SUMMARY
Embryonic stem cells (ESC) and induced pluripotent stem cells (iPSC) are characterized by their ability to self renew and differentiate into any cell type. The molecular mechanism behind this process is a complex interplay between the transcriptional factors with epigenetic regulators and signaling pathways. MicroRNAs are an integral part of this regulatory network with essential roles in pluripotent maintenance, proliferation and differentiation. MicroRNAs are a class of small non-coding RNAs that target protein-encoding mRNA to inhibit translation and protein synthesis. Discovered close to two decades ago, microRNAs have rapidly emerged as key regulatory molecules in several critical cellular processes across species. Recent studies have begun to clarify the specific role of microRNA in regulatory circuitries that control self renewal and pluripotency of both ESC and iPSC. These advances suggest a critical role for microRNAs in the process of reprogramming of somatic cells to pluripotent cells.
Keywords: microRNA, epigenetics, embryonic stem cells, induced pluripotent stem cells, Review
INTRODUCTION
Pluripotent stem cells hold immense promise for regenerative medicine due to their self renewal and potential for differentiation. Human embryonic stem cells (hESC) derived from the inner cell mass of a developing embryo can be propagated indefinitely in culture and have the ability to differentiate into virtually every cell type in the human body [1]. Despite this, their use in clinical applications remains a challenge owing to the potential rejection of cells following transplant and ethical concerns surrounding the use of discarded embryos [2]. Recently, the derivation of induced pluripotent stem cells (iPSC) via ectopic expression of defined factors in somatic cells [3–10] circumvents these issues. It offers a method to derive patient-specific pluripotent stem cells and also enables the creation of disease-specific stem cells to dissect the molecular mechanism underlying genetic disorders [11–13].
The molecular circuitry that maintains pluripotency is thought to comprise a complex network of transcription factors and epigenetic regulators externally connected via signaling pathways [14]. Several studies of gene expression analysis of pluripotent stem cells have led to the identification of a core regulatory network, composed of the transcription factors Oct4, Sox2 and Nanog, that governs pluripotency [15–18]. This core circuit includes mechanisms such as the epigenetic regulation of genes either by repressive chromatin marking of promoter regions by Polycomb family members or by promoter CpG methylation by Dnmt proteins [19–23].
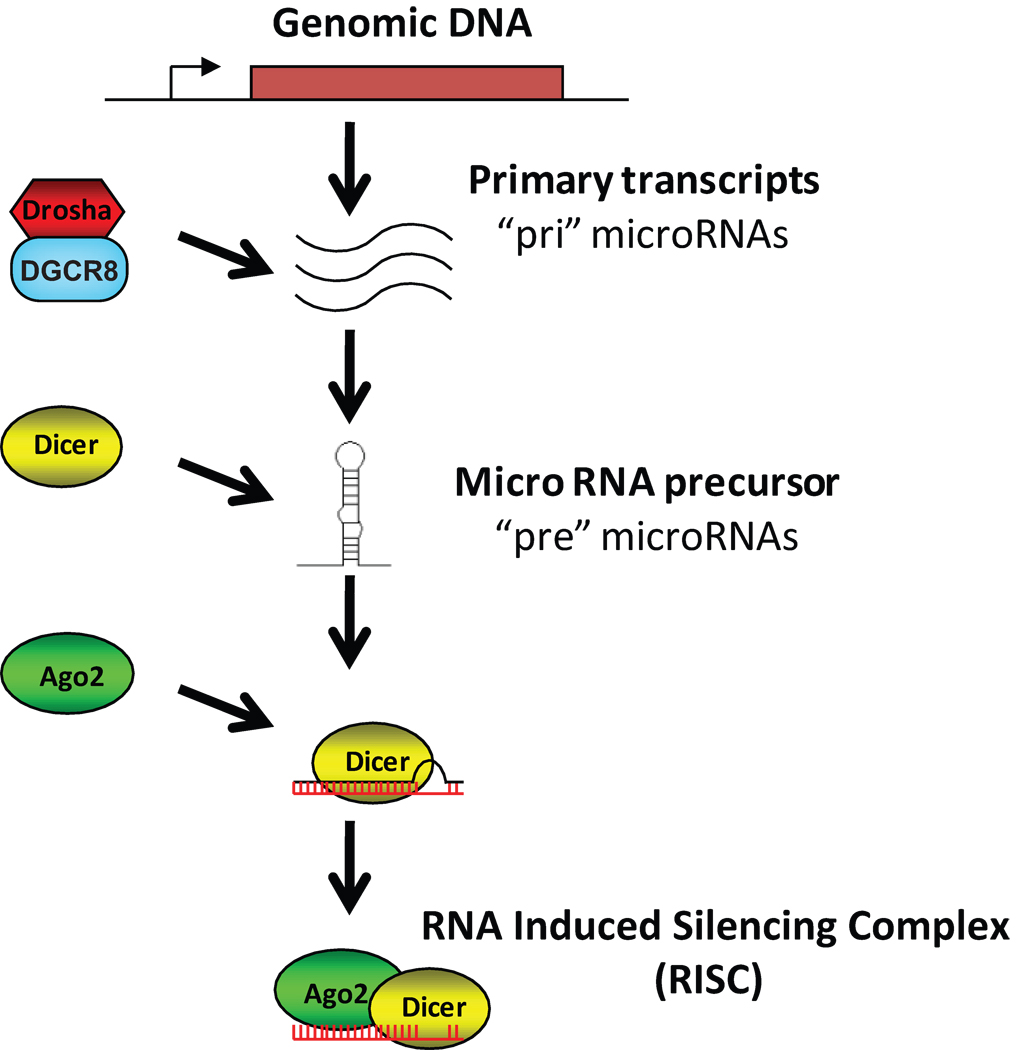
Recently, small noncoding RNAs molecules have emerged as posttranslational regulators of gene expression in diverse species. These small RNAs complex with the Argonaute (Ago) family of proteins as part of the RNA-induced silencing complex (RISC) to induce degradation or to prevent or activate translation of the gene transcript of target genes [24], ultimately regulating biological processes such as development, cell division, apoptosis, metabolism and cancer [25–27]. In mammals, the class of noncoding RNA that is predominantly associated with Ago proteins is termed microRNA (microRNA; Figure 1). The primary transcripts of microRNA (pri-microRNA) are processed by the Microprocessor (Drosha-DGCR8) complex to yield a stem-loop precursor microRNA (pre-microRNA) [28, 29]. In animals, pre-microRNA is exported from the nucleus to the cytoplasm by Exportin-5, where it is processed by Dicer to yield the double-stranded 19–25 nucleotide mature microRNA [30, 31]. One of the strands of the mature microRNA is incorporated into the RNA-induced silencing complex (RISC) and directed to 3’ untranslated region of mRNAs, targeting them for degradation, or by suppressing or activating translation [32–35]. Two newer families of noncoding RNA are endogenous small interfering RNA (Endo-siRNA) and Piwi-interacting RNA (PiRNA). The biogenesis of Endo-siRNA is similar to microRNA and is processed by Dicer and loaded into the RISC complex but target mRNA with complete complementarity to mediate their degradation [36]. PiRNA are 23–32 nucleotides long and are generated by a mechanism independent of Dicer by a less understood process involving the Piwi proteins [37, 38]. PiRNA are thought to play a key role in transposon silencing and epigenetic regulation [39].
