Abstract
Septins are conserved GTPases that form filaments and are required for cell division. During interphase, septin filaments associate with cellular membrane and cytoskeleton networks, yet the functional significance of these associations have, to our knowledge, remained unknown. We recently discovered that different septins, SEPT2 and SEPT11, regulate the InlB-mediated entry of Listeria monocytogenes into host cells. Here we address the role of SEPT2 and SEPT11 in the InlB-Met interactions underlying Listeria invasion to explore how septins modulate surface receptor function. We observed that differences in InlB-mediated Listeria entry correlated with differences in Met surface expression caused by septin depletion. Using atomic force microscopy on living cells, we show that septin depletion significantly reduced the unbinding force of InlB-Met interaction and the viscosity of membrane tethers at locations where the InlB-Met interaction occurs. Strikingly, the same order of difference was observed for cells in which the actin cytoskeleton was disrupted. Consistent with a proposed role of septins in association with the actin cytoskeleton, we show that cell elasticity is decreased upon septin or actin inactivation. Septins are therefore likely to participate in anchorage of the Met receptor to the actin cytoskeleton, and represent a critical determinant in surface receptor function.
Introduction
Originally identified in Saccharomyces cerevisiae as required for septum formation and cell division, septins form a family of GTPases among fungi and animals sharing a conserved role in cytokinesis (1). In mammals, septins are recognized for a variety of other essential cellular functions including roles in secretion, membrane remodeling, and cytoskeleton dynamics (1). Several human pathological conditions, including cancer (2), hereditary neuralgic amyotrophy (3), and Parkinson's disease (4), have been linked to septin dysfunction. Despite these important implications, the molecular functions of septins remain elusive, in part because the nature of septin complex assembly is only beginning to be understood (5,6). There are 14 human septin genes which are classified into four different groups based on sequence identity (6). Septins from different groups polymerize into heterooligomeric protein complexes and nonpolar filaments (7), and associate with cellular membranes, actin filaments, and microtubules (8,9). Thus, septins are increasingly regarded as an unconventional cytoskeletal element (10). Yet how septins function with the cytoskeleton remains to be determined.
The study of bacterial entry has been instrumental to uncover features of the cytoskeleton, and is one of the best examples of how understanding a bacterial-induced process can yield insight into basic cellular processes (11,12). A potential role for septins in the entry of bacteria originally emerged from a proteomic analysis of phagosomes containing latex beads coated with Listeria monocytogenes invasion proteins (13). Of the several bacterial surface proteins that participate in Listeria invasion (14), the two best characterized are InlA and InlB, either of which is independently sufficient to promote bacterial internalization into mammalian epithelial cells that express its respective receptor. InlA interacts with the surface protein E-cadherin (15), whereas InlB recognizes the receptor tyrosine kinase Met (16). These different interactions have well-described cell signaling consequences, including actin polymerization and membrane extension, which ultimately lead to bacterial uptake (17).
Septins have been implicated in a variety of membrane organization events including phagocytosis (8,18,19), and our previous work using septin siRNA has specifically shown that SEPT2 and SEPT11 regulate the InlB-mediated internalization process used by Listeria (20,21). Taken together, these data suggest a role for SEPT2 and SEPT11 on the properties of Met that, upon interaction with InlB, are critical for membrane extension and bacterial entry. In this study we use a combination of approaches, including flow cytometry, high content analysis (HCA) microscopy, and atomic force microscopy (AFM), to characterize the role of SEPT2 and SEPT11 in regulating properties of InlB-Met interaction, and reveal that septins coordinate Met receptor function at the surface of living cells.
Materials and Methods
Mammalian cells and culture conditions
HeLa human cervix carcinoma cells (ATCC CCL-2; American Type Culture Collection, Manassas, VA) were cultured in Modified Eagle's Medium plus GlutaMAX (GIBCO, Invitrogen, Carlsbad, CA) supplemented with 1 mM sodium pyruvate (GIBCO), 0.1 mM nonessential amino acid solution (GIBCO), and 10% fetal calf serum. Cells were grown at 37°C in 10% CO2 atmosphere.
Antibodies and reagents
Mouse monoclonal anti-human Met (clone DL-21) was purchased from Upstate Biotechnology (Lake Placid, NY). Secondary antibodies used were Cy5-conjugated (Jackson ImmunoResearch Laboratories, West Grove, PA) or Alexa 488-conjugated goat anti-mouse antibodies (Molecular Probes, Eugene, OR).
RNA interference and pharmacological inhibitors
HeLa cells (0.8 × 105) were plated in six-well plates (TPP, Trasadingen, Switzerland) and transfected the following day using oligofectamine (Invitrogen) following the manufacturer's instructions. The following scramble sequence, custom-designed from septin sequence,
AUAAGCGACGUCCGCGUGGtt (sense) and
CCACGCGGACGUCGCUUAUtt (antisense),
was applied as our control throughout experimentation. The custom scramble sequence, as well as predesigned siRNAs for SEPT2 (ID No. 14709) and SEPT11 (ID No. 125139), were all from Ambion (Naugatuck, CT) and taken from previous publications (20–22). Cells were tested 72 h after siRNA transfection.
Experiments involving latrunculin B or nocodazole were performed as described for other in vitro systems (21), and HeLa cells were treated for 60 min before interrogation with dimethyl sulfoxide, latrunculin B (100 nM), or nocodazole (10 μM). Latrunculin B and nocodazole were suspended in dimethyl sulfoxide and handled as suggested by the manufacturer (Sigma, St. Louis, MO).
Protein production and purification
InlB, coupled to a NH2-terminal 6× His tag, was purified as described previously (23). We used a soluble HGF/Met receptor generated by engineering a recombinant protein corresponding to the entire extracellular domain of Met, truncated before the transmembrane domain, i.e., decoy-Met (24). A myc-epitope tag and a poly-histidine tag were added at the C-terminus and the cDNA was subcloned into the pRRLsin.PPT.CMV.Wpre lentiviral vector (24). Lentivirus was produced using the pMD2G and pR87.4 plasmid Kit transfected in 293T cells (25). Proteins were recovered from the supernatant and purified on a NiNTA column (Qiagen, Valencia, CA) according to manufacturer's instructions, and protein quality was controlled by sodium-dodecyl-sulfate-polyacrylamide-gel-electrophoresis silver stain.
