Summary
Mycorrhizal species richness and host ranges were investigated in mixed deciduous stands composed of Fagus sylvatica, Tilia spp., Carpinus betulus, Acer spp., and Fraxinus excelsior. Acer and Fraxinus were colonized by arbuscular mycorrhizas and contributed 5% to total stand mycorrhizal fungal species richness. Tilia hosted similar and Carpinus half the number of ectomycorrhizal (EM) fungal taxa compared with Fagus (75 putative taxa). The relative abundance of the host tree the EM fungal richness decreased in the order Fagus > Tilia >> Carpinus. After correction for similar sampling intensities, EM fungal species richness of Carpinus was still about 30–40% lower than that of Fagus and Tilia. About 10% of the mycorrhizal species were shared among the EM forming trees; 29% were associated with two host tree species and 61% with only one of the hosts. The latter group consisted mainly of rare EM fungal species colonizing about 20% of the root tips and included known specialists but also putative non-host associations such as conifer or shrub mycorrhizas. Our data indicate that EM fungal species richness was associated with tree identity and suggest that Fagus secures EM fungal diversity in an ecosystem since it shared more common EM fungi with Tilia and Carpinus than the latter two among each other.
Electronic supplementary material
The online version of this article (doi:10.1007/s00572-010-0338-y) contains supplementary material, which is available to authorized users.
Keywords: Mycorrhizal community, Deciduous stand, Diversity, Temperate ecosystem
Introduction
In boreal and temperate forests of the palearctic realm, most tree species form ectomycorrhizal (EM) associations with a high number of fungal taxa. In these ecosystems, EM fungal species richness has been mainly studied with host trees of economic importance such as pine (Pinus sylvestris), spruce (Picea abies), oak (Quercus spp.), and beech (Fagus sylvatica). High-throughput sequencing revealed extremely high fungal species diversity in soils of these forests (Reich et al. 2009). Meta-analysis across ecosystems indicated that each of these tree species can be colonized by 160 to 226 different EM fungal taxa (De Roman et al. 2005). Within a given ecosystem, there is also considerable EM fungal diversity. For example, in beech forests, roots of old-growth trees are colonized by about 80 to 90 EM fungal taxa (Buée et al. 2005; Rumberger et al. 2005; Pena et al. 2010). By far less, i.e., only 10 to 15 EM fungal species have been identified for other deciduous European tree species that occur in mixed forests together with Fagus such as Tilia spp. (Tilia cordata Mill., Tilia platyphyllos Scop.) or Carpinus betulus (De Roman et al. 2005; Timonen and Kauppinen 2008). It is unknown whether Tilia and Carpinus are indeed associated with lower numbers of different mycorrhizal species than Fagus or whether these figures simply reflect differences in research intensity. There is also only limited information on the contribution of arbuscular mycorrhiza (AM)-forming tree species such as Fraxinus excelsior L. and Acer spp. (Acer pseudoplatanus L., Acer platanoides L.) to mycorrhizal species richness in mixed deciduous forests.
To date, tree species with a wide ecological amplitude such as Tilia, Carpinus, Fraxinus, and Acer (Ellenberg 1996; Marigo et al. 2000) are gaining importance for silvicultural management since mixed forests with these tree species may be more suitable to withstand climate change with lower summer precipitation anticipated for Central Europe (Gessler et al. 2007). It is expected that increasing tree species richness of forests will increase mycorrhizal fungal species richness due to host preferences of the fungi. This has been reported for boreal and temperate mixed coniferous–deciduous forests (Kernaghan et al. 2003; Ishida et al. 2007) as well as for wet, sclerophyllous forests in Australia (Tedersoo et al. 2008). Surprisingly, information on the importance of host species (i.e., root attachment) for mycorrhizal fungal taxa in mixed deciduous Central European forests is missing.
To uncover host–fungus interactions that shape mycorrhiza diversity in mixed deciduous forests, we characterized mycorrhizal species richness in a forest composed of members of five tree families (Fagaceae, Tiliaceae, Betulaceae, Oleaceae, and Aceraceae). We hypothesized that (1) multi-host fungal species are dominant with respect to root colonization, (2) increasing richness of EM-forming tree species increases EM fungal species richness because of fungal host specificity, and (3) AM-forming tree species contribute little to mycorrhizal fungal species richness in mixed forests. To test these hypotheses, we have chosen mixed deciduous forests containing Fagus, Tilia, Carpinus, Acer, and Fraxinus in the National Park Hainich (Thuringia, Germany). The National Park is covered with old-growth forests, which have not been managed for several decades (Meinen et al. 2009). Study plots identified in same climatic conditions with similar forest and edaphic structures (Leuschner et al. 2009; Meinen et al. 2009) were used for multiple samplings in different seasons to investigate mycorrhizal fungal species richness and their host preferences in this ecosystem.
Materials and methods
Site characteristics
The study was conducted in four deciduous forest stands in the northeastern part of National Park Hainich, Thuringa, Germany (51°05′28′′N, 10°31′24′′E). The forest has not been managed for at least four decades. Long-term annual sum of precipitation is 670 mm and annual mean temperature is 7.5°C (Leuschner et al. 2009). Four different forest plots (50 m × 50 m in the stands DL2b, DL2c, DL3b, and DL3c) at an altitude of 350 m above sea level within a radius of approximately 4 km were used for sampling. Mean tree density was 527 trees ha−1 with a total basal area of 38.8 m2 ha−1 (Online Resource 1). The plots contained F. sylvatica L., Tilia spp. (T. cordata Mill. or T. platyphyllos Scop.), C. betulus L., F. excelsior L., and Acer spp. (A. pseudoplatanus L. or A. platanoides L.) in varying proportions with mean contributions to the basal stem areas of 48%, 17%, 5%, 20%, and 7%, respectively (Online Resource 1). On two of the four plots (DL2b, DL2c), Carpinus was missing and Acer was rare. The plots were classified as Stellario-Carpinetum stachyetosum (DL2b, DL2b, DL3c) and as Hordelymo-Fagetum typicum (DL2c, Mölder et al. 2006). The plots were selected by the following criteria: low anthropogenic impact in the last decades, closed canopy, and homogeneous stand structure (Online Resource 1). All stands stocked on the soil type Luvisol that had developed from loess; the mean pH(H2O) was 5.3 (Guckland et al. 2009).
Sampling scheme
Soil cores were collected randomly using the following strategy: on each plot three 30-m-long lines were determined by choosing the starting point and direction randomly. On each line, five sampling points were randomly determined, thus defining 15 sampling points per plot. Soil cores (diameter 8 cm, depth 20 cm) were taken four times: 9th November 2006, 23rd April 2007, 5th July 2007, and 25th September 2007 adjacent to the defined 15 sampling points. The soil cores were stored at 4°C until analysis for a maximum of 4 weeks. A total of 240 soil cores were analyzed.
Root identification and morphotyping
Soil cores were soaked in tap water for 30 min. All roots were removed by careful washing and stored at 4°C between moist tissue papers. The mean fine root biomass per soil core was 2.3 g (±0.3). The roots of the different tree species were intermingled in the cores since root segregation was not observed (Meinen et al. 2009; Lang et al. 2010). The roots were sorted by C. Lang according to tree species with a stereomicroscope (Stemi SV 11; Zeiss, Jena, Germany) as described by Hölscher et al. (2002) and Korn (2004). Roots of grasses or herbs were rare and if present, they were removed. For the purpose of this study, T. cordata and T. platyphyllos were treated as one species (Tilia spp.) because their roots were indistinguishable. The same applied to Acer spp. (A. pseudoplatanus, A. platanoides). Of the 280 soil cores, 154 contained Fagus, 140 Tilia, 40 Carpinus, 199 Fraxinus, and 66 Acer roots, respectively.