Figure 1.
Schematic diagram of microRNA biogenesis.
The potential number of mRNA targets for the 700–1,000 cellular microRNAs currently known is yet to be defined, but has been estimated to reach >60% of all cellular mRNAs [40]. microRNA targets mRNA with a short ~7 nt “seed” sequence (corresponding to microRNA positions 2 through 8) using imperfect base-pair matching and therefore a single microRNA is predicted to match common binding sequences in the 3’ UTR or possibly other regions of mRNA. As mRNA targets were initially described, several bioinformatic algorithms using computational modeling of the seed-target interaction listed predictions from known 3’UTR sequences on publicly-available web sites [41, 42]. However, results among the target prediction algorithms vary and tend to show a high false positive rate [41, 43], leaving most investigators with huge lists of genes and very little confidence in the validity of the potential interactions. These predictions are, by their nature, probabilistic and so they normally require formal demonstration of action by biochemical methods.
microRNA analysis in stem cells
Novel methods
Methods for detection of microRNAs in hESC began with Northern blots but were quickly expanded to take advantage of quantitative real-time PCR (qPCR) [44] and microarrays [45, 46]. More recently, deep sequencing to detect small RNAs has proven to be a sensitive method [47], although careful comparisons have noted slight variations in results based on different sequencing strategies [48]. Using SOLiD sequencing, we observed a strong similarity between counted sequences and qPCR results [49] with a small number of exceptions. In one case, levels of expression of one microRNA disagreed between sequencing and qPCR results but it appears that variability in the 3’ terminus of the microRNA may skew the qPCR results, since TaqMan-based qPCR methods require high stringency matching at the 3’ end. This observation argues that qPCR assays may not always provide the most complete results—hybridization methods should detect all variants, with arrays summing all hybridization products into a single measurement, Northerns separating them by molecular weight, and deep sequencing identifying all sequence variations individually.
Identification of new microRNAs
Three groups have recently identified new microRNAs in hESC or early differentiation products (Table 1). Earlier identification of stem cell-specific microRNAs utilized classic cloning and sequencing approaches to identify the first stem cell-specific clusters [50, 51]. One deep sequencing study used high-throughput 454 pyrosequencing of concatamers built from small RNAs to detect more than 4 × 105 sequences [52]. From this starting point, alignments representing mature and “star” sequences were computed into hairpins using RNA folding algorithms. By these methods, Bar and colleagues identified 13 novel and 56 candidate microRNAs, using expression detection by a second method as validation.
Table 1.
Predictions of new microRNAs by deep sequencing in hESC.
| Reference | Sequencing Method |
Initial Sequences |
Prediction Method |
Number of New microRNA Genes |
|---|---|---|---|---|
| Bar et al. (2008) | 454 | 4 × 105 | Custom | 13 |
| Morin et al. (2008) | Illumina | 6 × 106 | miPred and custom | 104 |
| Goff et al. (2009) | SOLiD | 1.1 × 108 | miRDeep | 140* |
Goff originally reported 149 new genes but a revised analysis adjusts the number to 140.
Another group used Illumina sequencing and a custom microRNA prediction strategy to identify 104 novel microRNA genes in hESC [53]. These methods allowed the detection of 10-fold more RNA sequences than the Bar study (about 6 × 106 reads in hESC). Analyses of multiple aligned sequences also identified variability in processing, either by variation in cleavage site positions or by addition of a non-template nucleotide, as well as sequence modification by RNA editing. In some cases, most frequently variants were observed as isomers different from the reported sequences in miRBase [54], which could potentially confound prediction of seed sequences or construction of qPCR primers. Morin and colleagues employed several strategies for predicting novel microRNA genes among small RNA reads that did not align to known genes and ended up selecting 83 unique sequences from up to 104 distinct genes [53]. Not surprisingly, most of these sequences were expressed at moderate levels and were greatly regulated between ESC and embryoid body stages [53], consistent with their predicted role as regulators important in early differentiation events.
A more recent study combined deep sequencing, precursor structure modeling, and RNA immunoprecipitation (RIP) using Ago2-specific antibodies to identify 140 new microRNAs found in hESC and NSC [49]. Here, some 108 individual RNA sequence reads were aligned with human genome before matching the alignments with a computational model of Microprocessor and Dicer substrates. That is, the miRDeep algorithm [55] searched for adjacent RNA sequences, aligned along a hairpin sequence in genome, representing mature microRNA, star strand, and, in rare cases, loop strand that were likely produced by Dicer cleavage of a Microprocessor-produced precursor. This identified some 913 genomic loci that were consistent with encoding microRNA-like stem-loop structures, after filtering for previously-known microRNAs, other RNA genes, and highly repeated regions. These genomic loci were found to have chromatin modifications consistent with regulated gene expression [49]. To validate these predictions, RISC complexes were immunoprecipitated with an antibody against Ago2 and small RNAs prepared from the immunoprecipitations were sequenced. Of the 913 predictions, 140 RNAs could be identified in the Ago2 RIPs with confidence and were therefore considered to be microRNAs.
Some overlap was found among these three studies. Two of Bar’s predictions overlap Morin’s new microRNAs and 11 out of 13 were identified in at least one hESC line in Goff’s SOLiD expression dataset [49]. All but one of Bar’s 13 were Ago2-enriched in Goff’s data. Of the 83 distinct mature microRNA sequences identified by Morin, 70 could be identified in at least one hESC sample in the Goff data. However, only 31 of the Morin predicted microRNAs could be found to be enriched in Ago2-immunoprecipitated complexes. Note that each study specifically excluded the previously reported microRNAs identified in each earlier study since they became part of the “known” control collection as they were included in updated versions of miRBase. The overlaps listed here indicate that all three studies detected largely similar sequences. Perhaps the minor remaining differences can be ascribed to differences in each sequencing technology, particularly in the assembly of library mixtures, as has been noted previously [48].
Although these new microRNAs have not been demonstrated to exhibit regulatory function, there is some overlap among new microRNAs and existing microRNA families. In the Goff study it was noted that 10% of the 140 new microRNAs shared seed sequences with existing microRNAs [49]. Morin analyzed seed sequences enriched in hESC and found specific patterns of overlap between existing families and newly-described microRNAs [53]. Due to the large numbers of predictions in these reports, a new high-throughput biochemical approach for target identification may be required.
Discovery of microRNA targeting interactions
A novel high throughput targeting strategy is based on RNA immunoprecipitations (RIP) of RISC, most commonly one of the Ago proteins, and associated mRNAs and microRNAs. After extraction, associated RNAs can be identified by deep sequencing or microarrays. The major drawback of these techniques is the need to identify the mRNA and microRNA interactions bioinformatically since the precise targeting site is not detected directly.
To avoid this problem Nonne and coworkers have developed a tandem affinity purification method to sequentially pull down an Ago protein and then recover complexes binding a specific biotin labeled microRNA along with their associated mRNA targets, referred to as Tandem Affinity Purification of microRNA Target mRNAs (TAP-Tar) [56]. This approach eliminates the need to match all the microRNAs to their respective mRNA targets bioinformatically. The limitation of this approach is that the technique currently will only identify targets of a single tagged microRNA, thus limiting the high throughput identification of all microRNA targets under a specific condition. This technique also fails to identify the precise location of the microRNA response element on the recovered mRNAs.