Flow cytometry
siRNA-treated HeLa cells were washed twice in 1× phosphate-buffered saline (PBS) then detached with 0.05% EDTA in PBS. Single cell suspensions were adjusted at 106c/100 μL in PBS-staining buffer containing 2% bovine serum albumin, 0.05% NaN3, and 10% fetal calf serum, and incubated with anti-Met or isotype control (Molecular Probes) for 30 min at 4°C. After this time, cells were washed twice and incubated with Alexa 488-conjugated secondary antibodies for 30 min at 4°C. To quantify levels of total Met (intracellular plus cell surface), cells were permeabilized with Cytoperm buffer (BD Pharmingen, BD Biosciences, Franklin Lakes, NJ) before staining. Samples were analyzed using a FACSCalibur instrument (BD Biosciences). Dead cells were excluded based on forward and side scatter, and a minimum of 10,000 events was acquired per sample. The isotype control showed no positive staining on HeLa cells (data not shown). Results were analyzed using FlowJo software (http://www.flowjo.com/). Data represents at least three biological replicates per experiment. Results were analyzed for statistical significance using the Student's t-test.
High-content analysis microscopy
HeLa cells, transfected or not with the siRNA as described above, were plated on Falcon 96-wells plates. Cells were permeabilized or not, fixed, DAPI-stained, and incubated with anti-Met antibodies followed by Alexa 488-conjugated secondary antibodies. Data were acquired using the Operetta High-content screening system (PerkinElmer, Waltham, MA) and analyzed using the Acapella software (PerkinElmer). Cell area was analyzed on 96-wells plates, and Met expression was analyzed in quadruplicate/plate. Nine fields were recorded for quantitative analysis from each well. Raw values were obtained from a minimum of 45,000 cells per siRNA treatment for morphology assays, and 10,000 cells for immunocytochemistry assays. Results were analyzed for statistical significance using the Student's t-test.
Atomic force microscopy
Cells were grown on glass-bottom WillCo-dish plates (WillCo Wells, Amsterdam, The Netherlands) and when transfected oligofectamine (Invitrogen) was used. As a control for transfection, cells were cotransfected with the siGLO green indicator (Dharmacon, Lafayette, CO) and GFP positive cells were analyzed by AFM. All the experiments were performed at room temperature on a commercial stand-alone AFM (Bioscope II; Veeco, Santa Barbara, CA) combined with either an inverted optical microscope (Axiovert 200M; Zeiss, Jena, Germany) coupled to a Coolsnap ES camera (Roper Scientific, Tucson, AZ) driven under MetaMorph (Molecular Devices) software or an inverted optical microscope (TE 2000; Nikon, Melville, NY) with a DSQi1Mc camera (Nikon). AFM was operated in Modified Eagle's Medium buffer in contact mode or force-volume mode using Bio-Lever (imaging experiments) (Olympus, Melville, NY), Veeco DNP (force measurements), or Veeco MLCT (pulling and loading rate experiments).
Detailed descriptions of the AFM methods used for live cell imaging, surface Met quantification experiments, unbinding force and tether experiments, and elasticity experiments are provided in the Supporting Material. Where mentioned, statistical analysis was performed using R (Ver. 2.10, R-Project, http://cran.r-project.org/bin/windows/base/) and GraphPad Prism software (http://www.graphpad.com/prism/prism.htm).
Results
Septin depletion and cell shape change
To investigate how septins can regulate the bacterial internalization process, we performed our functional analysis on HeLa cells which do not express E-cadherin and thus allow the specific analysis of the InlB-Met entry pathway used by Listeria. We first considered the shape of noninfected HeLa cells upon siRNA treatment. HeLa cell shape can be differently affected depending on the septin targeted for depletion. Specifically, SEPT2-depleted cells appear compact and irregular in shape, whereas SEPT11-depleted cells appear large and flat (20,21). To obtain a three-dimensional representation of morphological changes upon septin depletion, the morphology of control, SEPT2-, and SEPT11-depleted cells was examined using AFM. AFM could define how siRNA-treated cells differed markedly in terms of cell shape and overall height distribution (Fig. 1 A). Most striking was the height variation in perinuclear space: compared to control cells, SEPT11-depleted cells were flat, whereas SEPT2-depleted cells were swollen throughout.
Figure 1.
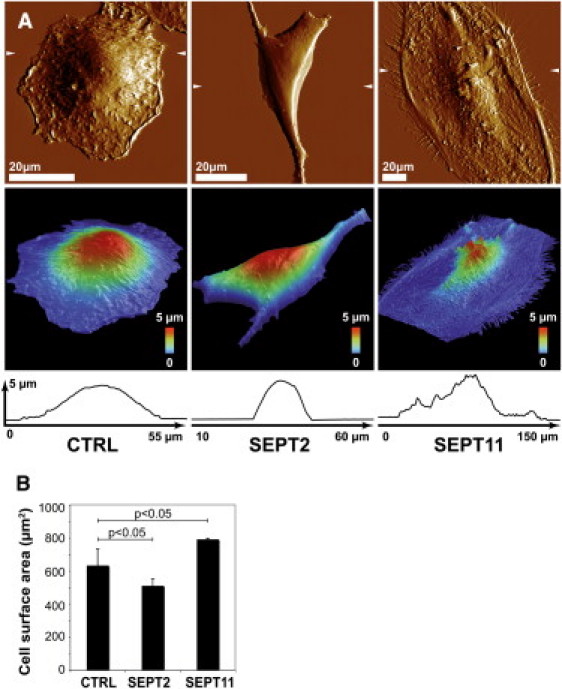
The impact of septin-depletion on cell shape. (A) AFM was used to examine cell shape and overall height of control (CTRL), SEPT2-, or SEPT11-depleted cells. (Top panels) AFM deflection images of cells recorded in contact mode at constant force. (Middle panels) Three-dimensional view of the height image with a pseudocolor scale ranging from low (blue) to high (red). (Arrows, top panels) Position of the cross-section profiles made on the height (bottom panels). One image representative of five is shown for each condition. (B) HCA microscopy was used to measure cell surface area of adherent CTRL (n = 98,356), SEPT2- (n = 156,823), or SEPT11- (n = 119,702) depleted cells. p-values, Student's t-test.
High content analysis (HCA) is an automated platform for performing fluorescence microscopy and quantitative image analysis that combines the efficiency of high-throughput techniques with multiplexed functional analysis applied to large samples of cells (26). To quantify the changes in cell shape upon septin-depletion, we performed HCA microscopy and measured the cell-projected surface area of adherent siRNA-treated cells. On average, the surface area for SEPT2-depleted cells was significantly smaller than control cells, whereas the surface area for SEPT11-depleted cells was significantly larger than control cells (Fig. 1 B). These results, obtained from a much larger sample of cells, are in accordance with previous results obtained by quantitative microscopy where the cell-occupied surface area of septin-depleted cells was measured after fixation (21).
It has been reported that septin depletions result in wider and flatter cells (27,28). Our data extend those findings, showing that septin depletions can affect overall cell shape, highlighting the particular role for septins in cell architecture.
