Abstract
Hypoxia inducible factor-1 (HIF-1) is a key transcription factor required for cellular adaptation to hypoxia, although its physiological roles and activation mechanisms during normoxia have not been studied sufficiently. The Warburg effect, which is a hallmark of malignant tumors that is characterized by increased activity of aerobic glycolysis, accompanies activation of HIF-1 during normoxia. Besides tumor cells that have multiple genetic and epigenetic alterations, normal macrophages also use glycolysis for ATP production by depending upon elevated HIF-1 activity even during normoxia. We recently found that activity of factor inhibiting HIF-1 (FIH-1) is specifically suppressed in macrophages by a nonproteolytic activity of membrane type-1 matrix metalloproteinase (MT1-MMP/MMP-14). Thus, MT1-MMP expressed in macrophages plays a significant role in regulating HIF-1 activity during normoxia. In the light of this finding, we examined here whether MT1-MMP contributes to the Warburg effect of tumor cells. All the tumor cell lines that express MT1-MMP exhibit increased glycolytic activity, and forced expression of MT1-MMP in MT1-MMP-negative tumor cells is sufficient to induce the Warburg effect. The cytoplasmic tail of MT1-MMP mediates the stimulation of aerobic glycolysis by increasing the expression of HIF-1 target genes. Specific intervention of the MT1-MMP-mediated activation of HIF-1 in tumor cells retarded tumor growth in mice. Systemic administration of a membrane-penetrating form of the cytoplasmic tail peptide in mice to inhibit HIF-1 activation competitively also exhibited a therapeutic effect on tumors.
Keywords: Cancer Therapy, Glycolysis, Hypoxia, Matrix Metalloproteinase, Tumor Metabolism, FIH-1, HIF-1, MT1-MMP, Mint3, Warburg Effect
Introduction
ATP is a key carrier of cellular energy generated via glycolysis and following oxidative phosphorylation (OXPHOS)2 in mitochondria. Oxygen levels regulate the relative contribution of glycolysis and OXPHOS to ATP production, through a process known as the Pasteur effect. OXPHOS is inefficient when the supply of oxygen is limited, and under such conditions cells produce ATP largely through glycolysis (anaerobic glycolysis). HIF-1 is a master transcription factor that mediates the adaptation of cells to hypoxia and directs cells to produce ATP via anaerobic glycolysis, and its activity is suppressed during normoxia by post-translational mechanisms, and this suppression is released by lowering of the oxygen levels (1–3). HIF-1 regulates the expression of multiple genes (4), and some of these encode glycolysis-related proteins as well as pyruvate dehydrogenase kinase-1 (PDK-1), which inhibits pyruvate consumption by OXPHOS (5, 6). Hypoxic conditions are frequently observed during inflammation and following tissue damage associated with various diseases. Growth of tumors beyond the capacity of the pre-existing capillary network gives rise to hypoxia within a tumor mass (1, 7, 8). HIF-1 is activated within the hypoxic area of tumor cells, and it switches the cellular ATP production system predominantly to anaerobic glycolysis, as long as glucose is available. At the same time, HIF-1 induces the expression of VEGF, which stimulates angiogenesis, leading to improved oxygen and nutrient supply (9).
Although much attention has been paid to the activity of HIF-1 during hypoxia, HIF-1 also plays a pivotal role in maintaining cellular activities in particular types of cells even during normoxia. One such example is macrophages in which ATP production depends largely on glycolysis even under oxygen-rich conditions (aerobic glycolysis), although the OXPHOS activity of the cells is negligible during normoxia (10–12). HIF-1 activity is responsible for aerobic glycolysis in macrophages. Indeed, HIF-1α knock-out mice demonstrated that HIF-1 is responsible for maintaining glycolytic activity during normoxia in macrophages, and knock-out of the HIF-1α subunit gene causes an 80% reduction in ATP levels of the cells (10).
Another example is malignant tumor cells where the Pasteur effect, which represents a physiological cellular response to oxygen levels, is disturbed, and the cells exhibit constitutively active glycolytic activity even under oxygen-rich conditions, via a phenomenon termed the “Warburg effect” (13, 14). HIF-1 is constitutively activated in such malignant tumor cells and regulates cellular glycolytic activity (15). Although cell growth and survival signals also modulate different steps of glycolysis (16–18), HIF-1 plays a major role in maintaining the basal activity of aerobic glycolysis. However, the mechanism activating HIF-1 activity as observed during the Warburg effect in tumor cells is not clear yet (7). The altered glucose metabolism and constitutive activation of HIF-1 that characterize malignant tumor cells have been thought to be possible targets for the development of anti-tumor therapy (8, 19). However, inhibition of HIF-1 activity not only suppresses the Warburg effect but also abrogates the physiological response of other normal cells to hypoxia and may worsen ischemic diseases. Therefore, it is important to clarify the specific mechanism that is boosting HIF-1 activity in tumor cells, particularly during normoxia, and to develop specific methods to intervene with this mechanism via HIF-1-targeted tumor therapy.
To understand the regulation of HIF-1 activity underlying the Warburg effect of tumors, we first turned to our previous study on HIF-1 regulation in macrophages. We have shown that macrophages lacking MT1-MMP exhibit reduced glycolytic activity and a 60% reduction in ATP levels (12). Because of the shortage of ATP, MT1-MMP-deficient macrophages exhibit multiple defects related to energy-dependent cell functions, such as motility, cytokine production, and phagocytosis (12, 20). MT1-MMP maintains ATP levels in macrophages through an activity encoded by its cytoplasmic tail (CPT) (12) and in a manner independent of the well established protease activity of the protein (21–23). The CPT binds factor inhibiting HIF-1 (FIH-1) (24) and guides FIH-1 to interact with Mint3, an inhibitor of FIH-1, and co-localizes within the same subcellular compartment (12, 25). Binding of Mint3 to FIH-1 abrogates the ability of FIH-1 to inhibit the transcriptional activity of HIF-1 (25). Mint3 co-localizes with MT1-MMP in the Golgi apparatus, where MT1-MMP is localized until it is exported to the surface of the cell (supplemental Fig. S1) (12, 25). The activation cascade of HIF-1 activity triggered by MT1-MMP is sufficient to maintain the aerobic glycolytic activity of macrophages despite the low protein levels of HIF-1 during normoxia.
MT1-MMP is a membrane protease that mediates pericellular proteolysis and thereby regulates various cellular functions in both physiological and pathological settings (21–23). Whereas MT1-MMP cleaves a variety of pericellular proteins, it is a potent cellular collagenase and thereby promotes invasion and proliferation of tumor cells in a collagen-rich environment (26–28). Although MT1-MMP is expressed almost exclusively in mesenchymal cells, it is expressed in carcinoma cells according to the activation of cellular programs related to the epithelium-mesenchyme transition (EMT) (29–32). Following the EMT of carcinoma cells resulting in an epithelial layer, MT1-MMP is expressed in the cells and digests the extracellular matrix (ECM), such as the basement membrane and collagen I-rich connective tissue. Cleavage products generated by proteolysis of components of the ECM, membrane proteins, and pericellular proteins can generate bioactive protein fragments that transduce a variety of signals that can further potentiate the malignant behavior of tumor cells (23, 33–36). Even though the importance of the proteolytic activity of MT1-MMP in tumor progression has been established (21, 37), we did not anticipate a link between this protein and the Warburg effect. Our recent studies on MT1-MMP in macrophages indicated a molecular link between MT1-MMP and aerobic glycolysis (12). Here, we show that MT1-MMP is also involved in the increased glycolytic activity observed in malignant tumor cells. We further evaluated the impact of specific intervention of the MT1-MMP-mediated activation of HIF-1 on tumor growth in vivo.
EXPERIMENTAL PROCEDURES
Cell Culture
Cells were cultured in DMEM (MDA-MB-231, MCF-7, T47D, T24, A431, and HT1080), DMEM high glucose (HepG2 and HEK293), or RPMI 1640 medium (Panc1, SW480, MKN7, KK47, PC9, and TMK1) containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Sigma) at 37 °C in humidified 5% CO2. For experiments conducted under hypoxic conditions, cells were cultured with 1% O2 and 5% CO2 in a model 9200 incubator (Wakenyaku) for 24 h.