Root tips of Fraxinus and Acer of about 5-mm length were immediately cut and stored frozen −80°C for analyses of arbuscular mycorrhizas (samples from 23rd April and 5th July 2007). For molecular analyses, the root tips were pooled per tree species and per sites and a total of 50 samples were analyzed. Subsamples per tree species, site, and sampling date were stored in 70% EtOH for the determination of AM fungal colonization. Root samples were cleared and stained with Lactophenol Blue after Phillips and Hayman (1970). AM fungal colonization was determined using the magnified intersect technique (McGonigle et al. 1990). The colonization with AM fungi (%) was calculated as: (intersects with AM fungal structures)/(all counted intersects with root tissue) × 100.
EM fungi of Fagus, Tilia, and Carpinus were subjected to morphological classification according to a simplified scheme after Agerer (1987–2006). The presence or absence of a hyphal mantle was recorded for vital root tips and the EM fungal (%) colonization was calculated as: EM root tips/(EM root tips + non-mycorrhizal root tips) × 100. Each morphotype was described by its color, the texture of the ectomycorrhizal mantel, branching, abundance of external hyphae, and rhizomorphs. Pictures were taken (Coolpix 4500; Nikon, Tokyo, Japan) and deposited together with the fungal description and molecular information (see below) under http://www.uni-goettingen.de/de/92389.html. Within each root sample, the following mean numbers of root tips were analyzed: 340 for Fagus, 217 for Tilia, and 280 for Carpinus. About 10 to 20 root tips per morphotype were pooled and stored frozen at −80°C for molecular analysis.
ITS sequencing and database searches
Frozen ectomycorrhizal root tips (a pool of 10 to 20 root tips per morphotype) were ground in a mill (Type MM2; Retsch, Haan, Germany). DNA was extracted with DNeasy Mini Plant Kit (Qiagen, Hilden, Germany) according to the manual. For ectomycorrhiza analyses, the internal transcribed spacer region of the fungal rDNA was amplified by using the primer ITS5 and ITS4 (MWG Biotech, Ebersberg, Germany) after White et al. (1990). The PCR mix was composed of 2.5 μl 10× PCR buffer, 1.5 μl 25 mM MgCl2, 0.5 μl 10 mM dNTPs (Fermentas, St-Leon-Rot, Germany), 2.0 μl 5 mM ITS5, 2.0 μl 5 mM ITS4, 0.1 μl (>10 U/μl) Taq-Polymerase (Thermus aquaticus expressed in Escherichia coli, courtesy of Dr. Patrick Hoegger, Büsgen-Institut, Abteilung Molekulare Holzbiotechnologie, Göttingen, Germany), 15.4 μl double deionized H2O (ddH2O), and 1 μl template DNA. The PCR was performed in a Mastercycler Gradient (Eppendorf, Hamburg, Germany) with the following settings: start 94°C for 60 s, 35 cycles at 94°C for 30 s and 55°C for 30 s, and 72°C for 45 s. After the final elongation step at 72°C for 10 min, the PCR was terminated.
To study AM fungal diversity in the field, a protocol was used that had been developed and successfully applied by Renker et al. (2003) for the detection of Glomeraceae, Archaeosporaceae, Gigasporaceae, Acaulosporaceae, Diversisporaceae, and Paraglomeraceae. Frozen root tips of Fraxinus and Acer were ground in a mill and DNA was extracted as above. DNA samples from each plot were pooled keeping tree species separately. PCR was conducted as above using the primers SSU Glom1 (5′-ATTACGTCCCTGCCCTTTGTACA-3′) and LSU Glom1 (5′-CTTCAATCGTTTCCCTTTCA-3′). Afterwards, the samples were digested with a restriction enzyme (AluI, 10 U/μl), and the resulting products were subjected to a second PCR step with ITS5 and ITS4 to amplify the ITS region as above. The PCR products were cloned. For ligation, the pGEM-T-System I (Promega, Madison, WI, USA) was used according to the instructions of the manufacturer. The plasmids were transformed into electrocompetent E. coli (TOP 10; Invitrogen, Carlsbad, CA, USA) and used for amplification of the ITS region as above.
Before sequencing, the DNA was purified. 2-Propanol (Roth, Karlsruhe, Germany) was added to the PCR products. DNA was precipitated for 1 h and centrifuged at room temperature (30 min, 17,900×g, Centrifuge 5417 R; Eppendorf, Hamburg, Germany). The pellet was air-dried and dissolved in ddH2O. The PCR products were labeled with the Big Dye Terminator Kit (Applied Biosystems, Foster City, CA, USA) and sequenced (ABI Prism 3100 Genetic Analyzer, 36 cm capillary, Matrix Pop 6; Applied Biosystems).
Analysis of the sequences
Sequences were aligned using Staden Package (4.10, http://staden.sourceforge.net). Sequences were compared with the databases NCBI GenBank (http://www.ncbi.nlm.nih.gov) and UNITE (http://unite.ut.ee) for fungal identification. If the homology was higher than 97% and the score higher than 900 bits, the species name suggested by the database was accepted. The name suggested by UNITE, a curated database for EM fungi (Kõljalg et al. 2005), was used preferentially and that of NCBI only if there was no entry in UNITE. The sequences were deposited in NCBI GenBank with the GenBank accession numbers: EU346870, EU346872, EU346875, EU350580–350582, EU816604–816688, EU826353–826355, and EU931248–EU931254. To obtain further information on the fungi, a phylogenetic tree (Online Resource 2) was created with Clustal X (version 1.83, http://bips.u-strasbg.fr/fr/Documentation/ClustalX/), Genedoc (version 2.6.002, http://www.nrb-sc.org/gfx/genedoc/index.html), and Mega (version 3.1, http://www.megasoftware.net/). To calculate the tree, the neighbor-joining method was used with the options: Bootstrap (2,500 replicates; seed = 70,189) and the model: Nucleotide with Kimura two-parameter settings.
Statistical analysis
The statistical analyses were done with STATGRAPHICS Plus for Windows 3.0. The abundance of EM fungi per plot, sampling date, and tree species were determined. Since these data were not normally distributed, the Kruskal–Wallis and Mann–Whitney tests were used to analyze differences in the abundance of fungal species on different tree species. Species accumulation curves (Chao 1) were calculated with EstimateS version 8.2.0 (Colwell 2006) and detrended correspondence analysis (DCA) was conducted with R 2.10.0 (R Development Core Team 2009).
Results
An inventory of mycorrhizal fungi in mixed deciduous stands
Mean EM fungal colonization of vital root tips of the EM-forming tree species Fagus, Tilia, and Carpinus was 96 ± 4%, and the relative abundance of AM fungal hyphae in roots of Fraxinus and Acer was 19 ± 9% regardless of the season and field plot. Fraxinus and Acer were strictly associated with AM fungi and Fagus, Tilia, and Carpinus with EM fungi. Molecular analysis of AM fungi in 50 samples of Fraxinus and Acer roots revealed seven different sequences for glomeromycota (Online Resource 2), of which two occurred only in Fraxinus roots and the others in Fraxinus and Acer.
