Abstract
Glycosylphosphatidylinositol (GPI) is a post-translational modification that anchors cell surface proteins to the plasma membrane, and GPI modifications occur in all eukaryotes. Biosynthesis of GPI starts on the cytoplasmic face of the endoplasmic reticulum (ER) membrane, and GPI precursors flip from the cytoplasmic side to the luminal side of the ER, where biosynthesis of GPI precursors is completed. Gwt1p and PIG-W are inositol acyltransferases that transfer fatty acyl chains to the inositol moiety of GPI precursors in yeast and mammalian cells, respectively. To ascertain whether flipping across the ER membrane occurs before or after inositol acylation of GPI precursors, we identified essential residues of PIG-W and Gwt1p and determined the membrane topology of Gwt1p. Guided by algorithm-based predictions of membrane topology, we experimentally identified 13 transmembrane domains in Gwt1p. We found that Gwt1p, PIG-W, and their orthologs shared four conserved regions and that these four regions in Gwt1p faced the luminal side of the ER membrane. Moreover, essential residues of Gwt1p and PIG-W faced the ER lumen or were near the luminal edge of transmembrane domains. The membrane topology of Gwt1p suggested that inositol acylation occurred on the luminal side of the ER membrane. Rather than stimulate flipping of the GPI precursor across the ER membrane, inositol acylation of GPI precursors may anchor the precursors to the luminal side of the ER membrane, preventing flip-flops.
Keywords: Endoplasmic Reticulum (ER), Glycosylphosphatidylinositol Anchors, Membrane Proteins, Post-translational Modification, Yeast Genetics, Inositol Acylation
Introduction
Many proteins attach to the cell surface via glycosylphosphatidylinositol (GPI)4 (1–3). GPI modification is conserved among eukaryotes. In the budding yeast Saccharomyces cerevisiae and in mammalian cells, ∼60 and ∼150 proteins, respectively, are predicted to be GPI-anchored (4, 5). The GPI-anchored proteins in yeast are mainly involved in cell wall integrity, and GPI biosynthesis is critical for growth of yeast cells (6); in mammals, GPI biosynthesis is essential for embryogenesis, but not for growth of individual cells (7, 8).
The biosynthesis of GPI and its attachment to target proteins occur on the endoplasmic reticulum (ER) membrane. GPIs, which have a conserved core structure, are preassembled in the ER via a multistep pathway before they are transferred to target proteins. The inositol moiety of GPI is often modified with a fatty acyl chain. In yeast and mammalian cells, the inositol acylation occurs at GlcN-PI to generate GlcN-(acyl)PI, and the acylation normally precedes the first mannosylation that generates Man-GlcN-(acyl)PI (9, 10). Once added, the inositol-linked acyl chain remains attached until the complete GPI precursor is transferred to a target protein in the ER. In many mammalian cells and in yeast, the acyl chain is removed from the inositol residue immediately after the transfer of the complete GPI precursor to a protein in the ER (11, 12).
Gwt1p and PIG-W have been identified as inositol acyltransferases in yeast and mammalian cells, respectively (13, 14). In S. cerevisiae, the compound, 1-[4-butylbenzyl]isoquinoline (BIQ) inhibits cell wall localization of GPI-anchored mannoproteins, and GWT1 was identified as a multicopy suppressor of BIQ sensitivity in S. cerevisiae (15). Gwt1p is the direct target of BIQ, and GWT1 null mutants grow very slowly and are defective in cell wall assembly (15). GWT1 is a homolog of PIG-W and can complement PIG-W mutant cells (14). GWT1 encodes a 490-amino acid protein that is predicted to have multiple membrane-spanning regions (15). Gwt1p localizes to the ER and is involved in inositol acylation, and gwt1 mutant cells produce fewer GPI-anchored proteins than wild-type cells, indicating that acylation is critical for the attachment of GPI to proteins (13). Characterization of temperature-sensitive gwt1 mutant cells also revealed the importance of GPI-anchored proteins for the transport of microdomain-associated membrane proteins, such as Tat2p and Fur4p, and their association with membrane microdomains (16).
GPIs are preassembled in association with the ER membrane via a multistep pathway. The two initial reactions of this pathway that generate GlcN-PI occur on the cytoplasmic side of the ER membrane (17–19); however, Gpi14p/PIG-M, which transfers the first mannose to GlcN-(acyl)PI, functions on the luminal side of the ER (20). Therefore, the GPI precursors must flip across the ER membrane before the first mannosylation. PIG-W encodes a 504-amino acid protein that spans the ER membrane and is predicted to have multiple transmembrane domains. The N terminus of PIG-W is oriented toward the ER lumen, and its C terminus is oriented toward the cytoplasm (14). Analysis with the TMpred program suggests that the PIG-W protein contains 13 transmembrane domains, and the conserved regions of PIG-W and its homologs are all predicted to orient toward the ER lumen, suggesting that the inositol acylation occurs on the luminal side of the ER (Fig. 1A) (14).
FIGURE 1.
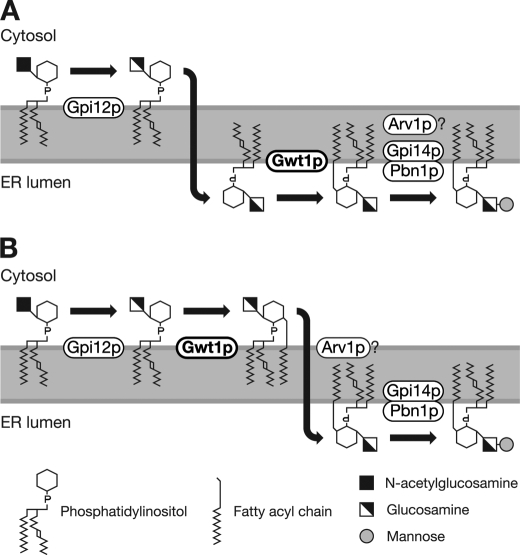
Two possible models for GPI biosynthetic pathway. A, if inositol acylation by Gwt1p occurs on the luminal side of the ER, GlcN-PI would flip from the cytoplasmic face to the luminal face of the ER membrane. B, if Arv1p is a flippase, GlcN-(acyl)PI, rather than GlcN-PI, would flip, and in this case, Gwt1p should function on the cytoplasmic face of the ER membrane.
However, a recent report suggests that yeast Arv1p is required for the delivery of GlcN-(acyl)PI to the first mannosyltransferase during GPI synthesis in the ER lumen (21). Arv1p contains six predicted transmembrane domains and seems to function as a multimer (21). These structural features of Arv1p are consistent with it being either a GPI flippase or an accessory protein that facilitates flipping of GPI (21); however, there is no experimental evidence that Arv1p is directly involved in the flipping of GPI precursors. If Arv1p were a GPI flippase or a factor that facilitates flipping of GPI, inositol acylation would precede the flipping of GPI precursors because GlcN-(acyl)PI accumulates in the arv1Δ mutant (21). If inositol acylation precedes flipping of GPI precursors, inositol acylation must occur on the cytoplasmic side of the ER (Fig. 1B).