Measurement of Lactic Acid Levels
Cells were seeded onto 24-well plates (1 × 105/well) in triplicate. Conditioned medium was collected after 6 h, and lactic acid was measured using an l-lactic acid kit (R-Biopharm), and values were normalized to the protein concentration determined using a Bradford assay kit (Bio-Rad). In some experiments, the medium was replaced with serum-free medium 64 h after siRNA treatment or adenoviral transduction, and the cells were incubated for another 6 h.
Measurement of ATP Concentrations
Cells were cultured in the presence or absence of the glycolysis inhibitor, 2-deoxyglucose (200 μg/ml; Sigma), for 4 h, and ATP levels were determined using the ATP bioluminescence assay kit CLS II (Roche Applied Science). ATP levels were normalized to the total protein concentration, determined using a Bradford assay kit (Bio-Rad).
Measurement of 2-NBDG Intake Levels
Cells were treated with 10 μm 2-NBDG (Invitrogen) for 6 h, washed with PBS three times, and lysed with Passive lysis buffer (Promega). After centrifugation, the fluorescence of 2-NBDG in clear cell lysates was measured using a fluorescence plate reader. 2-NBDG levels were normalized to the total protein concentration, determined using a Bradford assay kit (Bio-Rad).
Knockdown Experiments Using siRNA
The target sequences for siRNA were designed and then synthesized (B-bridge), and siRNAs were provided as a mixture consisting of three different siRNA target sequences. The siRNA sequences were as follows: Mint3, 5′-gauggaacuugaugaguca-3′, 5′-gggaggugcaccucgagaa-3′, and 5′-gguucuugguccuguauga-3′; FIH-1, 5′-cgacacaaaucuuguguau-3′, 5′-cagcaaacgcucaaugaca-3′, and 5′-gggaauggcuaccgggacu-3′; MT1-MMP, 5′-ggauggacacggagaauuu-3′, 5′-gcgaugaagucuucacuua-3′, and 5′-ggguagagacccugagaca-3′; and HIF-1α, 5′-cagaaauggccuugugaaa-3′, 5′-gauggaagcacuagacaaa-3′, and 5′-gcauauaucuagaagguau-3′. The control siRNAs were 5′-auccgcgcgauaguacgua-3′, 5′-uuacgcguagcguaauacg-3′, and 5′-uauucgcgcguauagcggu-3′. Cells were seeded (2.5 × 104/well onto 24-well plates) and transfected with a 10 nm siRNA mixture (containing the three sequences to the target) using LipofectamineTM RNAiMAX (Invitrogen) according to the manufacturer's instructions.
Knockdown Experiments Using shRNAs
The shRNA sequence used to knock down human Mint3 expression was 5′-gaugauggcgguggugacgggacgaaucccgucaccaccgccauca-3′, and the deoxyribose version of this gene sequence was subcloned into pENTR/U6 TOPO (Invitrogen) before being transferred via recombination into the lentivirus vector pLenti6 BlockIT. The shRNA-expressing lentiviral vectors were generated and used according to the manufacturer's instructions.
RNA Isolation, Reverse Transcription, and Real Time PCR
Total RNA was isolated from cells using TRIzol (Invitrogen) and subjected to reverse transcription (RT) using SuperScriptII (Invitrogen) and random primers. The RT products were then subjected to real time PCR in a Smart Cycler II System (Cepheid) using SYBR Green I (BioWhittaker Molecular Applications) and the following specific primers: ACTB (β-actin) sense, 5′-gggacgacatggagaaaatc-3′, and antisense, 5′-gggtgttgaaggtctcaaac-3′; VEGFA sense, 5′-ctccaccatgccaagtggtc-3′, and antisense, 5′-actcctggaagatgtccacc-3′; and SLC2A1 (GLUT-1) sense, 5′-gggcatgtgcttccagtatgt-3′, and antisense, 5′-accaggagcacagtgaagat-3′. The PCR products were sequenced, and their homogeneity was confirmed by dissociation temperature monitoring of SYBR Green I fluorescence.
Cell Growth Assay
Cells (1 × 104) were seeded onto a plastic dish and cultured at 37 °C in a humidified CO2 incubator. The cells were counted periodically using a hemocytometer.
Tumor Growth Assay
The tumorigenicity of the cells was examined using 6-week-old female BALB/c nude mice (Clea, Japan). Briefly, 1 × 106 (MDA-MB-231) or 1 × 107 (MCF-7) cells were injected subcutaneously into the dorsal side of mice, and the volumes of the implanted tumors were measured with a caliper and calculated using the formula V = (L × W2)/2, where V is volume (mm3); L is the biggest diameter (mm), and W is the smallest diameter (mm).
Treatment of Tumor Cells with Peptides in Vitro and Administration of Peptides to Tumor-bearing Mice
Tumor cells in culture were treated with 40 μg/ml synthesized (7R)-CPT (RRRRRRRGRRHGTPRRLLYCQRSLLDKV) or (7R)-SCR peptide (RRRRRRRGTLRPDRQVRHKLCLRGYLSR) (Invitrogen) for 48 h before being analyzed. For experimental treatment of tumors with the peptides, MDA-MB-231 cells were implanted as described above. (7R)-CPT or (7R)-SCR (50 mg/kg body weight) peptide was injected into the mice intraperitoneally at 0, 2, and 4 days after inoculation of the tumor cells and subsequently injected every 3 days. Tumor volume was monitored as described above.
Histopathology
The animals were sacrificed 7 (MCF-7) or 28 days (MDA-MB-231) after inoculation, and their tumors were embedded in O.C.T. compound (Sakura Finetek) and stored at −80 °C. Frozen sections (10 μm) were fixed with 4% paraformaldehyde/PBS and subjected to H&E staining or immunohistochemistry. Immunostaining was performed using rat anti-CD31 (Pharmingen) or rabbit anti-cleaved caspase-3 (Cell Signaling) antibodies. The nuclei were counter-stained with Hoechst 33342, and the sections were observed by CCD microscopy. The CD31-positive area was calculated using ImageJ software (National Institutes of Health).
Statistical Analysis
We compared two subject groups by the two-sided t test or the Mann-Whitney U test.
RESULTS
MT1-MMP Enhances Glycolytic Activity in MDA-MB-231 Breast Carcinoma Cells
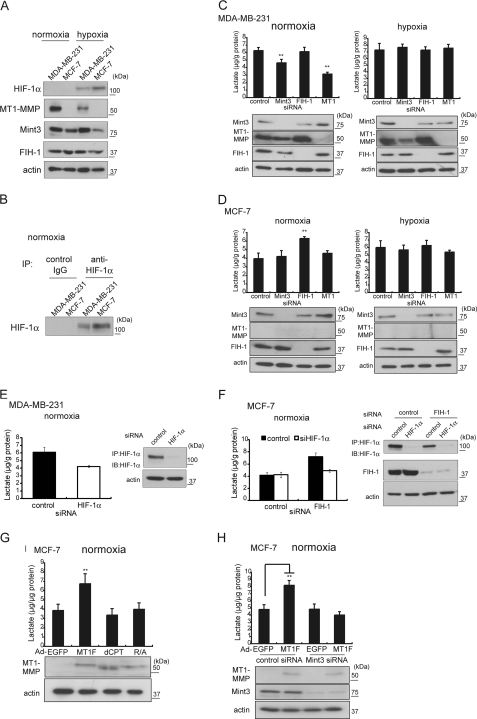
Human breast carcinoma cell lines MDA-MB-231 and MCF-7 have been used to study the Warburg effect. MDA-MB-231 is highly invasive in vitro, tumorigenic in mice, and exhibits elevated glycolytic activity, and MCF-7 is noninvasive, less tumorigenic, and exhibits less glycolytic activity (7). MDA-MB-231 cells express MT1-MMP, but MCF-7 cells do not (32). We confirmed these earlier findings by immunoblot analysis (Fig. 1A). We were, however, unable to detect HIF-1α protein in either cell line during normoxia by conventional immunoblot analysis (Fig. 1A). Detection of HIF-1α necessitated prior concentration of the protein by immunoprecipitation (Fig. 1B). Analysis of the production of lactate, the end product of glycolysis, in these cells confirmed that MDA-MB-231 cells exhibit higher glycolytic activity during normoxia than MCF-7 cells (Fig. 1, C and D, compare the control sample of each type of cell). Exposure of MDA-MB-231 cells to hypoxia gave rise to only a modest increase in the production of lactate, whereas exposure of MCF-7 cells to hypoxia increased the production of lactate by about 50% (Fig. 1, C and D, compare the control samples under normoxic and hypoxic conditions for each type of cell). The results indicate that the glycolytic activity of MCF-7 cells responds to hypoxic conditions, whereas the glycolytic activity of MDA-MB-231 cells during normoxia is already close to the upper limit of the capacity of the cell.
