Abstract
FoxO transcription factors have been implicated in lipid metabolism; however, the underlying mechanisms are not well understood. Here, in an effort to elucidate such mechanisms, we examined the phenotypic consequences of liver-specific deletion of three members of the FoxO family: FoxO1, FoxO3, and FoxO4. These liver-specific triply null mice, designated LTKO, exhibited elevated triglycerides in the liver on regular chow diet. More remarkably, LTKO mice developed severe hepatic steatosis following placement on a high fat diet. Further analyses revealed that hepatic NAD+ levels and Sirt1 activity were decreased in the liver of the LTKO mice relative to controls. At the mechanistic level, expression profile analyses showed that LTKO livers had significantly down-regulated expression of the nicotinamide phosphoribosyltransferase (Nampt) gene encoding the rate-limiting enzyme in the salvage pathway of NAD+ biosynthesis. Luciferase reporter assays and chromatin immunoprecipitation analyses demonstrated that Nampt is a transcriptional target gene of FoxOs. Significantly, overexpression of Nampt gene reduced, whereas knockdown increased, hepatic triglyceride levels in vitro and in vivo. Thus, FoxOs control the Nampt gene expression and the NAD+ signaling in the regulation of hepatic triglyceride homeostasis.
Keywords: Gene Regulation, Lipogenesis, Liver Metabolism, Metabolism, NAD, SIRT, Foxo1, Nampt
Introduction
The O family members of Forkhead transcription factors (FoxOs)3 are critical downstream effectors of the insulin/IGF-1 signaling pathway and play important roles in cellular growth and differentiation, protection against oxidative stress, and metabolic regulation (1–4). As a prototypical member of the FoxO family that includes four mammalian genes (FoxO1/3/4/6; FoxO2 is a pseudogene of FoxO3, and FoxO5 is the fish ortholog of FoxO3), FoxO1 has been implicated in nutrient and energy homeostasis (1–5). Although its function in glucose metabolism has been well documented (6–12), the role of FoxO1 in hepatic lipid metabolism remains an area of active investigation (6, 10, 12–17). Adenovirus-mediated overexpression of the constitutively nuclear FoxO1 mutant (FoxO1ADA) increases triglycerides in the mouse liver but decreases triglycerides in the circulation (15). In line with this observation, deletion of FoxO1 in the liver of systemic insulin receptor (IR) knock-out mice reverses the development of hepatic steatosis (10). However, liver-specific FoxO1 knock-out mice manifest increased secretion of VLDL triglycerides in the streptozotocin-induced diabetic state (17). Moreover, the results from two transgenic lines harboring constitutively active FoxO1 alleles are contradictory in regard to its role in hepatic lipid metabolism (6, 13, 14). One line engineered with an S253A mutation (mouse FoxO1) leads to elevated serum and liver triglycerides (8, 13), whereas a triple substitution FOXO1TSS-A allele (T24A, S256A, and S319A in human FOXO1) exhibits lower levels of plasma triglycerides and normal levels of liver triglycerides (6). The latter finding is consistent with the observations from liver-specific IR or IR substrate (Irs1/2) knock-out mice, which have constitutively active FoxOs and lower levels of serum triglycerides due to deficiency of hepatic insulin signaling in these mice (12, 18). In addition, liver-specific deletion of Akt2, the major inhibitory kinase for FoxOs, in the leptin-deficient ob/ob mice reduces fasted serum triglycerides and protects them from developing hepatic steatosis (19).
In addition to phosphorylation by numerous kinases, such as Akt2, FoxOs are also subject to acetylation by several protein acetylases (20–25). It is well known that FoxO acetylation can be reversed by NAD+-dependent deacetylases (sirtuins, such as Sirt1 and Sirt2) (20–25). Several sirtuins, including Sirt1/3/4/6, have been implicated in the regulation of lipid metabolism (26–41). The activity of sirtuins is regulated by intracellular NAD+ levels or the ratio of NAD+/NADH (42–45). In mammals, NAD+ biosynthesis is carried out by de novo and salvage pathways. The salvage pathway is catalyzed by nicotinamide phosphoribosyltransferase (Nampt) and nicotinamide-nucleotide adenylyltransferases (Nmnats), and Nampt has been shown to be the rate-limiting enzyme in this pathway (46). It has also been shown that overexpression of the Nampt gene can increase the Sirt1 activity (46).
Human NAMPT gene was initially characterized as a pre-B-cell colony-enhancing factor because it has a cytokine-like effect (47). Later, it was reported that Nampt (also called visfatin) could be secreted from adipocytes, hepatocytes, and leukocytes (48–51); however, the physiological function of the circulating visfatin is still elusive (52–54). The NAD+ biosynthetic function of Nampt has been clearly demonstrated by structural analysis and conditional knock-out mouse studies (55–57). Significantly, NAMPT mRNA is decreased in the liver of human subjects with nonalcoholic fatty liver disease, and NAMPT has been shown to protect hepatocytes against methyl methanesulfonate-induced apoptosis (58). However, whether Nampt may have protective functions against fatty liver disease is not yet clear.
Currently, it is not well understood how the Nampt gene is regulated at the molecular level in response to environmental cues. In differentiated 3T3-L1 adipocytes, expression of the Nampt gene can be induced by dexamethasone, a glucocorticoid receptor agonist, but suppressed by growth hormone, TNFα, and forskolin (59, 60); however, the underlying mechanisms are not clear. Additionally, glucose and the peroxisome proliferator-activated receptor α agonist WY-14643 have been shown to suppress the Nampt gene expression in hepatocytes and mouse liver, respectively (58), but it is unclear whether those are direct or indirect effects. Interestingly, the Nampt gene has been shown to be controlled by the circadian clock machinery involving Clock, Bmal1, and Sirt1 in mouse liver with a peak of the mRNA level at the beginning of the dark period and a peak of the protein level in the middle of the dark period during a 12-h light:12-h dark cycle (61, 62). The transcriptional profile of the Nampt gene in mice seems to have an inverse correlation with feeding-induced insulin secretion; however, the in vitro data regarding insulin effects on the Nampt gene expression are so far conflicting (58–60), and it is not yet known whether the Nampt gene can be regulated by FoxOs. In this study, we have identified that the Nampt gene is indeed a target of FoxO transcription factors. By regulating Nampt gene expression and subsequent sirtuin activity, FoxOs control lipid metabolism in the liver.
MATERIALS AND METHODS
Animals, Blood Chemistry, and Metabolic Analysis
FoxO1/3/4 floxed mice were generated and genotyped as described previously (63). Transgenic mice that carry a Cre coding sequence plus the albumin gene promoter were purchased from The Jackson Laboratory. To generate FoxO1/3/4 liver-specific triple knock-out mice, FoxO1/3/4 floxed mice were first crossed with the albumin-Cre mice, and the resultant FoxO1/3/4 heterozygotes were intercrossed to produce homozygotes. Sirt1 floxed mice were generated and genotyped as described previously (64). All procedures were performed in accordance with the Guide for Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the Institutional Animal Use and Care Committee of the Indiana University School of Medicine. High fat diet (45% of calories from fat) was purchased from Harlan Laboratories (Madison, WI). Blood glucose was measured using a glucose meter (Contour from Bayer). Serum insulin, free fatty acids, and triglycerides were measured using commercial assay kits (ALPCO and Wako USA). Tissue NAD+ and NADH were analyzed using an EnzyChrom NAD+/NADH assay kit from BioAssay Systems (Hayward, CA).