Thus, it is important to determine whether inositol acylation by Gwt1p/PIG-W occurs before or after the flipping of GPI, i.e. whether the inositol is acylated on the luminal or cytoplasmic side of the ER. However, it is difficult to predict which amino acid residues are essential for its O-acyltransferase activity because Gwt1p and PIG-W are not homologous to other known O-acyltransferases. Therefore, we first identified the essential amino acid residues of PIG-W and Gwt1p by changing highly conserved amino acid residues to alanine and determining whether these alanine mutants were functional. We then examined the membrane topology of Gwt1p using two strategies, glycosylation mapping and proteolytic cleavage analysis. We propose that inositol acylation occurred on the luminal side of the ER membrane based on the location of amino acid residues essential for Gwt1p/PIG-W function and determination of the membrane topology of Gwt1p.
EXPERIMENTAL PROCEDURES
Growth Media
S. cerevisiae cells were grown in rich medium containing 1% yeast extract, 2% Bacto-peptone, 0.005% adenine and supplemented with 2% glucose (YPAD) or 2% galactose (YPAG), or they were grown in synthetic medium containing 0.67% Bacto-yeast nitrogen base without amino acids supplemented with 2% glucose (SD) or 2% galactose (SG). Appropriate amino acids were added to the synthetic medium.
Construction of GWT1 Conditional Shutdown Strain (GC1)
The targeting vector for the GWT1 conditional shutdown was constructed by insertion of the following DNA fragments (in the order listed) into the pBluescript II SK+ vector (Stratagene Products Division, Agilent Technologies, La Jolla, CA): 220 bp of upstream sequences from the S. cerevisiae GWT1 (ScGWT1) ORF; a kanMX cassette (22); GAL10 promoter (23); Cryptococcus neoformans GWT1 (CnGWT1) ORF (13); and 237 bp of downstream sequence from the ScGWT1 ORF (see supplemental Fig. S3A). The linearized targeting vector was introduced into the W303-1B strain (MATα ade2-1 his3-11,-15 leu2-3,-112 trp1-1 ura3-1 can1-100) (24), and the transformants were selected on YPAG agar plate containing 200 μg/ml G418 (Sigma). The transformed clones were tested for growth suppression in YPAD medium and by genomic PCR analysis of the GWT1 locus. We named a transformed strain GC1 (GWT1 conditional shutdown 1) and used this strain in subsequent experiments.
Plasmid Construction, YEp352GHA
Plasmids and oligonucleotides used in this study are listed in supplemental Tables S1 and S2, respectively. For detection of recombinant proteins, a synthesized HA epitope sequence was inserted to the YEp352GAP-II expression vector (25) under the control of the TDH3 promoter. An adaptor containing the HA epitope sequence was synthesized by annealing two oligonucleotides (HA-EK-Fw and HA-EK-Rv), and this sequence was inserted into the EcoRI-KpnI restriction sites of the YEp352GAP-II vector. The resulting vector, designated YEp352GHA, had an HA epitope sequence followed by the restriction sites EcoRI, KpnI, XbaI, and SalI.
Plasmid Construction, pRS316-GWT1
Approximately 3 kb of the GWT1 locus containing the promoter, ORF, and terminator (NC_001142.8: chromosome X, 263947–260945) was inserted into a single copy vector pRS316 (26) to construct pRS316-GWT1.
Cloning of Mouse PIG-W ORF
The coding sequence of mouse PIG-W was cloned by genomic PCR because mouse PIG-W has no introns. We amplified a DNA fragment of ∼1500 bp from 100 ng of C57BL/6 mouse genomic DNA by 35 cycles of PCR using FastStart HiFi polymerase (Roche Diagnostics) and the mGW099FE and mGW104RS primers (supplemental Table S2). The PCR product was subjected to EcoRI and SalI digestion and subcloned into the YEp352GHA vector. The recombinant clone, YEp352GHA-musGWB, passed sequence verification and was used as the template in PCR-based mutagenesis when constructing the alanine replacement mutants (supplemental Table S1).
Plasmid Construction, YEp352GHA-scGWB and YEp352GHA-scGWA
The ScGWT1 ORF was amplified by PCR using S. cerevisiae genomic DNA as a template and primers sGW101FE and scGW96RK (supplemental Table S2), and this ORF was then subcloned into YEp352GHA vector. The resulting plasmid, YEp352GHA-scGWB, was used as the template in PCR-based mutagenesis when constructing the alanine replacement mutants (supplemental Table S1). The authentic ScGWT1 ORF has a SalI restriction site adjacent to the initiation codon, which caused difficulties in plasmid construction when using the SalI enzyme. Therefore, to remove the SalI site from the ScGWT1 ORF, we amplified the ScGWT1 ORF using primers 310FWE and 321R_Not (supplemental Table S2) and then subcloned the PCR product into the YEp352GHA vector. The resulting plasmid, named YEp352GHA-scGWA (supplemental Table S1), did not have a SalI site in the ScGWT1 ORF, and this modified ScGWT1 ORF was used as a template for PCR-based construction of the S2A constructs used in the glycosylation mapping.
Plasmid Construction for Alanine Replacement Mutagenesis
To introduce single amino acid replacements into ScGWT1 or the mouse PIG-W ORF, we performed overlap extension PCR, which is schematically diagrammed in the supplemental Fig. S1. The N-terminal fragment of ScGWT1 was amplified using the sGW101FE primer and the mutagenesis RV primer; the C-terminal fragment was amplified using the mutagenesis FW primer and the scGW96RK primer (supplemental Table S2). The overlapping N- and C-terminal fragments were mixed and subjected to the secondary PCR using primers sGW101FE and scGW96RK (supplemental Table S2). The resulting PCR fragment was subcloned into the YEp352GHA vector, and finally eight mutant GWT1 expression plasmids were constructed. Similarly, we constructed 17 different mutant PIG-W ORFs, and in each mutant, a single amino acid was replaced with an alanine.
Plasmid Construction for Glycosylation Mapping and Factor Xa Cleavage Analysis
To construct a series of insertional mutants, we ligated three DNA fragments and one vector together simultaneously (supplemental Fig. S2). For the glycosylation mapping, the S2A fragment, which encodes 53 residues of Suc2p containing three N-glycosylation sites, was amplified by PCR using primers SUC8FEK and SUC9RBgS (supplemental Table S2). The S2A fragment was digested with KpnI and BglII and subsequently used to construct all YEp352GHA-scGWT1(1-xx:S2A:xx-490) plasmids, where xx identifies the amino acid positions into which the S2A sequence was inserted (supplemental Table S1). The N- fragment was constructed by PCR amplification from the ScGWT1 ORF using the 310FWE primer and the position-specific RV primer (supplemental Table S2), followed by EcoRI and KpnI digestion. The C-fragment was constructed by PCR amplification from the ScGWT1 ORF using the position-specific FW primer and the 321R_Not primer (supplemental Table S2), followed by BglII and SalI digestion. Three fragments (N, S2A, and C) were mixed with the YEp352GHA vector, which had been digested with EcoRI and SalI, and all components were ligated together using T4 DNA ligase (Takara, Shiga, Japan). To generate the YEp352GHA-S2A-scGWT1(1–490) plasmid, the S2A fragment was excised by digestion with EcoRI and SalI and then inserted to EcoRI-SalI sites of YEp352GHA-scGWB. To construct the YEp352GAP-II-scGWT1(1–490)-S2A-HA plasmid, ScGWT1 ORF without a termination codon was amplified by PCR using the 310FWE primer and the 119RKsc primer (supplemental Table S2). The modified ScGWT1 fragment (EcoRI-KpnI), the S2A fragment (KpnI-BglII), and the Bgl-HA-stop-Sal-adaptor (mixture of oligonucleotides BgHASalF and BgHASalR) were inserted into EcoRI-SalI site of the YEp352GHA vector.