Septins regulate the level and distribution of surface Met
Our previous work has shown that septins differentially modulate the InlB-Met-mediated entry of L. monocytogenes. As compared to control cells, SEPT2-depleted cells are less permissive for entry (20), whereas SEPT11-depleted cells allow increased entry (21). We thus investigated the expression level of the InlB receptor Met in septin-depleted cells by flow cytometry. siRNA-treated cells were permeabilized or not before fluorescence labeling to quantify total cellular Met (i.e., intracellular plus cell surface) or only cell surface Met. As compared to control cells, SEPT11-depleted cells presented higher amounts of both total and cell surface Met, while SEPT2-depleted cells presented lower amounts (Fig. 2 A). These differences, as expected from the morphological studies presented above (Fig. 1 B), were confirmed using immunofluorescence assays analyzed by HCA microscopy (Fig. 2 B).
Figure 2.
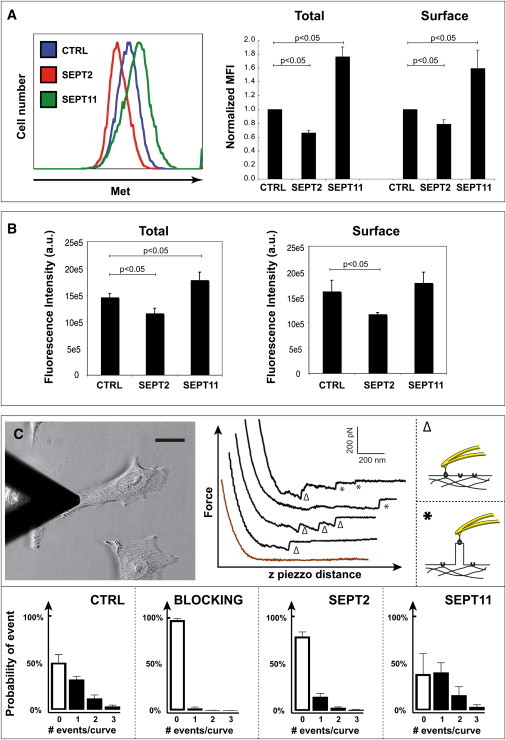
Septins regulate the level of Met protein. (A, left panel) Flow cytometry representative curves plot the intensity of total Met fluorescence (intracellular plus cell surface) for control (CTRL), SEPT2-, or SEPT11-depleted cells, where at least 10,000 events were acquired per sample. (Right panel) Median fluorescent intensity (MFI) ratios per sample were normalized by comparison with CTRL-transfected cells. (Graph) CTRL siRNA is figuratively presented as 1, and therefore does not have error bars, whereas the normalized MFI in septin-depleted cells are a relative value. Data represent the mean normalized MFI ratio mean ± SE from n ≥ 3 independent experiments for total (left histogram) and cell surface (right histogram) Met per siRNA treatment. p-values, Student's t-test. (B) HCA microscopy was used to measure the expression level of total (left panel) or surface (right panel) Met for CTRL, SEPT2-, or SEPT11-depleted cells. Raw values were obtained from four experiments per siRNA treatment. p-values, Student's t-test. (C) AFM force curve analysis of the InlB-Met interaction on living cells. (Top left panel) A tip functionalized with InlB is used to probe the cell surface (scale bar = 15 μm). (Top right panels) Representative examples of retraction force curves recorded at the surface of CTRL cells are shown. The curves displayed are (from bottom to top): no event (brown), single or multiple unbinding events, single and multiple tethers. Unbinding events are shown by Δ for V-shaped unbinding events and tethers are shown by asterisk (∗) for plateau events; probability of events are shown for cells treated with CTRL siRNA either with (i.e., blocking) or without InlB used as competitive inhibitor to demonstrate the specificity of the binding, and for cells treated with SEPT2 or SEPT11 siRNA. (Bar plots) Probability of having 0, 1, 2, or 3 events per retraction curve.
We used AFM to quantify the distribution of Met receptors at the surface of live siRNA-treated cells. An AFM piezo was applied to bring an InlB-coated cantilever into contact with living cells, and InlB-Met interaction events were detected by a specific software designed to analyze the force distance curves (see the Supporting Material). Here, 48 ± 7% of all the force curves presented a characteristic vertical force jump for cells treated with control siRNA (Fig. 2 C).
When competition experiments were performed using InlB added in the culture medium, 2.1 ± 1.3% of unbinding events were observed, demonstrating that the unbinding events analyzed were specific. Most of the force curves displayed single unbinding events, and multiple events were occasionally observed (Fig. 2 C). As compared to control cells, SEPT2-depleted cells presented a reduced number of InlB-Met interaction events (20 ± 4%), and SEPT11-depleted cells had an increased number of InlB-Met interaction events (60 ± 16%) (Fig. 2 C). The distribution maps did not reveal any impact of septin depletion on the clustering of Met (see Fig. S1 in the Supporting Material). Thus, AFM data obtained using live cells were in agreement with the flow cytometry and HCA data obtained using fixed cells (Fig. 2, A and B).
Taken together, different experimental techniques all converge to show that, as compared to control cells, SEPT11-depleted cells present higher amounts of Met available per cell surface unit, and SEPT2-depleted cells present lower amounts. These results can also be correlated to the macroscopic topology of cells as observed in Fig. 1 A, where SEPT2-depleted cells appear smoother than control or SEPT11-depleted cells. Therefore differences in Met availability at the surface of septin-depleted cells revealed by flow cytometry, HCA, and AFM correlate with patterns of Listeria infection (20,21). Because the Met receptor available at the cell surface is modulated by septin depletion, we focused on how septins could regulate the InlB-Met interaction.
Septin depletion reduces the unbinding force of InlB-Met interaction
We used AFM to characterize the interaction force between InlB and Met. As demonstrated from force profiles analyzed in other systems (29), AFM using functionalized tips allows measurements of the interaction forces between a ligand (i.e., InlB) and a receptor (i.e., Met). We measured unbinding forces between InlB coated on the cantilever tip and Met endogenously expressed at the surface of living cells at various loading rates ranging from 0.1 to 100 nN s−1. Dynamic force spectroscopy of the InlB-Met interaction was obtained by plotting the unbinding force as a function of the logarithm of the loading rate (Fig. 3 A). At each pulling velocity tested, a minimum of 1000 curves was recorded and analyzed.
Figure 3.