Lipogenesis and Fatty Acid Oxidation Analyses
De novo lipogenesis in mouse liver was measured essentially as described previously (6). Briefly, 2-month-old control and LTKO male mice were fasted overnight for 16 h before they were fed a high carbohydrate diet (Harlan Laboratories) for 4 h. Then animals were injected intraperitoneally with 3H2O (15 μCi/g; PerkinElmer Life Sciences), and they were sacrificed 1 h later for blood and tissue collection. Liver lipids were extracted and analyzed as described previously (6). De novo lipogenesis in mouse primary hepatocytes was analyzed using sodium [2-14C]acetate (PerkinElmer Life Sciences) as described previously (65). Lipogenic activity was presented as normalized radioactivity (counts per min (cpm)) to protein amount. Fatty acid oxidation in mouse primary hepatocytes was analyzed using [1-14C]palmitate (PerkinElmer Life Sciences) as described previously (66). Oxidation rates are presented as nmol of fatty acids oxidized/mg of protein/min.
Cell Culture
The human HEK293A cell line was purchased from Invitrogen, and cells were maintained in DMEM containing 100 units/ml penicillin, 100 μg/ml streptomycin, 4.5 g/liter glucose, and 10% FBS. Mouse primary hepatocytes were isolated and cultured as described previously (67). Briefly, primary hepatocytes were isolated from mice using collagenase perfusion under anesthesia. The viability of hepatocytes was assessed by the trypan blue exclusion method. Cells with viability >95% were used for the experiments.
Protein Analysis
Tissue or cells were homogenized in the lysis buffer (50 mm Hepes, pH 7.5, 150 mm NaCl, 10% glycerol, 1% Triton X-100, 1.5 mm MgCl2, 1 mm EGTA, 10 mm sodium pyrophosphate, 100 mm sodium fluoride, and freshly added 100 μm sodium vanadate, 1 mm PMSF, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). Protein extracts were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. Proteins were probed using the following antibodies: FoxO1/3/4 (Cell Signaling Technology), Sirt6 (Abcam), Sirt1, Sirt3, Nampt, PGC-1α, PGC-1β, and actinin (Santa Cruz Biotechnology). Detection of proteins was carried out by incubations with HRP-conjugated secondary antibodies followed by ECL detection reagents (Thermo Fisher Scientific). PGC-1α acetylation was analyzed as described previously (68). Briefly, PGC-1α was first immunoprecipitated from tissue or cell lysates and immunoblotted with anti-acetyl-lysine antibody (Cell Signaling Technology).
Histological Analysis
Liver histology was analyzed as described previously (69). Briefly, liver tissue was fixed in the buffered neutral formalin (10%) and embedded in paraffin prior to sectioning. Liver sections were stained with hematoxylin and eosin. Images were captured using a Leica DM750 microscope equipped with an EC3 digital camera and LAS EZ software.
Sirt1 Activity Assay
Sirt1 activities in liver extracts were assessed using a modified fluorescence assay kit (Endo Life Sciences, Plymouth Meeting, PA). Briefly, liver tissue was homogenized in the lysis buffer supplemented with 1 μm trichostatin A. Tissue extracts were added to the reaction mixture containing assay buffer and Fluor-de-Lys substrate (a p53 peptide), and the reaction took place for 20 min at room temperature. The reaction was stopped by adding 1× Developer II, 1 μm trichostatin A, and 10 mm nicotinamide. Fluorescence was measured in a fluorescence microplate reader with excitation at 360 nm and emission at 460 nm. Sirt1 activity was normalized to protein amount in the liver extracts.
Luciferase Reporter Assay
Mouse Nampt gene promoter was cloned by PCR using specific primers (see supplemental Table S1). The firefly luciferase reporter system (pGL4.10luc2 and pGL4.74hRluc/TK) was purchased from Promega (Madison, WI). DNA constructs were transfected into HEK293A cells, and luciferase activity was analyzed using the Dual-Luciferase Assay System from Promega.
Adenovirus Transduction
Adenoviruses carrying GFP, Sirt1, Nampt, and FoxO1 coding sequences were generated using the pAdEasy system (Agilent). Adenoviruses carrying GFP, Nampt, and FoxO1 shRNA sequences were generated using the BLOCK-iT system (Invitrogen). Generally, we used a multiplicity of infection of 100 for overexpression and a multiplicity of infection of 600 for shRNA knockdown experiments.
Real Time RT-PCR
RNA isolation was performed as described previously (12). Real time RT-PCR was performed in two steps. First, cDNA was synthesized using a cDNA synthesis kit (Applied Biosystems Inc.). Second, cDNA was analyzed by real time PCR using the SYBR Green Master Mix (Promega). Primer sequences are described in supplemental Table S1.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed as described previously (70). Briefly, mouse primary hepatocytes were seeded to 90% confluence before they were treated with 1% formaldehyde at 37 °C for 15 min. The cross-link reaction was stopped by adding glycine to a final concentration of 125 mm. Chromatin was sonicated to an average size of 150–300 bp. Immunoprecipitation was performed using the M2-FLAG antibody (Sigma) following the manufacturer's manual. ChIP DNA was analyzed by either regular PCR or real time PCR using the specific primers listed in supplemental Table S1.
Statistical Analysis
Data are presented as means ± S.E. The significance between two groups was assessed using a two-tailed unpaired Student's t test, and p < 0.05 was considered significant.
RESULTS
Hepatic Deficiency of FoxOs Leads to Triglyceride Accumulation in Mouse Liver
To study hepatic functions of FoxO transcription factors, we generated liver-specific FoxO1/3/4 knock-out mice (LTKO) using a Cre-loxP approach. We elected to focus on all three genes given the significant degree of functional redundancy in vivo (63). Deletion of these three genes in the mouse liver was confirmed by real time RT-PCR and immunoblot analyses (Fig. 1, A and B). The LTKO mice had normal body weight and fat mass compared with the control mice at ages of 3 and 5 months, respectively (Fig. 1, C and D). Whereas there was no difference in serum triglyceride (TG) levels between control and LTKO mice at 3 months of age (Fig. 1E), hepatic TG concentrations were significantly elevated in the LTKO mice (Fig. 1F). To further investigate the lipid phenotype, we challenged control and LTKO mice with a high fat diet. Strikingly, LTKO mice developed severe fatty liver in 3.5 months. Histologically, lipid droplets were larger and denser in liver sections of LTKO mice relative to control mice (Fig. 2A). Quantitatively, hepatic triglycerides were increased by 50% in LTKO mice compared with control mice (Fig. 2B). Additionally, LTKO mice also developed hypertriglyceridemia in the circulation after the high fat diet treatment (Fig. 2C).
FIGURE 1.