The KpnI-fXa-BglII adaptor, containing two factor Xa protease (fXa) cleavage sequences, was designed for fXa cleavage analysis (supplemental Fig. S2). Both KpnI and BglII restriction sites were unique in the YEp352GHA-scGWT1(1-xx:S2A:xx-490) plasmids (supplemental Table S1). Therefore, we were able to construct a series of YEp352GHA-scGWT1(1-xx:fXa:xx-490) plasmids easily, by removing the S2A fragment from the YEp352GHA-scGWT1(1-xx:S2A:xx-490) constructs by KpnI and BglII digestion and then inserting the KpnI-fXa-BglII adaptor into these digested constructs.
Complementation Assay
Plasmids to be tested for the ability to complement a GWT1 defect were introduced to GC1 cells, and the transformed cells were plated on SG−Ura agar plates. Several clones transformed with a single type of mutant (alanine or insertion) plasmid were mixed to generate pools of clones and then cultured in SG−Ura liquid medium for 2 days at room temperature before use in the complementation assay. In the liquid culture assay, cells from each pool were diluted 1000-fold with distilled water and added to assay medium, SD−Ura or SG−Ura. After incubation at 30 °C for 48 h in assay medium, the optical density of each pooled culture was measured at 620 nm using a Sunrise microplate reader (Tecan, Männedorf, Switzerland). In the agar plate assay, cells were diluted 1000-, 10,000-, or 100,000-fold with distilled water, spotted on SD−Ura or SG−Ura agar plates, and subsequently incubated at 30 °C for 48 h.
Immunoblot Analysis
Microsome fractions containing various types of HA-tagged Gwt1p were separated on 10% SDS-PAGE and transferred to a PVDF membrane (GE Healthcare). The blot was then incubated with anti-HA-HRP conjugate (clone 3F10; Roche Diagnostics) or anti-HA monoclonal antibody 16B12 (Covance, Princeton, NJ) and visualized with HRP-conjugated anti-mouse IgG and an ECL-Plus chemiluminescence detection system (GE Healthcare) according to the manufacturer's instructions. To detect Gwt1p, anti-Gwt1p mouse monoclonal antibody (epitope, amino acids 293–392 of Gwt1p) was also used as primary antibody. For chromogenic detection, One-step TMB-Blotting Reagent (Thermo Scientific, Waltham, MA) was used, according to the manufacturer's instructions.
Glycosylation Mapping
The GC1 cells, transformed with S2A-containing plasmids, were grown in 10 ml of SG−Ura liquid medium for 12 h at 30 °C and then collected by centrifugation. The supernatants were discarded, and the cell pellets were mixed with an approximately equal volume of glass beads and 250 μl of TC buffer (100 mm Tris-HCl, pH 8.0, complete protease inhibitor mixture (Roche Diagnostics)). These suspensions were subjected to vortex mixing for 4 min. After addition of 1000 μl of TC buffer, the suspensions were centrifuged at 500 × g to remove cell debris. The supernatants were moved to new tubes and centrifuged at 20,000 × g to concentrate the membrane fractions. The resulting pellets were suspended in 220 μl of DEN solution (1:1:8 mixture of 10× denaturing buffer; 10× G5 buffer for the endo-β-N-acetylglucosaminidase H enzyme (New England Biolabs, Ipswich, MA)/distilled water) and divided into two tubes. The endo-β-N-acetylglucosaminidase H enzyme (750 units) was added to one tube and control buffer to the other tube, and both tubes were incubated at 37 °C for 75 min. The samples were mixed with 50 μl of 2× SDS-PAGE sample buffer and heated at 95 °C for 3 min before loading to the gel and subjected to SDS-PAGE and immunoblot analysis.
Microsome Preparation and Factor Xa Protease Cleavage Analysis
Microsome preparation and fXa cleavage analysis were performed as described previously (27) with some modifications. The GC1 strain was transformed with plasmids carrying constructs that encode Gwt1p with fXa cleavage sites. Cell cultures were grown to the exponential phase in SGSCA−Ura medium (0.67% yeast nitrogen base, 5% galactose, 0.2% sucrose, 1% casamino acids, 40 μg/ml tryptophan, and 40 μg/ml adenine) and collected by centrifugation. The cells were washed, resuspended in Tris-Sorb buffer (1.0 m sorbitol, 50 mm Tris-HCl, pH 7.5, 10 mm dithiothreitol) containing Zymolyase 100T (Seikagaku, Tokyo, Japan), and incubated at 30 °C for 30 min. The resulting spheroplasts were suspended in lysis buffer (50 mm Tris-HCl, pH 7.5, 250 mm sorbitol, 1 mm phenylmethylsulfonyl fluoride) and homogenized by 20 strokes in a Dounce homogenizer. Cell debris was removed by centrifugation at 500 × g for 1 min. The resulting supernatant with the cell extracts were then centrifuged at 13,000 × g to sediment the ER-rich fraction. The ER-rich pellet was suspended in fXa buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 2 mm CaCl2). The samples were then digested with fXa protease (New England Biolabs) in the presence or absence of 0.3% digitonin at 4 °C for 2 h. After digestion, digitonin-free samples were centrifuged again at 13,000 × g and resuspended in fXa buffer. The samples were mixed with SDS-PAGE sample buffer and subjected to immunoblot analysis.
RESULTS
Construction of the GWT1 Conditional Shutdown Strain (GC1)
Previously, we demonstrated that the yeast GWT1 gene encodes a GlcN-PI-specific O-acyltransferase and that the acylation catalyzed by Gwt1p is critical for GPI synthesis in yeast (13). Sequence similarity searches indicate that Gwt1p does not share conserved motifs with other O-acyltransferases, suggesting that Gwt1p and the mammalian ortholog, PIG-W, have a novel mode of action. To elucidate the location of essential amino acids within Gwt1p and PIG-W, we first tried to determine the residues critical for protein function.
To determine whether the Gwt1p and PIG-W mutant proteins were functional, we used a complementation assay that involved the GWT1 shutdown strain, GC1. As shown in supplemental Fig. S3A, the authentic S. cerevisiae GWT1 was replaced by the kanMX-GAL10 promoter-C. neoformans GWT1 cassette. In this construct, we used the C. neoformans GWT1 gene (CnGWT1) as a Gwt1 protein donor to avoid homologous recombination between exogenous mutated GWT1/PIG-W constructs and the authentic GWT1 gene in S. cerevisiae genomic DNA. The strain GC1 was able to grow in media supplemented with galactose because CnGwt1p expressed under the galactose-inducible promoter complemented the ScGwt1p defect, but this strain was not able to grow in media supplemented with glucose because CnGwt1p expression was inhibited by glucose-induced repression of the galactose-inducible promoter (supplemental Fig. S3B, upper row). Overproduction of either ScGwt1p or the mouse PIG-W protein under the constitutive high expression promoter TDH3 of YEp352GHA rescued the lethality of the GC1 strain grown in the glucose media, confirming that these proteins are functional under these cultivation conditions (supplemental Fig. S3B, middle and lower rows).