For EM fungal analysis, a total of 53,322 of Fagus, 30,385 of Tilia, and 11,186 of Carpinus mycorrhizal tips were observed. These figures roughly reflect the fraction of soil cores of 0.55, 0.50, and 0.14 containing Fagus, Tilia, and Carpinus roots, respectively, thus exceeding the above-ground proportions of the latter two taxa. The sampling intensity of Carpinus was lower than that of the other tree species because it occurred on only half of the sampled plots. Taking all data together, a total of 130 different putative EM fungal species were recorded on root tips of Fagus, Tilia, and Carpinus. Of these species, 75 EM fungal were detected on Fagus, 68 on Tilia, and 43 on Carpinus root tips, respectively (Fig. 1a). If only samples from plots with Carpinus were considered, the number of root tips analyzed for Fagus and Tilia decreased to 19,969 and 18,230, but the number of EM fungal species would still be as high as 72 and 63 for Fagus and Tilia, respectively. Exponential fitting of the measured EM fungal species accumulation curves suggested that EM fungal species saturation was reached at 74 EM fungal taxa for Fagus, 66 for Tilia, and 43 for Carpinus, respectively (with R 2 ≥ 0.988, Fig. 1a). Species accumulation curves calculated with EstimateS suggested slightly higher than the measured values for species richness (Fig. 1b).
Fig. 1.
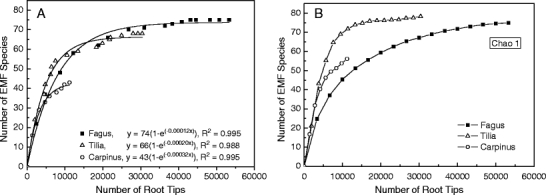
Species richness of ectomycorrhizal (EM) fungi at root tips of Fagus sylvatica, Tilia spp., and Carpinus betulus. Cumulative data from EM fungal analysis in 240 soil cores collected on four different plots and four sampling dates. a Number of root tips and measured species richness in our study. Data were fitted by Boltzmann functions. b Number of root tips and species richness estimated with Chao 1 (settings = 50 runs, without replacement)
All EM fungi were described according to their mantle properties (http://www.uni-goettingen.de/de/99297.html). The majority of these species (73%) was identified by ITS sequencing. These species colonized 97.7%, 93.7%, and 92.8% of the root tips of Fagus, Tilia, and Carpinus, respectively. Only very rare EM fungal species were not analyzed (Table 1, species with missing sequence information). Forty-seven percent of the EM fungal species were identified at the species level and most others at the level of the genus (Online Resource 2). Overall, EM fungal species richness contributed 95% to the total mycorrhizal species richness (EM + AM fungi) in this forest ecosystem.
Table 1.
Ectomycorrhizal fungi of Fagus sylvatica, Tilia spp., and Carpinus betulus roots and their relative abundance (1SE)
| Species (morphotype) | T | ACC | Name of best BLAST match | Abundance (%) | ||
|---|---|---|---|---|---|---|
| (Host) | ||||||
| Fagus | Tilia | Carpinus | ||||
| (Fagus + Tilia + Carpinus) | ||||||
| Amanita rubescens | BA | EU346872 | Amanita rubescens | 0.05 (0.03) | 0.07 (0.04) | 3.68 (2.30) |
| Cenococcum geophilum | AS | EU346870 | Cenococcum geophilum | 9.58a (1.74) | 14.60ab (2.13) | 19.96b (3.54) |
| Clavulina cristata | BA | EU816621 | Clavulina cristata | 14.16b (3.12) | 6.83a (2.34) | 2.38a (1.62) |
| Inocybe maculate | BA | EU816617 | Inocybe maculata | 1.01a (0.67) | 7.31b (1.24) | 15.28c (3.13) |
| Russula delica | BA | EU816643 | Russula delica | 4.85 (1.37) | 11.55 (3.37) | 6.23 (0.96) |
| Xerocomus chrysenteron | BA | EU350581 | Xerocomus chrysenteron | 1.57b (0.50) | 0.02a (0.02) | 1.70ab (1.04) |
| UECM (M73) | AS | EU816646 | Unc ECM (Pezizaceae) | 1.23a (0.67) | 0.63a (0.29) | 4.92b (1.80) |
| UECM (M83) | BA | EU816651 | Unc ECM (Tomentella) | 1.32b (0.63) | 0.08a (0.07) | 1.99ab (1.53) |
| UECM (M84) | BA | EU816652 | Unc ECM fungus | 1.22 (0.83) | 3.16 (1.55) | 0.74 (0.74) |
| UECM (M85) | BA | EU816653 | Unc ECM (Thelephoraceae) | 0.13 (0.10) | 1.70 (1.03) | 0.56 (0.56) |
| UECM (M87) | BA | EU816655 | Unc ECM (Thelephoraceae) | 0.37 (0.21) | 1.11 (0.51) | 1.24 (0.71) |
| (Fagus + Tilia) | ||||||
| Craterellus cornucopioides | BA | EU816605 | Craterellus cornucopioides | 2.49b (1.00) | 1.96a (1.26) | |
| Humaria hemisphaerica | AS | EU816610 | Humaria hemisphaerica | 1.58 (0.78) | 1.04 (0.66) | |
| Inocybe umbrina | BA | EU816641 | Inocybe umbrina | 1.67b (0.92) | 0.01a (0.01) | |
| Inocybe sp. (M88) | BA | EU816656 | Inocybe sp. | 0.15a (0.10) | 1.74b (0.68) | |
| Lactarius subdulcis | BA | EU346875 | Lactarius subdulcis | 12.69b (3.12) | 1.13a (0.44) | |
| Russula chloroides | BA | EU816642 | Russula chloroides | 1.61b (0.55) | 0.64a (0.45) | |
| Tomentella stuposa | BA | EU816618 | Tomentella stuposa | 0.33 (0.22) | 0.05 (0.05) | |
| Tomentella sublilacina | BA | EU816604 | Tomentella sublilacina | 11.11b (2.56) | 1.64a (0.59) | |
| Tomentella terrestris | BA | EU816649 | Tomentella terrestris | 0.93 (0.57) | 3.70 (2.55) | |
| Tomentella viridula | BA | EU816647 | Tomentella viridula | 1.10 (0.54) | 0.11 (0.11) | |
| Xerocomus pruinatus | BA | EU350582 | Xerocomus pruinatus | 0.60 (0.32) | 0.16 (0.12) | |
| UECM (M40) | BA | EU816627 | Unc ECM (Tomentella) | 1.22b (0.61) | 0.21a (0.16) | |
| UECM (M57) | BA | EU816638 | Tomentella sp. | 0.01 (0.01) | 0.62 (0.49) | |
| UECM (M63) | BA | EU816640 | Inocybe rimosa | 0.07 (0.04) | 0.33 (0.11) | |
| UECM (M82) | BA | EU816650 | Unc ECM fungus | 1.56 (0.88) | 0.13 (0.10) | |
| UECM (M86) | BA | EU816654 | Tomentella bryophila | 0.18a (0.17) | 1.27b (0.56) | |