Characterization of FoxO1/3/4 liver-specific knock-out mice (LTKO). A, FoxO1/3/4 mRNA levels in the liver of control and LTKO mice (n = 3) by real time RT-PCR. B, immunoblot analysis of FoxO1/3/4 proteins in the liver of control and LTKO mice. C, body weight of 3-month-old control and LTKO male mice (n = 6). D, relative weight of white adipose tissue (WAT) to body weight (BW) in 5-month-old control and LTKO male mice (n = 6–10). E, serum TG levels in 3-month-old control and LTKO male mice (n = 6). F, hepatic TG levels in 4-month-old control and LTKO male mice (n = 4–5). * indicates significance with p < 0.05 in control versus LTKO. Error bars indicate S.E.
FIGURE 2.
LTKO mice develop hepatic steatosis on high fat diet. A, H&E-stained liver sections (magnification, ×100). B and C, hepatic and serum triglycerides in control and LTKO male mice treated with a high fat diet for 3.5 months, respectively (n = 4–7). D, de novo lipogenesis in the liver of control and LTKO mice (n = 4). Data are presented as 3H incorporation (cpm) into lipids/g of liver tissue. E, expression of genes involved in lipogenesis was analyzed in the liver of control and LTKO male mice (n = 3) treated with a high fat diet for 3.5 months by real time RT-PCR. Acc1, acetyl-CoA carboxylase 1; Fasn, fatty-acid synthase; Gpam, glycerol-3-phosphate acyltransferase; Me1, malic enzyme 1. F and G, serum and liver FFAs in control and LTKO male mice treated with a high fat diet for 3.5 months, respectively (n = 4–7). H, expression of genes involved in fatty acid oxidation was analyzed in the liver of control and LTKO mice (n = 3) treated with a high fat diet for 3.5 months by real time RT-PCR. Ppara, peroxisome proliferator-activated receptor α; Cpt1a, carnitine palmitoyltransferase 1a; Acadsb, acyl-CoA dehydrogenase, short/branched chain; Acox1, acyl-CoA oxidase 1. * indicates significance with p < 0.05 in control versus LTKO. Error bars indicate S.E.
To elucidate the cause of triglyceride accumulation in the liver, we first measured hepatic de novo lipogenesis, and the results showed that lipid biosynthetic rates were up by 3.8-fold in the liver of LTKO mice compared with control mice (Fig. 2D). Then we analyzed expression of lipogenic genes. Although expression of Srebp-1c and PGC-1β, critical regulators for hepatic lipogenesis, was not altered (Figs. 2E and 3, A, H, and I), expression of fatty-acid synthase (Fasn), glycerol-3-phosphate acyltransferase (Gpam), and malic enzyme 1 (Me1) was significantly increased in the liver of LTKO mice relative to control mice (Fig. 2E). To examine whether fatty acid oxidation was impaired in LTKO mice, we measured free fatty acids (FFAs) in the serum and liver samples. Indeed, FFA levels were elevated by 26 and 37% in the circulation and liver of LTKO mice as compared with control mice, respectively (Fig. 2, F and G). Consistent with the FFA phenotype, expression of genes involved in fatty acid oxidation, including Lipin1 and acyl-CoA dehydrogenase, short/branched chain (Acadsb), was significantly decreased (Fig. 2H). These data suggest that lipogenesis might increase and fatty acid oxidation might decrease in the liver of LTKO mice.
FIGURE 3.
Protein levels of Sirt1/3/6 and PGC-1β in liver of control and LTKO mice. A, immunoblot analysis of Sirt1/3/6 and PGC-1β proteins in the liver of control and LTKO mice (n = 4) that were fed regular chow or a high fat diet (HFD) for 3.5 months. B–I, protein quantification analysis was performed on scanned Western blots from A using Quantity One software (Bio-Rad). Data are presented as values normalized to actinin (a loading control). A p value <0.05 was considered a statistical significant difference between control and LTKO samples. Error bars indicate S.E.
Nampt Gene Expression Is Down-regulated in FoxOs-deficient Liver
Because several sirtuins, including Sirt1/3/6, have been shown to regulate lipid metabolism (26–41), we were curious about whether FoxO deficiency could have any effect on sirtuin functions. First, we analyzed Sirt1/3/6 proteins by Western blots and observed no significant differences between control and LTKO mice on either chow or high fat diet (Fig. 3, A–G). Then we examined Sirt1 activity using two independent methods. An acetylation analysis of one of the Sirt1 substrates, PGC-1α, indicated an elevation of the PGC-1α acetylation in the liver of LTKO mice compared with control mice (Fig. 4A). Next, an enzymatic assay of Sirt1 using a fluorescence-labeled Sirt1 substrate (a peptide of p53) showed a 24% decrease in Sirt1 activity in the livers of LTKO mice relative to the control livers (Fig. 4B). These results suggest that sirtuin activities might be decreased in FoxO-deficient livers.
FIGURE 4.
Nampt gene is down-regulated in liver of LTKO mice. A and B, Sirt1 activity was assessed in the liver of control and LTKO mice by an acetylation analysis of PGC-1α and a fluorometric assay, respectively (n = 3). AFU, artificial fluorescence unit. C–E, NAD+ and NADH levels were measured in the liver of control and LTKO mice (n = 5). NAD+/NADH ratios were calculated according to the NAD+ and NADH values. F, expression of genes involved in NAD+ biosynthesis was analyzed in the liver of control and LTKO mice (n = 3) treated with a high fat diet for 3.5 months by real time RT-PCR. G, expression of heme decycling genes was analyzed in the liver of control and LTKO mice (n = 3) treated with a high fat diet for 3.5 months by real time RT-PCR. H, Nampt mRNA levels were analyzed in the liver of control and LTKO mice (n = 3) at ZT (under regular light/dark cycles, the time of lights on is ZT0 and the time of lights off is ZT12) 5, 11, and 16. I and J, Nampt protein was analyzed in the liver of control (Con) and LTKO (KO) mice at ZT 1, 6, 10, and 16. Both quantitative analyses (I) and immunoblots (J) are presented. * indicates significance with p < 0.05 in control versus LTKO. Error bars indicate S.E.
Because sirtuin activities are regulated by cellular NAD+ levels or NAD+/NADH ratios, we measured the concentrations of these molecules in the livers of control and LTKO mice. Although there was no significant change in NADH levels, NAD+ levels were decreased by 40% in LTKO livers relative to control livers (Fig. 4, C and D). Consequently, the NAD+/NADH ratio was also decreased by 44% in the LTKO liver (Fig. 4E). To determine what caused the decrease in NAD+ concentrations, we analyzed expression of a number of genes that are involved in NAD+ biosynthesis. No significant changes were observed in the expression of several genes involved in de novo biosynthesis of NAD+, including quinolinate phosphoribosyltransferase (Qprt), NAD synthetase 1 (Nadsyn1), and Nmnat1/2/3 (Fig. 4F). Additionally, heme oxygenase 1 (Hmox1) has been implicated previously in the regulation of the mitochondrial NAD+/NADH ratio (68); however, both Hmox1 and Hmox2 had no significant changes in their mRNA levels in the LTKO liver (Fig. 4G). In contrast, mRNA levels of the gene encoding the rate-limiting enzyme, Nampt, in the salvage pathway of NAD+ synthesis were significantly decreased at Zeitgeber times (ZT) 11 and 16 (Fig. 4H). Nampt protein was also decreased at most time points monitored, especially at ZT6 and ZT16 (Fig. 4, I and J), which is consistent with the previous report that Nampt protein levels peaked at about ZT6 and ZT18 in mouse liver, whereas Nampt mRNA peaks only at ZT12 during a 12:12-h light-dark cycle (61, 62). These results suggest that Nampt deficiency might lead to decreased NAD+ levels and impaired sirtuin functions in the liver of LTKO mice.