FIGURE 1.
MT1-MMP expression promotes aerobic glycolysis of MDA-MB-231 and MCF-7. A, expression of HIF-1α, MT1-MMP, FIH-1, and Mint3 in MDA-MB-231 and MCF-7 during normoxia and hypoxia. B, cell lysates from MDA-MB-231 and MCF-7 cells cultured during normoxia were subjected to immunoprecipitation (IP) using control IgG or anti-HIF-1α antibodies before immunoblot analysis. C and D, lactate production in MDA-MB-231 (C) and MCF-7 (D) cells during normoxia or hypoxia. The effect of knocking down of the expression of the indicated genes on lactate production (upper bar graphs) and protein expression (lower panels) is shown. E and F, effect of HIF-1α knockdown on lactate production (left panels) in MDA-MB-231 (E) and MCF-7 (F) cells. Protein expression was confirmed by immunoblot analysis (right panels). G and H, effect of forced expression of MT1-MMP or its mutants on lactate production in MCF-7 cells. G, cells were transduced with adenovirus vectors expressing EGFP (Ad-EGFP), FLAG-tagged MT1-MMP (Ad-MT1F), an MT1-MMP mutant lacking the CP tail (Ad-dCPT), or an MT1-MMP mutant unable to bind FIH-1 due to substitution of an Arg residue to an Ala (Ad-R/A). H, effect of forced expression of MT1-MMP on lactate production in MCF-7 cells treated with control siRNA or Mint3-targeted siRNA. The data in C–H were analyzed by the t test. **, p < 0.01; error bars indicate S.D. (n = 3).
The elevated glycolytic activity observed in the cells expressing MT1-MMP was reminiscent of macrophages, in which the expression of MT1-MMP leads to HIF-1 activation and sustained glycolytic activity during normoxia (12). In macrophages, the CPT of MT1-MMP mediates inactivation of FIH-1 via Mint3 and thereby activates HIF-1. We confirmed the expression of both Mint3 and FIH-1 in both types of cells by immunoblot analysis (Fig. 1A). We next tested whether knockdown of the expression of MT1-MMP, Mint3, or FIH-1 affected the glycolytic activity of MDA-MB-231 cells using specific siRNA sequences (Fig. 1, C and D). Depletion of Mint3 or MT1-MMP in MDA-MB-231 cells reduced lactate production by 25 and 50%, respectively, whereas depletion of FIH-1 had no effect. These results are similar to those observed in macrophages where FIH-1 activity is inhibited by Mint3 in an MT1-MMP-dependent manner (12). Under hypoxic conditions, depletion of neither MT1-MMP nor Mint3 affected lactate production in MDA-MB-231 cells (Fig. 1C, hypoxia). The results indicate that the effect of knockdown of MT1-MMP or Mint3 expression on glycolysis can only be observed during normoxia. Knockdown of their expression in hypoxic cells has no observable effect, as HIF-1 is already fully activated in such cells as a result of cellular response to hypoxia.
Unlike MDA-MB-231 cells, MCF-7 cells do not express MT1-MMP, although they express Mint3 and FIH-1 constitutively (Fig. 1A). In contrast to the results with MDA-MB-231 cells, transfection of MCF-7 cells with siRNA targeting Mint3 or MT1-MMP had no effect on lactate production during normoxia, whereas knockdown of FIH-1 expression in these cells increased lactate production by 1.6-fold, suggesting that FIH-1 is active in these cells, inhibits HIF-1 activity, and thereby suppresses glycolysis (Fig. 1D, normoxia). Exposure of MCF-7 cells to hypoxia increased lactate production by 1.5-fold, a value similar to the effect of FIH-1 depletion (Fig. 1D, hypoxia). These results indicate that FIH-1 is not inhibited by Mint3 in MCF-7 cells. We also confirmed the contribution of the HIF-1 activity of MDA-MB-231 cells and MCF-7 cells to the glycolytic activity of the cell during normoxia by knocking down expression of HIF-1α (Fig. 1, E and F).
We next evaluated whether MT1-MMP could stimulate glycolytic activity in MCF-7 cells by inhibiting FIH-1 via Mint3. Forced expression of MT1-MMP increased lactate production by 1.6-fold (Fig. 1E, Ad-MT1F). Thus, at least a part of the Warburg effect observed in MDA-MB-231 cells can be reproduced in MCF-7 cells by simply expressing MT1-MMP. We further observed that the CPT of MT1-MMP plays a vital role in stimulating glycolysis in MCF-7 cells; expression of a mutant MT1-MMP either lacking the entire CPT (Ad-dCPT) or having an amino acid substitution in the CPT that abrogates binding to FIH-1 (Ad-R/A) (12) failed to enhance glycolytic activity (Fig. 1E). In addition, the MT1-MMP-dependent stimulation of glycolysis in MCF-7 cells was dependent on the presence of Mint3 (Fig. 1F), indicating that tumor cells make use of the same MT1-MMP-dependent mechanism as macrophages for the constitutive maintenance of aerobic glycolysis.
Correlation between Expression of MT1-MMP and Production of Lactate in Tumor Cell Lines
MT1-MMP is frequently expressed in malignant tumor cells (33), suggesting a possibility that other tumor cell lines expressing MT1-MMP also exhibit enhanced glycolytic activity. We surveyed expression of HIF-1α, MT1-MMP, Mint3, and FIH-1 and glycolytic activity in 12 additional human tumor cell lines. HIF-1α, FIH-1, and Mint3 were expressed in all cell lines tested, although HIF-1α expression levels were low in T47D, MKN7, and HepG2 cells compared with the other cell lines (Fig. 2A). Nine cell lines expressed MT1-MMP and three others (T47D, TMK-1, and KK47) did not (Fig. 2A). The level of lactate production in the MT1-MMP-expressing cells (with the exception of Panc1) was generally greater than that observed in the three MT1-MMP-negative lines (Fig. 2B). Depletion of either MT1-MMP or Mint3 in the MT1-MMP-expressing cells, including Panc1, decreased lactate production, whereas depletion of FIH-1 had no effect (Fig. 2B and see supplemental Fig. S2 for protein expression). Thus, these results overall are similar to what we observed in MDA-MB-231 cells following the knockdown experiments. In contrast, the effects of gene knockdown upon lactate production in the three MT1-MMP-negative cell lines was similar to that observed in the parallel experiments in MCF-7 cells (Fig. 2C and supplemental Fig. S2 for protein expression). Thus, expression of MT1-MMP is a common feature of the tumor cell lines exhibiting increased glycolytic activity. Thus, we therefore will henceforth refer to the enhanced glycolytic activity correlated to expression of MT1-MMP as the MT1-MMP-dependent Warburg effect.
FIGURE 2.
MT1-MMP-expressing tumor cell lines promote aerobic glycolysis by Mint3. A, expression of HIF-1α, MT1-MMP, FIH-1, and Mint3 in MDA-MB-231, MCF-7, and 12 additional tumor cell lines. IP, immunoprecipitation; IB, immunoblot. B and C, analysis of lactate production in MT1-MMP-positive (B) and MT1-MMP-negative (C) tumor cell lines following knock down of Mint3, FIH-1, or MT1-MMP. The data in B and C were analyzed by the t test. *, p < 0.05; **, p < 0.01; error bars indicate S.D. (n = 3).
