Abstract
Fibrillar aggregation of the protein ataxin-3 is linked to the inherited neurodegenerative disorder Spinocerebellar ataxia type 3, a member of the polyQ expansion disease family. We previously reported that aggregation and stability of the nonpathological form of ataxin-3, carrying an unexpanded polyQ tract, are modulated by its N-terminal Josephin domain. It was also shown that expanded ataxin-3 aggregates via a two-stage mechanism initially involving Josephin self-association, followed by a polyQ-dependent step. Despite this recent progress, however, the exact mechanism of ataxin-3 fibrilization remains elusive. Here, we have used electron microscopy, atomic force microscopy, and other biophysical techniques to characterize the morphological and mechanical properties of nonexpanded ataxin-3 fibrils. By comparing aggregates of ataxin-3 and of the isolated Josephin domain, we show that the two proteins self-assemble into fibrils with markedly similar features over the temperature range 37–50°C. Estimates of persistence length and Young's modulus of the fibrils reveal a great flexibility. Our data indicate that, under physiological conditions, during early aggregation Josephin retains a nativelike secondary structure but loses its enzymatic activity. The results suggest a key role of Josephin in ataxin-3 fibrillar aggregation.
Introduction
The formation of organized protein fibrillar aggregates, often referred to as amyloid fibers, has been the object of extensive investigation, owing to its involvement in human disease and to the very unusual and somewhat ambiguous properties of fibrillar species (1). Several pathologies known as conformational disorders, which include Alzheimer's, Parkinson's, prion, and Huntington's diseases, have been associated with the formation of fibers formed by misfolded or abnormally self-assembled proteins and peptides. On the other hand, a positive functional role has also been recognized for amyloid or amyloidlike fibrils in bacteria and other organisms, including humans (2). The properties of these protein quaternary structures are exploited by cells and tissues for structural and protective functions, such as coat formation, stabilization, or scaffolding for catalytic reactions. These findings have inspired the design of artificial structures such as nanofibers for technical and medical applications (3). A detailed understanding of the mechanisms that regulate amyloid fiber assembly and of their mechanical properties is therefore critical to obtaining a more accurate picture of the dual role of amyloids and designing therapeutic interventions and novel biocompatible materials.
Here, we have investigated the properties of early aggregates formed by ataxin-3, the protein responsible for the neurodegenerative disorder Spinocerebellar ataxia type 3 (SCA3), otherwise known as Machado-Joseph disease. SCA3 belongs to the family of polyglutamine (polyQ) expansion diseases that includes Huntington's disease and several other spinocerebellar ataxias (4). These pathologies are caused by the abnormal expansion of trinucleotide repeats coding for glutamine, and protein aggregation and misfolding are indicated among the mechanisms responsible for disease initiation. Because of their link to misfolding and aggregation, the biophysical and structural properties of the clinically relevant polyQ proteins and their aggregation mechanisms at the molecular level have raised significant interest. PolyQ in particular has been the object of intense investigation, as it is insoluble in water and its tendency to aggregate depends on the number of consecutive residues within polyQ stretches (5). Although polyQ expansion is commonly accepted to be the main factor responsible for protein aggregation of polyQ proteins, consistent evidence indicates that this is not the only amyloidogenic element and that its properties are strongly modulated by the protein context surrounding the polyQ stretch (6–8).
This behavior is certainly true for ataxin-3, probably the best characterized of the polyQ containing proteins. Its aggregation properties are governed by the interplay of two regions: the enzymatically active N-terminal Josephin domain (the only structured portion of the molecule), and the polyQ stretch (present in the flexible C-terminus). PolyQ length influences the aggregation kinetics, which are faster for longer glutamine repeats, but aggregation is initiated and modulated by Josephin in a polyQ-independent step (9–13). Because the isolated Josephin domain is able to aggregate and form fibers (10), this region of the intact molecule is thought to modulate the aggregation properties of nonexpanded ataxin-3.
Interestingly, both full-length ataxin-3 and the isolated Josephin domain aggregate under physiological conditions. The formation at 37°C of amyloidlike fibrils with a beaded morphology has been reported for expanded and nonexpanded ataxin-3 (11,12,14). We have recently shown that, under these conditions, Josephin aggregation is promoted by hydrophobic surfaces exposed to solvent in the native protein structure (15). Although this observation could suggest that a structural rearrangement may not be necessary to induce aggregation, an α-to-β secondary structure transition is associated with fibril formation both for the intact protein and its N-terminal domain (11,12,15).
Despite its intensive characterization, the mechanism of ataxin-3 aggregation is still poorly understood and several important questions remain open:
How exactly does the globular Josephin domain contribute to ataxin-3 fiber formation and morphology?
What is the structure of ataxin-3 within the aggregates and at what stage during fibrilization do the conformational changes occur?
Does Josephin retain its cysteine protease activity within the fibrils?
Answers to these questions can only be provided by investigating further the early stages of aggregation of both ataxin-3 and Josephin and by comparing the species produced during fiber formation.
In this work, we characterized the morphological and mechanical properties of fibrils formed by ataxin-3 and by the isolated Josephin domain. Using a combination of different techniques, including electron microscopy (EM), atomic force microscopy (AFM), size exclusion chromatography (SEC), and circular dichroism (CD), we have monitored the formation of oligomeric aggregates and their subsequent assembly into protofibers. We show that these processes have remarkably similar properties for the full-length unexpanded protein and the Josephin domain. Based on our results, we suggest a leading role for the Josephin domain in ataxin-3 fiber formation.
Materials and Methods
Protein expression and purification
Human nonexpanded ataxin-3 (with a polyQ tract of 18 consecutive glutamines) and the isolated Josephin domain were expressed in the Escherichia coli strain BL21 and purified as GST fusion proteins using a Glutathione Sepharose affinity matrix (GE Healthcare, Chalfont St Giles, UK), as described in Masino et al. (10,16). The GST tags were removed using the recombinant Tobacco Etch Virus protease. The purity of all samples was >95%, as assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and mass spectrometry. Protein concentrations were determined using ultraviolet (UV) absorption with calculated extinction coefficients at 280 nm of 27,670 M−1 cm−1 for ataxin-3 and 25,440 M−1 cm−1 for the Josephin domain. No significant difference was observed when protein concentrations were measured in 6 M guanidinium hydrochloride or in pure buffer.