Nampt Is a Target Gene of FoxOs
To further examine whether Nampt is a transcriptional target of FoxO1, we overexpressed or knocked down FoxO1 in mouse primary hepatocytes. As expected, overexpression of FoxO1 increased Nampt mRNA levels by 62%, and knockdown of FoxO1 reduced Nampt transcripts by 30% (Fig. 5A). Similarly, Nampt protein levels also changed according to FoxO1 expression levels (Fig. 5B). To investigate whether the induction of Nampt is through FoxO1 interaction with the gene promoter, we first performed an in silico sequence analysis and found some FoxO putative binding sites (insulin-responsive element (IRE)) in the mouse, rat, and human NAMPT gene promoters in addition to previously identified E-box elements that can be bound by the Clock-Bmal1-Sirt1 complex (61, 62). One of the conserved IREs is located in the proximal upstream region to the E-box sequences (Fig. 5, C and D). To test whether this proximal IRE is functional, we performed luciferase reporter assays using a series of promoter constructs generated from the mouse Nampt gene. Co-transfection of FoxO1 with luciferase constructs induced the luciferase activity by 2–4-fold using longer Nampt gene promoter sequences (−444 to −1050 bp) (Fig. 5E). However, when shorter promoter sequences (−212 and −333 bp) were used for the luciferase reporter assays, there was no induction by FoxO1, whereas Clock and Bmal1 induced luciferase activities comparable with those of the longer promoter constructs (Fig. 5E). Similarly, the longer promoter sequences of the Nampt gene could also be up-regulated by FoxO3 (Fig. 5F), indicating a redundant role of FoxOs in the Nampt gene regulation. These results also suggest that a potential FoxO-binding site may be located between −333 and −444 bp in the Nampt gene promoter, and this is where the conserved IRE1 (−372 bp) is located. Moreover, association of FoxO1 with the Nampt gene promoter at this IRE1 site was also validated by ChIP-PCR, which showed an 8-fold enrichment in the conserved IRE1 sequence, whereas no enrichment was observed in another upstream IRE2 (−1373 bp) (Fig. 5, G and H). Because FoxO1 activity is regulated by insulin, we also observed that insulin suppressed the induction of the Nampt gene by FoxO1 in mouse primary hepatocytes (Fig. 5I).
FIGURE 5.
Nampt gene is a target of FoxOs. A and B, Nampt mRNA and protein analyses in mouse primary hepatocytes transduced with adenoviruses carrying GFP and FoxO1 overexpression or knockdown (shRNA) constructs (n = 3). C, a schematic diagram of the proximal promoter of the mouse Nampt gene. Previously identified E-box elements and a putative IRE (IRE1) are indicated. TSS, transcription start site. D, a sequence alignment among mouse, rat, and human NAMPT gene promoters at the putative IRE1 site. E and F, luciferase (Luc) reporter assays using serial deletion constructs of the mouse Nampt gene promoter and FoxO1/3-, Clock-, and Bmal1-overexpressing plasmids (n = 3). G and H, chromatin immunoprecipitation analysis of the putative IRE1 and an upstream IRE2 by regular and real time PCRs, respectively (n = 3). I, Nampt mRNA analysis in mouse primary hepatocytes transduced with GFP- or FoxO1-overexpressing adenoviruses in the absence or presence of insulin (n = 3). * indicates significance with p < 0.05. Error bars indicate S.E.
Nampt Regulates Lipid Metabolism
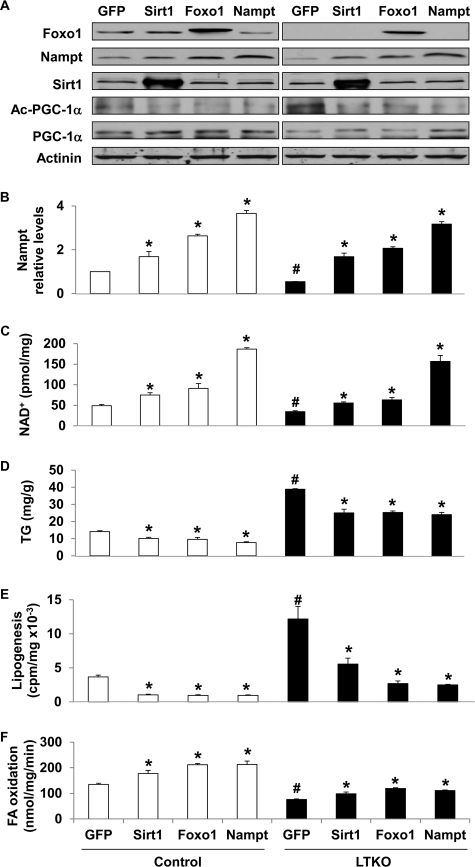
Next, we assessed the role of Nampt in lipid metabolism in both primary hepatocytes and liver. Overexpression of Sirt1, FoxO1, and Nampt was achieved by adenovirus transduction of mouse primary hepatocytes (Fig. 6, A and B), and this led to an increase in intracellular NAD+ concentrations by ∼50, 80, and 400%, respectively, in both control and LTKO hepatocytes (Fig. 6C). The elevation of NAD+ levels also led to an increased level of Sirt1 activity, which was indicated by lower acetylation levels in PGC-1α (Fig. 6A). As expected, intracellular TG concentrations were significantly decreased by ∼30–40% in both control and LTKO hepatocytes (Fig. 6D). To examine what contributed to the lower TG concentrations, we measured de novo lipogenesis and fatty acid oxidation after adenoviral transduction of mouse primary hepatocytes. The results showed that lipogenesis was decreased by about 70% in the control hepatocytes and 50–80% in the LTKO hepatocytes by overexpression of Sirt1, FoxO1, or Nampt (Fig. 6E), and fatty acid oxidation rates were increased by 30–50% in both control and LTKO hepatocytes overexpressing Sirt1, FoxO1, or Nampt (Fig. 6F).
FIGURE 6.
Nampt regulates lipid metabolism in mouse primary hepatocytes. A–D, protein, NAD+, and intracellular TG levels were analyzed in mouse primary hepatocytes that overexpressed GFP, Sirt1, Foxo1, and Nampt for 48 h using corresponding adenoviruses. E and F, de novo lipogenesis and fatty acid (FA) oxidation analyses in control and LTKO mouse primary hepatocytes that overexpressed GFP, Sirt1, Foxo1, and Nampt, respectively. * indicates significance with p < 0.05 between GFP and other gene overexpression in control or LTKO hepatocytes (n = 3). # indicates significance with p < 0.05 between control and LTKO hepatocytes (n = 3). Error bars indicate S.E.