Mutational Target Selection
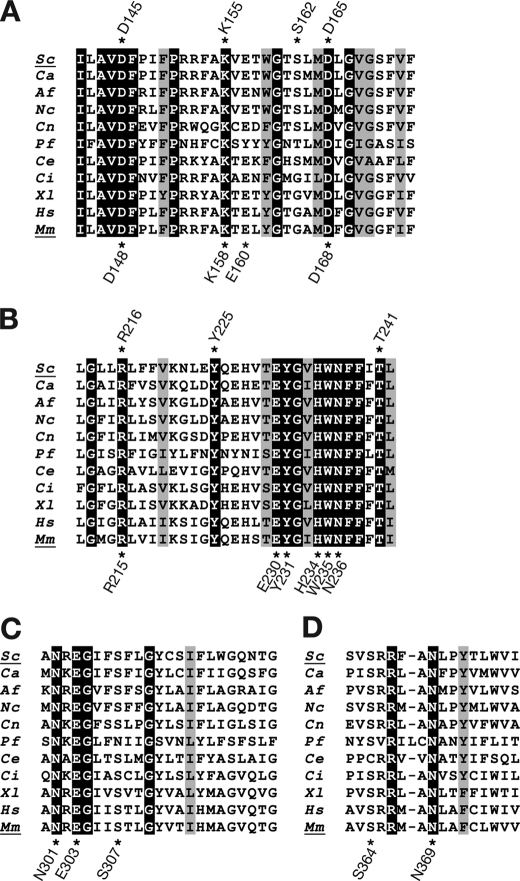
We identified target amino acids for mutagenesis based on sequence similarity information from several species. We aligned the Gwt1p/PIG-W amino acid sequences from five species (S. cerevisiae, Candida albicans, Aspergillus fumigatus, Mus musculus, and Homo sapiens) using the ClustalW algorithm and selected four highly conserved regions for further analysis (supplemental Fig. S4). We performed manual alignment of these four highly conserved regions from 11 species, S. cerevisiae, C. albicans, A. fumigatus, M. musculus, H. sapiens, Neurospora crassa, C. neoformans, Plasmodium falciparum, Caenorhabditis elegans, Ciona intestinalis, and Xenopus laevis (Fig. 2, A–D). We identified ∼30 residues that were identical in all 11 species and based on this level of conservation, we categorized these residues as potential targets for our mutational analysis. We prioritized among these candidate residues using two additional criteria; specifically, we focused on residues that were likely to participate directly in the O-acylation chemical reaction and on charged residues. Ser, His, and Asp residues are critical for activity of acyl-coenzyme A:sterol acyltransferase, which catalyzes a reaction similar to O-acylation (28, 29); therefore, these residues were selected as actual targets for mutagenesis. The charged amino acids, Lys, Arg, Glu, Asp, and His, were also selected as actual targets because they were reported to be important for catalytic activity of carnitine palmitoyltransferase I (30) and acyl CoA:diacylglycerol acyltransferase (31). Based on information on residues important to similar enzyme-catalyzed reactions, conserved sequences within Gwt1p/PIG-W homologs, and the chemical properties of individual residues, we selected residues in mouse PIG-W and ScGwt1p for replacement with alanine.
FIGURE 2.
Sequence alignment of S. cerevisiae Gwt1p, mouse PIG-W, and their orthologs. Four regions that were highly homologous among 11 species are depicted. Identical amino acid residues among all 11 species are highlighted. Conservative residues among all 11 species are shaded. Positions that were mutated to alanine in S. cerevisiae Gwt1p are marked with asterisks above the sequences; positions mutated in mouse PIG-W are marked below the sequence. Sc, S. cerevisiae; Ca, C. albicans; Af, A. fumigatus; Nc, N. crassa; Cn, C. neoformans; Pf, P. falciparum; Ce, C. elegans; Ci, Ciona intestinalis; Xl, X. laevis; Hs, H. sapiens; Mm, M. musculus.
Identification of the Essential Residues of Gwt1p/PIG-W by Alanine Replacement Mutagenesis
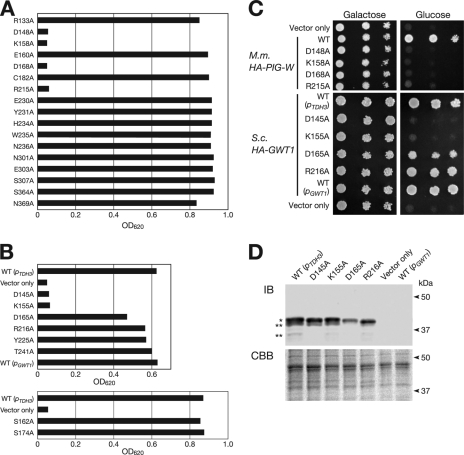
Initially, we selected 17 residues in mouse PIG-W and independently substituted each with an alanine using a PCR-based method; the resulting cDNAs were inserted to the high copy expression vector (YEp352GAP-II) with an HA epitope at the N terminus (supplemental Fig. S1). These mutant proteins were overproduced in the GC1 strain under the control of the TDH3 promoter; and their ability to complement the Gwt1p loss of function was evaluated by the growth of cells under CnGWT1 shutdown conditions. As shown in Fig. 3A, 4 of 17 mouse PIG-W mutant constructs did not rescue the growth defect of GC1 cells under shutdown conditions, suggesting that the residues replaced with alanine (D148A, K158A, D168A, and R215A) in these strains were essential for mouse PIG-W function. Unexpectedly, constructs with a mutation in any one of five residues (E230A, Y231A, H234A, W235A, or N236A) in the most highly conserved region (Fig. 2B) rescued the growth defect of GC1 cells (Fig. 3A), indicating that these residues were not essential for the PIG-W function. Based on the result of alanine replacement analysis of mouse PIG-W, we targeted eight residues in ScGwt1p for further alanine replacement analysis. Wild-type GWT1 was expressed under its own promoter or the TDH3 promoter. In the case of GWT1, only two of eight mutated proteins, D145A and K155A, did not rescue the growth defect of GC1 cells under shutdown conditions (Fig. 3, B and C). These residues corresponded to Asp-148 and Lys-158 of mouse PIG-W, respectively, indicating that these residues were essential for the function of these enzymes. In contrast, the residues Asp-165 and Arg-216 of yeast Gwt1p, which were homologous to the essential residues Asp-168 and Arg-215 of mouse PIG-W, respectively, were not essential for Gwt1p function, whereas the growth of GC1 cells producing D165A mutant Gwt1p was only slightly decreased (Fig. 3, B and C). We also checked the morphology of GC1 cells transformed with mutant GWT1 (supplemental Fig. S5). D145A and K155A mutant cells, unlike wild-type cells, were swollen and, in some cases, lysed. Temperature-sensitive gwt1-20 cells also have these same mutant phenotypes (13). Most D165A mutant cells were also swollen, although only a few of these cells lysed. R216A mutant cells were morphologically normal.
FIGURE 3.