Aggregation time course analysis
Ataxin-3 and Josephin samples were incubated at 37 or 50°C without shaking at a concentration of 14 μM, in 20 mM sodium phosphate, pH 6.5, 10 mM TCEP, and 0.025% NaN3. At different time points, aliquots were removed from stock solutions for SEC, SDS-PAGE, AFM, and EM measurements. Josephin samples treated at higher temperature were heated from 20°C to 80°C by increasing temperature in 10°C-steps and incubating samples at each temperature for ∼1 h. Monodispersion of the samples before aggregation was confirmed by SEC and EM data. SDS-PAGE showed that the proteins remained intact throughout the monitored incubation period. We observed that ataxin-3 and Josephin aggregation kinetics are strongly affected by buffer composition and in particular by the presence of aggregation inhibitor compounds. The Complete antiprotease cocktail (Roche USA, Nutley, NJ), for example, consistently increased Josephin aggregation rates.
AFM and EM measurements
AFM images were obtained at room temperature using a JPK Nanowizard II microscope combined with a TE2000 inverted microscope (Nikon, Melville, NY). Biolever cantilevers (RC800PSA, 200 × 20 μm; Olympus, Melville, NY) were employed, with an average tip-radius of 20 nm, a spring constant of 30 pN/nm (0.03 N/m), and resonant frequency of 37 kHz in air and ∼8 kHz in water. Ataxin-3 and Josephin samples were diluted in 20 mM Imidazole, 25 mM KCl, 2 mM EGTA, and 4 mM MgCl2, pH 7.4 to a final concentration of 0.1–0.2 μM and applied onto silanized glass coverslips or on freshly cleaved mica substrates treated with 150–200 mM MgCl2. Images of the fully hydrated specimens were recorded in intermittent contact mode in fluid, with a scan size between 300 nm and 2 μm and line scan rates between 0.3 and 1.4 Hz. Image analysis was performed using the JPK and WSxM software. Aggregate dimensions were measured from unprocessed topographical scans performed at the lowest possible scan force (<0.1 nN). Average height and apparent width values were obtained from image cross sections. Tip-sample dilation effects were modeled by assuming the tip to be a hemisphere with a radius larger than the radius of the measured specimen, also assumed to be spherical or cylindrical. The actual width (w) of the measured particles can then be calculated from the following relation between the tip radius (r) and the apparent particle width (wa): w = 2 (wa2/16 r) (17).
For EM measurements, ataxin-3 and Josephin samples were applied to carbon-coated, glow discharged grids and stained with 1% sodium silico-tungstate (pH 7). Protein concentration was diluted from 14 μM to 2–3 μM. The grids were viewed under minimum dose, accurate defocus conditions with a 1200EX instrument (JEOL, Peabody, MA) operated at 100 kV. Aggregate dimensions were measured using the ImageJ processing program (National Institutes of Health, Bethesda, MD). The fibril contour length was estimated both from EM and AFM images by approximating the fibril profile with consecutive segments. The width of Josephin and ataxin-3 fibrils was measured at the widest point along the fiber, i.e., in correspondence of the beads, and not of the linkers. Statistical analysis of particle dimensions (height, width, contour length, end-to-end distance, interbead distance) was performed by calculating average values on multiple observations.
Analysis of mechanical properties
Persistence length analysis was performed using two independent approaches. In the first, the persistence length Lp was estimated by evaluating the decay of tangent angle correlations along the fibrils, due to shape fluctuations caused by thermal motion (18); in the second, it was calculated using mean-square end-to-end distances 〈R2〉 (19).
SEC measurements
SEC experiments were performed on a Superdex 75 10/300 GL Tricorn column (GE Healthcare, Kings Park, NY) equilibrated in PBS (2 mM phosphate, pH 7.5, 5 mM KCl, and 140 mM NaCl). Aliquots of Josephin stock solutions removed at different time points were injected into the column and the signal was monitored at 215 and 280 nm. Peak areas were calculated after baseline subtraction using the GE Healthcare-supplied software.
CD measurements
Far-UV CD measurements were performed on a J-715 spectropolarimeter (JASCO, Oklahoma City, OK) equipped with a PTC-348 Peltier temperature control system. The spectra of Josephin samples incubated at 37°C were recorded at different time points using quartz cuvettes (Hellma, Plainview, NY) with pathlengths of 1 mm. Protein concentration was 14 μM. CD intensities are presented as the CD absorption coefficient calculated using the molar concentration of the proteins (ΔɛM). Far-UV CD spectral decomposition to obtain the secondary structure content was achieved with the CONTIN, SELCON, and CDSSTR methods (20). The possibility that artifacts may affect the CD spectra due to increased light scattering during aggregation could be excluded by recording the photomultiplier voltage during spectra acquisition.
Josephin enzymatic activity assays
The linear substrate GST-Ub52, comprising the human Ub precursor Ub52 fused to GST, was expressed in E. coli BL21 and purified using a Glutathione Sepharose affinity matrix (GE Healthcare), as described in Holowaty et al. (21). Reactions were performed at 37°C in 20 mM Tris-HCl (pH 8.0), 10 mM DTT. Josephin stock solutions (330 μM) were used fresh or after incubation at 37 or 50°C (to induce fibrilization) and then diluted into GST-Ub52 solutions to give a 10:1 enzyme/substrate molar ratio. The reactions were monitored by SDS-PAGE after incubation times of 3 and 24 h.
Results
Ataxin-3 aggregation proceeds through formation of spheroidal intermediates
We first characterized ataxin-3 samples after incubation at 37°C under native conditions for 40 h and analyzed their composition using EM. The micrographs show a heterogeneous population of aggregated species including spheroidal oligomers and, in higher proportion, elongated fibrils (Fig. 1, A and B). The filaments appear greatly flexible and with a beaded morphology. They can form annular structures (Fig. 1 B) and are sometimes branched. Although their contour lengths vary greatly, their width is consistently between 6 and 16 nm, with a large predominance of species (67%) 9–11-nm wide and an average width of 10 ± 2 nm (N, number of observations = 380) (Fig. 1 C) (Table S1 in the Supporting Material). The spheroidal species seem to assemble to form short structures of two or more linked globules (Fig. 1 B, inset). Their diameters vary between 7 and 16 nm, with an average of 11 ± 2 nm (N = 256) (Fig. 1 C). The correlation between the spheroidal dimensions and the fibril widths, together with the beaded morphology, suggests that the spheroidal particles could be building blocks that assemble into fibrils. Further analysis of negatively stained electron micrographs of longer fibrils also shows that their structure is irregular and is composed of repeating light- and dark-stained regions with an average spacing of ∼10–40 nm (Fig. S1 in the Supporting Material). Although these observations support a model of beads-on-string, they are also consistent with a ribbonlike morphology with twists at regular distances. The current resolution does not, however, allow us to discriminate between the possible models.
Figure 1.