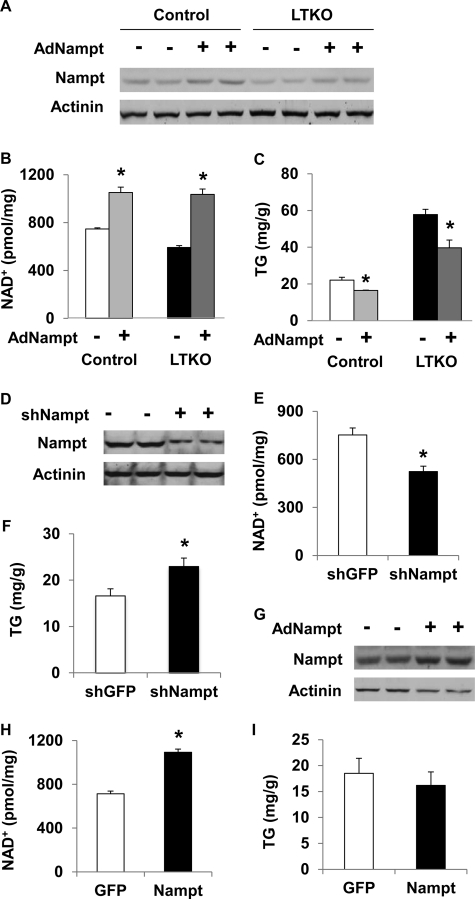
To further verify the TG-lowering effect of Nampt in vivo, we delivered Nampt-expressing adenoviruses to control and LTKO mice via tail vein injections. Nampt protein levels were increased by ∼63 and 69% in the liver of control and LTKO mice injected with Nampt adenoviruses compared with those animals injected with GFP adenoviruses, respectively (Fig. 7A). As a result, hepatic NAD+ concentrations were increased by 41 and 75% (Fig. 7B), and hepatic TG levels were decreased by 25 and 31% in control and LTKO livers (Fig. 7C), respectively. Conversely, knockdown of hepatic Nampt (Fig. 7D) reduced NAD+ concentrations by 30% and increased TG levels by 38% in the liver of wild-type mice (Fig. 7, E and F). To examine whether Sirt1 is required for the effect of Nampt on TG homeostasis, we overexpressed control GFP and Nampt genes in the liver of Sirt1-deficient mice. Nampt protein was increased by 71%, and NAD+ concentrations were increased by 53% in the liver of Nampt-overexpressing mice compared with GFP-overexpressing animals (Fig. 7, G and H); however, hepatic TG concentrations were not significantly different (Fig. 7I). These results suggest that Sirt1 might partly mediate the Nampt effects on hepatic lipid metabolism.
FIGURE 7.
Nampt regulates lipid metabolism in vivo. A–C, control and LTKO mice (n = 4) were injected with GFP- or Nampt-overexpressing adenoviruses via the tail vein, and Nampt protein, NAD+, and TG levels were analyzed in the liver of those mice 7 days postinjection. D–F, wild-type mice (n = 4) were injected with adenoviruses expressing GFP and Nampt shRNAs, and liver Nampt protein, NAD+, and TG levels were analyzed 7 days postinfection. G–I, Nampt-expressing adenoviruses were injected into mice that were deficient in hepatic Sirt1 (n = 4), and Nampt protein, NAD+, and TG levels in the livers were analyzed 7 days postinjection. * indicates significance with p < 0.05 between control and experimental groups. Error bars indicate S.E.
DISCUSSION
In this work, we have characterized FoxO1/3/4 liver-specific knock-out mice and provided evidence that these transcription factors play a critical role in lipid metabolism because LTKO mice exhibited elevated levels of hepatic TG and developed severe hepatic steatosis on a high fat diet. Gene expression analysis of the key regulators for lipogenesis and fatty acid oxidation, Srebp-1c, PGC-1β, and peroxisome proliferator-activated receptor α, did not reveal any significant changes in the liver of LTKO mice relative to control mice. Significantly, both elevated acetylation of PGC-1α and reduced deacetylation of p53 peptides indicate a decrease in Sirt1 activity in the LTKO liver. Because Sirt1 protein levels were not significantly changed, the deficiency of Sirt1 activity may be due to lower levels of NAD+, a co-substrate of the Sirt1 enzyme. Indeed, NAD+ measurements revealed a 40% decrease in the liver of LTKO mice compared with their littermates. The decrease in hepatic NAD+ concentrations is unlikely due to heme catabolism because there were no significant changes in expression of two heme decycling genes, Hmox1 and Hmox2. Rather, our results suggest that there might be a direct link between FoxOs and NAD+ homeostasis (for instance, regulation of NAD+ biosynthesis).
In mammals, cellular NAD+ biosynthesis is controlled by at least two pathways: de novo biosynthesis and the salvage pathway (71). We analyzed several genes that are involved in the de novo pathway, including Qprt, Nadsyn1, and Nmnat1/2/3, and found no significant differences between control and LTKO livers. In contrast, Nampt, the rate-limiting enzyme in the salvage pathway, was significantly decreased in the LTKO liver. Two isoforms of Nampt protein have been identified: intracellular and extracellular (52). The role of the extracellular form has not been resolved, although some reports suggest that it has insulin-sensitizing effects (52). The intracellular form of Nampt has been shown to play a critical role in the regulation of cellular NAD+ levels in lymphocytes using conditional knock-out mice because systemic knock-out of the Nampt gene is embryonically lethal (50, 56). Additionally, both extracellular and intracellular forms of Nampt have been shown to be important for regulation of insulin secretion in pancreatic β-cells, and the enzymatic activity is required for that function (50). Although there is an association of nonalcoholic fatty liver disease with low hepatic Nampt gene expression (58), it is not clear whether Nampt plays any role in the regulation of hepatic lipid metabolism.
Although the importance of Nampt in the regulation of NAD+ homeostasis and metabolism has been increasingly appreciated, it is not well understood how this gene is regulated in the first place. Interestingly, the Nampt gene has been found to be a circadian clock-controlled gene, and three E-box cis elements in the gene promoter have been shown to be involved in the regulation by Clock, Bmal1, and Sirt1 (61, 62). Whether other transcription factors also play a role in the transcriptional regulation of the Nampt gene has been poorly explored. By analyzing the mouse Nampt gene promoter, we have identified a conserved insulin-responsive element in the proximal promoter of human, mouse, and rat Nampt genes that is responsible for FoxO-regulated expression of the Nampt gene. These results have established a link between insulin signaling and NAD+ biosynthesis.