On the other hand, some groups have begun to footprint the exact binding location of RISC on the mRNAs in order to identify microRNA response elements. For example, Darnell and co-workers have developed a method called high-throughput sequencing-cross-linking immunoprecipitation (HITS-CLIP) [57–59] in which RNA-protein complexes are cross-linked in situ, partially digested with RNAase to trim away RNA that does not interact with the protein, and then immunoprecipitated with a specific antibody. The protein-bound RNA is then isolated and sequenced by deep sequencing to produce a map of every position on transcribed RNA where the RNA-binding protein is associated. This approach has been used successfully to identify Ago/microRNA associated mRNAs in the mouse brain [59]. The one caveat of this method is that even though all regulated response element sequences should be identified, investigators still need to match them to the targeting microRNAs by in silico analysis, since the precise location of the target is not determined by this method.
These techniques, even with their limitations, have the potential to verify computational targeting predictions as well as to provide input for improved targeting algorithms, but, more importantly, can provide direct evidence for microRNA interaction with specific isoforms of mRNA expressed in the current condition. It has been suggested that alternative splicing and alternative poly(A) site selection may contribute to affect microRNA targeting by changing the choice of 3’UTR sequences expressed [60]. Computational target predictions cannot include the variability in 3’UTR regulation if these variations have not been reported. Therefore, it will be important to interrogate the extent of microRNA regulatory events in ESCs or IPS cells directly using methods such as TAP-Tar or HITS-CLIP.
MicroRNA in ESC and their functional targets
The importance of microRNA function in stem cells has been established in C. elegans, Drosophila, Danio (zebrafish) and mice, where disruption of microRNA processing enzymes such as Dicer or DGCR8 leads to defects in cell proliferation and embryo development [61–67]. In mouse, Dicer-null animals are embryonic lethal demonstrating its critical role during early embryonic development [62]. However, dicer-null mouse ESC are viable albeit with marked defects in proliferation and differentiation. Notably, dicer-null ESCs show prolonged G0 and G1 phases of the cell cycle and this defective cell cycle progression could have an effect on other processes such as differentiation. Dicer-null ESC are defective in undergoing differentiation upon induction and fail to express differentiation markers such as HNF4a, BMP4 and GATA1 [64]. Further Dicer deficient cells show decreased levels of DNA methylation and methyltransferases (Dnmts) [68, 69] and increases telomerase recombination and elongation [68]. This defect in DNA methylation leads to incomplete and reversible silencing of the Oct4 pluripotent gene, thereby resulting in lack of differentiation [68, 69]. Since Dicer is needed for microRNA and endo-siRNA biogenesis, it can be argued that the phenotype may not be solely caused by lack of microRNA. One study reports that Dicer mutants had altered profiles of microRNA and not other small RNAs and half of the microRNAs detected were known regulators of cell cycle and oncogenesis [70].
More stringent evidence for microRNA requirement in ESC self-renewal and differentiation comes from the DGCR8-deficient ESC [65]. DGCR8 knock out mouse ESC show phenotypes similar to Dicer-deficient mouse ESC with reduced cell proliferation, abnormal cell cycle control and deficiencies in differentiation. DGCR8-null ESC arrest in the G1 phase, implicating a role for microRNA in ES cell cycle in promoting transition from G1 to S phase. These cells cannot fully silence the expression of self-renewal genes such as Oct4, Rex1, Nanog and Sox2 and subsequently show reduced expression of differentiation markers. DGCR8-mutant ESC when injected into host mice do not differentiate into the three germ layers to form teratomas, a features characteristic of normal embryonic stem cells [71–73]. Taking advantage of the simplified microRNA background in DGCR8-null mouse ESC, Blelloch and colleagues have discovered specific functions for families of microRNAs controlling cell cycle and self-renewal [74, 75], to be described in detail below.
However several differences were observed between the Dicer and DGCR8 mutant cells. While the Dicer-deficient ES cells did not expresses any differentiation markers [64], DGCR8 deficient ESC do express some differentiation markers and the defects in cell proliferation and cell cycle progression were less pronounced than Dicer-mutant cells [65]. These studies confirm the essential regulatory role of microRNAs in ESC proliferation, cell cycle and differentiation but also suggest that other small RNAs may play a role in this process as well.
Embryonic stem cells have been reported to express a small subset of unique microRNAs [50, 51, 53, 70, 76]. Most of these ES-specific microRNAs occur as two clusters. The human miR-371 cluster is located on chromosome 19 and is analogous to the mouse miR-290 cluster and the miR-302 cluster located on chromosome 4 is associated with both murine and human ESC [50, 51, 77]. Two additional clusters, miR-17 on chromosome 13 and the miR-106a cluster on chromosome X, have also been shown to be upregulated in ESC [78]. What is the evidence for these microRNAs having function in pluripotency and what are the targets of these microRNAs?
miR-290 and miR-371 Cluster
Transfection of Dicer-null cells, presumed to lack all microRNAs, with exogenous miR-290 family members can partially rescue a portion of their self-renewal phenotype, including Oct4 expression and reduced growth rate when [68, 69]. This miR-290 cluster comprises six microRNAs (miR-290 to miR-295) and all are expressed at high levels in undifferentiated cells, decreasing upon differentiation in mouse embryonic stem cells [50]. The human homolog of the miR-290 cluster on chromosome 19 includes miR-371, -372, -373 and -373*. These microRNAs have been identified as specific for human embryonic carcinoma and embryonic stem cells [51, 76, 79]. The cluster is transcribed as a single polycistronic transcript and therefore it is thought to be regulated by a common promoter [51]. Mouse mutants with a homozygous deletion of the miR 290–295 cluster are embryonic lethal, demonstrating its importance during embryo development [80]. Addition of miR-294 to a DGCR8-null ESC antagonizes the effect of added let-7, stabilizing the self-renewal phenotype [74]. This family of microRNAs plays a direct role in regulating ESC pluripotency and differentiation by down regulating Oct4 by targeting its epigenetic repressor DNA methyl transferases (DNMTs) such as retinoblastoma like 2 (RBL2) [68, 69]. The ES core transcriptional regulatory circuit, consisting of Oct4, Sox2 and Nanog, has recently been shown to be physically associated with promoters for microRNAs that are preferentially expressed in ESC such as the miR-290 cluster [81]. These findings demonstrate that the miR-290 (mouse) or miR-371 (human) cluster mediates the stabilization of pluripotency through de-repression of an Oct4 inhibitor, which in turn favors expression of this cluster through promoter binding. The miR-290–295 cluster has also been suggested to play a role in early germ cell development and spermatogenesis [82].