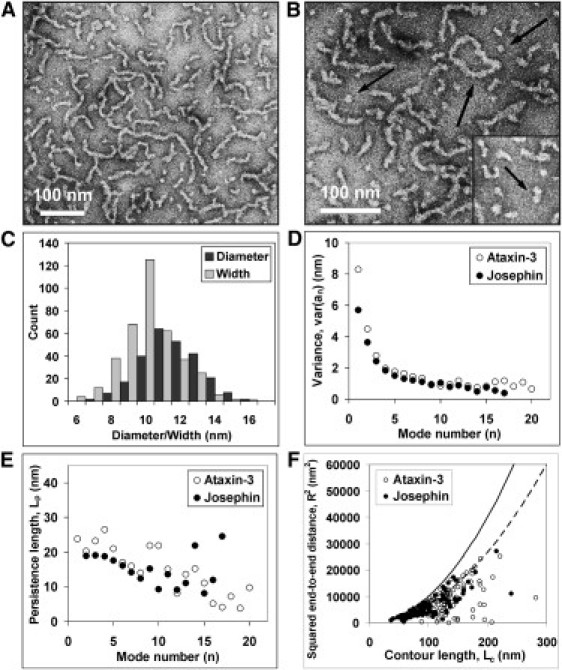
EM data of ataxin-3 samples incubated at 37°C and persistence length analysis. Sample concentration was 14 μM and the incubation time 4 h. (A and B) Micrographs of ataxin-3 oligomers and fibrils. (Arrows, B) Annular structure, isolated spheroidal oligomers, and oligomers that appear linked together (inset). (C) Distribution of widths of ataxin-3 fibrils and of diameters of spheroidal aggregates. (D and E) Persistence length analysis of ataxin-3 and Josephin fibrils performed using the decay of tangent correlations. The variance (D) and persistence length Lp (E) are plotted against mode number. Lp values reported in the text are the mean of the Lp estimates measured between modes 1–20. (F) Plot of squared end-to-end distances versus contour length. (Solid and dashed lines) Limit functions (for Lc << Lp) 〈R2〉2D = Lc2 and 〈R2〉3D proj = 2/3 Lc2, obtained from Eqs. 2 and 3, respectively.
AFM measurements on the same samples confirmed the presence of spheroidal oligomers (Fig. S2) with heights between 5 and 12 nm and an average value of 7 ± 2 nm (N = 143), in agreement with EM data (Table S1). The average apparent width of these spheroidal aggregates is 23 ± 4 nm (N = 39). This value is an overestimate of the actual diameter of the oligomers, owing to the lateral dilation caused by the AFM tip (17). After deconvolution, the calculated values for the actual width of the aggregates vary between 4 and 15 nm, with an average of 7 ± 3 nm. Although affected by larger errors, the width values are consistent with the more reliably measured height values, and confirm that the measured objects have an approximately spherical shape.
The difference between dimensions measured by EM and AFM may be due to the fact that negative staining EM is performed on samples adsorbed on carbon-coated grids and allowed to dry, whereas AFM images are acquired in solution on fully hydrated specimens. Nonetheless, the agreement between EM and AFM values is within the experimental error and the intrinsic instrumental differences.
Ataxin-3 fibrils have high flexibility and elasticity
We analyzed the mechanical properties of ataxin-3 fibrils obtained after 40 h of incubation at 37°C and imaged by EM to characterize their stiffness. The fibril persistence length Lp, a basic mechanical property that quantifies polymer flexibility, was calculated by decomposing the shape of the protofibrils into a sum of cosine waves of increasing frequency (or Fourier modes) (18). The images of protofibrils were digitized by subdividing the filaments into consecutive segments (Fig. S3 A). Their tangent angle was expressed as a sum of n cosine modes with amplitude an (Fig. S3, B and C). The amplitudes of the cosine waves for each mode are reported in Fig. S3 D. The fluctuation around the mean value of each mode amplitude, expressed as the variance var(an), provides an estimate for the persistence length Lp, which can be calculated as (18)
| (1) |
where Lc is the average contour length and n is the mode number. In the analysis, up to 22 modes were used for individual filaments. The variance, plotted against the mode number, has high values at low mode number and decreases for higher order modes (Fig. 1 D). This behavior is due to the fact that the variance is dominated by thermal fluctuations for lower order modes and by experimental noise for higher ones (18). As each mode provides an independent estimate of the persistence length, Lp values were calculated as an average of the values obtained for individual modes (Fig. 1 E). The analysis, carried out on 100 filaments, yielded a value for Lp of 15 ± 7 nm (Table S1).
Lp was also evaluated on the same subset of ataxin-3 fibrils by measuring end-to-end distances R (Fig. S4, A and B) and plotting the mean-squared end-to-end distances (〈R2〉) versus the corresponding Lc values (19) (Fig. 1 F). Two possible models were considered for data fitting, according to the following equations (19):
| (2) |
and
| (3) |
The model in Eq. 2 assumes that the fibrils reach conformational equilibrium in two dimensions when they are deposited on the substrate, whereas Eq. 3 assumes that the molecules do not equilibrate on the surface and adopt three-dimensional conformations that are orthogonally projected onto the substrate plane. Data fitting using Eqs. 2 and 3 yielded persistence length values of 21 ± 2 nm and 133 ± 33 nm, respectively. Because the latter Lp value exceeds the average contour length (126 ± 40 nm) of the fibrils analyzed, and this corresponds to the case of straight, rigid rods and does not describe flexible filaments, this is an indication that the three-dimensional model may not be suitable to fit our experimental data.
To validate this observation and conclusively discriminate between the two models, we considered the limit functions for Lc << Lp in each model: 〈R2〉2D = Lc2 and 〈R2〉3D proj = 2/3Lc2. These functions represent upper limits for 〈R2〉 values and are related to the limit case of straight fibrils, where end-to-end distances are equal to contour lengths. When the two limiting curves are compared with the squared end-to-end distance values obtained for ataxin-3 fibrils, the experimental values fall below the limit function of 〈R2〉2D but not that of 〈R2〉3Dproj (Fig. 1 F). We took this as a further indication that ataxin-3 fibrils reach equilibrium on the substrate and the data can be interpreted using the two-dimensional model but not the three-dimensional one, as also concluded for HypF-N fibrils (22).
The persistence lengths obtained using the decay of tangent correlations and end-to-end distances are thus in good agreement. If compared to Lp estimates obtained for other fibers, such as DNA (50 nm) (23), actin polymers (15 μm) (23), and insulin amyloid fibrils (42 μm) (24), the persistence length values measured for ataxin-3 fibrils indicate that they have great flexibility (Table S1). Importantly, these values show a good correlation with the dimensions of the spheroidal oligomers, thus suggesting that the oligomers are the precursors of elongated fibrils.