Because intracellular NAD+ levels regulate sirtuin activity, our findings also point to another layer of cross-talk between FoxOs and sirtuins in addition to deacetylation of FoxOs by Sirt1/2 as reported previously (22, 23, 25). Importantly, Sirt1 has been shown to regulate lipid metabolism through suppression of lipogenesis and promotion of fatty acid oxidation (27–33, 72). Deacetylation of SREBP-1 and PGC-1α by Sirt1 is critical for the corresponding regulations because deacetylated SREBP-1 is less stable than the acetylated form, whereas deacetylated PGC-1α is more active than the acetylated protein (26, 29, 32, 33). Overexpression of the Nampt gene not only increased NAD+ concentrations in mouse primary hepatocytes but also elevated the Sirt1 activity, which is consistent with the previous report (46). In addition to Sirt1, three other sirtuins (Sirt3/4/6) have also been shown to regulate lipid metabolism (36–41, 73). As a predominant sirtuin for deacetylation of mitochondrial proteins (74–76), Sirt3 plays a critical role in fatty acid oxidation, ketogenesis, and mitochondrial energy metabolism (35–38, 41, 74, 77–83). Sirt3 knock-out mice have increased levels of fatty acid oxidation intermediates and triglycerides and decreased levels of β-hydroxybutyrate (36, 37, 41). Sirt4, another mitochondrial sirtuin that has ADP-ribosyltransferase activity, has been reported to have an inhibitory function in the regulation of fatty acid oxidation and mitochondrial genes (40). Sirt6, a histone deacetylase, has been shown to suppress hepatic lipogenesis and promote fatty acid oxidation through deacetylation of lysine 9 of histone H3 in the promoter region of numerous lipogenic genes (39).
Through modulation of sirtuin functions, Nampt may play a critical role in hepatic lipid metabolism. This conjecture is supported by the data that Nampt overexpression reduces lipogenesis and increases fatty acid oxidation in primary hepatocytes. By activating Nampt gene expression, FoxOs may inhibit cellular triglyceride accumulation via the Nampt-sirtuin pathway. However, this molecular regulation might be disrupted under insulin resistance or diabetic conditions because FoxOs and the circadian clock may become dysregulated (8, 12, 14, 68, 84–87). As a result, NAD+/NADH ratios may drop below normal levels, and this leads to an impairment of sirtuin function (for example, dysregulated lipid metabolism).
It is possible that other FoxO-mediated mechanisms may also be involved in hepatic lipid metabolism. For example, overexpression of a constitutively nuclear form of FoxO1 (S253A) leads to up-regulation of the genes of apoC-III and microsomal triglyceride transfer protein in mouse liver, and this results in a 2-fold increase in serum and hepatic triglycerides (13, 14). However, overexpression of another constitutively active mutant of FoxO1 (TSS-A) causes a decrease in plasma triglycerides and down-regulation of hepatic lipogenesis (6). The inconsistency could be due to the difference in the levels of FoxO1 overexpression, activity, or experimental conditions. Nevertheless, our FoxO1/3/4 knock-out data share similar conclusion with the TSS-A FoxO1 transgenic results, although the underlying mechanisms might be distinct because Srebp-1c gene is suppressed in the TSS-A mouse liver but was not changed in the LTKO liver. Regardless of the detailed differences, our data suggest that FoxOs are required to maintain lipid homeostasis, and their deficiency makes the liver susceptible to the development of steatosis. The role of FoxOs in metabolic regulation can be better understood in the physiological context. During fasting, FoxOs become active and promote gluconeogenesis to maintain glucose homeostasis (6, 9–12, 88). Upon feeding, FoxOs are inhibited by insulin through Akt-mediated phosphorylation, and this may allow glucose metabolism and lipogenesis to proceed (6, 10, 12). However, prolonged activation of Akt may cause overaccumulation of lipid in the liver because hepatic deficiency of Akt2 protects mice from developing hepatic steatosis (19). It is possible that FoxOs might mediate part of the antisteatosis effect in the Akt2-deficient mouse model. In addition, FoxO1 has also been shown to protect type 1 diabetic mice from developing dyslipidemia, although a different mechanism, possibly through FGF21, might be involved (17).
In summary, we have identified a novel link between insulin signaling and NAD+ biosynthesis through FoxO-regulated Nampt gene expression. These findings have significant implications in both insulin signaling and sirtuin fields because a better understanding of the interactions between insulin and NAD+ signaling pathways may provide critical knowledge for developing novel therapeutics for metabolic syndrome, a rapidly growing health problem in the world (89, 90).
Acknowledgments
We thank Dr. Fred Alt for providing the Sirt1 floxed mice and Dr. Anna DePaoli-Roach for helping with animal import. We also thank Dr. Xiwen Xiong for the technical support.
This work was supported, in whole or in part, by National Institutes of Health Grant R00DK077505 from the NIDDK.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- FoxO
- O family member of Forkhead transcription factors
- Nampt
- nicotinamide phosphoribosyltransferase
- Sirt
- sirtuin
- ZT
- Zeitgeber time
- Srebp-1
- sterol regulatory element-binding protein 1
- PGC-1
- peroxisome proliferator-activated receptor γ coactivator 1
- Nmnat
- nicotinamide-nucleotide adenylyltransferase
- Qprt
- quinolinate phosphoribosyltransferase
- Nadsyn1
- NAD synthetase 1
- Hmox
- heme oxygenase
- IRE
- insulin-responsive element
- TG
- triglyceride
- IR
- insulin receptor.
REFERENCES
- 1. Accili D., Arden K. C. (2004) Cell 117, 421–426 [DOI] [PubMed] [Google Scholar]
- 2. Arden K. C. (2008) Oncogene 27, 2345–2350 [DOI] [PubMed] [Google Scholar]
- 3. Barthel A., Schmoll D., Unterman T. G. (2005) Trends Endocrinol. Metab. 16, 183–189 [DOI] [PubMed] [Google Scholar]
- 4. Gross D. N., van den Heuvel A. P., Birnbaum M. J. (2008) Oncogene 27, 2320–2336 [DOI] [PubMed] [Google Scholar]
- 5. Kamagate A., Kim D. H., Zhang T., Slusher S., Gramignoli R., Strom S. C., Bertera S., Ringquist S., Dong H. H. (2010) Endocrinology 151, 3521–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang W., Patil S., Chauhan B., Guo S., Powell D. R., Le J., Klotsas A., Matika R., Xiao X., Franks R., Heidenreich K. A., Sajan M. P., Farese R. V., Stolz D. B., Tso P., Koo S. H., Montminy M., Unterman T. G. (2006) J. Biol. Chem. 281, 10105–10117 [DOI] [PubMed] [Google Scholar]
- 7. Samuel V. T., Choi C. S., Phillips T. G., Romanelli A. J., Geisler J. G., Bhanot S., McKay R., Monia B., Shutter J. R., Lindberg R. A., Shulman G. I., Veniant M. M. (2006) Diabetes 55, 2042–2050 [DOI] [PubMed] [Google Scholar]