The expression of the miR-371 cluster is diverse and, in addition to ESC, has been shown to be expressed in embryonal carcinoma cells such as NTera 2 and 2102EP cells [76, 79, 83]. Expression of the miR-372/373 cluster associates with oncogenes involved in cellular transformation [84, 85]. Its expression and oncogenic properties correlate with p53 status of the cells and is thought to be inhibited by cisplatin [84, 86]. MiR-373 and miR-520 have also been shown to stimulate cancer cell migration and invasion in vitro and in vivo [87].
miR-302-367 Cluster
Another ESC-specific microRNA cluster is the miR302-367 cluster, shown to be highly and specifically expressed in undifferentiated ESC [53, 76, 79]. This cluster, encoded by human chromosome 4, consists of nine different microRNAs: miR-302a, -302a*, -302b, -302b*, -302c, -302c*, -302d, -367 and -367* [88]. Recently the putative promoter of miR-302-367 was identified and its transcriptional activity found to correlate with miR-302-367 levels [89]. Further, the ESC-specific transcription factors, Oct4, Nanog, Sox2 and Rex 1, were shown to be upstream regulators of the miR-302-367 promoter, based on effect of miR-302-367 promoter transcriptional activity with siRNA-mediated knockdown of these transcription factors and physical interaction of Oct4, Sox2 and Nanog with miR-302-367 promoter [89, 90]. In addition, miR-302-367 has been identified to post-transcriptionally regulate cyclin D1 and Cdk4, affecting cell cycle progression [90]. In addition to their role in self renewal and proliferation, miR-302-367 may indirectly induce the TGFβ/Nodal/Activin pathway via inhibition of intermediate negative regulators to maintain cells in the undifferentiated state [91].
miR-17-92 Cluster
This cluster, highly expressed in ESC and downregulated with differentiation, is located on chromosome 13 and a single polycistronic transcript generates six microRNAs: miR-17, -18a, -19a, -20a, -19b-1 and -92a-1 [50, 53, 92]. This cluster is a known OncomiR that promotes cell proliferation in several forms of cancer [93–97]. This cluster is activated by c-Myc, a repressor of other microRNAs such as miR-15 and let-7 [98]. The promoter region of this cluster contains putative transcription factor binding sites for E2F1, E2F2 and E2F3. miR-20a is thought to play a role cell proliferation and inhibition of apoptosis at the G1 to S transition via a feedback loop with E2F factors [97, 99–101]. The role of c-Myc in combination with Oct4, Sox2 and Klf4 in inducing somatic cells to induced pluripotent stem cells suggests that associated microRNAs play a key role in stem cell renewal and pluripotency. The down regulation of the miR-17-92 cluster with retinoic acid (RA)-induced differentiation suggests that down regulation of this microRNA maybe important for the process of stem cell differentiation [92]. Indeed, Melton and colleagues demonstrated that a let-7 inhibitor was able to replace exogenous c-Myc in the production of iPSC cultures, suggesting that Myc regulation of the miR-17-92 cluster may be a key component of achieving pluripotency, and that multiple stimuli (exogenous c-Myc or antagonism of let-7) can push the cell towards the same state [74].
let-7 Family
ESC are characterized by the presence of low levels of let-7, which is highly expressed in differentiating cell types and most, if not all, adult cell types [76, 92]. Dysregulated let-7 is also associated with multiple cancers [102–109], demonstrating its central role in stabilizing cell phenotype. Let-7g levels have been shown to be regulated by LIN28, a gene highly expressed in pluripotent cells, by inhibiting the Dicer-mediated processing of Pre-let-7 to mature let-7 [110]. The LIN28 gene is in turn targeted by let-7 via target sites in its 3’ UTR [110–113]. Both LIN28 and let-7g genes are associated with Oct4/Sox2/Nanog/Tcf3 binding in ESC, resulting in a regulatory loop where the core pluripotent transcription factors activate the LIN28 gene associated with maintenance of self-renewal and pluripotency while repressing let-7g to prevent differentiation [81]. The Lin28B promoter has been shown to be transcriptionally transactivated via direct association with Myc to mediate let-7 repression [114]. The let-7 family is also though to target RAS, a gene associated with cancer [115–118]; and HMGA2, a known regulator of differentiation and proliferation in mouse and human embryonic stem cells [119, 120]. A recent study reports that exogenous addition of let-7 microRNA can suppress self renewal in DGCR8 null mouse ESC but not wild type ESC and this suppression can be inhibited by introduction of ESC cell cycle regulating microRNAs such as miR-294, suggesting that they act via intersecting pathways to drive cell fate towards self renewal or differentiation [74]. There is however emerging evidence that Lin28 may alternate modes to sustain self renewal that does not involve regulation of let7 [121]
microRNA expression pattern in iPSC
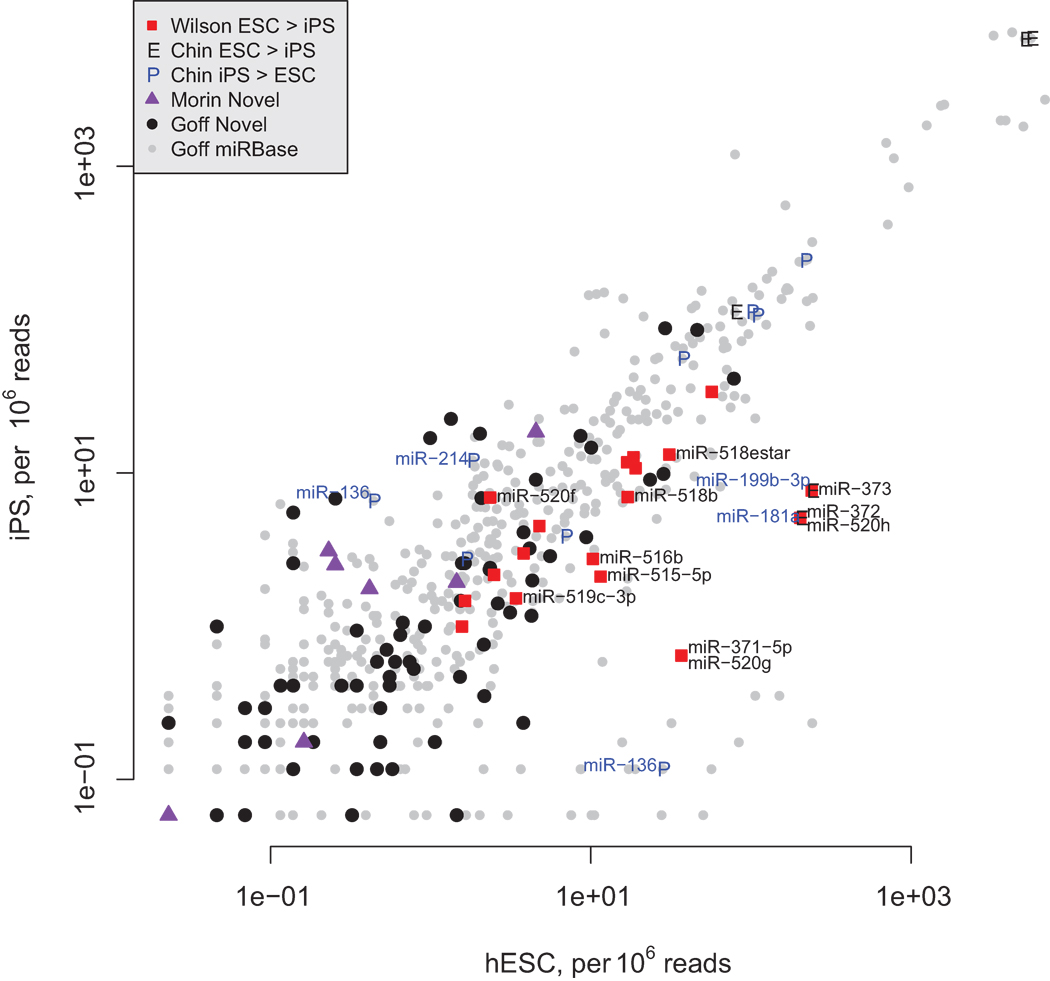
Induced pluripotent cells (iPSC) have been compared with embryonic-derived stem cells for gene expression by several groups, but only three published studies [49, 122, 123] compare microRNA expression with those found in human ESC (Figure 2). All three highlight differences between iPSC and hESC with microRNA expression patterns interpreted to show that iPSC do not appear to be as “undifferentiated” as hESC (Figure 3). For example, Wilson identified several members of the microRNA cluster encoded on chromosome 19, including miR-371/372/373 and many others, as more highly expressed in hESC compared to iPSC (Figure 2, red squares labeled in the lower right). Chin discovered several microRNAs that are predominantly expressed in either hESC or iPSC and several of these were confirmed by Goff, particularly the iPSC-specific microRNAs, including miR-181a, 199b-3p, and 214 (Figure 2, labeled in blue in upper left). These opposing groups of microRNAs that are differentially expressed between these two cell types indicate underlying differences in gene expression patterns between hESC and iPSC. The reduced expression of the ES-specific chromosome 19 cluster in iPSC predicts that regulatory signals are not sufficiently driving expression of microRNAs believed to help maintain self-renewal by stimulating cell cycle [74, 124]. This group of microRNAs, corresponding to the miR-294 cluster on mouse chromosome 7, has been shown to act in opposition to the let-7 family [74]. Reduced expression of this cluster can be predicted to be associated with reduced self-renewal and resistance to pluripotency.