Using the average of the two persistence length estimates (18 ± 5 nm), the bending rigidity of ataxin-3 fibrils, defined as CB = kBT Lp (19), is (8 ± 2) × 10−29 N m2 (Table S1). The Young's modulus (E), or modulus of elasticity, describes the tendency of an object to deform when force is applied and is a measure of its stiffness. Defined as the ratio of the linear stress to the linear strain, E is related to Lp by the equation E = kBT Lp/I, where I is the cross-sectional moment of inertia (19). As a first approximation, we calculated E for ataxin-3 fibrils by assuming that the fibrils have simple, isotropic homogenous cylindrical structure. In this case, I = πr4/4, where r is the fibrils radius (18). Using an average value of 5 nm for r, obtained from measurements of fibrils diameters by EM and AFM height measurements, we estimated a value of 0.16 ± 0.18 MPa for the Young's modulus, which indicates low stiffness, when compared to other biological materials (e.g., E is 2–14 GPa for fully formed, highly regular amyloid fibrils (25) and 1–10 GPa for silk (26)).
Ataxin-3 and Josephin fibrils have a similar morphology and mechanical properties
To further investigate how Josephin contributes to the aggregation pathway of ataxin-3, we incubated samples of the isolated Josephin domain at 37°C under the same conditions used for ataxin-3. EM micrographs recorded after 48 h of incubation show that dispersed spheroidal particles are the main species observed (Fig. 2 A, upper panel). As for ataxin-3, the shape of the globules is roughly spherical, although the contours are somehow irregular and the particles do not look completely isomorphic. Short assemblies of two or three oligomers that appear linked together are shown (Fig. 2 A, lower panel). The diameters of these particles vary between 6 and 17 nm, with an average of 10 ± 2 nm (N = 1051) (Fig. 2 B and Table S1). The predominant species (corresponding to 61% of the total measured particles) is 9–11-nm wide. In agreement with the dimensions measured by EM, AFM images show the presence of spheroidal aggregates 5–15 nm high, with an average height of 8 ± 2 nm (N = 111) (Fig. 2 C and Table S1). The oligomers' average apparent width is 28 ± 5 nm (N = 17), with actual widths after deconvolution varying between 4 and 18 nm and an average value of 10 ± 3 nm.
Figure 2.
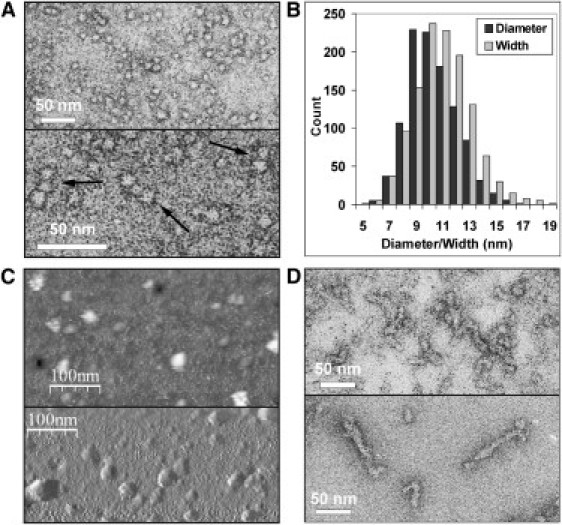
EM and AFM data of Josephin samples incubated at 37°C. Same concentration was 14 μM. (A) Electron micrographs of samples incubated for 48 h. (Arrows, lower panel) Assemblies of two or three globules that appear connected. (B) Distribution of Josephin spheroidal oligomer diameters and fibril widths measured in EM micrographs. The statistical analysis was performed on images obtained at all time points of incubation (2–15 days) in at least three separate experiments with different protein batches. (C) Similar morphology of Josephin oligomers, as observed in AFM images. The image in the lower panel was processed using a derivative filter to enhance edges. (D) Electron micrographs of samples incubated for three (upper panel) and six (lower panel) days.
After 72 h of incubation at 37°C, EM micrographs reveal a more heterogeneous composition of the samples, with spheroidal species present as well as elongated beaded assemblies that appear highly flexible (Fig. 2 D, upper panel). After six days, the fibrils elongate further, can form ringlike structures, and are sometimes branched (Fig. 2 D, lower panel).
Experiments performed using different Josephin batches showed some variability in protein aggregation kinetics and therefore in the species observed at different time points during incubation under the same conditions. Owing to the high tendency of Josephin to self-associate even at low temperature, small amounts of aggregated protein that are undetectable by SEC may be present at time zero in the Josephin stock solutions used. The differences observed in the kinetics can therefore be accounted for by seeding effects, as also reported for full-length ataxin-3 (11). Representative images of samples which aggregated with faster kinetics are shown in Fig. S5, A and B, recorded on samples incubated at 37°C for two and six days, respectively. Longer incubation times, up to 15 days, showed that no further changes are observed in fibril morphology and dimensions (see Fig. S5 C). Due to image resolution variability, a beaded morphology is more evident in certain subsets of images than in others. This could depend on the way the stain penetrates the samples on the EM grids.
Despite these differences, the aggregated species formed by all protein batches used in at least 15 separate incubation experiments have the same morphology and physical properties. In all analyzed samples, the average width of the elongated filaments is consistently 10 ± 2 nm (N = 1209), with values ranging from 5 to 19 nm and a predominance of species (51%) with granules 9–11 nm (Fig. 2 B and Table S1). In all cases, individual fibrils can appear tangled but do not assemble to form bundles with larger diameters. These observations support a model of ordered elongation at the two extremities only and correlate very well with those observed for ataxin-3 (Fig. 1 and Table S1), thus indicating that the isolated Josephin domain aggregates via a similar pathway to that of the full-length protein.
The distance between the bead centers in the fibers was measured in EM micrographs to obtain more detailed information on the fibrils structure. The values obtained for Josephin fibrils vary between 9 and 22 nm, with an average of 14 ± 2 nm (N = 251). Accordingly, the interbead average distances measured for ataxin-3 fibrils are between 9 and 23 nm, with an average value of 17 ± 4 nm (N = 40).
Analysis of the mechanical properties of Josephin fibrils was carried out on a representative subset of EM images obtained with samples incubated at 37°C for 48 h (Fig. S5 A). Distributions of contour lengths and end-to-end distances for 93 analyzed filaments are shown in Fig. S4, A and B. The persistence length values calculated using the decay of tangent correlations and end-to-end distances are, respectively, 15 ± 5 nm and 28 ± 3 nm (Fig. 1, D–F, Fig. S3 E, and Table S1). If we take the average of these values (22 ± 4 nm) and again assume that the fibrils have a simple, homogenous, cylindrical structure of average radius 5 nm (from EM measurements of fibril diameter and AFM height measurements), the estimate for the bending rigidity and Young's modulus of Josephin fibrils is CB = (9 ± 2) × 10−29 N m2 and E = 0.19 ± 0.18 MPa. These values indicate an unexpectedly high flexibility as observed for full-length ataxin-3. This close similarity is also evident from the plots of variance of mode amplitudes, persistence length, and squared end-to-end distances (Fig. 1, D–F).
