Abstract
Here, we report ion-helium collision cross sections (CCS) for a number of peptide, small protein, and peptide/protein ionic complexes. The CCS values reported here are compared to previously reported results.[1, 2] We also compare values for low charge state species, i.e., [M + H]+ and [M + 2H]2+, formed by MALDI with values for high charge state species formed by ESI, and the measured CCSs are compared with predicted CCS for solid-state and solution phase structures and calculated structures obtained by using a protein-protein structure algorithm generator, based on a combined Biomolecular complex Generation with Global Evaluation and Ranking[3] and Multi Dimensional Scaling[4].
Keywords: peptide and protein complex secondary structure, protein oligomers, ion-helium collision cross sections, Ion Mobility Spectrometry, protein docking, cluster analysis
Introduction
Protein folding and protein-protein interactions in a physiological environment depend on numerous factors (e.g., electrostatic interactions, intermolecular interactions with the solvent, entropic effects, intramolecular hydrogen bonding and Van der Waals interactions). In native environments protein folding and protein-protein interactions occur under a range of conditions including aqueous solutions as well as hydrophobic and low dielectric membranes.[5-7] Thus, solvent-free studies provide a means of separating solvation effects from intrinsic properties of the protein side-chain and protein-protein site interactions. For example, solvent-free studies may provide new approaches to studies of “non-native” structure that can be used to evaluate peptide and protein folding/unfolding models[8] as well as nucleation and growth[9].
Fundamental studies of gas-phase macromolecules, specifically studies of conformation and thermo-chemical properties of anhydrous peptides and proteins have grown rapidly since the introduction of ESI and MALDI.[10-13] For example, hydrogen/deuterium (H/D) exchange reactions are frequently used to probe the number of exchangeable hydrogen atoms as a measure of the folding of a targeted protein.[14-19] IR spectroscopic techniques have been developed to study secondary structural elements, based on comparisons of theoretically estimated vibrational frequencies of candidate structures, but this approach is often limited to the study of small peptides.[20, 21] Ion Mobility Spectrometry (IMS) combined with theoretical simulations has proven to be the most versatile technique for conformational analysis of intermediate and equilibrium structures of biomolecules by measuring the ion-neutral, collision cross-section (CCS) of molecular ions.[1, 2, 22] IMS combined with theoretical calculations is a powerful technique for probing the equilibrium between conformational states of the macromolecular ions. [23-26] A major difficulty in simulating processes occurring in IMS experiments is the long timescale over which structural rearrangements may occur (~ few milliseconds) whereas molecular dynamics simulations are typically limited to nanoseconds.[27-29] IMS measured CCSs correspond to an average of all thermodynamically available conformations accessible during the experimental measuring time or drift time. Although a number of theoretical methods have been developed to reduce the computation time,[30-38] further improvements are needed to efficiently correlate the theoretical results with available experimental data for peptide systems.[39] The theoretical limitation becomes even more pronounced as the molecule of interest increases from small model peptide ions to large ionic proteins or protein complexes; the later systems demand the use of non-dynamic, theoretical strategies to effectively sample the conformational space.[40-43]
The goal of this study is to provide a high confidence level ion-helium CCS database of peptides, proteins and peptide/protein complexes. These new data will expand the region of reference conformational space to protein and protein complexes, and complement previous reports on CCS databases for peptide systems.[1, 2] The advantages of a combined theoretical and experimental analysis of the molecular complex conformational space is illustrated for the insulin oligomer (n = 1 - 7) using a protein-protein structure algorithm generator, based on a combined Biomolecular complex Generation with Global Evaluation and Ranking[3] and Multi Dimensional Scaling[4] analysis.