Impact of the MT1-MMP-dependent Warburg Effect on Tumor Growth
The HIF-1α subunit of the heterodimeric HIF-1 is regulated negatively by two types of oxygen-dependent modifications, whereas the HIF-1β subunit is constitutively active (2, 3). FIH-1 hydroxylates an asparagine residue within the transcriptional activation domain of HIF-1α and thereby inhibits the binding of HIF-1α to p300/CBP (24, 38). In addition, prolyl hydroxylases hydroxylate two proline residues within HIF-1α, thereby initiating its degradation by the proteasome (39, 40).
To evaluate the impact of the MT1-MMP-dependent Warburg effect on tumor growth, we established stable Mint3-depleted cells by expressing a specific shRNA targeting Mint3 in MDA-MB-231 cells. The depletion of Mint3 did not affect the level of HIF-1α during normoxia (Fig. 3, A and B), the level of expression of prolyl hydroxylases (supplemental Fig. S3), or the prolyl hydroxylase-dependent regulation of HIF-1α during hypoxia (Fig. 3A). However, depletion of Mint3 markedly reduced expression of the mRNAs encoding the HIF-1 target genes, VEGFA (9) and SLC2A1 which encodes glucose transporter 1 (GLUT1) (41) (Fig. 3C). Depletion of Mint3 also reduced the expression of proteins involved in glycolysis (supplemental Fig. 4A). Consequently, the depleted cells exhibited a decrease in glucose transporter activity measured by uptake of a fluorescent glucose analog 2-NBDG, lactate production, and ATP production by 42, 27, and 23%, respectively (Fig. 2C). Thus, the glycolytic activity of Mint3-depleted MDA-MB-231 cells was suppressed to the level observed in MCF-7 cells (Fig. 3D, compare lactate/shMint3 with Fig. 1D, MCF-7/normoxia/control). The rate of growth of the depleted cells in culture during normoxia or hypoxia was unaffected (Fig. 3E).
FIGURE 3.
Tumor-promoting role of the MT1-MMP-induced Warburg effect in MDA-MB-231 cells, as determined by in vitro experiments. Mint3 expression was knocked down in MDA-MB-231 cells using Mint3 shRNA (shMint3). Control cells were transfected with LacZ shRNA (shLacZ). The effect on the following parameters was assessed. A, Mint3 protein expression (left panel) and HIF-1α protein expression in response to hypoxia (right panel); B, HIF-1α protein expression during normoxia; C, VEGF and GLUT1 mRNA expression; D, 2-NBDG intake (left panel), lactate production (middle panel), and ATP production in the presence or absence of the glycolysis inhibitor, 2-deoxyglucose (right panel); E, in vitro cell proliferation during normoxia (left panel) and hypoxia (right panel). The error bars indicate S.D. (n = 3), and the data were analyzed by the t test. IP, immunoprecipitation; IB, immunoblot. **, p < 0.01.
2-Deoxyglucose (2-DG) is a glucose analog that inhibits glycolysis. 2-DG treatment decreased ATP production in control MDA-MB-231 cells to 40% of the value observed in cells receiving vehicle alone, and under such conditions the effect of Mint3 depletion was not evident (Fig. 3D, far right). In a previous study, we observed that 2-DG treatment inhibited ATP production nearly completely in macrophages, in which OXPHOS activity is negligible, whereas 2-DG did not inhibit ATP production in MEFs, which produce ATP by OXPHOS (12). Therefore, MDA-MB-231 cells appear to have an intermediate phenotype between macrophages and MEFs.
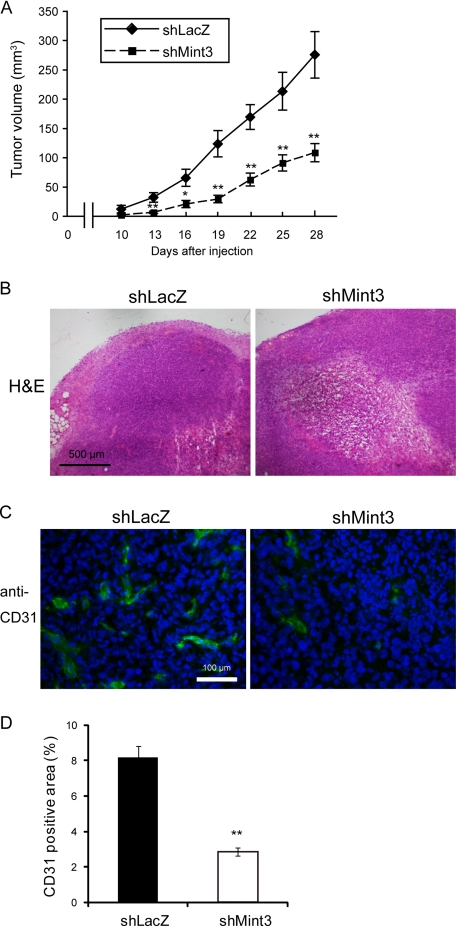
We next monitored tumor growth of Mint3-depleted MDA-MB-231 cells (shMint3) implanted into immunodeficient mice. The Mint3-depleted cells formed tumors that were 40% smaller than those produced by the control cells by day 28 (Fig. 4A). Tumors formed by the Mint3-depleted cells frequently showed necrotic areas (Fig. 4B) and had fewer vessels within the tumor tissue as evaluated by CD31 staining (Fig. 4, C and D). The results are consistent with the observed marked reduction of VEGFA expression in the depleted cells (Fig. 3C). Because depletion of Mint3 does not affect the rate of growth of the cells in culture (Fig. 3E), the effect of Mint3 depletion on tumor growth presumably reflects the pleiotropic effect of HIF-1 activity rather than a simple consequence of decreased glucose metabolism. Because HIF-1 regulates a number of genes encoding paracrine factors and ECM regulators (42), the ability of HIF-1 to regulate the tumor microenvironment is presumably important in suppression of tumor growth observed following Mint3 depletion.
FIGURE 4.
Tumor-promoting role of the MT1-MMP-induced Warburg effect in MDA-MB-231 cells, as determined by in vivo experiments. A, tumor growth upon subcutaneous implantation in immunodeficient mice. The tumors were dissected 28 days after implantation and analyzed as follows: B, H&E staining; C, immunostaining for CD31. D, CD31-positive area in C was calculated using ImageJ software. A and D, error bars indicate S.E. (n = 10), and the data were analyzed by the Mann-Whitney U test. *, p < 0.05; **, p < 0.01.
Construction of a Mutant Mint3 That Cannot Inhibit FIH-1
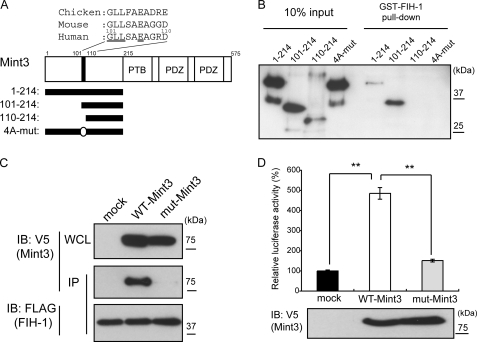
Three members of the Mint/X11/APBA family of proteins share a common C-terminal domain structure containing PTB and PDZ domains, whereas the N-terminal portion of each protein is unique (43). Because Mint3 is an adaptor protein, it can bind multiple proteins. For example, Mint3 binds amyloid precursor protein, Arf3 and Arf4, and furin via the C-terminal PTB and PDZ domains (44–46), whereas binding to FIH-1 is mediated by the N-terminal region (25). Therefore, the effect of Mint3 depletion on tumor growth might be mediated by proteins other than FIH-1. To confirm the importance of FIH-1 as the target of the effect of Mint3-depletion on tumor growth, we first generated a mutant Mint3 that specifically lacks the binding site for FIH-1. The N-terminal half of Mint3 (amino acids 1–214) contains no known protein motif, but it is sufficient to bind FIH-1 fused to GST (Fig. 5, A and B), as reported previously (25). A Mint3 fragment spanning 101–214 amino acids retains FIH-1 binding activity, but further deletion of an additional 10 amino acids generated a fragment spanning 110–214 amino acids (Fig. 5A) that failed to bind FIH-1 (Fig. 5B). We observed that some residues within the 101–110-amino acid region are conserved among vertebrate homologs (Fig. 5A, underlined residues, and supplemental Fig. S5). Based on this knowledge, we introduced alanine substitutions into the conserved amino acid residues in the N-terminal fragment (Fig. 5A, 4A-mut) and observed that the mutant protein could not bind GST-FIH-1 (Fig. 5B). To confirm the lack of an interaction between FIH-1 and the mutant Mint3 with the four alanine substitutions (mut-Mint3) within cells, FIH-1 was expressed in HEK293 cells as a FLAG-tagged form along with either wild-type Mint3 (WT-Mint3) or mut-Mint3 expressed as V5-tagged forms (Fig. 5C). Immunoprecipitation of FLAG-tagged FIH-1 co-precipitated V5-tagged Mint3 but not mut-Mint3. Thus, the conserved amino acids of Mint3 are indispensable for binding to FIH-1.