Taken together, these data indicate that the morphological and mechanical features of ataxin-3 fibrils are largely influenced by the Josephin domain.
Effects of temperature on Josephin and ataxin-3 aggregate morphology
To investigate the effects of temperature on the morphological and mechanical properties of the aggregates, we also characterized ataxin-3 and Josephin samples incubated at 50°C. As expected, the increase of incubation temperature from 37 to 50°C has dramatic effects on self-association kinetics, with complete disappearance of monomeric proteins within 6 h, at a concentration of 14 μM (data not shown). Also at this temperature, however, EM micrographs recorded after 6 h of incubation show that ataxin-3 and Josephin samples are composed by aggregated species ranging from oligomers to elongated fibrils with a beaded morphology (Fig. 3, A and B). The diameters of the oligomers in EM micrographs vary between 7 and 16 nm (average 11 ± 2 nm, N = 102) for ataxin-3 and between 6 and 17 nm (average 11 ± 2 nm, N = 261) for Josephin, whereas the average width of the fibrils is 10 ± 2 nm (N = 296) and 11 ± 2 nm (N = 587) for ataxin-3 and Josephin, respectively (Table S2).
Figure 3.
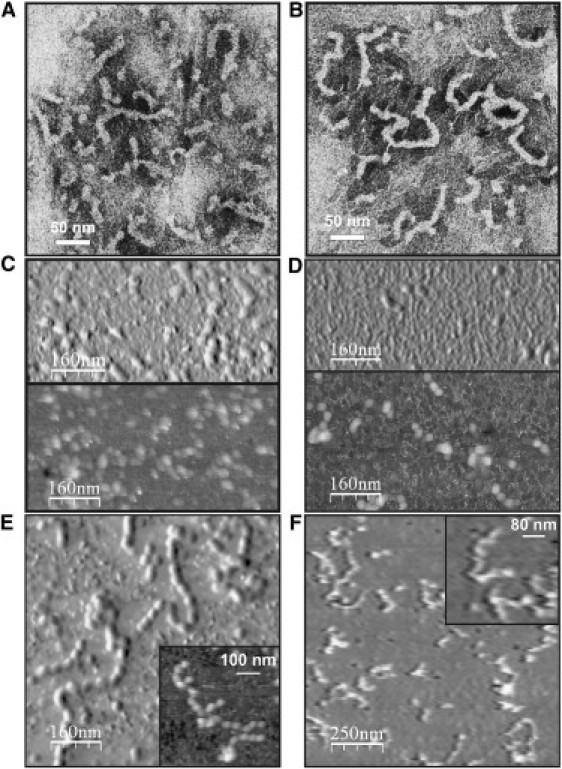
Fiber formation at 50°C. (A) Electron micrograph of ataxin-3 incubated at 50°C for 6 h. (B) Electron micrograph of Josephin incubated at 50°C for 6 h. (C) AFM images of ataxin-3 incubated at 50°C for 6 h. (D) AFM images of Josephin incubated at 50°C for 11 h. (E) AFM images of ataxin-3 incubated at 50°C for 11 h. (F) AFM images of Josephin heated to 80°C. (Upper panels in images C–E were processed with a derivative filter to enhance the edges.)
AFM measurements of samples incubated for 6–11 h at 50°C show the presence of spheroidal oligomers as well as flexible fibrils (Fig. 3, C–E). Height measurements yielded values of 6–12 nm (average 8 ± 1 nm, N = 61) for ataxin-3 oligomers, 4–10 nm (average 7 ± 1 nm, N = 45) for Josephin oligomers, and average values of 8 ± 1 nm (N = 71) and 7 ± 1 nm (N = 70) for ataxin-3 and Josephin fibrils, respectively, in agreement with EM observations (Table S2). The average interbead distances measured on AFM images, in which the beaded morphology is more easily observed, is 37 ± 7 nm (N = 47) and 29 ± 5 nm (N = 13) for ataxin-3 and Josephin fibrils, respectively. If we interpreted the data with the bead-on-string model, the fact that the measured interbead distances vary within wide ranges (24–50 nm for ataxin-3 and 24–36 nm for Josephin) could suggest the presence of a flexible linker between individual beads.
Josephin samples gradually heated to 80°C were also analyzed using AFM. In agreement with our previously reported EM data (10), Josephin forms fibrils also under these conditions (Fig. 3 F). The morphology and dimensions of these fibrils are closely similar to those of fibrils obtained at 37 and 50°C. The persistence length values estimated for 58 filaments using tangent correlations and end-to-end distances are, respectively, 19 ± 8 nm and 32 ± 4 nm, again in agreement with the values measured at 37°C.
Thus, temperature affects the kinetics of aggregation, but not the morphology of ataxin-3 and Josephin fibrils, that retain strikingly similar properties also when formed at 50°C and at higher temperature.
In the initial stages of aggregation, Josephin retains a nativelike secondary structure
To study how the secondary structure of Josephin evolves during aggregation, we recorded far-UV CD spectra of Josephin samples incubated at 37°C and analyzed the same samples using SEC and EM. SEC experiments, that monitor the conversion of monomeric Josephin into higher order species, were performed by measuring elution profiles of sample aliquots at different time points (Fig. 4 A). We observed a progressive decrease of the peak corresponding to monomeric Josephin (with a retention volume ∼11.4 mL) and the appearance of a peak corresponding to the column void volume (retention volume ∼7.7 mL), as previously described (15). According to the separation range of the column used, the aggregated species are estimated to have a molecular mass of ≥80 kDa and thus to be formed by four or more Josephin monomers, which have a molecular mass of 21.2 kDa.
Figure 4.