| (Fagus + Carpinus) | ||||||
| Melanogaster broomeianus | BA | EU816648 | Melanogaster broomeianus | 0.23 (0.14) | 0.56 (0.56) | |
| Russula puellaris | BA | EU816628 | Russula puellaris | 0.86 (0.36) | 1.29 (0.93) | |
| Thelephoraceae sp. (M5) | BA | EU816607 | Thelephoraceae sp. | 1.15b (0.31) | 7.91a (0.87) | |
| Tomentella pilosa | BA | EU816644 | Tomentella pilosa | 1.13 (0.87) | 0.06 (0.06) | |
| Tuber puberulum | AS | EU816619 | Tuber puberulum | 0.46a (0.18) | 2.17b (1.02) | |
| UECM (M6) | BA | EU816608 | Unc ECM (Tomentella) | 1.79b (0.64) | 0.57a (0.57) | |
| UECM (M42) | AS | EU816629 | Unc ECM (Trichocomaceae) | 0.15 (0.11) | 1.16 (0.86) | |
| M25 | 0.04 (0.03) | 0.40 (0.34) | ||||
| M33 | 0.02 (0.02) | 0.09 (0.09) | ||||
| M58 | 0.06 (0.04) | 0.25 (0.20) | ||||
| (Tilia + Carpinus) | ||||||
| Cortinarius infractus | BA | EU816664 | Cortinarius infractus | 0.06 (0.06) | 1.00 (0.99) | |
| Piloderma lanatum | BA | EU816674 | Piloderma lanatum | 0.16a (0.12) | 1.78b (1.04) | |
| Rhizopogon sp. (M103) | BA | EU816666 | Rhizopogon sp. | 0.06 (0.06) | 0.18 (0.18) | |
| Sebacina sp. (M90) | BA | EU826353 | Sebacina sp. | 0.97b (0.37) | 2.51a (0.61) | |
| UECM (M99) | BA | EU816663 | Tomentella coerulea | 0.40 (0.29) | 1.15 (0.47) | |
| UECM (M102) | AS | EU816665 | Unc ECM (Trichocomaceae) | 6.09b (1.53) | 0.50a (0.50) | |
| UECM (M125) | BA | EU816679 | Unc ECM (Sebacinaceae) | 0.33 (0.24) | 0.07 (0.07) | |
| M130 | 0.76 (0.34) | 2.76 (2.76) | ||||
| (Fagus) | ||||||
| Cortinarius anomalus | BA | EU816625 | Cortinarius anomalus | 0.21 (0.19) | ||
| Genea hispidula | AS | EU816611 | Genea hispidula | 0.95 (0.40) | ||
| Inocybe asterospora | BA | EU816612 | Inocybe asterospora | 0.89 (0.57) | ||
| Tomentella sp. (M61) | BA | EU816639 | Tomentella sp. | 0.84 (0.45) | ||
| Laccaria maritima | BA | EU816633 | Laccaria maritima | 0.26 (0.16) | ||
| Lactarius blennius | BA | EU816609 | Lactarius blennius | 0.32 (0.15) | ||
| Lactarius fluens | BA | EU816606 | Lactarius fluens | 0.48 (0.25) | ||
| Russula fellea | BA | EU816623 | Russula fellea | 1.41 (0.67) | ||
| Russula ochroleuca | BA | EU350580 | Russula ochroleuca | 5.54 (2.28) | ||
| Russula raoultii | BA | EU816634 | Russula raoultii | 2.30 (1.44) | ||
| Russula solaris | BA | EU816636 | Russula solaris | 0.37 (0.16) | ||
| Xerocomus badius | BA | EU816626 | Xerocomus badius | 0.27 (0.25) | ||
| UECM (M13) | BA | EU816613 | Unc ECM (Russula) | 2.02 (0.77) | ||
| UECM (M14) | AS | EU816614 | Hydnotrya tulasnei | 0.05 (0.03) | ||
| UECM (M18) | BA | EU816615 | Unc ECM (Atheliaceae) | 0.14 (0.10) | ||
| UECM (M28) | AS | EU816622 | Unc ECM fungus | 1.18 (0.59) | ||
| UECM (M44) | AS | EU816630 | Unc ECM (Pezizaceae) | 0.14 (0.10) | ||
| UECM (M45) | AS | EU816631 | Unc ECM | 0.14 (0.06) | ||
| UECM (M52) | AS | EU816635 | Uncultured soil fungus clone | 0.14 (0.14) | ||
| UECM (M56) | AS | EU816637 | Uncultured (Pezizomycotina) | 0.41 (0.18) | ||
| UECM (M70) | BA | EU816645 | Inocybe glabripes | 0.60 (0.46) | ||
| M7* | 0.10 (0.10) | |||||
| M16 | 0.17 (0.11) | |||||
| M20 | 0.20 (0.20) | |||||
| M31 | 0.06 (0.04) | |||||
| M34 | 0.21 (0.19) | |||||
| M38* | 0.12 (0.12) | |||||
| M46 | 0.36 (0.20) | |||||
| M47 | 0.06 (0.04) | |||||
| M50 | 0.09 (0.06) | |||||
| M55 | 0.30 (0.11) | |||||
| M64 | 0.01 (0.01) | |||||
| M65* | 0.06 (0.06) | |||||
| M71 | 0.33 (0.29) | |||||
| M72 | 0.29 (0.21) | |||||
| M76 | 0.06 (0.05) | |||||
| M78* | 0.05 (0.05) | |||||
| M81* | 0.02 (0.02) | |||||
| (Tilia) | ||||||
| Inocybe cookie | BA | EU816677 | Inocybe cookie | 1.64 (1.48) | ||
| Inocybe geophylla | BA | EU816657 | Inocybe geophylla | 2.11 (0.70) | ||
| Russula cyanoxantha | BA | EU816662 | Russula cyanoxantha | 0.37 (0.20) | ||
| Russula pectinatoides | BA | EU816667 | Russula pectinatoides | 4.02 (1.67) | ||
| Sebacina aff. epigaea | BA | EU816673 | Sebacina aff. Epigaea | 1.58 (1.05) | ||
| Peziza michelii | AS | EU816678 | Peziza michelii | 0.57 (0.30) | ||
| Peziza succosa* | AS | EU816676 | Peziza succosa | 0.21 (0.21) | ||
| Tuber borchii | AS | EU816671 | Tuber borchii | 0.22 (0.12) | ||
| UECM (M19) | AS | EU816616 | Unc ECM (Pezizaceae) | 3.92 (1.58) | ||
| UECM (M91) | BA | EU816658 | Unc ECM (Inocybe) | 2.04 (0.77) | ||
| UECM (M92) | AS | EU816659 | Unc ECM (Pezizales) | 0.03 (0.03) | ||
| UECM (M93) | BA | EU816660 | Uncultured Thelephoraceae | 0.46 (0.32) | ||
| UECM (M95) | BA | EU816661 | Inocybe actuella | 1.06 (0.64) | ||
| UECM (M106) | AS | EU816668 | Unc ECM (Hydnobolites) | 0.82 (0.61) | ||
| UECM (M108) | BA | EU816669 | Unc ECM fungus | 0.27 (0.15) | ||
| UECM (M109)* | AS | EU816670 | Unc ECM (Terfeziaceae) | 0.32 (0.32) | ||
| UECM (M115) | BA | EU816672 | Unc ECM (Thelephoraceae) | 0.43 (0.35) | ||
| UECM (M119)* | AS | EU816675 | Unc ECM (Pezizaceae) | 1.23 (1.23) | ||
| UECM (M126) | BA | EU816680 | Inocybe sp. | 1.87 (0.79) | ||
| UECM (M127) | BA | EU816681 | Tomentella sp. | 2.01 (0.86) | ||
| UECM (M128) | BA | EU816682 | Tomentella sp. | 0.42 (0.30) | ||
| M104* | 0.14 (0.14) | |||||
| M107 | 0.03 (0.02) | |||||
| M110 | 0.58 (0.29) | |||||
| M112 | 0.30 (0.22) | |||||
| M114 | 0.22 (0.12) | |||||
| M117 | 1.04 (0.55) | |||||
| M120 | 0.31 (0.19) | |||||
| M122 | 0.42 (0.25) | |||||
| M129 | 0.04 (0.03) | |||||
| M131 | 0.13 (0.10) | |||||
| M132 | 0.16 (0 09) | |||||