FIGURE 5.
Mutant Mint3 unable to bind FIH-1 fails to activate HIF-1. A, structure of Mint3. The amino acid sequences of the 101–110-amino acid region in the human, mouse, and chicken homologs of Mint3 are shown. The N-terminal His6-Mint3 polypeptides 1–214, 101–214, and 110–214 used for pulldown assays with GST-FIH-1 are indicated below. In the 4A-mut peptide, the underlined residues in HGLLSAEAGRD110 were substituted with alanine. B, pulldown assays with GST-FIH-1 and the His6-Mint3 peptides. C, ability of FLAG-FIH-1 expressed in HEK293 cells to co-immunoprecipitate with wild-type (WT-Mint3) or the 4A substitution mutant (mut-Mint3) of Mint3. FIH-1 was immunoprecipitated using anti-FLAG antibody and Mint3, and its mutant were detected by immunoblot (IB) using anti-V5 antibody. D, effect of mut-Mint3 and WT-Mint3 on the transcriptional activity of the HIF-1α CAD reporter in HEK293 cells. The reporter assay system is depicted in supplemental Fig. S5. Error bars indicate S.D. (n = 3).
To confirm the effect of the Mint3 mutation on the regulation of HIF-1α, we used a luciferase reporter assay that can specifically monitor FIH-1-mediated regulation of HIF-1α activity (supplemental Fig. S6) as described previously (25, 47). Expression of WT-Mint3 in HEK293 cells together with the reporter gave rise to increased reporter gene expression, indicating that Mint3 inhibits the endogenous FIH-1 and thereby activates HIF-1α activity (Fig. 5, C and D). However, the mut-Mint3 failed to stimulate the expression of the reporter gene.
Mint3 Mutant Cannot Enhance Glycolytic Activity or Tumor Growth in Vivo
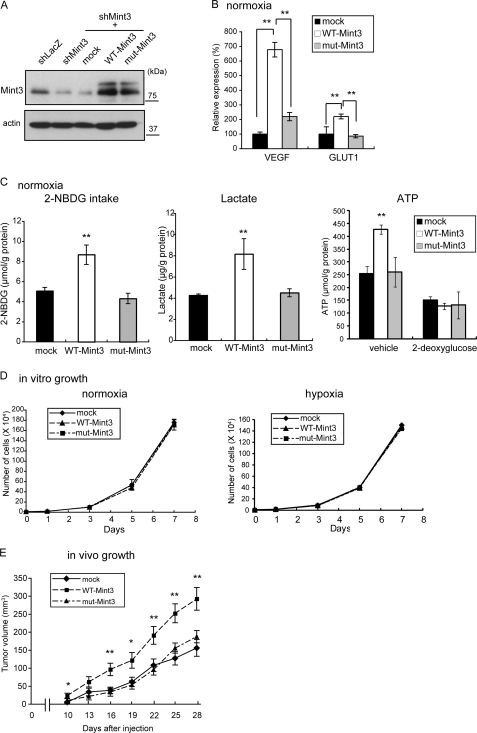
To confirm that the ability of Mint3 to bind FIH-1 is essential for the regulation of HIF-1 and its target genes, we expressed shRNA-resistant forms of the mRNAs encoding WT-Mint3 or mut-Mint3 in Mint3-depleted MDA-MB-231 derivative cells (MDA-KD) (Fig. 6A). Expression of WT-Mint3, but not mut-Mint3, enhanced the expression of HIF-1 target genes (Fig. 6B) and glycolysis-related proteins (supplemental Fig. 4, B and C) and augmented 2-NBDG intake, lactate production, and ATP production (Fig. 6C) in MDA-KD cells. Although the expression of WT- and mut-Mint3 did not affect the proliferation of MDA-KD cells in culture during normoxia or hypoxia (Fig. 6D), expression of WT-Mint3 but not mut-Mint3 enhanced the growth of tumors by MDA-KD cells in mice (Fig. 6E). Taken together, these results indicate that FIH-1 is the major target of Mint3 depletion leading to suppression of tumor growth in mice.
FIGURE 6.
Mutant Mint3 unable to bind FIH-1 fails to restore the Warburg effect in Mint3 knocked down MDA-MB-231 cells. WT-Mint3 or mut-Mint3 was expressed in Mint3-knockdown MDA-MB-231 cells using shMint3-resistant expression constructs and the following parameters were analyzed: A, Mint3 levels; B, VEGF and GLUT1 mRNA levels; C, 2-NBDG intake (left panel), lactate production (middle panel), and ATP production in the presence or absence of the glycolysis inhibitor, 2-deoxyglucose (right panel); D, cell growth in culture during normoxia (left panel) and hypoxia (right panel); E, growth in immunodeficient mice. B–D, error bars indicate S.D. (n = 3), and the data were analyzed by the t test. E, error bars indicate S.E. (n = 12), and the data were analyzed by the Mann-Whitney U test. *, p < 0.05; **, p < 0.01.
CPT of MT1-MMP Is Sufficient to Induce the Warburg Effect in MCF-7 Cells and Increases Survival of Tumor Cells in Vivo
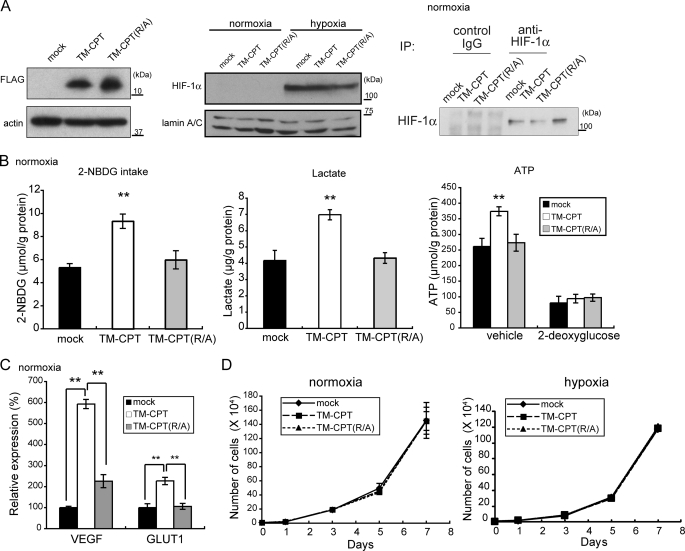
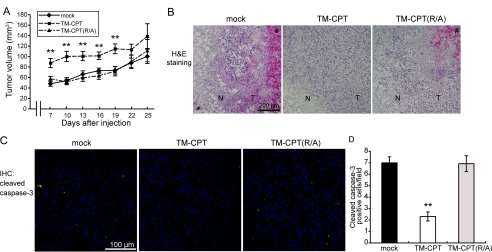
The CPT of MT1-MMP is indispensable for the MT1-MMP-dependent Warburg effect in MCF-7 cells (Fig. 1E). Because the membrane-anchored form of the CP tail (TM-CPT) is sufficient for augmenting HIF-1 activity in macrophages (12), we examined whether expression of TM-CPT or its substitution mutant TM-CPT(R/A) could enhance glycolysis in MCF-7 cells (Fig. 7A). Expression of these TM-CPT constructs did not alter expression of HIF-1α (Fig. 7A) or prolyl hydroxylases (supplemental Fig. S7) during normoxia, and hypoxia induced the accumulation of HIF-1α protein in cells expressing these TM-CPT constructs (Fig. 7A). However, the expression of TM-CPT but not TM-CPT(R/A) enhanced 2-NBDG intake, lactate production, and ATP production in MCF-7 cells (Fig. 7B) and also stimulated the expression of HIF-1 target genes (Fig. 7C and supplemental Fig. 4D). Cell growth in culture was unaffected by the expression of either the TM-CPT or the (Arg/Ala) mutant (Fig. 7D). The parental MCF-7 cells are weakly tumorigenic in mice, whereas cells expressing the TM-CPT formed larger tumors than those expressing TM-CPT(R/A) or the control cells, particularly at early time points after implantation (Fig. 8A, days 7–18). The difference in size of the tumors became less statistically significant after 22 days due to increased variation in each group. Thus, the TM-CPT-mediated Warburg effect appears to increase the ability of MCF-7 cells to adapt and survive in the implanted tissue. To confirm this further, we analyzed the tissues of 7-day-old tumors by subjecting them to H&E staining (Fig. 8B) and immunohistochemical analysis to detect the cleaved protein products produced by caspase-3 activity (Fig. 8C). Necrotic areas were commonly observed in tumors derived from parental cells (mock) or TM-CPT(R/A)-expressing cells but not in those expressing TM-CPT (Fig. 8B). The tumors expressing TM-CPT had fewer apoptotic cells than those derived from mock cells or TM-CPT (R/A)-expressing cells (Fig. 8, C and D). Thus, the artificially induced Warburg effect in MCF-7 cells appears to promote cell survival in vivo.