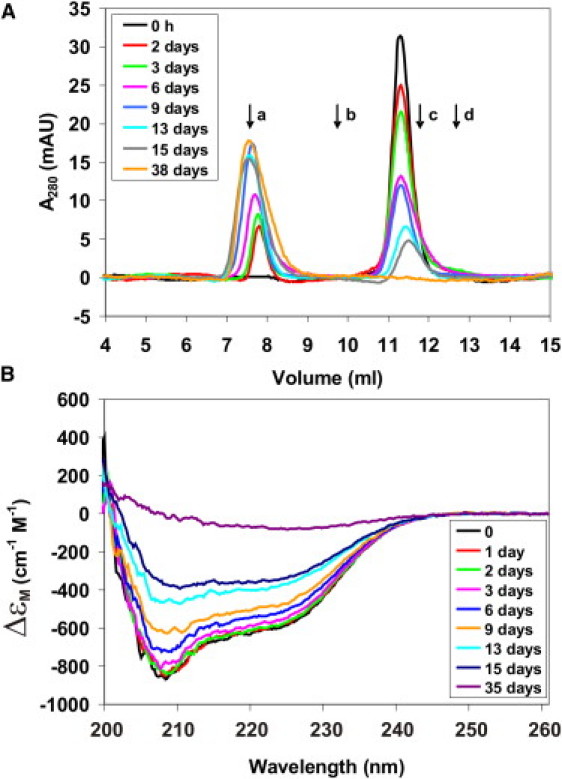
Josephin secondary structure changes upon aggregation. (A) SEC profiles and (B) far-UV CD spectra of 14 μM Josephin recorded at different time points during incubation at 37°C. (Arrows in A) Position of size markers: (a) Blue Dextran (7.79 mL); (b) Ovalbumin, 43 kDa (9.80 mL); (c) Chymotrypsinogen, 25 kDa (11.77 mL); and (d) Ribonuclease A, 13.7 kDa (12.81 mL). Note that Josephin (21.2 kDa) elutes as a protein with a slightly larger molecular mass because of its oblong shape.
Under the conditions used, ∼15% of the monodispersed protein has converted into aggregated species after two days of incubation as estimated from the SEC data. Far-UV CD data recorded within these two days, however, show virtually no differences in secondary structure (Fig. 4 B). After three days, small spectral variations are observed, that become progressively more evident in samples incubated for six days or longer. The changes consist in a decrease in signal intensity and in variations in the spectrum shape similar to those measured during Josephin aggregation at high temperature (10). The decrease in CD signal intensity is both attributable to secondary structure changes and to a loss of signal due to protein precipitation during aggregation. The CD signal results from the contribution of monomeric protein as well as soluble aggregates. This is confirmed by the observation that, after 15 days of incubation, SEC data report a loss of ∼78% of monomeric protein, whereas the reduction in CD signal total intensity at this time point corresponds to <50%. Spectral deconvolution (20) at different time points indicates a progressive loss of helical conformation and a concomitant increase in β-sheet structure. After 15 days of incubation, a decrease of up to 20% of helical content and an increase of up to 13% of β-sheet conformation are observed. EM micrographs reveal the presence of elongated, flexible fibrils (Fig. S5 C).
Comparison of far-UV CD, SEC, and EM data thus confirms that a significant structural α-to-β rearrangement occurs during fibril formation. However, during the initial stages of self-assembly, Josephin seems to retain a native or nativelike secondary structure, with significant changes occurring only at later aggregation stages.
Josephin fibrillar aggregates do not retain enzymatic function
To test whether Josephin enzymatic activity is retained during aggregation, we performed Ub cleavage assays using the linear substrate GST-Ub52, as previously described (27,28). Enzymatic reactions, monitored by SDS-PAGE, were performed at 37°C with two separate sets of Josephin samples: the first contained fresh protein, and the second, Josephin samples incubated at 37°C for 39 h. SEC profiles showed that, under the conditions used, fresh Josephin is entirely monomeric before the enzymatic reactions, whereas the protein incubated for 39 h completely aggregates into higher-molecular-weight species (Fig. S6 A). The fractions containing aggregated Josephin samples that eluted in the void volume of the column were collected. They did not show precipitation but analysis by EM confirmed the presence of short protofibrils. SDS-PAGE data indicate that the cleavage reaction with fresh Josephin samples is ∼20–30% complete after 3 h, under the conditions used (Fig. S6 B). The amount of cleaved substrate remained unchanged after 24 h at 37°C. A substantial proportion of the Josephin sample aggregated within this time lag, as also indicated by the significant amount of protein retained in the stacking gel.
In contrast, enzymatic reactions performed with Josephin samples incubated at 37°C for 39 h show hardly any cleavage after 3 h or 24 h and thus that activity is mostly lost (Fig. S6 B). Similarly, no substrate cleavage was observed in reactions performed with Josephin samples previously incubated at 50°C for 15 h, and shown by SEC to have completely converted into aggregated species (data not shown).
Discussion
The information currently available on the properties of the fibrillar aggregates formed by ataxin-3 is limited, despite the biological and medical interest of this protein. Here, we have carried out a systematic analysis under different experimental conditions of the morphological and mechanical properties of fibrils formed by an ataxin-3 isoform carrying a nonexpanded polyQ tract (18 glutamines) and compared them to those of the isolated Josephin domain. This is not the first time that ataxin-3 fibers both of expanded and nonexpanded ataxin-3 isoforms have been described (11,12,14), and our data are in excellent agreement with these reports. However, here we go much beyond these observations.
Using EM and AFM, we show that at 37°C ataxin-3 forms oligomers with diameters of ∼7–11 nm, which seem to further assemble into filaments. The fibrils retain a constant width of ∼10 nm and do not assemble further to form larger, mature fibers. Ataxin-3 fibrils formed at 50°C show remarkably similar properties. Although aggregation kinetics at this temperature are faster than at 37°C, no significant differences are observed in the morphology and dimensions of the aggregates. This suggests that aggregation at these two temperatures may occur via a similar assembly mechanism.
Here, we show for the first time, to our knowledge, that the features of the aggregated species of intact ataxin-3 are qualitatively and quantitatively reproduced by the isolated Josephin domain. The morphologies of the oligomers and fibrils formed by the full-length protein and by Josephin alone are remarkably similar both at 37 and 50°C. EM and AFM images show broadly consistent results, within the technical differences between the two methods and despite the fact that measurements of surface-immobilized fibrils do not necessarily reflect fibril structures solely determined by thermal fluctuations in solution. The identical trend observed when measuring all particle dimensions, with values determined by AFM consistently slightly lower than those estimated by EM, further demonstrates that the species analyzed have the same dimensions. The mechanical parameters describing the bending rigidity of the fibrils are also almost identical. Although we cannot exclude the possibility that adhesion of the particles to the different substrates (carbon-coated grids for EM and glass or mica for AFM) may introduce systematic artifacts, the consistency between EM micrographs of dried specimens and AFM images of fully hydrated samples supports our conclusions. These data are in agreement with our previous observations that ataxin-3 and isolated Josephin undergo analogous conformational changes coupled with aggregation, when incubated at different temperatures, from 25 to 85°C (data not shown and (10)). We conclude that the Josephin domain has a strong influence on the ataxin-3 aggregation pathway and on the shape and structure of ataxin-3 fibrils.