Figure 2.
Scatterplot of microRNA sequence frequencies (per 106 reads) in iPS or hESC taken from Goff et al. (2009) and highlighting overlapping microRNAs from Wilson et al. (2009), Chin et al. (2009) and Morin et al. (2008). MicroRNAs found to be enriched in hESC over iPS by Wilson are noted as red squares and those found to be at least 2-fold different in Goff are labeled in black. Among those microRNAs found to be ESC enriched by Chin, labeled with the character “E,” none was confirmed by Goff. However, several of those microRNAs found by Chin to be enriched in iPS (labeled “P”) are confirmed by Goff (labeled in blue to the left of the “P” character if they were at least 2-fold different). Note also the number of novel microRNAs found by Goff (black dots) that are enriched in iPS cells.
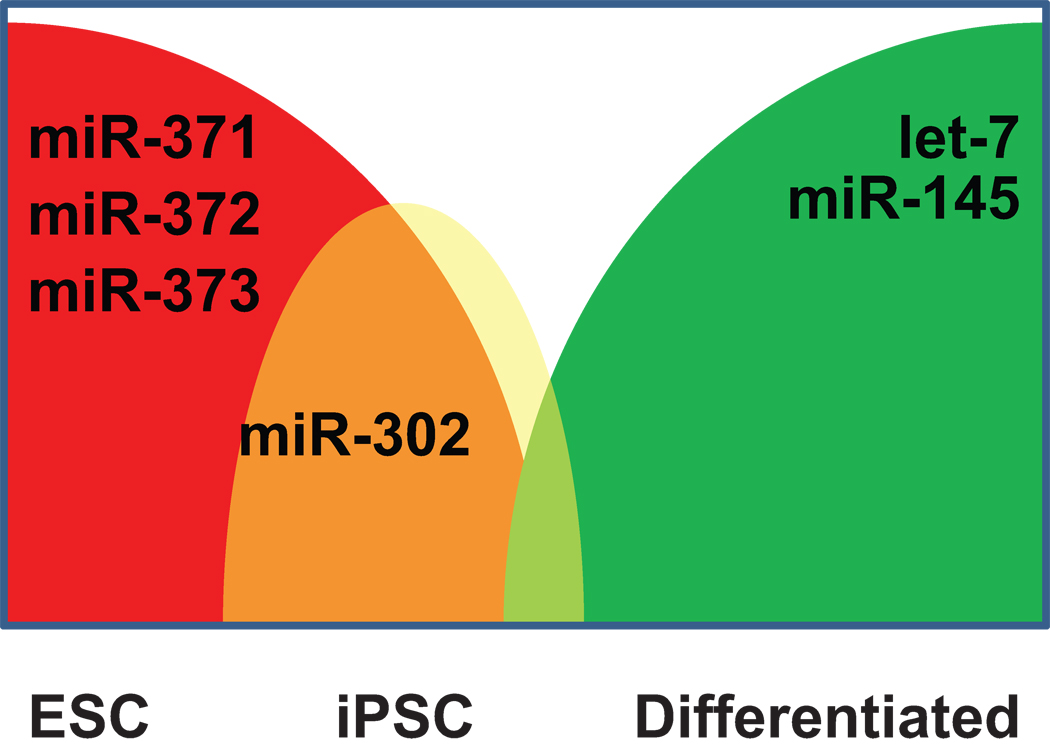
Figure 3.
Schematic of microRNA expression characterizing differentiation status. Representative microRNAs are shown that are characteristic of each stage, with miR-302 being strongly expressed in both ESC and iPSC, but the miR-371 cluster more highly expressed in ESC than iPSC.
Relevance of microRNA regulation in ESC/iPSC
Hornstein and Shomron propose that the interactions between microRNAs and the network of protein-coding genes evolved in order to buffer stochastic perturbations and thereby confer robustness to developmental genetic programs [125]. The concept of this proposed role for microRNAs arose from Waddington’s original canalization hypothesis [126], originally utilized to describe evolution of populations. This hypothesis can be extrapolated beyond its original vision to explain cellular processes such as pluripotency or differentiation in ESC.
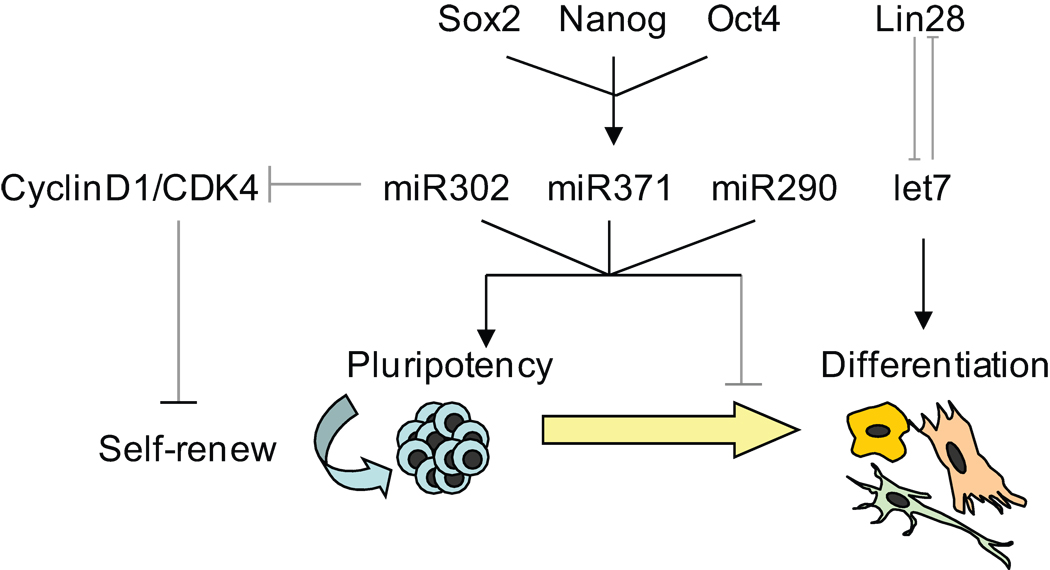
The core stem cell circuitry, composed of pluripotent transcription factors, clearly interacts with groups of microRNAs [81] which coordinate to de-repress the genes involved in pluripotent cell maintenance and suppress genes involved in development and differentiation [74]. Indeed, the pluripotent-specific microRNAs serve to maintain ESCs in their pluripotent state by inhibiting genes that would favor differentiation or cause cells to leave the cell cycle, usually considered to be a trigger for differentiation (Figure 4).