FIGURE 7.
Ability of MT1-MMP CP tail expression to induce the Warburg effect in MCF-7 cells. The membrane-anchored form of the CP tail (TM-CPT) and its mutant TM-CPT(R/A) were expressed in MCF-7 cells as FLAG-tagged forms and their effects on the following MCF-7 parameters were evaluated. A, TM-CPT and TM-CPT(R/A) protein expression, as determined by immunoblot using anti-FLAG antibody (left panel), HIF-1α protein expression in response to hypoxia (middle panel), and HIF-1α protein expression during normoxia (right panel). B, 2-NBDG intake (right panel), lactate production (middle panel), and ATP production in the presence or absence of the glycolysis inhibitor, 2-deoxyglucose (right panel). C, VEGF and GLUT1 mRNA levels. D, cell growth in culture. B–D, error bars indicate S.D. (n = 3) and the data were analyzed by the t test. **, p < 0.01; IP, immunoprecipitation.
FIGURE 8.
Ability of MT1-MMP CP tail expression to induce the Warburg effect in MCF-7 cells. A, tumor growth in immunodeficient mice. B, mock-, TM-CPT-, and TM-CPT(R/A)-expressing tumors were dissected 7 days after implantation and examined by H&E staining. N, normal tissue; T, tumor tissue; #, necrotic area. C and D, tumor tissues were also subjected to immunohistochemical (IHC) analysis to determine cleaved caspase-3 levels. A and D, error bars indicate S.E. (n = 12), and the data were analyzed by the Mann-Whitney U test. **, p < 0.01.
Experimental Therapy of MDA-MB-231 Tumors by Systemic Administration of a Synthetic CPT Peptide
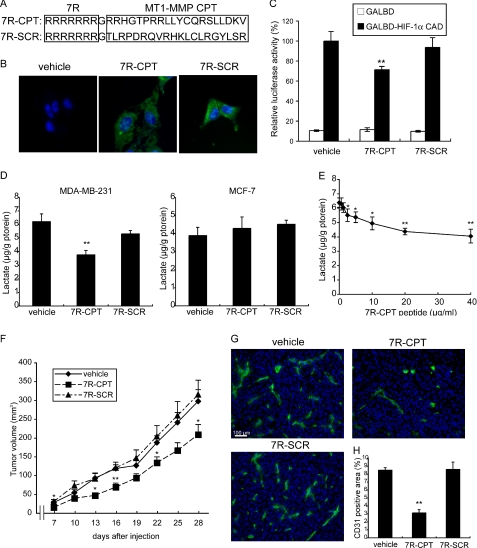
Because the interaction between the CPT of MT1-MMP and FIH-1 plays a pivotal role in the Mint3-mediated inhibition of FIH-1 (12), we next evaluated the therapeutic effect of interrupting this interaction in vivo using a soluble synthetic CPT peptide as a competitor. The CPT peptide ((7R)-CPT) includes seven Arg residues at the N terminus to allow penetration of the cell membrane (48), and the (7R)-SCR (control) peptide is made up of a scrambled sequence of the CPT amino acids (Fig. 9A). FITC-labeled peptides were incorporated into cultured MDA-MB231 cells after incubation for 3 h (Fig. 9B), and the (7R)-CPT peptide significantly inhibited the luciferase activity used to monitor HIF-1α activity regulated by FIH-1 in the cells (Fig. 9C). Lactate production by MDA-MB-231 cells was suppressed by (7R)-CPT but not by (7R)-SCR, and treatment of MCF-7 cells with (7R)-CPT had no effect, indicating the specificity of the activity of the peptide toward cells expressing MT1-MMP (Fig. 9D). The level of lactate production by MDA-MB-231 cells was reduced to the levels observed in MCF-7 cells when the MDA-MB-231 cells were treated with 40 μg/ml (7R)-CPT (Fig. 9, D and E). The (7R)-CPT also suppressed lactate production in other tumor cell lines expressing MT1-MMP (supplemental Fig. S8).
FIGURE 9.
Membrane-penetrating MT1-MMP CP tail peptides suppress aerobic glycolysis and tumor growth in MDA-MB-231 cells. A, sequences of membrane-penetrating MT1-MMP CP tail peptides ((7R)-CPT) and control peptides with a scrambled CP tail sequence ((7R)-SCR). B, photographs of MDA-MB-231 cells treated with vehicle, FITC-labeled (7R)-CPT, or (7R)-SCR peptides for 3 h. C, effect of (7R)-CPT and (7R)-SCR peptides on the transcriptional activity of the HIF-1α CAD reporter in MDA-MB-231 cells. D, effect of (7R)-CPT and (7R)-SCR peptides on lactate production in MDA-MB-231 (left panel) and MCF-7 cells (right panel). E, effect of the (7R)-CPT peptide concentration on lactate production in MDA-MB-231 cells. F, effect of the intraperitoneal injection of (7R)-CPT or (7R)-SCR peptides on the tumor growth of MDA-MB-231 cells in immunodeficient mice. G and H, tumors in F were dissected 28 days after implantation and analyzed by immunostaining for CD31 (G) and the CD31-positive area was calculated using ImageJ software (H). C–E, error bars indicate S.D. (n = 3), and the data were analyzed by the t test. F and H, error bars indicate S.E. (n = 12), and the data were analyzed by the Mann-Whitney U test. *, p < 0.05; **, p < 0.01.
The peptides exhibited no apparent toxicity toward MDA-MB-231 cells or noncancerous cells such as NIH3T3 and HEK293 during cell growth in vitro (supplemental Fig. S9 and data not shown). MDA-MB-231 cells were subsequently implanted subcutaneously in mice, and the peptides (50 mg/kg body weight) were injected into the intraperitoneal cavity at 0, 2, and 4 days after implantation of the tumor cells and every 3 days thereafter according to the in vivo protein transduction method (49) by which the peptides were delivered into the tumor tissue (supplemental Fig. S10). Tumor growth was significantly retarded by (7R)-CPT peptide but not by (7R)-SCR (Fig. 9F). As was observed following Mint3 depletion in MDA-MB-231 cells (Fig. 4, C and D), administration of (7R)-CPT but not (7R)-SCR reduced the number of tumor blood vessels (Fig. 9, G and H).