Determination of essential residues of mouse PIG-W and S. cerevisiae Gwt1p. A, growth of GC1 cells transformed with mutant PIG-W. In this figure, “R133A,” for example, means that the arginine at the 133rd residue position of PIG-W was substituted with alanine. In GC1 cells, wild-type or mutant PIG-W was expressed under the control of TDH3 promoter from a YEp plasmid. Samples of each strain were precultured for 2 days in SG−Ura medium, diluted 1000-fold, cultured in SD−Ura at 30 °C for 2 days, and measured for absorbance at 620 nm. B, growth of GC1 cells transformed with wild-type or mutant forms of GWT1. The experimental procedure was the same as that described for A. Wild-type GWT1 was expressed under its own promoter (pGWT1) or the TDH3 promoter (pTDH3). All mutant GWT1 were expressed under the control of TDH3 promoter. Results on the upper and lower panels were obtained from different sets of experiments. C, growth of GC1 cells transformed with mutant S. cerevisiae GWT1 or mouse PIG-W on plates. The cells were spotted on SG−Ura (Galactose) or SD−Ura (Glucose) plates after making a 10× serial dilution (left to right) and then incubated at 30 °C for 2 days. M.m., M. musculus; S.c., S. cerevisiae. D, amount of mutant Gwt1p from GC1 cells was estimated by immunoblotting. Membrane fractions of the GC1 cells expressing wild-type or mutant Gwt1p were prepared as described under “Experimental Procedure” for glycosylation mapping. Each sample was split into two equal portions, which were separately subjected to SDS-PAGE. One gel (upper panel) was used for immunoblot analysis using anti-Gwt1p monoclonal antibody, and single and double asterisks indicate an intact and partially degraded Gwt1p, respectively. The other gel (lower panel) was subjected to Coomassie Brilliant Blue R-250 (CBB) staining as a loading control to assess relative protein content between samples.
We suspected that the decrease in complementation activity observed for Gwt1p mutants D145A, K155A, and D165A was caused by a decrease in protein stability. Therefore, we examined the amount of mutant Gwt1p produced in the GC1 cells by immunoblot analysis. Notably, we did not detect wild-type Gwt1p expressed under its own promoter (Fig. 3D), although it could rescue the growth of GC1 cells (Fig. 3, B and C). This observation indicated that a small amount of Gwt1p was sufficient for the growth of GC1 cells. As shown in the Fig. 3D, comparable amounts of D145A and K155A mutant Gwt1p were detected, suggesting that the inability of these mutants to rescue the growth defect was not caused by the protein instability but by the loss of function. Although D165A mutant Gwt1p seemed to be slightly unstable, the amount of the mutant D165A protein was much larger than the amount of wild-type Gwt1p expressed from its own promoter (Fig. 3D), suggesting that the slow growth phenotype of D165A was not caused by protein instability but by the loss of function. The R216A mutation apparently did not influence function or protein stability of Gwt1p (Fig. 3D). These results indicated that two residues in Gwt1p, Asp-145 and Lys-155, which were conserved in a wide range of organisms, were essential for the function of GlcN-PI O-acyltransferase.
Hydropathic Profile Analysis of Gwt1p
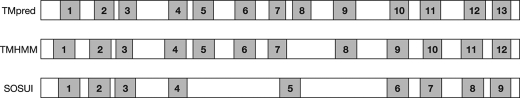
The precise membrane topology of Gwt1p has not been determined. Three well known algorithms for the prediction of membrane-spanning segments, TMpred, TMHMM, and SOSUI, suggested different numbers of the transmembrane domains (TMDs) (Fig. 4), indicating that a simple estimation of membrane topology of Gwt1p was difficult. Therefore, biological experiments were essential to determine the number of TMDs in Gwt1p. We designed biological topology-mapping experiments based on the TMpred prediction because it proposed the maximum number of TMDs, which is 13.
FIGURE 4.
Prediction for the transmembrane domain of Gwt1p using three different prediction programs. TMpred, TMHMM, and SOSUI were used to identify putative transmembrane domains in Gwt1p. Shaded areas indicate putative transmembrane domains.
Glycosylation Mapping
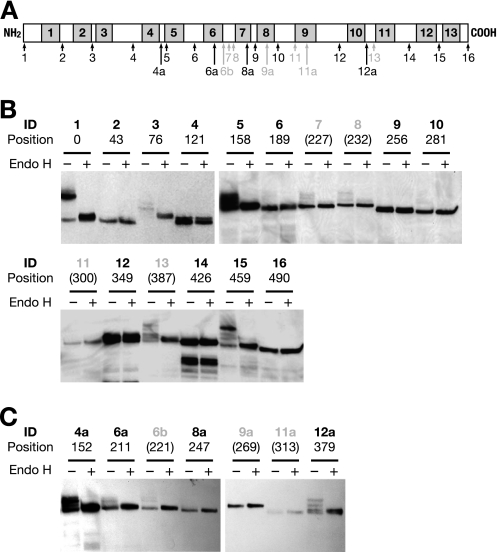
The S2A cassette, consisting of a 53-amino acid sequence from Suc2p, contains three NX(S/T) sites for N-linked glycosylation (27). This reporter cassette has been used to determine the topology of several membrane proteins, including Lcb1p, Shr3p, and Lag1p (27, 32, 33), demonstrating the usefulness of this method. To determine the topology of Gwt1p, we first constructed 16 Gwt1p-S2A fusion protein expression plasmids; in each construct, an S2A cassette was inserted into a predicted loop (Fig. 5A, 1–16). These plasmids were introduced to the GC1 cells, and their ability to rescue growth under shutdown conditions was examined to assess whether they accurately reflect native Gwt1p topology. Of the 16 fusion proteins, 12 supported growth of GC1 cells under CnGWT1 shutdown conditions and were therefore assumed to be functional and to retain their native topology (Fig. 5A, 1–6, 9, 10, 12, and 14–16). We constructed seven additional Gwt1p-S2A fusions (Fig. 5A, 4a, 6a, 6b, 8a, 9a, 11a, and 12a) to cover all possible loops. However, we could not obtain any functional Gwt1p-S2A fusion proteins containing an insertion between putative TMDs 6 and 7.
FIGURE 5.
Determination of membrane topology of Gwt1p by S2A glycosylation mapping. A, diagram of the positions of S2A sequence insertions. Shaded areas indicate putative transmembrane domains predicted by TMpred. Black numbers below arrows indicate the identification (ID) numbers of the fusion proteins in which Gwt1p remained functional after S2A insertion. Gray numbers below gray arrows indicate identification numbers in which Gwt1p lost its function after S2A insertion. B and C, immunoblot analysis of HA-tagged and S2A sequence-inserted Gwt1p, respectively. Endo-β-N-acetylglucosaminidase H (Endo H)-treated (+) and untreated (−) microsome fractions were subjected to SDS-PAGE and immunoblotting using an anti-HA monoclonal antibody. Identification numbers of the fusion proteins and their amino acid positions are shown above each lane. Gray identification numbers and amino acid positions in parentheses indicate that S2A sequence-inserted Gwt1p was not functional.
The state of protein glycosylation in these Gwt1p-S2A proteins was examined (Fig. 5, B and C). We confirmed that the N and C termini of Gwt1p were oriented on the luminal and cytosolic side, respectively (Fig. 5B, 1 and 16), as Murakami et al. (14) demonstrated in the case for mammalian PIG-W protein. In addition, we detected glycosylation of several fusion constructs, 3, 4a, 5, 12a, and 15 (Fig. 5, B and C), suggesting that the loops between TMD 2 and TMD 3 (hereafter loop 2–3), loop 4–5, loop 10–11, and loop 12–13, were oriented on the luminal side. We saw a trace of glycosylation for the construct 10 (loop 8–9), but the orientation of this loop was ambiguous. We did not obtain any functional insertion mutants into loop 6–7, although we did see a trace of the glycosylated form of Gwt1p-S2A using the constructs 6b, 7, and 8.