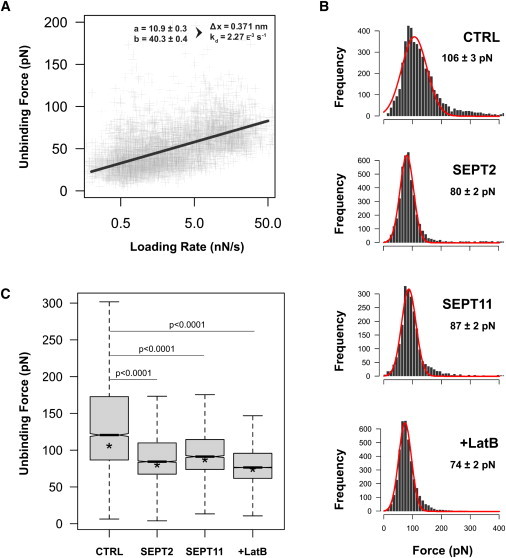
Septins regulate the interaction of InlB-Met. (A) The unbinding force (pN) increases linearly with the loading rate (nN s−1). Force and loading rates are calculated automatically for each unbinding event. The fit gives a width for the energy barrier Δx = 0.371 nm and a dissociation rate constant of k0d = 2.3 × 10−3 s−1. (B, Force histograms) Control (CTRL), SEPT2-depleted, SEPT11-depleted, or latrunculin B-treated unbinding forces. Force distribution is fitted by a Gaussian curve with peak and error values indicated. (C, Box plot) Unbinding forces. Septin-depletion, or actin inhibition by latrunculin B, reduces the interaction force between InlB and Met. (Lines) Medians and asterisk (∗) shows peak positions of Fig. 3B. p-values, Mann-Whitney test.
Unbinding of a ligand from a receptor has been widely analyzed using the Bell model and then by the Bell-Evans model (30,31) (see the Supporting Material). We investigated the energy involved in the InlB-Met complex by performing a dynamic force spectroscopy experiment recording unbinding forces as a function of the loading rate. The loading rate defines the rate at which two molecules are pulled away, in N s−1. It is calculated as
Strikingly, one regime was detected for the InlB-Met interaction with the mean unbinding force depending logarithmically on the loading rate. From these data, we could determine a dissociation rate constant
(Fig. 3 A, and see the Supporting Material). Using previous work that has reported a 20–30 nM dissociation constant (KD) for the InlB-Met interaction (32,33), we could calculate an association rate constant
on living cells.
To analyze directly the InlB-Met interaction, we performed intermolecular dynamic force spectroscopy using purified Met. We purified the extracellular domain of Met that carried a poly-histidine tag on the C-terminus, i.e., decoy-Met (24), and coated the purified protein on the substratum using covalent oriented linkage (34) (see Materials and Methods). To ensure that correct coating by the protein was obtained, we scanned 3 μm2 of the surface using 0.06 N/m sharp nitride lever cantilevers (Veeco) in contact mode (Fig. S2 A). The 1-μm2 surface was then scoured with the AFM tip by increasing speed and force. Height profiles showed that the protein layer reached ∼8 nm, which is in agreement with the structure information known for Met (35). Force curves were then acquired using the force-volume mode (Fig. S2 B). To control the specificity of the interaction, InlB was added to the medium. We found a dramatic decrease of the interactions from 80 ± 5% to 8 ± 2% of the curves. Fig. S2 C shows dynamic force spectroscopy analysis of the InlB-Met interaction by varying the loading rate. We found a dissociation rate constant
(Fig. S2 C). We thus found a decrease of the KD for the InlB-Met interaction when experiments were performed in cells versus experiments performed with purified molecules, a feature that has already been noticed (36). Our results from experiments performed with cells and with purified molecules are compatible with the unbinding force required to overcome one energy barrier (Fig. S2 D).
We then explored the InlB-Met interaction force in siRNA-treated cells. A quantity of 25 × 105 curves were analyzed per conditions and specific events were collected, analyzed, and plotted in a force histogram (Fig. 3 B). A clear peak arose from this distribution, and results were analyzed to yield a mean value ± SE. (Fig. 3 C). Based on these profiles, control cells presented a strong population of Met receptors with a defined rupture force peak of 106 ± 3 pN. For SEPT2-depleted cells, a clear rupture force peak centered significantly lower at 80 ± 2 pN (Fig. 3 B). Similar results were obtained for SEPT11-depleted cells (87 ± 2 pN). These results show that septin-depletion dramatically decreases the mean interaction force between the InlB-coated tip and cell surface Met.
Because these AFM measurements were obtained from living cells, the impact of septin depletion on the interaction between InlB and Met could reflect several variables including receptor anchorage to the plasma membrane, membrane tension and bending, and possible regulation by the cytoskeleton. The individual contribution of these variables is currently difficult to determine. To address the role of actin in the regulation of InlB-Met interaction, we performed AFM experiments in which cells were treated with latrunculin B, an inhibitor of actin polymerization. Treatment of cells by latrunculin B significantly decreased the unbinding force of InlB-Met interaction to the same level observed from septin depletion (74 ± 2 pN; Fig. 3, B and C). These data are in agreement with the proposed role for septins in modulation of the actin network (9,10). Thus, results performed using AFM clearly demonstrate that septin-depletion, like inactivation of the actin cytoskeleton, significantly reduces the unbinding force of InlB-Met interaction.
Septin depletion reduces membrane tethers
To investigate the role of septins and actin in InlB-Met interaction, we exploited an AFM approach that measures the formation of membrane tethers, i.e., long, thin (tens of nm), extendable cylindrical membrane tubes that stretch from the cell surface to the AFM tip as it retracts (37). Living-cell membrane tethering has been investigated using different techniques including micropipettes, laser optical tweezers, and AFM (37,38), and the dilatation extension of the membrane that is likely to occur under the AFM tip can be compared to the dilatation of the cytoskeleton during micropipette aspiration (39). AFM has been used recently to follow membrane tether formation with another bacterial adhesin-cell surface receptor pair, i.e., the heparin-binding hemagglutinin-heparan sulfate proteoglycan on A549 cells (40). Here we used a similar approach considering the plateau events occurring from 400 nm distance and the InlB-Met unbinding when applying an extraction force (see asterisk in Fig. 2 C).
We analyzed whether septin-depletion could affect the detachment of the tensed Met receptor-associated membrane from the cytoskeleton, leading to the extraction of membrane tethers. The threshold force for tether extraction corresponds to the minimal force required to extract a membrane tether from the plasma membrane, and is mainly determined by the adhesion energy between the membrane and cytoskeleton. This is defined by the membrane tension, the membrane-binding stiffness, and the membrane-cytoskeleton association energy (41,42) (Fig. 4 A, and see the Supporting Material). The threshold force was determined, considering 400–600 events per time point, as 46 ± 2 pN for control cells (Fig. 4 B). We did not observe significant differences in cells depleted for SEPT2 or SEPT11 (Fig. 4 B). Our threshold values were comparable to those obtained from previous reports of ligand-receptor analysis performed on a variety of cell types (40,43). Also in agreement with previous reports (43), we observed a net decrease in the threshold force after latrunculin B treatment (Fig. 4 B), highlighting the role of actin in the adhesion energy between the membrane and the cytoskeleton.
Figure 4.