- 8. Qu S., Altomonte J., Perdomo G., He J., Fan Y., Kamagate A., Meseck M., Dong H. H. (2006) Endocrinology 147, 5641–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B. M. (2003) Nature 423, 550–555 [DOI] [PubMed] [Google Scholar]
- 10. Matsumoto M., Pocai A., Rossetti L., Depinho R. A., Accili D. (2007) Cell Metab. 6, 208–216 [DOI] [PubMed] [Google Scholar]
- 11. Liu Y., Dentin R., Chen D., Hedrick S., Ravnskjaer K., Schenk S., Milne J., Meyers D. J., Cole P., Yates J., 3rd, Olefsky J., Guarente L., Montminy M. (2008) Nature 456, 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dong X. C., Copps K. D., Guo S., Li Y., Kollipara R., DePinho R. A., White M. F. (2008) Cell Metab. 8, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Altomonte J., Cong L., Harbaran S., Richter A., Xu J., Meseck M., Dong H. H. (2004) J. Clin. Investig. 114, 1493–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kamagate A., Qu S., Perdomo G., Su D., Kim D. H., Slusher S., Meseck M., Dong H. H. (2008) J. Clin. Investig. 118, 2347–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsumoto M., Han S., Kitamura T., Accili D. (2006) J. Clin. Investig. 116, 2464–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu Z., Jiao P., Huang X., Feng B., Feng Y., Yang S., Hwang P., Du J., Nie Y., Xiao G., Xu H. (2010) J. Clin. Investig. 120, 3901–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haeusler R. A., Han S., Accili D. (2010) J. Biol. Chem. 285, 26861–26868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Biddinger S. B., Hernandez-Ono A., Rask-Madsen C., Haas J. T., Alemán J. O., Suzuki R., Scapa E. F., Agarwal C., Carey M. C., Stephanopoulos G., Cohen D. E., King G. L., Ginsberg H. N., Kahn C. R. (2008) Cell Metab. 7, 125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leavens K. F., Easton R. M., Shulman G. I., Previs S. F., Birnbaum M. J. (2009) Cell Metab. 10, 405–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 21. Jing E., Gesta S., Kahn C. R. (2007) Cell Metab. 6, 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kitamura Y. I., Kitamura T., Kruse J. P., Raum J. C., Stein R., Gu W., Accili D. (2005) Cell Metab. 2, 153–163 [DOI] [PubMed] [Google Scholar]
- 23. Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. (2004) Cell 116, 551–563 [DOI] [PubMed] [Google Scholar]
- 24. Wang F., Tong Q. (2009) Mol. Biol. Cell 20, 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Y., Hou H., Haller E. M., Nicosia S. V., Bai W. (2005) EMBO J. 24, 1021–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feige J. N., Lagouge M., Canto C., Strehle A., Houten S. M., Milne J. C., Lambert P. D., Mataki C., Elliott P. J., Auwerx J. (2008) Cell Metab. 8, 347–358 [DOI] [PubMed] [Google Scholar]
- 27. Hou X., Xu S., Maitland-Toolan K. A., Sato K., Jiang B., Ido Y., Lan F., Walsh K., Wierzbicki M., Verbeuren T. J., Cohen R. A., Zang M. (2008) J. Biol. Chem. 283, 20015–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pfluger P. T., Herranz D., Velasco-Miguel S., Serrano M., Tschöp M. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9793–9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Purushotham A., Schug T. T., Xu Q., Surapureddi S., Guo X., Li X. (2009) Cell Metab. 9, 327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen Z., Liang X., Rogers C. Q., Rideout D., You M. (2010) Am. J. Physiol. Gastrointest. Liver Physiol. 298, G364–G374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu F., Gao Z., Zhang J., Rivera C. A., Yin J., Weng J., Ye J. (2010) Endocrinology 151, 2504–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walker A. K., Yang F., Jiang K., Ji J. Y., Watts J. L., Purushotham A., Boss O., Hirsch M. L., Ribich S., Smith J. J., Israelian K., Westphal C. H., Rodgers J. T., Shioda T., Elson S. L., Mulligan P., Najafi-Shoushtari H., Black J. C., Thakur J. K., Kadyk L. C., Whetstine J. R., Mostoslavsky R., Puigserver P., Li X., Dyson N. J., Hart A. C., Näär A. M. (2010) Genes Dev. 24, 1403–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ponugoti B., Kim D. H., Xiao Z., Smith Z., Miao J., Zang M., Wu S. Y., Chiang C. M., Veenstra T. D., Kemper J. K. (2010) J. Biol. Chem. 285, 33959–33970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang R. H., Li C., Deng C. X. (2010) Int. J. Biol. Sci. 6, 682–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bao J., Scott I., Lu Z., Pang L., Dimond C. C., Gius D., Sack M. N. (2010) Free Radic. Biol. Med. 49, 1230–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hallows W. C., Yu W., Smith B. C., Devires M. K., Ellinger J. J., Someya S., Shortreed M. R., Prolla T., Markley J. L., Smith L. M., Zhao S., Guan K. L., Denu J. M. (2011) Mol. Cell 41, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., Newgard C. B., Farese R. V., Jr., Alt F. W., Kahn C. R., Verdin E. (2010) Nature 464, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kendrick A. A., Choudhury M., Rahman S. M., McCurdy C. E., Friederich M., Van Hove J. L., Watson P. A., Birdsey N., Bao J., Gius D., Sack M. N., Jing E., Kahn C. R., Friedman J. E., Jonscher K. R. (2011) Biochem. J. 433, 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim H. S., Xiao C., Wang R. H., Lahusen T., Xu X., Vassilopoulos A., Vazquez-Ortiz G., Jeong W. I., Park O., Ki S. H., Gao B., Deng C. X. (2010) Cell Metab. 12, 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nasrin N., Wu X., Fortier E., Feng Y., Bare' O. C., Chen S., Ren X., Wu Z., Streeper R. S., Bordone L. (2010) J. Biol. Chem. 285, 31995–32002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shimazu T., Hirschey M. D., Hua L., Dittenhafer-Reed K. E., Schwer B., Lombard D. B., Li Y., Bunkenborg J., Alt F. W., Denu J. M., Jacobson M. P., Verdin E. (2010) Cell Metab. 12, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 44. Smith J. S., Brachmann C. B., Celic I., Kenna M. A., Muhammad S., Starai V. J., Avalos J. L., Escalante-Semerena J. C., Grubmeyer C., Wolberger C., Boeke J. D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Van Gool F., Gallí M., Gueydan C., Kruys V., Prevot P. P., Bedalov A., Mostoslavsky R., Alt F. W., De Smedt T., Leo O. (2009) Nat. Med. 15, 206–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Revollo J. R., Grimm A. A., Imai S. (2004) J. Biol. Chem. 279, 50754–50763 [DOI] [PubMed] [Google Scholar]