| M133 | 0.41 (0.22) | |||||
| (Carpinus) | ||||||
| Inocybe corydalina | BA | EU816683 | Inocybe corydalina | 0.99 (0.74) | ||
| Inocybe hirtella | BA | EU826355 | Inocybe hirtella | 0.29 (0.21) | ||
| UECM (M135) | BA | EU816684 | Unc ECM (Sebacinaceae) | 1.35 (0.82) | ||
| UECM (M136) | AS | EU816685 | Vouchered mycorrhizae (Humaria) | 0.57 (0.27) | ||
| UECM (M137) | BA | EU816686 | Unc ECM (Agaricales) | 9.20 (3.73) | ||
| UECM (M138) | AS | EU816687 | Unc ECM (Tuber) | 0.87 (0.41) | ||
| UECM (M142)* | AS | EU816688 | Hymenoscyphus ericae | 0.11 (0.11) | ||
| UECM (M143) | AS | EU826354 | Uncultured soil fungus | 0.45 (0.30) | ||
| M15* | 0.22 (0.22) | |||||
| M39* | 0.36 (0.36) | |||||
| M139* | 0.04 (0.04) | |||||
| M140* | 0.14 (0.14) | |||||
| M141 | 0.91 (0.94) | |||||
| M145 | 1.41 (0.84) | |||||
EM fungi were identified by ITS sequencing. If the sequence homology was higher than 97% and the score higher than 900 bits, the name suggested by the database (UNITE, NCBI) was accepted. Unknown ectomycorrhizas were called UECM if sequence information was available. The genus or family name is indicated under best BLAST match (for further details, see Online Resource 2). Species for which sequence information was not available were denominated by an internal morphotype number (M). Morphotypes can be viewed under http://www.uni-goettingen.de/de/goe-fungi/92389.html. For statistical analysis, data per plot (n = 4), date (n = 4), and tree species were used (n = 3). Since the data were not normally distributed, a non-parametric statistical test (Mann–Whitney test) was used and significant differences at p ≤ 0.05 between tree species were indicated by different letters
T taxonomic classification according to basidiomycota (BA) or ascomycota (AS), ACC accession number in NCBI databank, Unc ECM uncultured ectomycorrhiza, AS ascomycota, BA basidiomycota, M morphotype, UECM unknown ectomycorrhizal fungus. *Singleton found only in one sample
Phylogenetic analysis indicated that the EM fungal species were from all major fungal families known to form EM (Online Resource 3). Approximately 25% belonged to the ascomycota (Online Resource 3). Among all genera, Tomentella sp. and Inocybe sp. were most abundant and contributed 24–26% and 11–16%, respectively, to species richness roots of Fagus, Tilia, and Carpinus.
Host range and preferences of EM fungal species
To disentangle possible effects of season and site from host effects, a DCA was conducted (Fig. 2). Prior to DCA, morphotypes that only occurred in one sample (Table 1) were removed. The first two axes of the DCA, which explained 37.6% (DCA1) and 28.7% (DCA2) of the variance, clearly separated fungi according to tree species (Fig. 2). This shows that the most important factor for the EM fungal community composition was tree species identity and neither sampling site nor sampling date (Fig. 2).
Fig. 2.
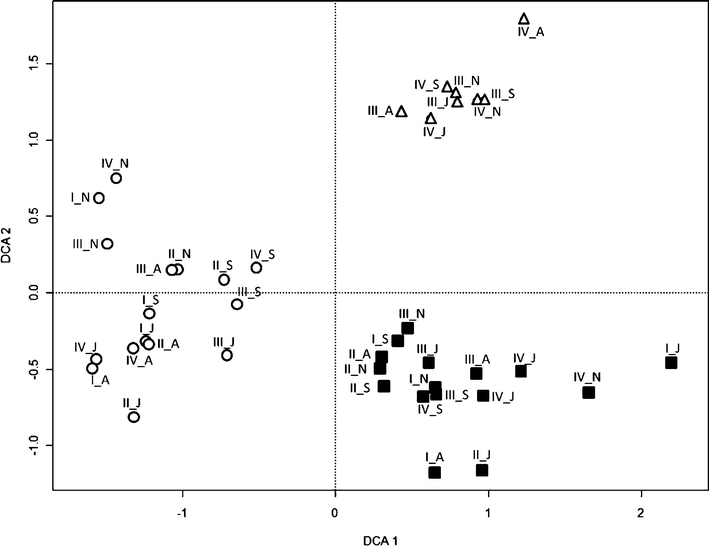
Detrended correspondence analysis (DCA) for the EM fungal community structures per plot, sampling date, and tree species. The analysis was based on the relative abundance of EM fungi. One hundred percent of the mycorrhizal root tips are all EM fungi for each sampling date, plot, and tree species. I–IV plot numbers, N November 2006, A April 2007, J July 2007, S September 2007, circles Fagus sylvatica, triangles Carpinus betulus, closed squares Tilia spp.
Among all EM fungal taxa, 11 species were commonly found on root tips of Fagus, Tilia, and Carpinus and were therefore classified as species with broad host range. These multi-host EM fungi colonized about 35% (Fagus) to 60% (Carpinus) of the root tips (Fig. 3). The most abundant species were Clavulina cristata, Cenococcum geophilum, Russula delica, and Inocybe maculata (Table 1). In spite of their ability to colonize the different host trees studied here, about half of the multi-host EM fungal species showed significantly higher abundances on one or two tree species than on the remaining ones pointing to pronounced host preferences in this ecological context (Table 1).
Fig. 3.
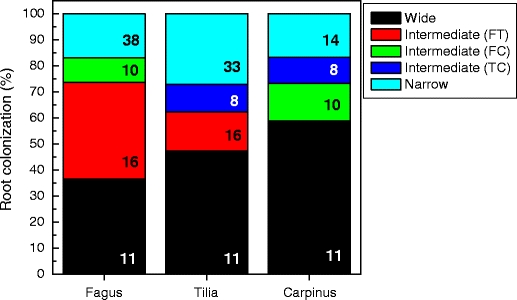
Overview on relative abundance and species richness of ectomycorrhizal taxa on roots of Fagus sylvatica, Tilia spp., and Carpinus betulus. Figures in the bars indicate the number of EM fungal species, which were classified as W = EM fungi with a wide host range found on Fagus, Tilia, and Carpinus (black); I = EM fungi with an intermediate host range occurring on roots of two tree taxa (red Fagus and Tilia, green Fagus and Carpinus, marine Tilia and Carpinus); and N = EM fungi with a narrow host range occurring only on one tree taxon (turquoise)
Thirty-four EM fungal species were found on two host tree species and were therefore classified as EM fungi with intermediate host range (Fig. 3). Most of these EM fungal species showed significantly higher abundance on root tips of one of the two host trees, indicating host preferences (Table 1). For example, Lactarius subdulcis and Tomentella sublilacina were abundant on Fagus, scarce on Tilia, and absent on Carpinus roots. Similarly, a Thelephoraceae species was abundant on Carpinus, and a Trichocomaceae species (UECM-M102) was abundant on Tilia and infrequent on Fagus and Carpinus roots, respectively. The EM fungi with an intermediate and broad host range colonized together about 75% to 85% of the root tips (Fig. 3).