The morphology, dimensions, and persistence lengths measured for ataxin-3 and Josephin filaments are comparable to those of protofibrils formed by several other amyloidogenic proteins or peptides, such as α-lactalbumin (25), αB-crystallin (25), HypF-N (22), β-lactoglobulin (29), huntingtin (30,31), and β2-microglobulin (32,33). However, if compared to these systems, the stiffness measured for ataxin-3 and Josephin fibrils through our wormlike chain (WLC) analysis is surprisingly low, indicating a higher flexibility of the ataxin-3 fibers. For comparison, the values obtained for the Young's moduli (0.16–0.19 MPa) are roughly 2–3 orders-of-magnitude-lower than those measured for some of the protofibrils mentioned above (e.g., 60 MPa for HypF-N (22) and 0.14 GPa for α-lactalbumin (25)). These differences may reflect the implicit assumption of the WLC model that the filament is a homogeneous material. In contrast, based on our EM and AFM images, the filaments have an irregular morphology; the WLC model may thus not give an accurate-enough description of the system.
The observed structures could be interpreted either as a string of spheroidal beads or as a twisted planar sheet. The fact that we observe isolated spheroidal particles and small chains of these particles at early aggregation stages, and that the dimensions of the spheroids correlate with the width of the elongated fibrils make, for the time being, our preferred interpretation that the fibrils are composed of beads-on-a-string with mechanically stiff regions connected by flexible linkages. Such structures might then be better described by a freely-jointed-chain model, with the additional constraint that the connecting linkages exhibit steric hindrance in which each joint in the chain is not free to adopt all angular orientations. Under this assumption, we expect that the mechanical properties will be dominated by the length of the rigid elements and the angular freedom of the linker. This is consistent with what we observe: the filaments may be interpreted as being formed by relatively stiff protein beads with a diameter of ∼8–10 nm, and linkers to the next subunit with an angular freedom of ∼30–45°.
Josephin, the only structured region of ataxin-3, has been suggested to be the triggering element of ataxin-3 aggregation even in the presence of a polyQ-expansion (12,13): aggregation of both nonexpanded and expanded ataxin-3 has been shown to proceed via the same first step which involves the Josephin domain (11,12). Although nonexpanded ataxin-3 forms SDS-soluble fibrils, polyQ expansion induces a subsequent aggregation step in which these fibrils further assemble into large, highly stable, SDS-insoluble fibers (13). Recent evidence further supports these findings by showing that inhibition of Josephin self-association by a small heat-shock protein significantly slows down expanded ataxin-3 aggregation (34). These observations suggest that self-assembly of the Josephin domain within ataxin-3 plays a role in pathogenesis, and thus a better understanding of these processes is crucial to the rationalization of SCA3 disease mechanisms.
Analysis of the conformational changes coupled to Josephin aggregation indicates that, at the early stages of the aggregation pathway, Josephin retains a high secondary structure content and that a conversion of α-helical structure into a β-sheet-enriched conformation occurs only after aggregation starts. Similar structural changes were previously reported also during aggregation of ataxin-3 at 37°C (11). Enzymatic assays performed with samples that have undergone aggregation indicate that Josephin fibrils do not retain the proteolytic activity. This result is consistent with our previous data showing that the surface areas involved in ubiquitin binding include two hydrophobic patches that promote self-association (28,35). It can be expected that, when these regions are involved in the process of self-association, they become unavailable for target interaction.
Although further evidence is required to establish the exact temporal steps in which structural rearrangements take place along Josephin aggregation pathway, we can use the information available to suggest a working model for Josephin fibril formation and extend it to ataxin-3: the intrinsic tendency of Josephin to self-associate promotes the formation of the first intermediate species, involving the hydrophobic patches on its surface (Fig. 5). This is likely to be a dimeric form, because studies of the ataxin-3 aggregation mechanisms suggested that dimerization through the Josephin domain may be the first step in the process of self-assembly (11).
Figure 5.
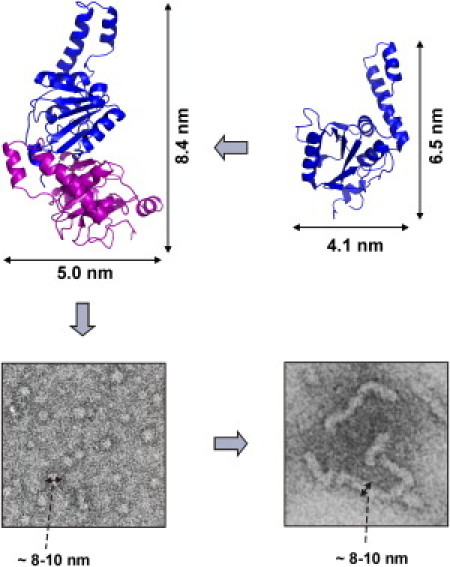
Schematic representation of Josephin fibril formation pathway. Monomeric Josephin (PDB entry: 1YZB) self-associates into dimeric structures (11), which further aggregate into spheroidal oligomers ∼8–10-nm wide. We propose that the globules could then assemble linearly to give rise to elongated, flexible fibrils. A conformational transition to a β-rich structure is observed after the initial aggregation stages. The dimer model displayed here is only representative, but was formulated to accommodate the notion that aggregation involves enzymatically important regions. Increase in β-structure content could be initially achieved by pairing-up β-strands from different monomers. Approximate dimensions of the Josephin monomer and of the dimer are indicated.
A Josephin dimer or a small multiple of it is of a size compatible with the oligomers we observe by EM and AFM. The oligomers may then further assemble to form the elongated fibril. As in full-length ataxin-3 Josephin accounts approximately for half of the molecule; the remaining flexible C-terminal portion (16) may pack against the rest of the molecule within the oligomers, decorate the fibrils by sticking out in solution, or contribute to the formation of flexible linkers. Further progress in characterizing the fibrilization pathway of ataxin-3 will provide important clues for the rationalization of drugs that are able to inhibit the aggregation process.
Acknowledgments
This work was supported by the Medical Research Council, UK (grant No. U117584256).
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Contributor Information
Justin Molloy, Email: jmolloy@nimr.mrc.ac.uk.
Annalisa Pastore, Email: apastor@nimr.mrc.ac.uk.