Factor Xa Protease Cleavage Analysis
To confirm the results from the glycosylation mapping and to determine precisely the orientation of each loop, we analyzed Gwt1p fusion proteins using microsomal fXa cleavage. fXa cleaves proteins containing the Ile-Glu-Gly-Arg (IEGR) tetrapeptide at the C-terminal side of the arginine. The two copies of fXa cleavage site, IEGRIEGR, were inserted into Gwt1p at the same positions where the individual S2A cassettes were inserted to generate Gwt1p-IEGR fusion proteins. The complementation analysis of Gwt1p-fXa fusion proteins revealed that complementation activity was dependent on the position of the cassette regardless of the cassette sequences; specifically, we could not obtain a functional Gwt1p-fXa fusion protein at loop 6–7, as was the case for the Gwt1p-S2A fusions (supplemental Fig. S6).
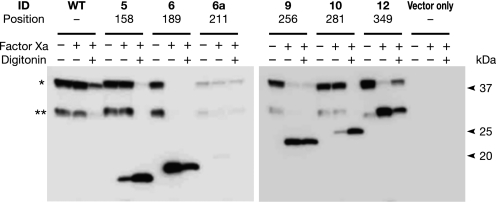
GC1 cells expressing Gwt1p-fXa fusion proteins from a plasmid were grown in culture, and microsomal membranes were prepared from these cells. The sensitivity of Gwt1p-fXa fusion proteins to fXa protease cleavage was assessed by comparing digestion of the fusion proteins in the absence or presence of 0.3% digitonin, which solubilizes membranes (Fig. 6). When fXa cleavage sites were inserted into loop 5–6 (6), loop 7–8 (9), or loop 9–10 (12) of Gwt1p (see also Fig. 5A), the fXa protease was accessible to the fusion proteins in the presence or absence of digitonin, suggesting that these positions were located on the cytosolic side of the ER membrane. In contrast, when fXa cleavage sites were inserted into loop 4–5 (5) and loop 8–9 (10) of Gwt1p, the fusion proteins were resistant to fXa cleavage in the absence of digitonin treatment; specifically, these fusion proteins were sensitive to fXa cleavage only after digitonin treatment of microsomes, suggesting that these residues do not face the cytosol. It is conceivable that these residues were located either in the lumen of the ER or in the ER membrane. The proposed orientation of loop 4–5 (5), loop 5–6 (6), loop 7–8 (9), and loop 9–10 (12) based on fXa cleavage analysis was identical to the proposed orientation based on the glycosylation mapping, suggesting that these results were consistent and reliable. A summary of the results by the glycosylation mapping and fXa cleavage analysis is shown in Fig. 7A.
FIGURE 6.
Determination of membrane topology of Gwt1p by factor Xa protease cleavage analysis. Membrane fractions from GC1 cells expressing wild-type (WT) or Gwt1p fusion proteins with fXa cleavage sequences, tagged with an HA epitope, were prepared and digested with fXa in the presence (+) or absence (−) of 0.3% digitonin at 4 °C for 2 h. After digestion, digitonin-free samples were centrifuged and resuspended in fXa buffer. The samples were mixed with SDS-PAGE sample buffer and subjected to immunoblot analysis. An asterisk indicates intact Gwt1p detected with anti-HA antibody. Double asterisk indicates a partially degraded Gwt1p.
FIGURE 7.
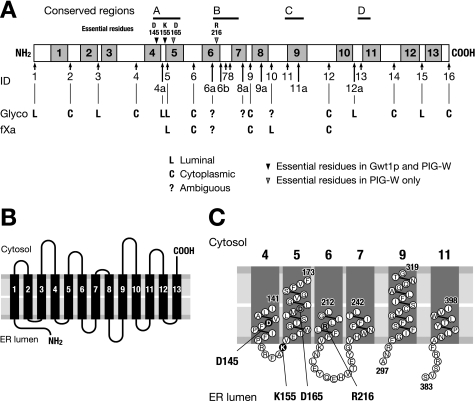
Schematic diagram of the topology of Gwt1p. A, summary of the results of glycosylation (Glyco) mapping and fXa cleavage analysis. Four conserved regions (A–D) and essential residues (Asp-145, Lys-155, Asp-165, and Arg-216) are also shown. Asp-145 and Lys-155 were essential for both Gwt1p and PIG-W, but Asp-165 and Arg-216 were essential for PIG-W function alone. B, model of the membrane topology of Gwt1p. C, possible structure of four conserved regions. Numbers indicate amino acid positions.
Although we detected only a trace of glycosylation in loop 8–9 (10) of Gwt1p in the S2A-reporter assay, we could see marked protection from fXa cleavage of the fXa-target cassette inserted into loop 8–9 (10), indicating that loop 8–9 was located on the luminal side of the ER membrane. Although we obtained a “glycosylation-negative” result in the S2A-reporter assay, the oligosaccharyltransferase enzymes may not have been accessible to the S2A sequence due to an inappropriate conformation of the substrate. In contrast, the accessibility of the fXa enzyme to the substrate was guaranteed in the fXa cleavage assay after digitonin treatment. Therefore, we assigned loop 8–9 of Gwt1p to the luminal side of the ER based on the more reliable fXa cleavage assay result.
No clear evidence indicated that fXa cleaved the 6a fusion protein even after digitonin treatment. One possibility was that position 6a may be located in the transmembrane domain. If position 6a was in a transmembrane domain, then TMD 6 and TMD 7 were present because positions 6 and 9 faced the cytoplasmic side. The other possibility was that fXa could not access position 6a due to some solid structure around this position. If so, neither TMD 6 nor TMD 7 exists. However, we think that this latter case is unlikely for two reasons as follows: 1) all three programs used for the prediction of TMDs suggested the existence of TMD 7, and if TMD 7 exists, TMD 6 should also be present based on the results of the glycosylation mapping of positions 6 and 9; 2) if TMD 6 or TMD 7 did not exist, the positions 6b, 7, and 8 should all face the cytoplasmic side. However, the glycosylation mapping showed a faint but reproducible N-glycosylation band at these three sites, suggesting that these sites faced the ER lumen. Therefore, we proposed that TMD 6 and TMD 7 were both present and favor the hypothesis that Gwt1p is a membrane protein containing 13 transmembrane domains, as shown in Fig. 7B.
DISCUSSION
It was proposed that mammalian PIG-W was a multiple membrane-spanning protein with 13 TMDs, and the conserved regions among PIG-W and its homologs were predicted to face the luminal side of the ER, suggesting that inositol acylation occurred at the luminal face of the ER (14). After that, Arv1p was reported to be a candidate GPI flippase, although its enzymatic activity was not demonstrated directly (21). If Arv1p is a flippase, inositol acylation should occur on the cytoplasmic face of the ER because GlcN-(acyl)PI accumulates in arv1 mutant cells (21). These circumstances led us to experimentally identify essential residues in the Gwt1p and PIG-W proteins and the actual membrane topology of Gwt1p. The GWT1 conditional shutdown strain, GC1, allowed us to pursue both objectives. An advantage of using the GC1 strain was that whether an individual residue of Gwt1p/PIG-W was essential for function could be determined by measuring absorbance at 620 nm of cell cultures (i.e. cell growth), without assaying Gwt1p/PIG-W enzymatic activity directly.