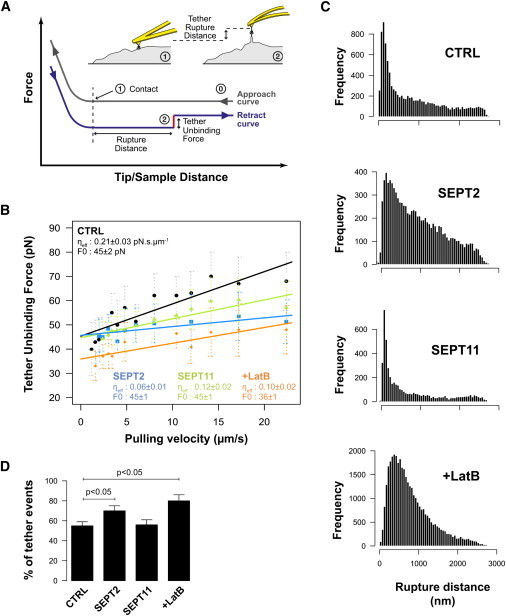
Impact of cytoskeleton disruption on membrane tethers. (A) Membrane tethers can be seen as force plateaus on retraction force curves. We measured the force of these plateaus (pN, red segment), as well as distance from the point of contact (nm, horizontal distance). (B) Tether unbinding force versus pulling velocity of the cantilever. Unbinding force increases linearly with the pulling velocity. SEPT2- and SEPT11-depleted cells show a smaller effective viscosity (ηeff) than control (CTRL) cells. Latrunculin B (LatB)-treated cells show a smaller effective viscosity as well as a smaller tether extraction force (F0) (n > 100 events/condition, mean ± SD). SEPT2- and SEPT11-depleted cell viscosities were significantly different (p-value = 0.012, GraphPad Prism two-tailed slope test). (C, Frequency histograms) Unbinding distances at 12 μm s−1 pulling velocity. CTRL and SEPT11-depleted cells show a clear peak soon after the point of contact whereas SEPT2-depleted cells show longer tethers in the same manner as LatB-treated cells. (D, Bar plots) Probability of having tether events. Data show mean ± SD. p-values, Student's t-test.
As the pulling velocity of tether elongation was increased, there was an increase in the force of the tether from the viscous components of membrane movement into the tether (Fig. 5 B). There are several different analyses of the viscous components contributing to the overall viscous force. These contributions are mainly determined by the membrane curvature and the resulting interbilayer sliding, the membrane viscosity, and membrane sliding over the cytoskeleton (41,42). This represents the effective viscosity, but it is possible to estimate an effective membrane viscosity neglecting contributions of the viscous shear between the halves of the bilayer (44–46). Strikingly, and unlike what was observed with the threshold force, we observed a significant reduction in the effective viscosity of septin-depleted cells as compared to control cells (Fig. 4 B).
Figure 5.
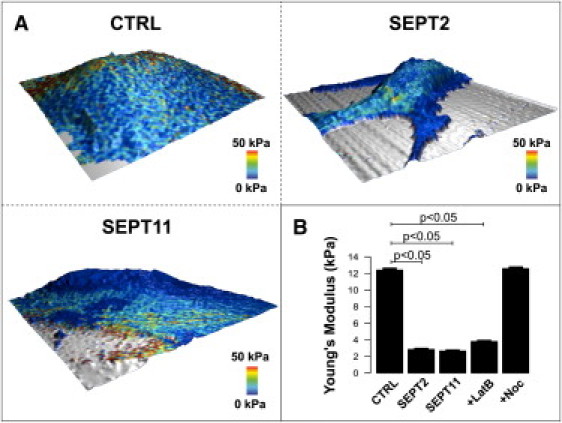
Cortical cell elasticity. (A) Representative three-dimensional elasticity heat maps of control (CTRL), SEPT2-, or SEPT11-depleted cells. Scan size is 50 μm2. In addition to topography is mapped the elasticity (Young's modulus) at 50 nm indentation. (Blue) Soft areas; (red) harder parts of the cell. (B) Mean elasticity values for CTRL, SEPT2-, or SEPT11-depleted cells, or latrunculin B (LatB)- or nocodazole (Noc)-treated cells (n = 20/condition with 1024 measurements per cell). Analysis was performed using 20,000 approach curves obtained from scan frames selected neither on the cell soma nor at the edge of the cell, and are summarized in a bar plot with mean ± SE error bars. p-values, Student's t-test.
Interestingly, we also observed significantly different effective viscosities between SEPT2- and SEPT11-depleted cells, with the former having a more pronounced effect (Fig. 4 B). The molecular reasons underlying changes in effective viscosity are not fully understood, although it is assumed to be due to subcortical cytoskeleton network organization (43). Smaller effective viscosity implies that less energy is consumed during tether extraction, possibly reflecting a change in the actin network packing and anchoring to the membrane. Indeed, previous studies have reported a smaller effective viscosity upon actin disruption (41,43,45,47,48), and we also observed a reduced effective viscosity in latrunculin B-treated cells (Fig. 4 B). The rupture distance is due to the specific InlB-Met unbinding which depends on membrane mechanical properties.
We obtained frequency histograms of the rupture distances of control, septin-depleted, or latrunculin B-treated cells (Fig. 4 C). The distribution profile of SEPT2-depleted or latrunculin B-treated cells displayed more unbinding events at longer distances versus the profiles obtained for control or SEPT11-depleted cells. As compared to 560 nm for control cells, the median rupture distance was 330 nm, 770 nm, or 650 nm for SEPT11-depleted, SEPT2-depleted, or latrunculin B-treated cells, respectively. Differences in median rupture distance (Fig. 4 C) and percentage of events associated with tethers (Fig. 4 D) between SEPT2- and SEPT11-depleted cells are likely due to the functional redundancy described for SEPT6 group members that has been reported to compensate for SEPT11 depletion (21,49,50), whereas SEPT2 is reported to be less dispensable (7,20,22) (see Discussion and the Supporting Material). The enhanced effect observed with latrunculin B treatment is in agreement with previous reports that disruption of the actin cytoskeleton can increase tether length (48,51).
Given these results, we hypothesized that septins are likely to play a role in linking the membrane to the cytoskeleton.
Septin inactivation decreases cortical cell elasticity
To test the role of septins in linking membrane to the cytoskeleton, we examined the specific contribution of SEPT2 or SEPT11 to cell elasticity using the AFM force-mapping method (52) (Fig. 5 A). Previous AFM reports have demonstrated that after disruption of the actin network, the elastic (i.e., Young's) modulus is reduced (53,54). We confirmed these observations in our experimental system (Fig. 5 B). In contrast to actin, cell elasticity does not depend on microtubule dynamics, because microtubule networks are localized deeper in the cell than cortical actin (54,55). In agreement with this, we found that perturbation of the microtubule networks by nocodazole did not affect the cell stiffness analyzed in the first 50 nm of tip indentation (Fig. 5 B).