DISCUSSION
In this study, we demonstrated that tumor cells expressing MT1-MMP exhibited enhanced glycolytic activity that is similar to the level observed when cells are exposed to hypoxia in culture. The mechanism responsible for the increased glycolytic activity of tumor cells is the same as that used by macrophages; the CPT of MT1-MMP binds FIH-1 and thereby suppresses the activity of the latter by promoting its binding to Mint3, an inhibitor of FIH-1 (12). Thus, expression of MT1-MMP contributes to the Warburg effect of tumors by activating HIF-1. Based on this understanding of the mechanism mediating the Warburg effect in tumors, we used Mint3 depletion or expression of a membrane-penetrating CPT peptide to mitigate the MT1-MMP-dependent suppression of FIH-1. The re-activation of FIH-1 in cells treated by these methods results in inhibition of HIF-1 activity specifically during normoxia, whereas the treated cells responded to hypoxia in a normal fashion and exhibited increased expression of HIF-1 protein and lactate production. The specific impact of MT1-MMP-mediated HIF-1 activation was also manifested as enhanced tumor growth in mice, increased survival of tumor cells, and increased tumor blood vessel formation. The effect of MT1-MMP-mediated HIF-1 activation on tumor cell behavior in vivo is not restricted to MDA-MB-231 cells, because knockdown of Mint3 reduced survival of Panc1 cells and TM-CPT expression increased survival of T47D cells (supplemental Fig. S11). Systemic administration of the soluble CPT peptide intraperitoneally also exhibited therapeutic effect against MDA-MB-231 cells implanted subcutaneously in mice. In this case, tumor-associated macrophages (50) and endothelial cells (51) that also express MT1-MMP in the tumor microenvironment could conceivably also be targeted by the peptide and might contribute to the therapeutic effect.
In our previous study, we observed that 2-DG treatment of macrophages abolished ATP production almost completely, although it showed no effect on MEFs that use OXPHOS in mitochondria for ATP production (12). Unlike the results with MEFs, treatment of MDA-MB-231 cells with 2-DG reduced ATP levels by 60% (Fig. 3D), suggesting that OXPHOS is not fully active in the latter cells. This partial dysfunction of OXPHOS activity in MDA-MB-231 cells may increase the cell's dependence upon the MT1-MMP-dependent Warburg effect for ATP production, and indeed Mint3 depletion in these cells caused a 23% reduction in ATP levels (Fig. 3D). In addition to its effect on glycolysis, HIF-1 affects multiple cellular functions by regulating the expression of a variety of genes that regulate cell growth (cyclin D1 and IGF2), motility (Met, SDF-1α, and CXCR4), angiogenesis (VEGF and PDGF), and the ECM environment (fibronectin, collagen VI, and MMP-2) (42). Indeed, Mint3 depletion in MDA-MB-231 cells affects the motility and invasiveness of the cell (supplemental Fig. S12). Therefore, it is most likely that the inhibition of tumor growth of Mint3-depleted MDA-MB-231 cells in mice is a consequence of pleiotropic effects upon HIF-1 signaling during normoxia.
The MT1-MMP-induced activation of HIF-1 presumably cooperates with many genetic and epigenetic alterations associated with tumors that have recently been reported to contribute to the Warburg effect (16–19). For example, constitutive expression of an embryonic isoform of pyruvate kinase M2 (PKM2) in tumor cells is responsible for enhanced lactate production (52). Switching of isoform expression is regulated by alternative splicing of the mRNA. The Myc oncoprotein is known to regulate alternative splicing, so as to suppress the expression of the adult isoform (PKM1) and promote the expression of the PKM2 isoform, through an effect upon heterogeneous nuclear ribonucleoprotein activity (53). Unlike PKM1, the activity of PKM2 can be regulated by tyrosine kinases such as FGFR to provide a metabolic advantage to tumor cells (19). A lack of the tumor suppressor p53 also contributes to the Warburg effect, as demonstrated by the observation that knock-out of the p53 gene suppresses OXPHOS and increases lactate production (54). Synthesis of cytochrome c oxidase 2 (SCO2) was identified as the downstream mediator of the suppression of OXPHOS by p53. Glycolytic activity is also suppressed by a p53-inducible gene product called TIGAR that inhibits phosphofructokinase and suppresses GLUT3 expression via modulation of the IKK-NF-κB pathway (55, 56). The Ras-PI3K-Akt pathway also regulates glucose uptake and early steps of glucose metabolism (57). Constitutive defects in the TCA cycle due to mutations in succinate dehydrogenase or fumarate dehydratase genes are associated with certain types of tumor development and activation of HIF-1 activity (58). However, activation of HIF-1 alone is insufficient to render cancer cells addictive to aerobic glycolysis for energy generation. The effects of genetic and epigenetic alterations on OXPHOS are presumably at the core of the addiction of cancer cells to the Warburg effect. It is of note that embryonic fibroblasts express MT1-MMP accompanying activation of HIF-1. Treatment of cells with a glucose analog, 2DG, decreases cellular ATP levels in cancer cells and macrophages that have lower OXPHOS activity. In contrast, 2DG treatment does not affect ATP levels of fibroblasts that depend upon OXPHOS for ATP generation during normoxia.
Expression of MT1-MMP is usually absent in normal epithelial cells, and its expression is closely linked to cellular programs regulating EMT (32). Indeed, Snail induces expression of MT1-MMP (29), and the promoter activity of the MT1-MMP gene is controlled by β-catenin, which is a downstream mediator of Wnt signals (31). The protease activity of MT1-MMP is important for the invasive and proliferative nature of tumor cells following the EMT. However, this study additionally sheds light on a new link between the EMT and activation of HIF-1 via the expression of MT1-MMP. Therefore, the induction of MT1-MMP expression during the EMT may also induce the metabolic reprogramming of tumor cells such that they gain a proliferative advantage by better utilizing the available carbon sources.
The MT1-MMP/FIH-1/Mint3 pathway is a mechanism regulating HIF-1 activity independently of oxygen levels. Cancer cells exposed to hypoxia employ the well established hypoxia response mechanism to activate HIF-1, and contribution of the MT1-MMP/FIH-1/Mint3 pathway becomes negligible. Therefore, the MT1-MMP/FIH-1/Mint3 pathway will not affect the behavior of cancer cells within hypoxic areas of a tumor. We believe that HIF-1 activation by the MT1-MMP/FIH-1/Mint3 pathway affords an advantage to those cancer cells that disseminate from the primary tumor mass and metastasize to secondary sites. The results of our xenograft model experiments support this idea by demonstrating that the MT1-MMP/FIH-1/Mint3 pathway contributes to tumor survival and growth in metastatic foci rather than in the primary lesion. Taken together, we believe that the regulation of HIF-1 by the CPT of MT1-MMP reveals a promising new molecular target for the development of therapeutics to treat cancer that would act to abrogate the MT1-MMP-dependent Warburg effect of tumor cells as well as the energy-dependent functions of tumor-associated macrophages that promote tumor progression (50).
Acknowledgments
We thank Naohiko Koshikawa, Ikuo Yana, Makoto Nagano, Daisuke Hoshino, Nagayasu Egawa, and Dr. Mitsuaki Yoshida for helpful discussions. We also thank Dr. Robert Whittier for help in preparing the manuscript.
This work was supported by the Specific Coordination Fund for Promoting Science (to T. S.) and by a grant-in-aid for scientific research (S) on priority areas, the “Integrative Research toward the Conquest of Cancer,” from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Figs. 1–12 and additional reference.
- OXPHOS
- oxidative phosphorylation
- MT1-MMP
- membrane type-1 matrix metalloproteinase
- CPT
- cytoplasmic tail
- EMT
- epithelium-mesenchyme transition
- ECM
- extracellular matrix
- 2-DG
- 2-deoxyglucose
- MEF
- mouse embryo fibroblast
- 2-NBDG
- 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy-d-glucose.