Based on our experimental results, we concluded that S. cerevisiae Gwt1p was a membrane protein with 13 TMDs and that the N and C termini of Gwt1p faced the luminal and cytoplasmic side of the ER, respectively. This conclusion is consistent with the membrane topology of PIG-W, predicted by Murakami et al. (14). In addition, we demonstrated that four highly conserved regions of the protein were in luminal loops or near the luminal face of transmembrane domains and that essential amino acid residues, specifically aspartic acid at position 145 (Asp-145) and lysine at position 155 (Lys-155), are near the luminal face of TMD 4 and in loop 4–5, respectively (Fig. 7C); these results strongly indicate that the luminal amino acids are important for the function of Gwt1p.
To assess whether the loss of function in D145A and K155A mutant Gwt1p was caused by the loss of acyltransferase activity, we assessed the morphology of cells expressing these mutant proteins. Previously, we demonstrated that gwt1-20 temperature-sensitive mutant cells that are defective in acylation of inositol during GPI biosynthesis have a swollen morphology (13). Although we did not assay the acyltransferase activity of D145A or K155A mutant Gwt1p, it is plausible, even likely, that these mutant Gwt1p were also defective in acylation of inositol because GC1 cells expressing D145A or K155A mutant Gwt1p had the swollen phenotype observed in gwt1-20 cells.
In addition to Asp-145 and Lys-155 in Gwt1p, we identified four amino acid residues, aspartic acid at position 148 (Asp-148), lysine at position 158 (Lys-158), aspartic acid at position 168 (Asp-168), and arginine at position 215 (Arg-215), that are necessary for the function of mouse PIG-W. Asp-145 and Lys-155 of ScGwt1p correspond to Asp-148 and Lys-158, respectively, of PIG-W. Asp-168 and Arg-215 of PIG-W correspond to Asp-165 and Arg-216 of ScGwt1p. In Fig. 7C, Arg-216 also mapped near the luminal face of TMD 6, indicating again the importance of the luminal amino acid residues. Notably, all the essential residues identified by the alanine replacement method were charged amino acids, suggesting that the charges may have some important role in transferring a fatty acyl chain to the inositol ring of GPI, although the catalytic mechanism is not known.
In S. cerevisiae, neither Asp-165 nor Arg-216 was essential for Gwt1p function; the growth of the R216A mutant was comparable with cells expressing wild-type Gwt1p, although the growth rate of the D165A mutant was slightly decreased relative to wild type (Fig. 3, B and C). Currently, we do not understand why Asp-165 and Arg-216 of Gwt1p were not essential, but the corresponding residues in PIG-W (Asp-168 and Arg-215, respectively) were essential. It may be that yeast Gwt1p differs from mammalian PIG-W in some molecular characteristics, although the membrane topology is apparently the same. For instance, binding efficiency of Gwt1p/PIG-W to a compound, BIQ, shows species specificity (34). In fact, we selected these two different inositol acyltransferases, yeast Gwt1p and mammalian PIG-W, in the hope of developing species-specific inhibitors of inositol acyltransferase. Results from these types of experiments are described elsewhere (34). Further alanine substitution analysis may provide clues that can lead to the development of antifungal reagents that may be useful in treating mycoses.
Alanine replacement experiments revealed that the conserved region A (Fig. 2A) was important for Gwt1p/PIG-W function. However, the alanine substitution in the conserved region B (Fig. 2B) had little effect on Gwt1p/PIG-W function, although the amino acid sequence, EYG(I/L/V)HWNFF, was highly conserved among inositol acyltransferases from various species. Nevertheless, we suppose that the conserved region B was also important for the Gwt1p/PIG-W function, because the insertion of the S2A or fXa sequence into the conserved region B caused Gwt1p to lose the ability to rescue the growth of GC1 cells. In contrast, Gwt1p remained functional after the insertion of S2A and fXa sequences into the conserved region A. It is conceivable that the conserved regions A and B contribute to Gwt1p/PIG-W function in different ways because two experimental procedures, alanine replacement and S2A/fXa insertion, caused different effects on these regions. The insertion of the S2A or fXa sequence into the conserved region C (11) or D (13) impaired the function of Gwt1p. These regions seem to have similar characteristics to region B, in that Gwt1p remained functional after alanine mutagenesis, but lost its function after insertion of amino acid sequences.
Our results strongly support the conclusion that Gwt1p of S. cerevisiae is a membrane protein with 13 transmembrane domains and that the N and C termini of Gwt1p faced the luminal and cytoplasmic sides, respectively, of the ER. We propose that inositol acylation occurs on the luminal side of the ER membrane based on our mapping of functionally essential residues of the enzyme and of membrane topology, as shown in Fig. 1A. Acyl-CoA, a substrate for the inositol acyltransferase, is synthesized in the cytosol or on the cytoplasmic face of organelle membranes (35). If inositol acylation occurs on the luminal face of the ER membrane, acyl-CoA must be delivered to the luminal side of the ER membrane. However, the presence of acyl-CoA on the luminal side of the ER membrane has not been demonstrated. The lipid remodeling pathway of GPI-anchored proteins may offer insight into this issue. The lipid moieties of GPI-anchored proteins are modified in the ER. This process is called lipid remodeling (36), and an unsaturated fatty acyl chain at the sn-2 position of phosphatidylinositol is removed by Per1p (37), and a C26 fatty acid is transferred to the sn-2 position by Gup1p instead (38). The lipid remodeling process in yeast occurs on the luminal side of the ER membrane and the donor of the C26 fatty acid is C26-CoA. Therefore, it is highly possible that acyl-CoA does exist in the lumen of the ER.
If flipping of GPI precursors occurs as shown in Fig. 1A, alternative hypotheses for the function of Arv1p must be developed. Arv1p should function after Gwt1p, because the arv1Δ mutant accumulates GlcN-(acyl)PI and has a defect in synthesis of Man-GlcN-(acyl)PI (21). Moreover, the defect of Man-GlcN-(acyl)PI synthesis in the arv1Δ mutant is neither due to the defect in GPI-mannosyltransferase activity nor to synthesis of dolichol phosphomannose, which is the donor substrate for GPI-mannosyltransferase (21). Thus, the molecular function of Arv1p remains unclear, although Arv1p is required for the delivery of GlcN-(acyl)PI to the GPI-mannosyltransferase I Gpi14p-Pbn1p complex (21).
Another important question is as follows. What is the physiological role of the inositol acylation? Our results indicate that functional residues in Gwt1p reside on the luminal face of the ER membrane, and this finding is consistent with the hypothesis that inositol acylation occurs on the luminal face of the ER membrane. The inositol acylation is followed by the transfer of mannose and phosphoethanolamine to the GPI precursor, and these processes occur on the luminal side of the ER membrane. Thus, if inositol acylation occurs on the luminal face of the ER membrane, then it may inhibit flip-flop of GPI precursors and “fix” them on the luminal side. The flip-flop of GlcN-PI was recently demonstrated using fluorescent derivatives of GPI precursors and a biochemical reconstitution approach (39). Although fluorescent probes for GlcN-(acyl)PI have not been developed yet, such methods may significantly advance our understanding of the mechanism of flipping of GPI precursors across the ER membrane and our understanding of the precise function of Arv1p.