Experimental Method
The experimental details of the MALDI-IM-MS instrumentation and data acquisition used in this study have been described elsewhere.[44-47] Briefly, ions are formed in an IM drift cell by matrix-assisted laser desorption ionization (MALDI) using a microcrystal Nd:YAG laser (355 nm, Powerchip Nanolaser, JDS Uniphase Corp.), operating at a pulse rate of 200-300 Hz. Ions entering the drift cell are separated on the basis of drift time through the cell maintained at room temperature, a helium pressure of ~3.0 torr and field strength/pressure ratio (E/p) of 10 - 40 V cm−1 torr−1. Ions exiting the IM drift cell are focused (by a multi-element Einzel lens) into an orthogonal-TOF ion source. The TOF is biased at a potential of −6 kV and the ions are extracted by applying a voltage pulse to the TOF push/pull electrodes (+675V/−675 V, respectively); the TOF extraction potential is pulsed at a rate ~2 kHz, and typical mass resolution for the reflectron TOF is 1500 - 3000. In all experiments, the temperature of the bath gas is ca. 300 K. The experiments were performed using a laser power near the ion desorption threshold to minimize peak broadening of the arrival time distribution (ATD) due to space charge effects.[48]
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. The analytes consist of neuropeptide Y 1-24 (Human), melittin (honey bee venom), insulin β-chain (bovine), β-amyloid 1-40, insulin (bovine pancreas), ubiquitin (bovine red blood cells), cytochrome C (horse heart), RNase A (bovine pancreas), lysozyme (chicken egg white), and apomyoglobin (horse heart). All analytes were dissolved in distilled water at a concentration of 1.0 mg/ml and mixed (1:1 v:v) with 5 mg/ml α-cyano-4-hydroxycinnamic acid (CHCA), dissolved in 60% acetonitrile, 38% distilled water, 1% of 10% trifluoroacetic acid solution (TFA), and 1% ammonium phosphate solution. The mixture of matrix and analyte was spotted onto a 96 well MALDI target plate. The sample spots were prepared in quadruplicate to guarantee spot-to-spot reproducibility and triplicate CCS measurements were performed as independent experiments.
The ion-helium collision cross-sections (CCSs) were determined by using the method described by Mason and McDaniel.[49] Briefly, CCSs are obtained by recording the ATD at different applied voltages (V) to the IM cell and plotting the mean ATD vs 1/V. The ATD-intercept of this linear plot equals the time the ions spend outside the IM cell (to), which permits the determination of the CCS values as a function of the drift time (td = <ATD> −to) from the equation:
| (1) |
where z and e are number of charges and elementary charge, kb is Boltzmann's constant, T is drift gas temperature, mg and mi are the bath gas and ion masses, E is the electric field, L is drift length, p is the helium bath gas pressure, and No is bath gas number density. The CCSs are reported including the propagated error of ATD, to determination, and standard deviation of CCS at different E/p values. A fullerene mixture was used as a CCS reference.[50, 51]
Theoretical and Computational Details
The structures of protein complexes were predicted using a Biomolecular complex Generation with Global Evaluation and Ranking (BiGGER) program[3]. Briefly, BiGGER uses an efficient algorithm to search and select a set of candidate complexes. Each candidate is then evaluated according to the estimated probability of being an accurate model of the complex. This allows the user to rank all models according to a single criterion and focus on the more likely ones if no additional information is available. In the first stage, a step-by-step search through all the possible docking configurations is performed, followed by a stringent filtering criterion that will retain only a small fraction of the possible structures. The maximum number of docking candidates was set to 5000; the docking search was performed using angular steps of 15 degrees, and docking resolution and added radius of 1.0 Å. In the second stage, each solution is evaluated using a neural network to estimate its likelihood of being an accurate model of the complex. The overall evaluation of docked solutions is given by the Interaction Global Score. This is the estimated probability (in percentage) that a docking solution with that Neural Network response is an accurate model of the complex as a function of the surface docking score, side chain contact filter, electrostatics interaction score, hydrophobic score and side chain contact score.[52, 53] Despite modeling some side chain flexibility of the individual docking units, BiGGER assumes that there are no significant conformational changes during complex formation. To overcome this problem, the top 100 configurations were further optimized using the Open Force Field program in Accelrys Cerius2, by Accelrys Inc[54]. The Consistent Force Field (version 2002) was employed in the geometry optimization[55]. Following geometry optimization, the theoretical ion-neutral, collision cross-sections were calculated using the Trajectory Method implemented in the MOBCAL software, with all the interaction potentials taken as the default values. [50, 51]