We then analyzed whether septin-depletion could affect the cell elasticity of living cells. Similar to results obtained upon disruption of the actin cytoskeleton by latrunculin B, SEPT2- or SEPT11-depletion significantly decreased cell elasticity by approximately threefold (Fig. 5 B). Considering that the depletion of SEPT2 or SEPT11 does not affect the expression of actin (20,21) (Fig. S3), these results strongly suggest that septin inactivation, like actin inactivation, decreases cell cortical elasticity. These results are consistent with septin function being more closely associated to the actin cytoskeleton than microtubule networks, and strongly suggest that septins play a role in membrane anchorage to the actin cytoskeleton. In doing so, septins possibly regulate Met-based signal transduction, the precise molecular basis of which remains to be determined.
Altogether, our results clearly establish that septins modulate the biophysical properties of membrane domains and their association to the actin cytoskeleton.
Discussion
Septins are increasingly recognized as an unconventional cytoskeleton component different from actin, microtubules, and intermediate filaments (6,10). Because the study of host-microbial interaction has been instrumental in investigating the function of cytoskeleton components (11,12), we used this approach here to analyze septin function. Using AFM to specifically establish the role of SEPT2 and SEPT11, we reveal that septins orchestrate functional properties of the Met receptor. This strongly introduces the septin cytoskeleton as a novel regulator (to our knowledge) of surface receptor dynamics.
Biological processes, such as bacterial entry, rely on molecular interactions that can be directly measured using AFM. AFM can investigate surfaces of living cells to single-molecule resolution (37), and the probing of individual ligand-receptor interactions can map cell surface receptors and assay receptor functionality (29). By analyzing nanotube extension forces, plasma membrane properties such as the extent of anchoring to the cytoskeleton and viscosity can also be characterized (37). Herein, we used AFM to investigate interactions between InlB and Met at the molecular level with purified molecules and in a cellular context (Fig. 3 and Fig. S2). In both cases, specificity was ensured by competitive inhibition experiments. The KD indicated that the dissociation of the InlB-Met complex involves one energy barrier in both cases, and we obtained values for the KD comparable to what has been obtained using the same approach for other protein pairs (e.g., transferrin and transferrin receptor) (36).
When interpreting intermolecular interaction measurements on living cells, it is critical to consider these interactions as measured using purified proteins (Fig. S2). Other important parameters that are likely to also play a role in modulating the ligand-receptor interaction include membrane elasticity, lipid bilayer sliding, membrane sliding over the cytoskeleton, and cytoskeleton anchorage of the membrane receptor. We thus investigated membrane tether formation by AFM to give insights into these points (37). Septin depletion significantly reduced the effective viscosity (Fig. 4 B), a property that depends on the subcortical cytoskeleton network organization (43). These results are consistent with the association of septins to actin filaments, and particularly reinforces the proposed role for septins in controlling the rigidity of the plasma membrane (56).
A link between the Met receptor and the actin cytoskeleton has recently been established based on tyrosine kinase activity (57,58), and based on interactions with integrin (59) and ezrin-radixin-moesin surface proteins (60). Although the link of Met to actin remains to be deciphered fully, our results show that it can be modulated by septins. Another important point to consider when examining ligand-receptor interaction is the cell stiffness, a property that is dependent upon the cytoskeleton, cytoplasm viscosity, and organelles encountered during tip indentation into the cell. We investigated the role of septins in cell stiffness and observed that septin depletion, like actin disruption, significantly reduced cell elasticity (Fig. 5, A and B).
These results are compatible with those showing that unbinding forces measured with our InlB-functionalized tip involved septin-modulated Met-actin interactions (Fig. 3 C). Interestingly, we observed differences in the mechanical properties between SEPT2- and SEPT-11 depleted cells. These differences can potentially be explained by the proposed redundancy of SEPT6 group members (e.g., SEPT11) and/or the distribution of different septin complexes inside the cell (see the Supporting Material). It will next be important to determine how the Met-actin link is achieved, and whether different septin complexes are involved in the regulation of actin networks located at the subcortical level and deeper in the cell.
Septin filament assembly is stimulated by septin interaction with the membrane component phosphatidyl inositol polyphosphate and affects the organization of the underlying membranes (61). Septins have been implicated in a variety of membrane transformation/organization events such as bud neck morphogenesis and asymmetric compartmentalization between dividing yeast cells, dendritic spine morphogenesis of neurons, cortical organization in mammalian spermatozoa and lymphocytes, and phagocytosis (6,8,9,18,19), yet the biophysical basis for the septin-membrane interplay remains largely unknown. To explore septin function in membrane transformations during bacterial entry, we addressed the role of SEPT2 and SEPT11 in InlB-Met interactions.
Although the underpinning cytoskeletal organization remains to be determined, our data predict that ligand-receptor interactions that result in membrane extension are stabilized by the presence of septin assemblies. Previous work has shown that the raft-association of Met can modulate the efficiency of Listeria entry into the host cell (62). Interestingly, the regulation of the raft-associated receptor dynamics upon ligand-mediated clustering and bacteria-induced activation has been proposed to depend on the membrane-actin skeleton “fences” and the anchored-protein “pickets” (63). Should septins play a role in the organization of the membrane skeleton as limiting the cell surface area where receptors partition, our study (to our knowledge) initiates the concept that functional dynamics of activated receptors depend on the presence of septins in the subcortical cytoskeleton meshwork.
Our results highlight a function for septins in regulating the dynamics of the Met receptor at the cell surface, and possibly its linkage to the underlying cytoskeleton. Future work will investigate the direct or indirect molecular basis of Met anchorage to the cytoskeleton, as well as the putative involvement of septins in regulating membrane-trafficking events.
Acknowledgments
We thank Daniel Navajas and members of the P.C. and F.L. laboratories for helpful discussions. J. Warein is acknowledged for expert technical assistance. We are grateful to Y. Ciczora for the production and purification of the Met receptor. We thank A. Ndoye (PerkinElmer) for help in handling the Operetta System. We thank P. Michieli for the decoy-Met and D. Trono for the lentivirus-based expression system.
This work was supported by grants from the Fondation pour la Recherche Médicale (No. INE20041102821), French Minister for Research (No. ACI-CE-042466), from the Centre National de la Recherche Scientifique (Projet International de Collaboration Scientifique No. 2010-5221) and ERDS funds (No. OBJ 2-2006/1-4.1-261 No. 8078)/Région Nord-Pas-de-Calais (No. 05480182) to F.L., and from the Agence Nationale pour la Recherche (No. ANR-05-MIM-PMIIM-013-01) to F.L. and P.C. S.M. is supported by a long-term fellowship from the Canadian Institutes of Health Research. P.C. is an international research scholar from the Howard Hughes Medical Institute and recipient of a European Research Council Advanced Grant Award (No. 233348).