REFERENCES
- 1. Denko N. C. (2008) Nat. Rev. Cancer 8, 705–713 [DOI] [PubMed] [Google Scholar]
- 2. Kaelin W. G., Jr., Ratcliffe P. J. (2008) Mol. Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 3. Semenza G. L. (2001) Curr. Opin. Cell Biol. 13, 167–171 [DOI] [PubMed] [Google Scholar]
- 4. Lee J. W., Bae S. H., Jeong J. W., Kim S. H., Kim K. W. (2004) Exp. Mol. Med. 36, 1–12 [DOI] [PubMed] [Google Scholar]
- 5. Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V. (2006) Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 6. Papandreou I., Cairns R. A., Fontana L., Lim A. L., Denko N. C. (2006) Cell Metab. 3, 187–197 [DOI] [PubMed] [Google Scholar]
- 7. Gatenby R. A., Gillies R. J. (2004) Nat. Rev. Cancer 4, 891–899 [DOI] [PubMed] [Google Scholar]
- 8. Semenza G. L. (2003) Nat. Rev. Cancer 3, 721–732 [DOI] [PubMed] [Google Scholar]
- 9. Forsythe J. A., Jiang B. H., Iyer N. V., Agani F., Leung S. W., Koos R. D., Semenza G. L. (1996) Mol. Cell. Biol. 16, 4604–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cramer T., Yamanishi Y., Clausen B. E., Förster I., Pawlinski R., Mackman N., Haase V. H., Jaenisch R., Corr M., Nizet V., Firestein G. S., Gerber H. P., Ferrara N., Johnson R. S. (2003) Cell 112, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Michl J., Ohlbaum D. J., Silverstein S. C. (1976) J. Exp. Med. 144, 1465–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sakamoto T., Seiki M. (2010) J. Biol. Chem. 285, 29951–29964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Warburg O. (1930) Metabolism of Tumors, p. 327, Constable and Co., Ltd., London [Google Scholar]
- 14. Warburg O. (1956) Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 15. Semenza G. L. (2010) Curr. Opin. Genet. Dev. 20, 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dang C. V. (2009) Sci. Signal. 2, pe75. [DOI] [PubMed] [Google Scholar]
- 17. Kim J. W., Dang C. V. (2006) Cancer Res. 66, 8927–8930 [DOI] [PubMed] [Google Scholar]
- 18. Kroemer G., Pouyssegur J. (2008) Cancer Cell 13, 472–482 [DOI] [PubMed] [Google Scholar]
- 19. Hitosugi T., Kang S., Vander Heiden M. G., Chung T. W., Elf S., Lythgoe K., Dong S., Lonial S., Wang X., Chen G. Z., Xie J., Gu T. L., Polakiewicz R. D., Roesel J. L., Boggon T. J., Khuri F. R., Gilliland D. G., Cantley L. C., Kaufman J., Chen J. (2009) Sci. Signal. 2, ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sakamoto T., Seiki M. (2009) Genes Cells 14, 617–626 [DOI] [PubMed] [Google Scholar]
- 21. Itoh Y., Seiki M. (2006) J. Cell. Physiol. 206, 1–8 [DOI] [PubMed] [Google Scholar]
- 22. Rowe R. G., Weiss S. J. (2009) Annu. Rev. Cell Dev. Biol. 25, 567–595 [DOI] [PubMed] [Google Scholar]
- 23. Kessenbrock K., Plaks V., Werb Z. (2010) Cell 141, 52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mahon P. C., Hirota K., Semenza G. L. (2001) Genes Dev. 15, 2675–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakamoto T., Seiki M. (2009) J. Biol. Chem. 284, 30350–30359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hotary K., Allen E., Punturieri A., Yana I., Weiss S. J. (2000) J. Cell Biol. 149, 1309–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hotary K. B., Allen E. D., Brooks P. C., Datta N. S., Long M. W., Weiss S. J. (2003) Cell 114, 33–45 [DOI] [PubMed] [Google Scholar]
- 28. Wolf K., Wu Y. I., Liu Y., Geiger J., Tam E., Overall C., Stack M. S., Friedl P. (2007) Nat. Cell Biol. 9, 893–904 [DOI] [PubMed] [Google Scholar]
- 29. Ota I., Li X. Y., Hu Y., Weiss S. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20318–20323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Polette M., Gilles C., Nawrocki-Raby B., Lohi J., Hunziker W., Foidart J. M., Birembaut P. (2005) Cancer Res. 65, 7691–7698 [DOI] [PubMed] [Google Scholar]
- 31. Takahashi M., Tsunoda T., Seiki M., Nakamura Y., Furukawa Y. (2002) Oncogene 21, 5861–5867 [DOI] [PubMed] [Google Scholar]
- 32. Pulyaeva H., Bueno J., Polette M., Birembaut P., Sato H., Seiki M., Thompson E. W. (1997) Clin. Exp. Metastasis 15, 111–120 [DOI] [PubMed] [Google Scholar]
- 33. Seiki M., Yana I. (2003) Cancer Sci. 94, 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koshikawa N., Mizushima H., Minegishi T., Iwamoto R., Mekada E., Seiki M. (2010) Cancer Res. 70, 6093–6103 [DOI] [PubMed] [Google Scholar]
- 35. Guess C. M., Quaranta V. (2009) Matrix Biol. 28, 445–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morrison C. J., Butler G. S., Rodríguez D., Overall C. M. (2009) Curr. Opin. Cell Biol. 21, 645–653 [DOI] [PubMed] [Google Scholar]
- 37. Rowe R. G., Weiss S. J. (2008) Trends Cell Biol. 18, 560–574 [DOI] [PubMed] [Google Scholar]
- 38. Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. (2003) EMBO J. 22, 4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang J., Zhao Q., Mooney S. M., Lee F. S. (2002) J. Biol. Chem. 277, 39792–39800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ebert B. L., Firth J. D., Ratcliffe P. J. (1995) J. Biol. Chem. 270, 29083–29089 [DOI] [PubMed] [Google Scholar]
- 42. Pouysségur J., Dayan F., Mazure N. M. (2006) Nature 441, 437–443 [DOI] [PubMed] [Google Scholar]
- 43. Rogelj B., Mitchell J. C., Miller C. C., McLoughlin D. M. (2006) Brain Res. Rev. 52, 305–315 [DOI] [PubMed] [Google Scholar]
- 44. Han J., Wang Y., Wang S., Chi C. (2008) J. Cell Sci. 121, 2217–2223 [DOI] [PubMed] [Google Scholar]
- 45. Hill K., Li Y., Bennett M., McKay M., Zhu X., Shern J., Torre E., Lah J. J., Levey A. I., Kahn R. A. (2003) J. Biol. Chem. 278, 36032–36040 [DOI] [PubMed] [Google Scholar]
- 46. Tanahashi H., Tabira T. (1999) Biochem. Biophys. Res. Commun. 255, 663–667 [DOI] [PubMed] [Google Scholar]
- 47. Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. (2002) Science 295, 858–861 [DOI] [PubMed] [Google Scholar]
- 48. Futaki S., Suzuki T., Ohashi W., Yagami T., Tanaka S., Ueda K., Sugiura Y. (2001) J. Biol. Chem. 276, 5836–5840 [DOI] [PubMed] [Google Scholar]
- 49. Schwarze S. R., Ho A., Vocero-Akbani A., Dowdy S. F. (1999) Science 285, 1569–1572 [DOI] [PubMed] [Google Scholar]
- 50. Joyce J. A., Pollard J. W. (2009) Nat. Rev. Cancer 9, 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yana I., Sagara H., Takaki S., Takatsu K., Nakamura K., Nakao K., Katsuki M., Taniguchi S., Aoki T., Sato H., Weiss S. J., Seiki M. (2007) J. Cell Sci. 120, 1607–1614 [DOI] [PubMed] [Google Scholar]
- 52. Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., Cantley L. C. (2008) Nature 452, 230–233 [DOI] [PubMed] [Google Scholar]
- 53. David C. J., Chen M., Assanah M., Canoll P., Manley J. L. (2010) Nature 463, 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Matoba S., Kang J. G., Patino W. D., Wragg A., Boehm M., Gavrilova O., Hurley P. J., Bunz F., Hwang P. M. (2006) Science 312, 1650–1653 [DOI] [PubMed] [Google Scholar]
- 55. Bensaad K., Tsuruta A., Selak M. A., Vidal M. N., Nakano K., Bartrons R., Gottlieb E., Vousden K. H. (2006) Cell 126, 107–120 [DOI] [PubMed] [Google Scholar]
- 56. Kawauchi K., Araki K., Tobiume K., Tanaka N. (2008) Nat. Cell Biol. 10, 611–618 [DOI] [PubMed] [Google Scholar]
- 57. Manning B. D., Cantley L. C. (2007) Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gottlieb E., Tomlinson I. P. (2005) Nat. Rev. Cancer 5, 857–866 [DOI] [PubMed] [Google Scholar]