Supporting Material
References
- 1.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Fowler D.M., Koulov A.V., Kelly J.W. Functional amyloid—from bacteria to humans. Trends Biochem. Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Scheibel T. Protein fibers as performance proteins: new technologies and applications. Curr. Opin. Biotechnol. 2005;16:427–433. doi: 10.1016/j.copbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Cummings C.J., Zoghbi H.Y. Trinucleotide repeats: mechanisms and pathophysiology. Annu. Rev. Genomics Hum. Genet. 2000;1:281–328. doi: 10.1146/annurev.genom.1.1.281. [DOI] [PubMed] [Google Scholar]
- 5.Chen S., Berthelier V., Wetzel R. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J. Mol. Biol. 2001;311:173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- 6.Bulone D., Masino L., Pastore A. The interplay between PolyQ and protein context delays aggregation by forming a reservoir of protofibrils. PLoS ONE. 2006;1:e111. doi: 10.1371/journal.pone.0000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.La Spada A.R., Taylor J.P. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson A.L., Bottomley S.P. Towards the treatment of polyglutamine diseases: the modulatory role of protein context. Curr. Med. Chem. 2010;17:3058–3068. doi: 10.2174/092986710791959800. [DOI] [PubMed] [Google Scholar]
- 9.Marchal S., Shehi E., Lange R. Structural instability and fibrillar aggregation of non-expanded human ataxin-3 revealed under high pressure and temperature. J. Biol. Chem. 2003;278:31554–31563. doi: 10.1074/jbc.M304205200. [DOI] [PubMed] [Google Scholar]
- 10.Masino L., Nicastro G., Pastore A. Characterization of the structure and the amyloidogenic properties of the Josephin domain of the polyglutamine-containing protein ataxin-3. J. Mol. Biol. 2004;344:1021–1035. doi: 10.1016/j.jmb.2004.09.065. [DOI] [PubMed] [Google Scholar]
- 11.Gales L., Cortes L., Macedo-Ribeiro S. Towards a structural understanding of the fibrilization pathway in Machado-Joseph's disease: trapping early oligomers of non-expanded ataxin-3. J. Mol. Biol. 2005;353:642–654. doi: 10.1016/j.jmb.2005.08.061. [DOI] [PubMed] [Google Scholar]
- 12.Ellisdon A.M., Thomas B., Bottomley S.P. The two-stage pathway of ataxin-3 fibrillogenesis involves a polyglutamine-independent step. J. Biol. Chem. 2006;281:16888–16896. doi: 10.1074/jbc.M601470200. [DOI] [PubMed] [Google Scholar]
- 13.Ellisdon A.M., Pearce M.C., Bottomley S.P. Mechanisms of ataxin-3 misfolding and fibril formation: kinetic analysis of a disease-associated polyglutamine protein. J. Mol. Biol. 2007;368:595–605. doi: 10.1016/j.jmb.2007.02.058. [DOI] [PubMed] [Google Scholar]
- 14.Bevivino A.E., Loll P.J. An expanded glutamine repeat destabilizes native ataxin-3 structure and mediates formation of parallel β-fibrils. Proc. Natl. Acad. Sci. USA. 2001;98:11955–11960. doi: 10.1073/pnas.211305198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masino L., Nicastro G., Pastore A. Functional interactions as a survival strategy against abnormal aggregation. FASEB J. 2011;25:45–54. doi: 10.1096/fj.10-161208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masino L., Musi V., Pastore A. Domain architecture of the polyglutamine protein ataxin-3: a globular domain followed by a flexible tail. FEBS Lett. 2003;549:21–25. doi: 10.1016/s0014-5793(03)00748-8. [DOI] [PubMed] [Google Scholar]
- 17.Graveland-Bikker J.F., Schaap I.A., de Kruif C.G. Structural and mechanical study of a self-assembling protein nanotube. Nano Lett. 2006;6:616–621. doi: 10.1021/nl052205h. [DOI] [PubMed] [Google Scholar]
- 18.Gittes F., Mickey B., Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivetti C., Guthold M., Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 20.Sreerama N., Woody R.W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 21.Holowaty M.N., Sheng Y., Frappier L. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J. Biol. Chem. 2003;278:47753–47761. doi: 10.1074/jbc.M307200200. [DOI] [PubMed] [Google Scholar]
- 22.Relini A., Torrassa S., Gliozzi A. Detection of populations of amyloid-like protofibrils with different physical properties. Biophys. J. 2010;98:1277–1284. doi: 10.1016/j.bpj.2009.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howard J. Sinauer Associates; Sunderland, MA: 2001. Mechanics of Motor Proteins and the Cytoskeleton. [Google Scholar]
- 24.Smith J.F., Knowles T.P., Welland M.E. Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2006;103:15806–15811. doi: 10.1073/pnas.0604035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knowles T.P., Fitzpatrick A.W., Welland M.E. Role of intermolecular forces in defining material properties of protein nanofibrils. Science. 2007;318:1900–1903. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- 26.Vollrath F., Knight D.P. Liquid crystalline spinning of spider silk. Nature. 2001;410:541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- 27.Chow M.K., Mackay J.P., Bottomley S.P. Structural and functional analysis of the Josephin domain of the polyglutamine protein ataxin-3. Biochem. Biophys. Res. Commun. 2004;322:387–394. doi: 10.1016/j.bbrc.2004.07.131. [DOI] [PubMed] [Google Scholar]
- 28.Nicastro G., Menon R.P., Pastore A. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc. Natl. Acad. Sci. USA. 2005;102:10493–10498. doi: 10.1073/pnas.0501732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gosal W.S., Clark A.H., Ross-Murphy S.B. Fibrillar β-lactoglobulin gels. Part 1. Fibril formation and structure. Biomacromolecules. 2004;5:2408–2419. doi: 10.1021/bm049659d. [DOI] [PubMed] [Google Scholar]
- 30.Poirier M.A., Li H., Ross C.A. Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization. J. Biol. Chem. 2002;277:41032–41037. doi: 10.1074/jbc.M205809200. [DOI] [PubMed] [Google Scholar]
- 31.Díaz-Hernández M., Moreno-Herrero F., Lucas J.J. Biochemical, ultrastructural, and reversibility studies on huntingtin filaments isolated from mouse and human brain. J. Neurosci. 2004;24:9361–9371. doi: 10.1523/JNEUROSCI.2365-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gosal W.S., Morten I.J., Radford S.E. Competing pathways determine fibril morphology in the self-assembly of β2-microglobulin into amyloid. J. Mol. Biol. 2005;351:850–864. doi: 10.1016/j.jmb.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 33.Kad N.M., Myers S.L., Thomson N.H. Hierarchical assembly of β2-microglobulin amyloid in vitro revealed by atomic force microscopy. J. Mol. Biol. 2003;330:785–797. doi: 10.1016/s0022-2836(03)00583-7. [DOI] [PubMed] [Google Scholar]
- 34.Robertson A.L., Headey S.J., Bottomley S.P. Small heat-shock proteins interact with a flanking domain to suppress polyglutamine aggregation. Proc. Natl. Acad. Sci. USA. 2010;107:10424–10429. doi: 10.1073/pnas.0914773107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicastro G., Masino L., Pastore A. Josephin domain of ataxin-3 contains two distinct ubiquitin-binding sites. Biopolymers. 2009;91:1203–1214. doi: 10.1002/bip.21210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