Eighty-five EM fungal species were found only on roots of one host tree and were therefore classified as species with a narrow host range. Among the identified EM fungal species, 21 were unique for Fagus, 21 for Tilia, and eight for Carpinus in our study (Table 1). The EM fungi with narrow host range contributed 65% to species richness, but colonized only 15–25% of the root tips (Fig. 3).
Discussion
A comparison of mycorrhizal species richness of different host taxa in a mixed deciduous temperate forest
This study provides a comprehensive analysis of mycorrhizal community species richness in beech forests mixed with Tilia, Carpinus, Fraxinus, and Acer. Tilia has occasionally been reported to form AM (Harley and Harley 1987 and references therein), but this was not observed here. Both Tilia and Carpinus shared more common EM fungal taxa with Fagus than among each other. Therefore, we suggest that Fagus, which is the potentially dominant tree species in most Central European forests (Ellenberg 1996), secures EM fungal species richness and is therefore ecologically important as a warrantor of EM fungal diversity. Tilia was colonized by the same set of abundant EM fungi and moreover hosted a high number of unique EM fungal species with low abundance as did Fagus. Tilia was, thus, ecologically equivalent in fostering high EM fungal community species richness. This result is important because, in the light of the current debate on prevention of biodiversity erosion, our data suggest that Tilia is recommendable as a host taxon able to maintain high mycorrhizal diversity.
Based on the data of our study, Carpinus appears to be less useful in this respect. If we corrected for the higher sampling intensities of Tilia and Fagus, their EM fungal species richness would decrease only marginally (−5 and −3 EM fungal taxa) and, thus, would still be 32% and 40% higher than that of Carpinus. However, on our study plots Carpinus was a subordinate tree species (Jacob et al. 2010). Overstory plants can influence EM fungal diversity of understory plants (Kennedy et al. 2003). Since EM fungal species richness, especially that of rare species, is strongly dependent on plant carbon productivity and supply with recent photoassimilates (Druebert et al. 2009; Pena et al. 2010), it is possible that carbohydrate allocation to the below-ground compartment was too limited for Carpinus to maintain high EM fungal species richness. This would suggest that, in addition to the host tree species composition, the stand structure might also have influenced mycorrhizal community richness. To unravel the factors controlling mycorrhizal fungal species richness, this aspect will deserve further attention in future studies.
In contrast to EM fungi, the contribution of AM fungal taxa to mycorrhizal species richness was low (5%). Their host tree species Fraxinus and Acer were as abundant as Tilia and Carpinus, respectively, in this ecosystem (Meinen et al. 2009) and were found here in 66% and 23% of the soil cores. We can, therefore, assume that AM fungi have ample opportunities for root colonization. Employing pyrosequencing for the detection of fungi in different forest soils, Bueé et al. (2009) also found only one operational taxonomic unit (OTU) belonging to the glomeromycota compared to 33 OTUs classified as potential EM fungal species. These observations support earlier notions that the species richness of AM fungi is generally lower than that of EM fungi (Smith and Read 2008).
Host ranges and preferences of EM fungi
Host range and host specificity are important determinants of EM fungal community composition in mixed forests. In our study, the number of multi-host EM fungal species, i.e., fungi associated with Fagus, Carpinus, and Tilia, constituted only a small fraction (8%) of the total mycorrhizal species richness. Since these multi-host EM fungi colonized the largest fraction of the root tips of EM-forming host trees, our data support that mycorrhizal species with a large host range are strong competitors (Horton and Bruns 1998; Cullings et al. 2001; Kennedy et al. 2003; Richard et al. 2005; Walker et al. 2005; Ishida et al. 2007; Twieg et al. 2007; Hubert and Gehring 2008; Tedersoo et al. 2008). The advantage may be that plants in different environments can always find suitable fungal associates (Bruns et al. 2002). But the differences in colonization found here for different tree taxa suggest that multi-host EM fungal species still exhibit host preferences in a given ecological context or that their competitiveness differs on different host trees.
The majority of EM fungi in this ecosystem showed clear host preferences. The category of fungi with narrow host range contained specialists, for example fungi typically associated with Fagaceae such as Lactarius blennius, Lactarius fluens, Russula fellea, Russula raoultii, and Russula solaris (Brand 1991; Beenken 2004, Agerer in http://www.deemy.de/). An advantage of specialized associations may be improved adaptation to host physiology for nutrient exchange (Baxter and Dighton 2001; Hobbie et al. 2005) or other host or fungal benefits. Since the “host range” categories used in this study were based on colonization patterns, they reflect realization of ecological niches on the background of genetic affinities or barriers to certain plant–fungus interactions (Molina and Trappe 1982; Molina et al. 1992; Dickie 2007). Therefore, they are flexible rather than fixed entities. For example, various EM fungal species which have previously been documented only on Fagus or Quercus (Inocybe umbrina; UNITE, http://www.unite.ut.ee), Peziza michelii (Tedersoo et al. 2006), Russula pectinatoides (Agerer, http://www.deemy.de; Dickie and Reich 2005), Tomentella terrestris (Kjøller 2006), and C. cristata (Buée et al. 2005; 2007; Kjøller 2006) were found here for the first time on Tilia or Carpinus. Therefore, our study shows that the host range of these fungi is greater than previously known. Otherwise, fungi species known from the literature to associate with Fagus, e.g., Russula cyanoxantha (Agerer in http://www.deemy.de; DeRoman et al. 2005; Grebenc and Kraigher 2007) and Cortinarius infractus (Garnica et al. 2003) were not colonizing Fagus roots but Tilia and Carpinus. This suggests that their host preferences are also strongly affected by ecological factors. The category of fungi with narrow host range furthermore included taxa previously not known as colonizers of Fagus: Cortinarius anomalus is a documented associate of shrubs of maqui in semi-arid climate (Cistus sp.) and of early succession tree species such as Salix sp. (Watling 2005; Barden 2007); Xerocomus badius is a typical EM fungus of spruce (Gronbach 1988) and Inocybe asterospora of the orchid Cephalathera longifolia (Leake 2004). Colonization of Fagus roots with putative non-host EM fungal taxa has been reported previously (Pena et al. 2010). These non-host EM fungal associations occurred only at low frequency and were very labile when the photoassimilate supply was interrupted (Pena et al. 2010). It has been suggested that Fagus provides ecosystem services by maintaining non-host fungi which may constitute the insurance for future forest development and adaptation to changing environmental conditions (Pena et al. 2010).
