) Trends Genet. 22, 627–636 [DOI] [PubMed] [Google Scholar]
- 4. Hulpiau P., van Roy F. (2009) Int. J. Biochem. Cell Biol. 41, 349–369 [DOI] [PubMed] [Google Scholar]
- 5. Anders J., Kjar S., Ibáñez C. F. (2001) J. Biol. Chem. 276, 35808–35817 [DOI] [PubMed] [Google Scholar]
- 6. Kjaer S., Hanrahan S., Totty N., McDonald N. Q. (2010) Nat. Struct. Mol. Biol. 17, 726–731 [DOI] [PubMed] [Google Scholar]
- 7. Schalm S. S., Ballif B. A., Buchanan S. M., Phillips G. R., Maniatis T. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13894–13899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bordeaux M. C., Forcet C., Granger L., Corset V., Bidaud C., Billaud M., Bredesen D. E., Edery P., Mehlen P. (2000) EMBO J. 19, 4056–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cañibano C., Rodriguez N. L., Saez C., Tovar S., Garcia-Lavandeira M., Borrello M. G., Vidal A., Costantini F., Japon M., Dieguez C., Alvarez C. V. (2007) EMBO J. 26, 2015–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thibert C., Teillet M. A., Lapointe F., Mazelin L., Le Douarin N. M., Mehlen P. (2003) Science 301, 843–846 [DOI] [PubMed] [Google Scholar]
- 11. Furne C., Rama N., Corset V., Chédotal A., Mehlen P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14465–14470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsunaga E., Tauszig-Delamasure S., Monnier P. P., Mueller B. K., Strittmatter S. M., Mehlen P., Chédotal A. (2004) Nat. Cell Biol. 6, 749–755 [DOI] [PubMed] [Google Scholar]
- 13. Mehlen P., Puisieux A. (2006) Nat. Rev. Cancer 6, 449–458 [DOI] [PubMed] [Google Scholar]
- 14. Goldschneider D., Mehlen P. (2010) Oncogene 29, 1865–1882 [DOI] [PubMed] [Google Scholar]
- 15. Mille F., Thibert C., Fombonne J., Rama N., Guix C., Hayashi H., Corset V., Reed J. C., Mehlen P. (2009) Nat. Cell Biol. 11, 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Encinas M., Rozen E. J., Dolcet X., Jain S., Comella J. X., Milbrandt J., Johnson E. M., Jr. (2008) Cell Death Differ. 15, 1510–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takeichi M., Nakagawa S. (2001) Curr. Protoc. Cell Biol. 9, Unit 9.3 [DOI] [PubMed] [Google Scholar]
- 18. Tsui C. C., Pierchala B. A. (2010) J. Neurosci. 30, 5149–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takahashi M., Asai N., Iwashita T., Isomura T., Miyazaki K., Matsuyama M. (1993) Oncogene 8, 2925–2929 [PubMed] [Google Scholar]
- 20. Trupp M., Raynoschek C., Belluardo N., Ibáñez C. F. (1998) Mol. Cell. Neurosci. 11, 47–63 [DOI] [PubMed] [Google Scholar]
- 21. Takeichi M. (2007) Nat. Rev. Neurosci. 8, 11–20 [DOI] [PubMed] [Google Scholar]
- 22. Yap A. S., Niessen C. M., Gumbiner B. M. (1998) J. Cell Biol. 141, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thoreson M. A., Anastasiadis P. Z., Daniel J. M., Ireton R. C., Wheelock M. J., Johnson K. R., Hummingbird D. K., Reynolds A. B. (2000) J. Cell Biol. 148, 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kasemeier-Kulesa J. C., Bradley R., Pasquale E. B., Lefcort F., Kulesa P. M. (2006) Development 133, 4839–4847 [DOI] [PubMed] [Google Scholar]
- 25. McCarthy K. D., Partlow L. M. (1976) Brain Res. 114, 391–414 [DOI] [PubMed] [Google Scholar]
- 26. Gould T. W., Yonemura S., Oppenheim R. W., Ohmori S., Enomoto H. (2008) J. Neurosci. 28, 2131–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Enomoto H., Crawford P. A., Gorodinsky A., Heuckeroth R. O., Johnson E. M., Jr., Milbrandt J. (2001) Development 128, 3963–3974 [DOI] [PubMed] [Google Scholar]
- 28. Li J., Yuan J. (2008) Oncogene 27, 6194–6206 [DOI] [PubMed] [Google Scholar]
- 29. Dix M. M., Simon G. M., Cravatt B. F. (2008) Cell 134, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Philippova M., Joshi M. B., Kyriakakis E., Pfaff D., Erne P., Resink T. J. (2009) Cell Signal 21, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 31. Anastasiadis P. Z. (2007) Biochim. Biophys. Acta 1773, 34–46 [DOI] [PubMed] [Google Scholar]
- 32. Paratcha G., Ledda F., Ibáñez C. F. (2003) Cell 113, 867–879 [DOI] [PubMed] [Google Scholar]
- 33. Ledda F., Paratcha G., Sandoval-Guzmán T., Ibáñez C. F. (2007) Nat. Neurosci. 10, 293–300 [DOI] [PubMed] [Google Scholar]