- 47. Samal B., Sun Y., Stearns G., Xie C., Suggs S., McNiece I. (1994) Mol. Cell. Biol. 14, 1431–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Friebe D., Neef M., Kratzsch J., Erbs S., Dittrich K., Garten A., Petzold-Quinque S., Bluher S., Reinehr T., Stumvoll M., Bluher M., Kiess W., Korner A. (2011) Diabetologia, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garten A., Petzold S., Barnikol-Oettler A., Körner A., Thasler W. E., Kratzsch J., Kiess W., Gebhardt R. (2010) Biochem. Biophys. Res. Commun. 391, 376–381 [DOI] [PubMed] [Google Scholar]
- 50. Revollo J. R., Körner A., Mills K. F., Satoh A., Wang T., Garten A., Dasgupta B., Sasaki Y., Wolberger C., Townsend R. R., Milbrandt J., Kiess W., Imai S. (2007) Cell Metab. 6, 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tanaka M., Nozaki M., Fukuhara A., Segawa K., Aoki N., Matsuda M., Komuro R., Shimomura I. (2007) Biochem. Biophys. Res. Commun. 359, 194–201 [DOI] [PubMed] [Google Scholar]
- 52. Garten A., Petzold S., Körner A., Imai S., Kiess W. (2009) Trends Endocrinol. Metab. 20, 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Imai S., Kiess W. (2009) Front. Biosci. 14, 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moschen A. R., Gerner R. R., Tilg H. (2010) Curr. Pharm. Des. 16, 1913–1920 [DOI] [PubMed] [Google Scholar]
- 55. Khan J. A., Tao X., Tong L. (2006) Nat. Struct. Mol. Biol. 13, 582–588 [DOI] [PubMed] [Google Scholar]
- 56. Rongvaux A., Galli M., Denanglaire S., Van Gool F., Drèze P. L., Szpirer C., Bureau F., Andris F., Leo O. (2008) J. Immunol. 181, 4685–4695 [DOI] [PubMed] [Google Scholar]
- 57. Wang T., Zhang X., Bheda P., Revollo J. R., Imai S., Wolberger C. (2006) Nat. Struct. Mol. Biol. 13, 661–662 [DOI] [PubMed] [Google Scholar]
- 58. Dahl T. B., Haukeland J. W., Yndestad A., Ranheim T., Gladhaug I. P., Damås J. K., Haaland T., Løberg E. M., Arntsen B., Birkeland K., Bjøro K., Ulven S. M., Konopski Z., Nebb H. I., Aukrust P., Halvorsen B. (2010) J. Clin. Endocrinol. Metab. 95, 3039–3047 [DOI] [PubMed] [Google Scholar]
- 59. Kralisch S., Klein J., Lossner U., Bluher M., Paschke R., Stumvoll M., Fasshauer M. (2005) J. Endocrinol. 185, R1–R8 [DOI] [PubMed] [Google Scholar]
- 60. MacLaren R., Cui W., Cianflone K. (2007) Diabetes Obes. Metab. 9, 490–497 [DOI] [PubMed] [Google Scholar]
- 61. Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone-Corsi P. (2009) Science 324, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ramsey K. M., Yoshino J., Brace C. S., Abrassart D., Kobayashi Y., Marcheva B., Hong H. K., Chong J. L., Buhr E. D., Lee C., Takahashi J. S., Imai S., Bass J. (2009) Science 324, 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Paik J. H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J. W., Carrasco D. R., Jiang S., Gilliland D. G., Chin L., Wong W. H., Castrillon D. H., DePinho R. A. (2007) Cell 128, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cheng H. L., Mostoslavsky R., Saito S., Manis J. P., Gu Y., Patel P., Bronson R., Appella E., Alt F. W., Chua K. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Harada N., Oda Z., Hara Y., Fujinami K., Okawa M., Ohbuchi K., Yonemoto M., Ikeda Y., Ohwaki K., Aragane K., Tamai Y., Kusunoki J. (2007) Mol. Cell. Biol. 27, 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mao X., Kikani C. K., Riojas R. A., Langlais P., Wang L., Ramos F. J., Fang Q., Christ-Roberts C. Y., Hong J. Y., Kim R. Y., Liu F., Dong L. Q. (2006) Nat. Cell Biol. 8, 516–523 [DOI] [PubMed] [Google Scholar]
- 67. Xiong Y., Collins Q. F., An J., Lupo E., Jr., Liu H. Y., Liu D., Robidoux J., Liu Z., Cao W. (2007) J. Biol. Chem. 282, 4975–4982 [DOI] [PubMed] [Google Scholar]
- 68. Cheng Z., Guo S., Copps K., Dong X., Kollipara R., Rodgers J. T., Depinho R. A., Puigserver P., White M. F. (2009) Nat. Med. 15, 1307–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dong X., Park S., Lin X., Copps K., Yi X., White M. F. (2006) J. Clin. Investig. 116, 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ghoshal K., Datta J., Majumder S., Bai S., Dong X., Parthun M., Jacob S. T. (2002) Mol. Cell. Biol. 22, 8302–8319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bogan K. L., Brenner C. (2008) Annu. Rev. Nutr. 28, 115–130 [DOI] [PubMed] [Google Scholar]
- 72. Rodgers J. T., Puigserver P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shi T., Fan G. Q., Xiao S. D. (2010) J. Dig. Dis. 11, 55–62 [DOI] [PubMed] [Google Scholar]
- 74. Ahn B. H., Kim H. S., Song S., Lee I. H., Liu J., Vassilopoulos A., Deng C. X., Finkel T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14447–14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lombard D. B., Alt F. W., Cheng H. L., Bunkenborg J., Streeper R. S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., Yang Y., Chen Y., Hirschey M. D., Bronson R. T., Haigis M., Guarente L. P., Farese R. V., Jr., Weissman S., Verdin E., Schwer B. (2007) Mol. Cell. Biol. 27, 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Verdin E., Hirschey M. D., Finley L. W., Haigis M. C. (2010) Trends Biochem. Sci. 35, 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cimen H., Han M. J., Yang Y., Tong Q., Koc H., Koc E. C. (2010) Biochemistry 49, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hafner A. V., Dai J., Gomes A. P., Xiao C. Y., Palmeira C. M., Rosenzweig A., Sinclair D. A. (2010) Aging 2, 914–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kong X., Wang R., Xue Y., Liu X., Zhang H., Chen Y., Fang F., Chang Y. (2010) PLoS One 5, e11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Palacios O. M., Carmona J. J., Michan S., Chen K. Y., Manabe Y., Ward J. L., 3rd, Goodyear L. J., Tong Q. (2009) Aging 1, 771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schlicker C., Gertz M., Papatheodorou P., Kachholz B., Becker C. F., Steegborn C. (2008) J. Mol. Biol. 382, 790–801 [DOI] [PubMed] [Google Scholar]
- 82. Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shi T., Wang F., Stieren E., Tong Q. (2005) J. Biol. Chem. 280, 13560–13567 [DOI] [PubMed] [Google Scholar]
- 84. Bass J., Takahashi J. S. (2010) Science 330, 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hsieh M. C., Yang S. C., Tseng H. L., Hwang L. L., Chen C. T., Shieh K. R. (2010) Int. J. Obes. 34, 227–239 [DOI] [PubMed] [Google Scholar]
- 86. Kim J. J., Li P., Huntley J., Chang J. P., Arden K. C., Olefsky J. M. (2009) Diabetes 58, 1275–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kohsaka A., Laposky A. D., Ramsey K. M., Estrada C., Joshu C., Kobayashi Y., Turek F. W., Bass J. (2007) Cell Metab. 6, 414–421 [DOI] [PubMed] [Google Scholar]
- 88. Altomonte J., Richter A., Harbaran S., Suriawinata J., Nakae J., Thung S. N., Meseck M., Accili D., Dong H. (2003) Am. J. Physiol. Endocrinol. Metab. 285, E718–E728 [DOI] [PubMed] [Google Scholar]
- 89. Bruce K. D., Hanson M. A. (2010) J. Nutr. 140, 648–652 [DOI] [PubMed] [Google Scholar]
- 90. Guarente L. (2006) Nature 444, 868–874 [DOI] [PubMed] [Google Scholar]