Figure 4.
Regulatory network in pluripotent stem cells. Key genes are represented in italics and microRNAs in bold.
Specifically, miR-302, which has been shown to maintain “stemness” in ESCs, is a negative regulator of G1 and promotes entry into S phase by regulating cyclin D family members [90, 127]. On the other hand, upon hESC differentiation, miR-145 is induced, which in turn serves to inhibit OCT4, SOX2, and KLF4 [128]. Upregulation of miR-145 expression caused a significant diminution of the self-renewal marker SSEA4 and an increase in multiple differentiation markers associated with all three germ layers [128]. These few examples indicate how microRNAs can be understood to stabilize the cellular state endogenously —either as self-renewing/pluripotent or differentiated cultures.
In the case of iPSC, one can propose that by activating ES specific microRNAs in differentiated cells one would induce de-differentiation and revert cells to a pluripotent state. This is certainly the case. MiR-291-3p, -294 and -295 have been shown to increase the efficiency of reprogramming mouse embryonic fibroblast when exogenously expressed with Oct4, Sox2 and Klf4 [129]. In addition, exogenous addition of members of the miR-302 cluster has also been reported to enhance reprogramming of human skin cancer cells into a pluripotent, ESC-like state [130]. These cell lines, constitutively expressing miR-302, began to express several ES cell markers, such as Oct3/4, SSEA-3, SSEA-4, Sox2, and Nanog [130]. Most importantly is that teratoma assays of EBs derived from these cells gave rise to all three embryonic germ layers [130]. Finally, antagonism of the differentiation-specific let-7 family can replace one of the four classic iPSC-inducing transcription factors (c-Myc) in the production of iPSC cultures [74].
As these examples prove, microRNAs integrally participate in the switch between pluripotency and differentiation. Regulation of microRNA levels is required for a full transition from one state to the other. Stable expression of specific classes of microRNAs stabilizes either pluripotency or differentiation. Slight differences in microRNA levels help to distinguish pluripotent cells derived from embryos (ESC) from cells generated by exogenous expression of pluripotency transcription factors (iPSC). As the full regulatory network controlling the entry into or the exit from the pluripotent state is identified, microRNAs clearly represent important stabilizers or actuators that cannot be ignored.
Executive summary.
Small, noncoding, regulatory RNAs known as miRNA have a critical role in regulatory networks that maintain pluripotency in embryonic stem cells (ESCs) and induced pluripotent stem cells.
miRNA expression analyses in stem cells utilize traditional tools to confirm expression of known miRNAs as well as novel technologies such as deep sequencing to identify new miRNAs.
Cells defective in the miRNA processing machinery such as Dicer- and DGCR8-knockout ESCs have been utilized to identify specific miRNA function in regulating pluripotency as well as downstream targets.
These studies have led to the identification of distinct clusters of miRNAs specific for ESCs.
Recently, comparative miRNA studies between ESCs and induced pluripotent stem cells have highlighted slight differences between the two.
These differences in miRNA expression patterns have an effect on the pluripotency maintenance networks.
It is proposed that miRNAs interact with epigenetic regulatory mechanisms to stabilize pluripotency or to switch to a differentiated state.
ACKNOWLEDGEMENTS
Supported by grants to RPH from NIH (R21MH085088, RC1CA147187), the New Jersey Commission on Science & Technology, the New Jersey Commission on Spinal Cord Research, and Invitrogen, Inc.
REFERENCES
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science (New York, N.Y. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Choumerianou DM, Dimitriou H, Kalmanti M. Stem cells: Promises versus limitations. Tissue engineering. 2008;14(1):53–60. doi: 10.1089/teb.2007.0216. [DOI] [PubMed] [Google Scholar]
- 3.Aoi T, Yae K, Nakagawa M, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science (New York, N.Y. 2008;321(5889):699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nature protocols. 2007;2(12):3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa M, Koyanagi M, Tanabe K, et al. Generation of induced pluripotent stem cells without myc from mouse and human fibroblasts. Nature biotechnology. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 7.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451(7175):141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 8.Maherali N, Sridharan R, Xie W, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell stem cell. 2007;1(1):55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science (New York, N.Y. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 10.Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent es-cell-like state. Nature. 2007;448(7151):318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 11.Muller R, Lengerke C. Patient-specific pluripotent stem cells: Promises and challenges. Nat Rev Endocrinol. 2009;5(4):195–203. doi: 10.1038/nrendo.2009.18. [DOI] [PubMed] [Google Scholar]
- 12.Lengerke C, Daley GQ. Disease models from pluripotent stem cells. Annals of the New York Academy of Sciences. 2009;1176:191–196. doi: 10.1111/j.1749-6632.2009.04962.x. [DOI] [PubMed] [Google Scholar]
- 13.Lengerke C, Daley GQ. Autologous blood cell therapies from pluripotent stem cells. Blood reviews. 2009 doi: 10.1016/j.blre.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 15.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132(4):567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Daley GQ. Molecular basis of pluripotency. Human molecular genetics. 2008;17(R1):R23–R27. doi: 10.1093/hmg/ddn050. [DOI] [PubMed] [Google Scholar]
- 18.Loh YH, Wu Q, Chew JL, et al. The oct4 and nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature genetics. 2006;38(4):431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 19.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagarkova MA, Volchkov PY, Lyakisheva AV, Philonenko ES, Kiselev SL. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell cycle (Georgetown, Tex. 2006;5(4):416–420. doi: 10.4161/cc.5.4.2440. [DOI] [PubMed] [Google Scholar]
- 21.Yeo S, Jeong S, Kim J, Han JS, Han YM, Kang YK. Characterization of DNA methylation change in stem cell marker genes during differentiation of human embryonic stem cells. Biochemical and biophysical research communications. 2007;359(3):536–542. doi: 10.1016/j.bbrc.2007.05.120. [DOI] [PubMed] [Google Scholar]
- 22.Fouse SD, Shen Y, Pellegrini M, et al. Promoter cpg methylation contributes to es cell gene regulation in parallel with oct4/nanog, pcg complex, and histone h3 k4/k27 trimethylation. Cell stem cell. 2008;2(2):160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohn F, Weber M, Rebhan M, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Molecular cell. 2008;30(6):755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nature chemical biology. 2007;3(1):36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 25.Kloosterman WP, Plasterk RH. The diverse functions of micrornas in animal development and disease. Developmental cell. 2006;11(4):441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Kato M, Slack FJ. Micrornas: Small molecules with big roles - c. Elegans to human cancer. Biology of the cell / under the auspices of the European Cell Biology Organization. 2008;100(2):71–81. doi: 10.1042/BC20070078. [DOI] [PubMed] [Google Scholar]