Concluding remarks
It is well known that soil properties, climatic conditions, and physiological factors such as tree age affect EM fungal species richness in a stand (Conn and Dighton 2000; Wardle 2006). In accordance with other studies (Kernaghan et al. 2003; Ishida et al. 2007; Tedersoo et al. 2008), we found that the number of mycorrhizal species increased with increasing number of host tree species. Since the contributions of different tree taxa to mycorrhizal species numbers varied considerably, our study highlights that the increment in fungal species numbers depended on tree species identity or tree social status but was not simply a function of tree species numbers (Dickie 2007). Therefore, stand composition is important for below-ground mycorrhizal community species richness. We found clear host preferences. However, overall generalist fungi colonized the major fraction of root tips.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(XLS 29 kb)
(DOC 254 kb)
(DOC 44 kb)
Acknowledgments
We are grateful to T. Klein, R. Schulz, and P. Hawighorst for help with sample collection and to M. Franke-Klein, M. Fastenrath, and R. Pena for help with morphotyping. We thank the DFG (German Science Foundation) for funding this work in the context of the graduate school “Die Bedeutung der Biodiversität für Stoffkreisläufe und biotische Interaktionen in temperaten Laubwäldern (GRK 1086)” and two anonymous reviewers for helpful comments.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Agerer R (1987–2006) Colour atlas of Ectomycorrhizae. Einhorn Verlag + Druck GmbH, Schwäbisch Gmünd
- Barden N. Helianthemum grasslands of the peak district and their possible mycorrhizal associates. Field Mycol. 2007;8:119–126. doi: 10.1016/S1468-1641(10)60384-2. [DOI] [Google Scholar]
- Baxter JW, Dighton J. Ectomycorrhizal diversity alters the growth of gray birch Betula populifolia (Marshall) seedlings in experimental host-symbiont culture conditions. New Phytol. 2001;152:139–149. doi: 10.1046/j.0028-646x.2001.00245.x. [DOI] [PubMed] [Google Scholar]
- Beenken L (2004) Die Gattung Russula. Untersuchungen zu ihrer Systematik anhand von Ektomykorrhizen. Dissertation, Universität München. Available at http://edoc.ub.uni-muenchen.de/archive/00003175/01/Beenken_Ludwig.pdf
- Brand F. Ektomykorrhizen an Fagus sylvatica. Charakterisierung und Identifizierung, ökologische Kennzeichnung und unsterile Kultivierung. Libri Botanici. 1991;2:1–229. [Google Scholar]
- Bruns TD, Bidartondo MI, Taylor DL. Host specificity in ectomycorrhizal communities: what do the exceptions tell us? Integ Comp Biol. 2002;42:352–359. doi: 10.1093/icb/42.2.352. [DOI] [PubMed] [Google Scholar]
- Bueé M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, Martin F. 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 2009;184:449–456. doi: 10.1111/j.1469-8137.2009.03003.x. [DOI] [PubMed] [Google Scholar]
- Buée M, Vairelles D, Garbaye J. Year-round monitoring of diversity and potential metabolic activity of the ectomycorrhizal community in a beech (Fagus sylvatica) forest subjected to two thinning regimes. Mycorrhiza. 2005;15:235–245. doi: 10.1007/s00572-004-0313-6. [DOI] [PubMed] [Google Scholar]
- Buée M, Courty PE, Mignot D, Garbaye J. Soil niche effect on species diversity and catabolic activities in an ectomycorrhizal fungal community. Soil Biol Biochem. 2007;39:1947–1955. doi: 10.1016/j.soilbio.2007.02.016. [DOI] [Google Scholar]
- Colwell RK (2006) EstimateS: statistical estimation of species richness and shared species from samples, version 8. Available at purl.oclc.org/estimates
- Conn C, Dighton J. Litter quality influences on decomposition, ectomycorrhizal community structure and mycorrhizal root surface acid phosphatase activity. Soil Biol Biochem. 2000;32:489–496. doi: 10.1016/S0038-0717(99)00178-9. [DOI] [Google Scholar]
- Cullings KW, Vogler DV, Parker VT, Makhija S. Defoliation effects on the ectomycorrhizal community of a mixed Pinus contorta/Picea engelmannii stand in Yellowstone Park. Oecologia. 2001;127:533–539. doi: 10.1007/s004420000610. [DOI] [PubMed] [Google Scholar]
- De Roman M, Claveria V, DeMiguel AM. A revision of the descriptions of ectomycorrhizas published since 1961. Mycol Res. 2005;109:1063–1104. doi: 10.1017/S0953756205003564. [DOI] [PubMed] [Google Scholar]
- Dickie IA. Host preference, niches and fungal diversity. New Phytol. 2007;174:230–233. doi: 10.1111/j.1469-8137.2007.02055.x. [DOI] [PubMed] [Google Scholar]
- Dickie IA, Reich PB. Ectomycorrhizal fungal communities at forest edges. J Ecol. 2005;93:244–255. doi: 10.1111/j.1365-2745.2005.00977.x. [DOI] [Google Scholar]
- Druebert C, Lang C, Valtanen K, Polle A. Beech carbon productivity as driver of ectomycorrhizal abundance and diversity. Plant Cell Environ. 2009;32:992–1003. doi: 10.1111/j.1365-3040.2009.01983.x. [DOI] [PubMed] [Google Scholar]
- Ellenberg H. Vegetation Mitteleuropas mit den Alpen in ökologischer, dynamischer und historischer Sicht. 5. Stuttgart: Ulmer; 1996. [Google Scholar]
- Garnica S, Weiß M, Oertel B, Oberwinkler F. Phylogenetic relationships of European Phlegmacium species (Cortinarius, Agaricales) Mycol. 2003;95:1155–1170. doi: 10.2307/3761917. [DOI] [PubMed] [Google Scholar]
- Gessler A, Keitel C, Kreuzwieser J, Matyssek R, Seiler W, Rennenberg H. Potential risks for European beech (Fagus sylvatica L.) in a changing climate. Trees. 2007;21:1–11. doi: 10.1007/s00468-006-0107-x. [DOI] [Google Scholar]
- Grebenc T, Kraigher H. Types of ectomycorrhiza of mature beech and spruce at ozone-fumigated and control forest plots. Environ Monit Assess. 2007;127:47–59. doi: 10.1007/s10661-006-9414-3. [DOI] [PubMed] [Google Scholar]
- Gronbach E. Charakterisierung und Identifizierung von Ektomykorrhizen in einem Fichtenbestand mit Untersuchungen zur Merkmalsvariabilität in sauer beregneten Flächen. Bibl Mycol. 1988;125:1–217. [Google Scholar]
- Guckland A, Brauns M, Flessa H, Thomas F, Leuschner C. Acidity, nutrient stocks and organic matter content in soils of a temperate deciduous forest with different abundance of European beech (Fagus sylvatica L.) J Plant Nutr Soil Sci. 2009;172:500–511. doi: 10.1002/jpln.200800072. [DOI] [Google Scholar]
- Harley JL, Harley EL. A check-list of mycorrhiza in the British flora. New Phytol. 1987;105:1–102. doi: 10.1111/j.1469-8137.1987.tb00674.x. [DOI] [Google Scholar]
- Hobbie EA, Jumpponen A, Trappe J. Foliar and fungal 15N:14N ratios reflect development of mycorrhizae and nitrogen supply during primary succession: testing analytical models. Oecologia. 2005;146:258–268. doi: 10.1007/s00442-005-0208-z. [DOI] [PubMed] [Google Scholar]
- Hölscher D, Hertel D, Leuschner C, Hottkowitz M. Tree species diversity and soil patchiness in a temperate broad-leaved forest with limited rooting space. Flora. 2002;197:118–125. [Google Scholar]
- Horton TR, Bruns TD. Multiple-host fungi are the most frequent and abundant ectomycorrhizal types in a mixed stand of Douglas fir (Pseudotsuga menziesii) and bishop pine (Pinus muricata) New Phytol. 1998;139:331–339. doi: 10.1046/j.1469-8137.1998.00185.x. [DOI] [Google Scholar]
- Hubert NA, Gehring CA. Neighboring trees affect ectomycorrhizal fungal community composition in a woodland-forest ecotone. Mycorrhiza. 2008;18:363–374. doi: 10.1007/s00572-008-0185-2. [DOI] [PubMed] [Google Scholar]