Supporting Material
References
- 1.Hall P.A., Russell H.S.E., Pringle J.R. Wiley InterScience; New York: 2009. The Septins. [Google Scholar]
- 2.Hall P.A., Russell S.E. The pathobiology of the septin gene family. J. Pathol. 2004;204:489–505. doi: 10.1002/path.1654. [DOI] [PubMed] [Google Scholar]
- 3.Kuhlenbäumer G., Hannibal M.C., Chance P.F. Mutations in SEPT9 cause hereditary neuralgic amyotrophy. Nat. Genet. 2005;37:1044–1046. doi: 10.1038/ng1649. [DOI] [PubMed] [Google Scholar]
- 4.Ihara M., Yamasaki N., Kinoshita M. Sept4, a component of presynaptic scaffold and Lewy bodies, is required for the suppression of α-synuclein neurotoxicity. Neuron. 2007;53:519–533. doi: 10.1016/j.neuron.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Barral Y., Kinoshita M. Structural insights shed light onto septin assemblies and function. Curr. Opin. Cell Biol. 2008;20:12–18. doi: 10.1016/j.ceb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Weirich C.S., Erzberger J.P., Barral Y. The septin family of GTPases: architecture and dynamics. Nat. Rev. Mol. Cell Biol. 2008;9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- 7.Sirajuddin M., Farkasovsky M., Wittinghofer A. Structural insight into filament formation by mammalian septins. Nature. 2007;449:311–315. doi: 10.1038/nature06052. [DOI] [PubMed] [Google Scholar]
- 8.Spiliotis E.T., Nelson W.J. Here come the septins: novel polymers that coordinate intracellular functions and organization. J. Cell Sci. 2006;119:4–10. doi: 10.1242/jcs.02746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinoshita M. Diversity of septin scaffolds. Curr. Opin. Cell Biol. 2006;18:54–60. doi: 10.1016/j.ceb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Spiliotis E.T., Nelson W.J. Septin functions in the mammalian cytoskeleton. In: Hall P.A., Russell H.S.E., Pringle J.R., editors. The Septins. Wiley-Blackwell; Hoboken, NJ: 2008. pp. 229–246. [Google Scholar]
- 11.Mostowy S., Cossart P. From pathogenesis to cell biology and back. Cell Host Microbe. 2009;5:510–513. doi: 10.1016/j.chom.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Cossart P., Sansonetti P.J. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 13.Pizarro-Cerdá J., Jonquières R., Cossart P. Distinct protein patterns associated with Listeria monocytogenes InlA- or InlB-phagosomes. Cell. Microbiol. 2002;4:101–115. doi: 10.1046/j.1462-5822.2002.00169.x. [DOI] [PubMed] [Google Scholar]
- 14.Bierne H., Cossart P. Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol. Mol. Biol. Rev. 2007;71:377–397. doi: 10.1128/MMBR.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mengaud J., Ohayon H., Mege R.-M. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell. 1996;84:923–932. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y., Naujokas M., Ireton K. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell. 2000;103:501–510. doi: 10.1016/s0092-8674(00)00141-0. [DOI] [PubMed] [Google Scholar]
- 17.Hamon M., Bierne H., Cossart P. Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 2006;4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- 18.Caudron F., Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev. Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Huang Y.-W., Yan M., Trimble W.S. Mammalian septins are required for phagosome formation. Mol. Biol. Cell. 2008;19:1717–1726. doi: 10.1091/mbc.E07-07-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mostowy S., Nam Tham T., Cossart P. Septins regulate bacterial entry into host cells. PLoS ONE. 2009;4:e4196. doi: 10.1371/journal.pone.0004196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mostowy S., Danckaert A., Cossart P. Septin 11 restricts InlB-mediated invasion by Listeria. J. Biol. Chem. 2009;284:11613–11621. doi: 10.1074/jbc.M900231200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mostowy S., Bonazzi M., Cossart P. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe. 2010;8:433–444. doi: 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Pizarro-Cerdá J., Lecuit M., Cossart P. Measuring and analyzing invasion of mammalian cells by bacterial pathogens: the Listeria monocytogenes system. In: Sansonetti P.J., Zychlinsky A., editors. Molecular Cellular Microbiology, Methods in Microbiology. Academic Press; London, UK: 2002. pp. 161–177. [Google Scholar]
- 24.Michieli P., Mazzone M., Comoglio P.M. Targeting the tumor and its microenvironment by a dual-function decoy Met receptor. Cancer Cell. 2004;6:61–73. doi: 10.1016/j.ccr.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 25.Salmon P., Trono D. John Wiley & Sons; Hoboken, NJ: 2006. Production and Titration of Lentiviral Vectors. [DOI] [PubMed] [Google Scholar]
- 26.Conrad C., Gerlich D.W. Automated microscopy for high-content RNAi screening. J. Cell Biol. 2010;188:453–461. doi: 10.1083/jcb.200910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kremer B.E., Adang L.A., Macara I.G. Septins regulate actin organization and cell-cycle arrest through nuclear accumulation of NCK mediated by SOCS7. Cell. 2007;130:837–850. doi: 10.1016/j.cell.2007.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita M., Field C.M., Mitchison T.J. Self- and actin-templated assembly of mammalian septins. Dev. Cell. 2002;3:791–802. doi: 10.1016/s1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- 29.Hinterdorfer P., Dufrêne Y.F. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods. 2006;3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 30.Bell G.I. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 31.Merkel R., Nassoy P., Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 32.Machner M.P., Frese S., Heinz D.W. Aromatic amino acids at the surface of InlB are essential for host cell invasion by Listeria monocytogenes. Mol. Microbiol. 2003;48:1525–1536. doi: 10.1046/j.1365-2958.2003.03532.x. [DOI] [PubMed] [Google Scholar]
- 33.Schubert W.D., Heinz D.W. Structural aspects of adhesion to and invasion of host cells by the human pathogen Listeria monocytogenes. ChemBioChem. 2003;4:1285–1291. doi: 10.1002/cbic.200300624. [DOI] [PubMed] [Google Scholar]
- 34.Chevalier S., Cuestas-Ayllon C., de la Fuente J.M. Creating biomimetic surfaces through covalent and oriented binding of proteins. Langmuir. 2010;26:14707–14715. doi: 10.1021/la103086b. [DOI] [PubMed] [Google Scholar]