- 27.Stefani G. Slack FJ: Small non-coding rnas in animal development. Nature reviews. 2008;9(3):219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 28.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microrna precursors by the nuclear processing enzyme drosha. The EMBO journal. 2005;24(1):138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The drosha-dgcr8 complex in primary microrna processing. Genes & development. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Y, Cullen BR. Structural requirements for pre-microrna binding and nuclear export by exportin 5. Nucleic acids research. 2004;32(16):4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartel DP. Micrornas: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Carrington JC, Ambros V. Role of micrornas in plant and animal development. Science (New York, N.Y. 2003;301(5631):336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 33.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: Short rnas that silence gene expression. Nature reviews. 2003;4(6):457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 34.Pickford AS, Cogoni C. Rna-mediated gene silencing. Cell Mol Life Sci. 2003;60(5):871–882. doi: 10.1007/s00018-003-2245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: Micrornas can up-regulate translation. Science (New York, N.Y. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 36.Okamura K, Lai EC. Endogenous small interfering rnas in animals. Nature reviews. 2008;9(9):673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin H, Yin H. A novel epigenetic mechanism in drosophila somatic cells mediated by piwi and pirnas. Cold Spring Harbor symposia on quantitative biology. 2008;73:273–281. doi: 10.1101/sqb.2008.73.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomson T, Lin H. The biogenesis and function of piwi proteins and pirnas: Progress and prospect. Annual review of cell and developmental biology. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin H, Lin H. An epigenetic activation role of piwi and a piwi-associated pirna in drosophila melanogaster. Nature. 2007;450(7167):304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 40.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mrnas are conserved targets of micrornas. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microrna targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. Mirbase: Tools for microrna genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microrna targets. Nat Methods. 2006;3(11):881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 44.Aravin A, Tuschl T. Identification and characterization of small rnas involved in rna silencing. FEBS Lett. 2005;579(26):5830–5840. doi: 10.1016/j.febslet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 45.Lakshmipathy U, Hart RP. Concise review: Microrna expression in multipotent mesenchymal stromal cells. Stem Cells. 2008;26(2):356–363. doi: 10.1634/stemcells.2007-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W, Ruan K. Microrna detection by microarray. Anal Bioanal Chem. 2009;394(4):1117–1124. doi: 10.1007/s00216-008-2570-2. [DOI] [PubMed] [Google Scholar]
- 47.Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of micrornas by deep sequencing. Brief Bioinform. 2009 doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linsen SE, de Wit E, Janssens G, et al. Limitations and possibilities of small rna digital gene expression profiling. Nat Methods. 2009;6(7):474–476. doi: 10.1038/nmeth0709-474. [DOI] [PubMed] [Google Scholar]
- 49.Goff LA, Davila J, Swerdel MR, et al. Ago2 immunoprecipitation identifies predicted micrornas in human embryonic stem cells and neural precursors. PLoS ONE. 2009;4(9):e7192. doi: 10.1371/journal.pone.0007192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific micrornas. Dev Cell. 2003;5(2):351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 51.Suh MR, Lee Y, Kim JY, et al. Human embryonic stem cells express a unique set of micrornas. Dev.Biol. 2004;270(2):488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 52.Bar M, Wyman SK, Fritz BR, et al. Microrna discovery and profiling in human embryonic stem cells by deep sequencing of small rna libraries. Stem Cells. 2008;26(10):2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morin RD, O'Connor MD, Griffith M, et al. Application of massively parallel sequencing to microrna profiling and discovery in human embryonic stem cells. Genome Res. 2008;18(4):610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. Mirbase: Microrna sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Friedlander MR, Chen W, Adamidi C, et al. Discovering micrornas from deep sequencing data using mirdeep. Nat Biotechnol. 2008;26(4):407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 56.Nonne N, Ameyar-Zazoua M, Souidi M, Harel-Bellan A. Tandem affinity purification of mirna target mrnas (tap-tar) Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jensen KB, Darnell RB. Clip: Crosslinking and immunoprecipitation of in vivo rna targets of rna-binding proteins. Methods Mol Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Licatalosi DD, Mele A, Fak JJ, et al. Hits-clip yields genome-wide insights into brain alternative rna processing. Nature. 2008;456(7221):464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute hits-clip decodes microrna-mrna interaction maps. Nature. 2009;460(7254):479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mrnas with shortened 3' untranslated regions and fewer microrna target sites. Science. 2008;320(5883):1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of dicer-deficient murine embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nature genetics. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 63.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microrna pathway. Nature. 2005;435(7044):974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 64.Kanellopoulou C, Muljo SA, Kung AL, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & development. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. Dgcr8 is essential for microrna biogenesis and silencing of embryonic stem cell self-renewal. Nature genetics. 2007;39(3):380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microrna-producing enzyme dicer1 is essential for zebrafish development. Nature genetics. 2003;35(3):217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 67.Nimmo RA, Slack FJ. An elegant mirror: Micrornas in stem cells, developmental timing and cancer. Chromosoma. 2009;118(4):405–418. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benetti R, Gonzalo S, Jaco I, et al. A mammalian microrna cluster controls DNA methylation and telomere recombination via rbl2-dependent regulation of DNA methyltransferases. Nature structural & molecular biology. 2008;15(3):268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sinkkonen L, Hugenschmidt T, Berninger P, et al. Micrornas control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nature structural & molecular biology. 2008;15(3):259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 70.Calabrese JM, Seila AC, Yeo GW, Sharp PA. Rna sequence analysis defines dicer's role in mouse embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(46):18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bodnar MS, Meneses JJ, Rodriguez RT, Firpo MT. Propagation and maintenance of undifferentiated human embryonic stem cells. Stem cells and development. 2004;13(3):243–253. doi: 10.1089/154732804323099172. [DOI] [PubMed] [Google Scholar]
- 72.Keller G. Embryonic stem cell differentiation: Emergence of a new era in biology and medicine. Genes & development. 2005;19(10):1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 73.Menendez P, Bueno C, Wang L. Human embryonic stem cells: A journey beyond cell replacement therapies. Cytotherapy. 2006;8(6):530–541. doi: 10.1080/14653240601026654. [DOI] [PubMed] [Google Scholar]
- 74.Melton C, Judson RL, Blelloch R. Opposing microrna families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463(7281):621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y, Blelloch R. Cell cycle regulation by micrornas in embryonic stem cells. Cancer Res. 2009;69(10):4093–4096. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lakshmipathy U, Love B, Goff LA, et al. Microrna expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem cells and development. 2007;16(6):1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strauss WM, Chen C, Lee CT, Ridzon D. Nonrestrictive developmental regulation of microrna gene expression. Mamm Genome. 2006;17(8):833–840. doi: 10.1007/s00335-006-0025-7. [DOI] [PubMed] [Google Scholar]
- 78.Laurent LC, Chen J, Ulitsky I, et al. Comprehensive microrna profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem cells (Dayton, Ohio) 2008;26(6):1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 79.Josephson R, Ording CJ, Liu Y, et al. Qualification of embryonal carcinoma 2102ep as a reference for human embryonic stem cell research. Stem cells (Dayton, Ohio) 2007;25(2):437–446. doi: 10.1634/stemcells.2006-0236. [DOI] [PubMed] [Google Scholar]
- 80.Ambros V, Chen X. The regulation of genes and genomes by small rnas. Development (Cambridge, England) 2007;134(9):1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 81.Marson A, Levine SS, Cole MF, et al. Connecting microrna genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134(3):521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, et al. Microrna biogenesis is required for mouse primordial germ cell development and spermatogenesis. PloS one. 2008;3(3):e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Voorhoeve PM. Micrornas: Oncogenes, tumor suppressors or master regulators of cancer heterogeneity? Biochimica et biophysica acta. 2009 doi: 10.1016/j.bbcan.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 84.Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates mirna-372 and mirna-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124(6):1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 85.Voorhoeve PM, le Sage C, Schrier M, et al. A genetic screen implicates mirna-372 and mirna-373 as oncogenes in testicular germ cell tumors. Advances in experimental medicine and biology. 2007;604:17–46. doi: 10.1007/978-0-387-69116-9_2. [DOI] [PubMed] [Google Scholar]