- Ishida TA, Nara K, Hogetsu T. Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer–broadleaf forests. New Phytol. 2007;174:430–440. doi: 10.1111/j.1469-8137.2007.02016.x. [DOI] [PubMed] [Google Scholar]
- Jacob M, Leuschner C, Thomas FM. Productivity of temperate broad-leaved forest stands differing in tree species diversity. Ann For Sci. 2010;67:503. doi: 10.1051/forest/2010005. [DOI] [Google Scholar]
- Kennedy PG, Izzo AD, Bruns TD. There is high potential for the formation of common mycorrhizal networks between understorey and canopy trees in a mixed evergreen forest. J Ecol. 2003;91:1071–1080. doi: 10.1046/j.1365-2745.2003.00829.x. [DOI] [Google Scholar]
- Kernaghan G, Widden P, Bergeron Y, Légaré S, Paré D. Biotic and abiotic factor affecting ectomycorrhizal diversity in boreal mixed-woods. Oikos. 2003;102:497–504. doi: 10.1034/j.1600-0706.2003.12415.x. [DOI] [Google Scholar]
- Kjøller R. Disproportionate abundance between ectomycorrhizal root tips and their associated mycelia. FEMS Microbiol Ecol. 2006;58:214–224. doi: 10.1111/j.1574-6941.2006.00166.x. [DOI] [PubMed] [Google Scholar]
- Kõljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Hoiland K, Kjoller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Vrålstad T, Ursing BM. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005;166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- Korn S (2004) Experimentelle Untersuchung der Wasseraufnahme und der hydraulischen Eigenschaften des Wurzelsystems von sechs heimischen Baumarten, Dissertation, Universität zu Göttingen. Available at http://webdoc.sub.gwdg.de/diss/2004/korn/korn.pdf
- Lang C, Dolynska A, Finkeldey R, Polle A. Are beech roots territorial? For Ecol Manage. 2010;260:1212–1217. doi: 10.1016/j.foreco.2010.07.014. [DOI] [Google Scholar]
- Leake JR. Myco-heterotroph/epiparasitic plant interactions with ectomycorrhizal and arbuscular mycorrhizal fungi. Curr Op Plant Biol. 2004;7:422–428. doi: 10.1016/j.pbi.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Leuschner C, Jungkunst HF, Fleck S. Functional role of forest diversity: pros and cons of synthetic stands and across-site comparisons in established forests. Basic Appl Ecol. 2009;10:1–9. doi: 10.1016/j.baae.2008.06.001. [DOI] [Google Scholar]
- Marigo G, Peltier JP, Girel J, Pautou G. Success in the demographic expansion of Fraxinus excelsior L. Trees. 2000;15:1–13. doi: 10.1007/s004680000061. [DOI] [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. A new method which gives an objective measure of colonization of roots by vesicular–arbuscular mycorrhizal fungi. New Phytol. 1990;115:495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x. [DOI] [PubMed] [Google Scholar]
- Meinen C, Leuschner Ch, Ryan NT, Hertel D. No evidence of spatial root system segregation and elevated fine root biomass in multi-species temperate broad-leaved forests. Trees. 2009;23:941–950. doi: 10.1007/s00468-009-0336-x. [DOI] [Google Scholar]
- Mölder A, Bernhardt-Römermann M, Schmidt W. Forest ecosystem research in Hainich National Park (Thuringia): first results on flora and vegetation in stands with contrasting tree species diversity. Waldoekologie online. 2006;3:83–99. [Google Scholar]
- Molina R, Trappe J. Patterns of ectomycorrhizal host specificity and potential among Pacific Northwest conifers and fungi. For Sci. 1982;28:423–458. [Google Scholar]
- Molina R, Massicotte H, Trappe JM. Specificity phenomenon in mycorrhizal symbiosis: community ecological consequences and practical implications. In: Allen MF, editor. Mycorrhizal functioning. London: Chapman and Hall; 1992. pp. 357–423. [Google Scholar]
- Pena R, Offermann C, Simon J, Naumann PS, Geßler A, Holst J, Mayer H, Kögel-Knabner I, Rennenberg H, Polle A. Girdling affects ectomycorrhizal diversity and reveals functional differences of EM community composition in a mature beech forest (Fagus sylvatica) Appl Environ Microbiol. 2010;76:1831–1841. doi: 10.1128/AEM.01703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JM, Hayman DS. Improved procedures for clearing roots and staining parasitic and vesicular–arbuscular mycorrhizal fungi for rapid assessment of infection. Trans Brit Mycol Soc. 1970;55:158–160. doi: 10.1016/S0007-1536(70)80110-3. [DOI] [Google Scholar]
- R Development Core Team (2009). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. Available at http://www.R-project.org
- Reich M, Kohler A, Martin F, Buée M. Development and validation of an oligonucleotide microarray to characterise ectomycorrhizal fungal communities. BMC Microbiology. 2009;9:241. doi: 10.1186/1471-2180-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renker C, Heinrichs J, Kaldorf M, Buscot F. Combining nested PCR and restriction digest of the internal transcribed spacer region to characterize arbuscular mycorrhizal fungi on roots from the field. Mycorrhiza. 2003;13:191–198. doi: 10.1007/s00572-002-0214-5. [DOI] [PubMed] [Google Scholar]
- Richard F, Millot S, Gardes M, Selosse MA. Diversity and specificity of ectomycorrhizal fungi retrieved from an old-growth Mediterranean forest dominated by Quercus ilex. New Phytol. 2005;166:1011–1023. doi: 10.1111/j.1469-8137.2005.01382.x. [DOI] [PubMed] [Google Scholar]
- Rumberger MD, Lentzsch P, Münzenberger B, Hüttl RF. Nutrient amounts of ectomycorrhizae analysed by EDX using ESEM and ICP. Mycorrhiza. 2005;15:307–312. doi: 10.1007/s00572-005-0350-9. [DOI] [PubMed] [Google Scholar]
- Smith SE, Read D. Mycorrhizal symbiosis. 3. London: Academic; 2008. [Google Scholar]
- Tedersoo L, Hansen K, Perry BA, Kjøller R. Molecular and morphological diversity of pezizalean ectomycorrhiza. New Phytol. 2006;170:581–596. doi: 10.1111/j.1469-8137.2006.01678.x. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Jairus T, Horton BM, Abarenkov K, Suvi T, Saar I, Kõljalg U. Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol. 2008;180:479–490. doi: 10.1111/j.1469-8137.2008.02561.x. [DOI] [PubMed] [Google Scholar]
- Timonen S, Kauppinen P. Mycorrhizal colonisation patterns of Tilia trees in street, nursery and forest habitats in southern Finland. Urban For Urban Green. 2008;7:265–276. doi: 10.1016/j.ufug.2008.08.001. [DOI] [Google Scholar]
- Twieg BD, Durall DM, Simard SW. Ectomycorrhizal fungal succession in mixed temperate forests. New Phytol. 2007;176:437–447. doi: 10.1111/j.1469-8137.2007.02173.x. [DOI] [PubMed] [Google Scholar]
- Walker JF, Miller OK, Horton JL. Hyperdiversity of ectomycorrhizal fungus assemblages on oak seedlings in mixed forests in the southern Appalachian Mountains. Mol Ecol. 2005;14:829–838. doi: 10.1111/j.1365-294X.2005.02455.x. [DOI] [PubMed] [Google Scholar]
- Wardle DA. The influence of biotic interactions on soil biodiversity. Ecol Lett. 2006;9:870–886. doi: 10.1111/j.1461-0248.2006.00931.x. [DOI] [PubMed] [Google Scholar]
- Watling R. Fungal associates of Salix repens in northern oceanic Britain and their conservation significance. Mycol Res. 2005;109:1418–1424. doi: 10.1017/S0953756205004004. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelgard DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic; 1990. pp. 315–322. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS 29 kb)
(DOC 254 kb)
(DOC 44 kb)