Acknowledgments
We are grateful to Ken-ichi Nakayama, Yoh-ichi Shimma, Yasunori Chiba, Xiao-dong Gao, and Michiyo Okamoto at National Institute of Advanced Industrial Science and Technology and Junro Kuromitsu, Takeshi Nagasu, Katsura Hata, Naoaki Watanabe, and Mamiko Miyazaki at Eisai Co., Ltd., for helpful discussions. We thank Nozomi Kumagai, Ikuko Seiki, and Kayoko Ebihara (Eisai Co., Ltd.) for technical assistance.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S6.
- GPI
- glycosylphosphatidylinositol
- BIQ
- 1-[4-butylbenzyl]isoquinoline
- ER
- endoplasmic reticulum
- fXa
- factor Xa protease
- PI
- phosphatidylinositol
- TMD
- transmembrane domain.
REFERENCES
- 1. Kinoshita T., Inoue N. (2000) Curr. Opin. Chem. Biol. 4, 632–638 [DOI] [PubMed] [Google Scholar]
- 2. Pittet M., Conzelmann A. (2007) Biochim. Biophys. Acta 1771, 405–420 [DOI] [PubMed] [Google Scholar]
- 3. Orlean P., Menon A. K. (2007) J. Lipid Res. 48, 993–1011 [DOI] [PubMed] [Google Scholar]
- 4. Caro L. H., Tettelin H., Vossen J. H., Ram A. F., van den Ende H., Klis F. M. (1997) Yeast 13, 1477–1489 [DOI] [PubMed] [Google Scholar]
- 5. Kinoshita T., Inoue N., Takeda J. (1995) Adv. Immunol. 60, 57–103 [DOI] [PubMed] [Google Scholar]
- 6. Leidich S. D., Drapp D. A., Orlean P. (1994) J. Biol. Chem. 269, 10193–10196 [PubMed] [Google Scholar]
- 7. Hyman R. (1988) Trends Genet. 4, 5–8 [DOI] [PubMed] [Google Scholar]
- 8. Hirose S., Mohney R. P., Mutka S. C., Ravi L., Singleton D. R., Perry G., Tartakoff A. M., Medof M. E. (1992) J. Biol. Chem. 267, 5272–5278 [PubMed] [Google Scholar]
- 9. Costello L. C., Orlean P. (1992) J. Biol. Chem. 267, 8599–8603 [PubMed] [Google Scholar]
- 10. Urakaze M., Kamitani T., DeGasperi R., Sugiyama E., Chang H. M., Warren C. D., Yeh E. T. (1992) J. Biol. Chem. 267, 6459–6462 [PubMed] [Google Scholar]
- 11. Sipos G., Puoti A., Conzelmann A. (1994) EMBO J. 13, 2789–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen R., Walter E. I., Parker G., Lapurga J. P., Millan J. L., Ikehara Y., Udenfriend S., Medof M. E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9512–9517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Umemura M., Okamoto M., Nakayama K., Sagane K., Tsukahara K., Hata K., Jigami Y. (2003) J. Biol. Chem. 278, 23639–23647 [DOI] [PubMed] [Google Scholar]
- 14. Murakami Y., Siripanyapinyo U., Hong Y., Kang J. Y., Ishihara S., Nakakuma H., Maeda Y., Kinoshita T. (2003) Mol. Biol. Cell 14, 4285–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsukahara K., Hata K., Nakamoto K., Sagane K., Watanabe N. A., Kuromitsu J., Kai J., Tsuchiya M., Ohba F., Jigami Y., Yoshimatsu K., Nagasu T. (2003) Mol. Microbiol. 48, 1029–1042 [DOI] [PubMed] [Google Scholar]
- 16. Okamoto M., Yoko-o T., Umemura M., Nakayama K., Jigami Y. (2006) J. Biol. Chem. 281, 4013–4023 [DOI] [PubMed] [Google Scholar]
- 17. Vidugiriene J., Menon A. K. (1993) J. Cell Biol. 121, 987–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Watanabe R., Kinoshita T., Masaki R., Yamamoto A., Takeda J., Inoue N. (1996) J. Biol. Chem. 271, 26868–26875 [DOI] [PubMed] [Google Scholar]
- 19. Nakamura N., Inoue N., Watanabe R., Takahashi M., Takeda J., Stevens V. L., Kinoshita T. (1997) J. Biol. Chem. 272, 15834–15840 [DOI] [PubMed] [Google Scholar]
- 20. Maeda Y., Watanabe R., Harris C. L., Hong Y., Ohishi K., Kinoshita K., Kinoshita T. (2001) EMBO J. 20, 250–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kajiwara K., Watanabe R., Pichler H., Ihara K., Murakami S., Riezman H., Funato K. (2008) Mol. Biol. Cell 19, 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wach A., Brachat A., Pöhlmann R., Philippsen P. (1994) Yeast 10, 1793–1808 [DOI] [PubMed] [Google Scholar]
- 23. Ogawa-Mitsuhashi K., Sagane K., Kuromitsu J., Takagi H., Tsukahara K. (2009) Yeast 26, 587–593 [DOI] [PubMed] [Google Scholar]
- 24. Thomas B. J., Rothstein R. (1989) Cell 56, 619–630 [DOI] [PubMed] [Google Scholar]
- 25. Abe H., Shimma Y., Jigami Y. (2003) Glycobiology 13, 87–95 [DOI] [PubMed] [Google Scholar]
- 26. Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilstring C. F., Ljungdahl P. O. (2000) J. Biol. Chem. 275, 31488–31495 [DOI] [PubMed] [Google Scholar]
- 28. Guo Z., Cromley D., Billheimer J. T., Sturley S. L. (2001) J. Lipid Res. 42, 1282–1291 [PubMed] [Google Scholar]
- 29. Lu X., Lin S., Chang C. C., Chang T. Y. (2002) J. Biol. Chem. 277, 711–718 [DOI] [PubMed] [Google Scholar]
- 30. Treber M., Dai J., Woldegiorgis G. (2003) J. Biol. Chem. 278, 11145–11149 [DOI] [PubMed] [Google Scholar]
- 31. McFie P. J., Stone S. L., Banman S. L., Stone S. J. (2010) J. Biol. Chem. 285, 37377–37387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han G., Gable K., Yan L., Natarajan M., Krishnamurthy J., Gupta S. D., Borovitskaya A., Harmon J. M., Dunn T. M. (2004) J. Biol. Chem. 279, 53707–53716 [DOI] [PubMed] [Google Scholar]
- 33. Kageyama-Yahara N., Riezman H. (2006) Biochem. J. 398, 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakamoto K., Tsukada I., Tanaka K., Matsukura M., Haneda T., Inoue S., Murai N., Abe S., Ueda N., Miyazaki M., Watanabe N., Asada M., Yoshimatsu K., Hata K. (2010) Bioorg. Med. Chem. Lett. 20, 4624–4626 [DOI] [PubMed] [Google Scholar]
- 35. Black P. N., DiRusso C. C. (2007) Biochim. Biophys. Acta 1771, 286–298 [DOI] [PubMed] [Google Scholar]
- 36. Fujita M., Jigami Y. (2008) Biochim. Biophys. Acta 1780, 410–420 [DOI] [PubMed] [Google Scholar]
- 37. Fujita M., Umemura M., Yoko-o T., Jigami Y. (2006) Mol. Biol. Cell 17, 5253–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bosson R., Jaquenoud M., Conzelmann A. (2006) Mol. Biol. Cell 17, 2636–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vishwakarma R. A., Menon A. K. (2005) Chem. Commun. 2005, 453–455 [DOI] [PubMed] [Google Scholar]