- 86.Duale N, Lindeman B, Komada M, et al. Molecular portrait of cisplatin induced response in human testis cancer cell lines based on gene expression profiles. Molecular cancer. 2007;6:53. doi: 10.1186/1476-4598-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang Q, Gumireddy K, Schrier M, et al. The micrornas mir-373 and mir-520c promote tumour invasion and metastasis. Nature cell biology. 2008;10(2):202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 88.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microrna expression atlas based on small rna library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barroso-delJesus A, Romero-Lopez C, Lucena-Aguilar G, et al. Embryonic stem cell-specific mir302-367 cluster: Human gene structure and functional characterization of its core promoter. Molecular and cellular biology. 2008;28(21):6609–6619. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Card DA, Hebbar PB, Li L, et al. Oct4/sox2-regulated mir-302 targets cyclin d1 in human embryonic stem cells. Molecular and cellular biology. 2008;28(20):6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barroso-del Jesus A, Lucena-Aguilar G, Menendez P. The mir-302-367 cluster as a potential stemness regulator in escs. Cell cycle (Georgetown, Tex. 2009;8(3):394–398. doi: 10.4161/cc.8.3.7554. [DOI] [PubMed] [Google Scholar]
- 92.Gu P, Reid JG, Gao X, et al. Novel microrna candidates and mirna-mrna pairs in embryonic stem (es) cells. PloS one. 2008;3(7):e2548. doi: 10.1371/journal.pone.0002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microrna cluster, mir-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer research. 2005;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 94.Venturini L, Battmer K, Castoldi M, et al. Expression of the mir-17-92 polycistron in chronic myeloid leukemia (cml) cd34+ cells. Blood. 2007;109(10):4399–4405. doi: 10.1182/blood-2006-09-045104. [DOI] [PubMed] [Google Scholar]
- 95.Kutay H, Bai S, Datta J, et al. Downregulation of mir-122 in the rodent and human hepatocellular carcinomas. Journal of cellular biochemistry. 2006;99(3):671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rinaldi A, Poretti G, Kwee I, et al. Concomitant myc and microrna cluster mir-17-92 (c13orf25) amplification in human mantle cell lymphoma. Leukemia & lymphoma. 2007;48(2):410–412. doi: 10.1080/10428190601059738. [DOI] [PubMed] [Google Scholar]
- 97.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. C-myc-regulated micrornas modulate e2f1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 98.Chang TC, Yu D, Lee YS, et al. Widespread microrna repression by myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sylvestre Y, De Guire V, Querido E, et al. An e2f/mir-20a autoregulatory feedback loop. The Journal of biological chemistry. 2007;282(4):2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 100.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-rna cluster by e2f transcription factors. The Journal of biological chemistry. 2007;282(4):2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 101.Trimarchi JM, Lees JA. Sibling rivalry in the e2f family. Nature reviews. 2002;3(1):11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 102.Yu F, Yao H, Zhu P, et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 103.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 micrornas in human lung cancers in association with shortened postoperative survival. Cancer research. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 104.Akao Y, Nakagawa Y, Naoe T. Let-7 microrna functions as a potential growth suppressor in human colon cancer cells. Biological & pharmaceutical bulletin. 2006;29(5):903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 105.Calin GA, Sevignani C, Dumitru CD, et al. Human microrna genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microrna signature in pancreatic cancer. International journal of cancer. 2007;120(5):1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Szafranska AE, Davison TS, John J, et al. Microrna expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26(30):4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 108.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microrna reduces tumor growth in mouse models of lung cancer. Cell cycle (Georgetown, Tex. 2008;7(6):759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 109.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microrna family. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microrna processing by lin28. Science (New York, N.Y. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rybak A, Fuchs H, Smirnova L, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature cell biology. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 112.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the let-7 precursor loop mediates regulated microrna processing. RNA (New York, N.Y. 2008;14(8):1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Piskounova E, Viswanathan SR, Janas M, et al. Determinants of microrna processing inhibition by the developmentally regulated rna-binding protein lin28. The Journal of biological chemistry. 2008;283(31):21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 114.Chang TC, Zeitels LR, Hwang HW, et al. Lin-28b transactivation is necessary for myc-mediated let-7 repression and proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(9):3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johnson SM, Grosshans H, Shingara J, et al. Ras is regulated by the let-7 microrna family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 116.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microrna represses cell proliferation pathways in human cells. Cancer research. 2007;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 117.Morris JPt, McManus MT. Slowing down the ras lane: Mirnas as tumor suppressors? Sci STKE. 2005;2005(297):pe41. doi: 10.1126/stke.2972005pe41. [DOI] [PubMed] [Google Scholar]
- 118.Stahlhut Espinosa CE, Slack FJ. The role of micrornas in cancer. The Yale journal of biology and medicine. 2006;79(3–4):131–140. [PMC free article] [PubMed] [Google Scholar]
- 119.Lee YS, Kim HK, Chung S, Kim KS, Dutta A. Depletion of human micro-rna mir-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. The Journal of biological chemistry. 2005;280(17):16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 120.Li O, Vasudevan D, Davey CA, Droge P. High-level expression of DNA architectural factor hmga2 and its association with nucleosomes in human embryonic stem cells. Genesis. 2006;44(11):523–529. doi: 10.1002/dvg.20242. [DOI] [PubMed] [Google Scholar]
- 121.Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. Lin28 alters cell fate succession and acts independently of the let-7 microrna during neurogliogenesis in vitro. Development (Cambridge, England) 137(6):891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- 122.Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC. Microrna profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18(5):749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chin M, Mason M, Xie W, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5(1):111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Y, Baskerville S, Shenoy A, Babiarz J, Baehner L, Blelloch R. Embryonic stem cell|[ndash]|specific micrornas regulate the g1-s transition and promote rapid proliferation. Nature Genetics. 2008;40(12):1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hornstein E, Shomron N. Canalization of development by micrornas. Nat Genet. 2006;38 Suppl:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 126.Waddington CH. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183(4676):1654–1655. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 127.Lee NS, Kim JS, Cho WJ, et al. Mir-302b maintains "Stemness" Of human embryonal carcinoma cells by post-transcriptional regulation of cyclin d2 expression. Biochem Biophys Res Commun. 2008;377(2):434–440. doi: 10.1016/j.bbrc.2008.09.159. [DOI] [PubMed] [Google Scholar]
- 128.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. Microrna-145 regulates oct4, sox2, and klf4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 129.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific micrornas promote induced pluripotency. Nat Biotechnol. 2009 doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lin SL, Chang DC, Chang-Lin S, et al. Mir-302 reprograms human skin cancer cells into a pluripotent es-cell-like state. RNA. 2008;14(10):2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]