- 35.Tolbert W.D., Daugherty J., Xu H.E. A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist. Proc. Natl. Acad. Sci. USA. 2007;104:14592–14597. doi: 10.1073/pnas.0704290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yersin A., Osada T., Ikai A. Exploring transferrin-receptor interactions at the single-molecule level. Biophys. J. 2008;94:230–240. doi: 10.1529/biophysj.107.114637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller D.J., Helenius J., Dufrêne Y.F. Force probing surfaces of living cells to molecular resolution. Nat. Chem. Biol. 2009;5:383–390. doi: 10.1038/nchembio.181. [DOI] [PubMed] [Google Scholar]
- 38.Sheetz M.P. Cell control by membrane-cytoskeleton adhesion. Nat. Rev. Mol. Cell Biol. 2001;2:392–396. doi: 10.1038/35073095. [DOI] [PubMed] [Google Scholar]
- 39.Discher D.E., Mohandas N., Evans E.A. Molecular maps of red cell deformation: hidden elasticity and in situ connectivity. Science. 1994;266:1032–1035. doi: 10.1126/science.7973655. [DOI] [PubMed] [Google Scholar]
- 40.Dupres V., Verbelen C., Dufrêne Y.F. Force spectroscopy of the interaction between mycobacterial adhesins and heparan sulphate proteoglycan receptors. ChemPhysChem. 2009;10:1672–1675. doi: 10.1002/cphc.200900208. [DOI] [PubMed] [Google Scholar]
- 41.Dai J., Sheetz M.P. Mechanical properties of neuronal growth cone membranes studied by tether formation with laser optical tweezers. Biophys. J. 1995;68:988–996. doi: 10.1016/S0006-3495(95)80274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao J.Y., Hochmuth R.M. Micropipette suction for measuring picoNewton forces of adhesion and tether formation from neutrophil membranes. Biophys. J. 1996;71:2892–2901. doi: 10.1016/S0006-3495(96)79486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y., Girdhar G., Shao J.-Y. Single membrane tether extraction from adult and neonatal dermal microvascular endothelial cells. Am. J. Physiol. Cell Physiol. 2007;292:C1272–C1279. doi: 10.1152/ajpcell.00251.2006. [DOI] [PubMed] [Google Scholar]
- 44.Waugh R.E. Surface viscosity measurements from large bilayer vesicle tether formation. I. Analysis. Biophys. J. 1982;38:19–27. doi: 10.1016/S0006-3495(82)84526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hochmuth F.M., Shao J.Y., Sheetz M.P. Deformation and flow of membrane into tethers extracted from neuronal growth cones. Biophys. J. 1996;70:358–369. doi: 10.1016/S0006-3495(96)79577-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun M., Graham J.S., Grandbois M. Multiple membrane tethers probed by atomic force microscopy. Biophys. J. 2005;89:4320–4329. doi: 10.1529/biophysj.104.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Girdhar G., Shao J.Y. Membrane tether extraction from human umbilical vein endothelial cells and its implication in leukocyte rolling. Biophys. J. 2004;87:3561–3568. doi: 10.1529/biophysj.104.047514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edmondson K.E., Denney W.S., Diamond S.L. Neutrophil-bead collision assay: pharmacologically induced changes in membrane mechanics regulate the PSGL-1/P-selectin adhesion lifetime. Biophys. J. 2005;89:3603–3614. doi: 10.1529/biophysj.105.066134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ono R., Ihara M., Nosaka T. Disruption of Sept6, a fusion partner gene of MLL, does not affect ontogeny, leukemogenesis induced by MLL-SEPT6, or phenotype induced by the loss of Sept4. Mol. Cell. Biol. 2005;25:10965–10978. doi: 10.1128/MCB.25.24.10965-10978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanai N., Nagata K., Inagaki M. Biochemical and cell biological characterization of a mammalian septin, Sept11. FEBS Lett. 2004;568:83–88. doi: 10.1016/j.febslet.2004.05.030. [DOI] [PubMed] [Google Scholar]
- 51.Finger E.B., Bruehl R.E., Springer T.A. A differential role for cell shape in neutrophil tethering and rolling on endothelial selectins under flow. J. Immunol. 1996;157:5085–5096. [PubMed] [Google Scholar]
- 52.Radmacher M., Cleveland J.P., Hansma P.K. Mapping interaction forces with the atomic force microscope. Biophys. J. 1994;66:2159–2165. doi: 10.1016/S0006-3495(94)81011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rotsch C., Radmacher M. Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: an atomic force microscopy study. Biophys. J. 2000;78:520–535. doi: 10.1016/S0006-3495(00)76614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu H.W., Kuhn T., Moy V.T. Mechanical properties of L929 cells measured by atomic force microscopy: effects of anticytoskeletal drugs and membrane crosslinking. Scanning. 1998;20:389–397. doi: 10.1002/sca.1998.4950200504. [DOI] [PubMed] [Google Scholar]
- 55.Kasas S., Wang X., Catsicas S. Superficial and deep changes of cellular mechanical properties following cytoskeleton disassembly. Cell Motil. Cytoskeleton. 2005;62:124–132. doi: 10.1002/cm.20086. [DOI] [PubMed] [Google Scholar]
- 56.Gilden J., Krummel M.F. Control of cortical rigidity by the cytoskeleton: emerging roles for septins. Cytoskeleton (Hoboken) 2010;67:477–486. doi: 10.1002/cm.20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abella J.V., Vaillancourt R., Park M. The Gab1 scaffold regulates RTK-dependent dorsal ruffle formation through the adaptor Nck. J. Cell Sci. 2010;123:1306–1319. doi: 10.1242/jcs.062570. [DOI] [PubMed] [Google Scholar]
- 58.Abella J.V., Parachoniak C.A., Park M. Dorsal ruffle microdomains potentiate Met receptor tyrosine kinase signaling and down-regulation. J. Biol. Chem. 2010;285:24956–24967. doi: 10.1074/jbc.M110.127985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hui A.Y., Meens J.A., Elliott B.E. Src and FAK mediate cell-matrix adhesion-dependent activation of Met during transformation of breast epithelial cells. J. Cell. Biochem. 2009;107:1168–1181. doi: 10.1002/jcb.22219. [DOI] [PubMed] [Google Scholar]
- 60.Orian-Rousseau V., Morrison H., Ponta H. Hepatocyte growth factor-induced Ras activation requires ERM proteins linked to both CD44v6 and F-actin. Mol. Biol. Cell. 2007;18:76–83. doi: 10.1091/mbc.E06-08-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanaka-Takiguchi Y., Kinoshita M., Takiguchi K. Septin-mediated uniform bracing of phospholipid membranes. Curr. Biol. 2009;19:140–145. doi: 10.1016/j.cub.2008.12.030. [DOI] [PubMed] [Google Scholar]
- 62.Seveau S., Bierne H., Cossart P. Role of lipid rafts in E-cadherin- and HGF-R/Met-mediated entry of Listeria monocytogenes into host cells. J. Cell Biol. 2004;166:743–753. doi: 10.1083/jcb.200406078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morone N., Nakada C., Kusumi A. Three-dimensional molecular architecture of the plasma-membrane-associated cytoskeleton as reconstructed by freeze-etch electron tomography. Methods Cell Biol. 2008;88:207–236. doi: 10.1016/S0091-679X(08)00412-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