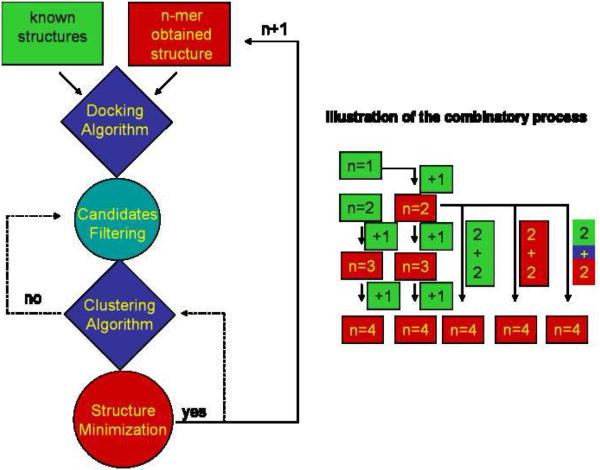
The search for candidate structures of the protein-protein complexes was performed following the scheme contained in Figure 1. An initial set of candidate structures measured experimentally is pre-defined as docking input elements. After each cycle, the highest score structure is selected as a potential docking input for the next cluster size. All potential combinations of docking pre-defined inputs are considered. For example, the scheme contained in Figure 1 considers the structure of a protein monomer (n1) and dimer (n2) as pre-defined docking elements. For n = 2, the candidature structure pool is composed of the dimer structure and the top 100 (n1+n1) structures. The highest score structure from the 100 n1+n1 structures is then defined as the n1n1 docking element and used in the search for larger cluster sizes. That is, the search for trimer (n3) candidates will consider n1n1, n2, and n1 as docking elements, which yield n1n1+n1 and n2+n1 as potential combinations. In the insulin example, since the number of potential combinations increases geometrically with the cluster size, the number of new defined docking elements after the optimization cycle for a given combination was simplified to the highest score candidate (i.e., only 1 structure); nevertheless, this algorithm has the potential to set the number of new defined elements by the number of structurally different cluster families. The docking elements for the insulin monomer (n1) and dimer (n2) were obtained from X-ray diffraction data.[56]
Figure 1.
Scheme of the protein-protein candidate structure generation algorithm. On the right, the combinatory process is illustrated for the case of pre-defined protein monomer and dimer structures as docking elements (green). New docking elements are obtained from each combinatory output (red).
A user-developed algorithm was used to classify the resulting optimized structures in clusters. The cluster analysis is based on the comparison of all structures and subsequent classifications in clusters, i.e., structures are classified such that a given cluster contains a set of structures that are more similar to one another than to members of other clusters. Comparison between the similarities of two structures is made using the average root mean square deviation (RMSD) using the SUPPOSE program.[57] Average RMSD values are computed from the pair-wise RMSD measured between corresponding backbone atoms of two structures, once the corresponding atoms have been identified and the molecules have been rotated and translated as rigid bodies to the best match. A multidimensional scaling (MDS) method was used to generate an n-dimensional projection of all the structures, where the inter-point distances are “close” to their average RMSD values.[4] Briefly, the dimension, n, of the MDS space was chosen such that the inter-point distances and the RMSD values correspond within a stress value below 5%.[4] The structures were classified in the n-dimensional MDS space using a hierarchical clustering algorithm (HCA).[58] The HCA classifies the structures without a predetermined number of clusters, which permits users to determine the natural grouping with interactive visual feedback (e.g., dendrograms, 3-D projections and/or color mosaic). The optimum number of clusters can be defined by the user or can be determined using an iterative algorithm from a user-defined RMSD tolerance criteria within a cluster.
Results and Discussion
Ion Mobility – Mass Spectrometry separates gas-phase ions on the basis of ion-neutral collision cross section (CCS) and m/z ratios. One of the objectives of the present study is to compile a database of CCS values for single-charged peptide and protein ionic complexes, which can be used as calibration standards for CCS values (Table 1). Establishing a CCS database of macromolecules and molecular complexes is especially important for techniques such as Differential Mobility Analyzers (DMA),[59] Field Asymmetric waveform Ion Mobility Spectrometers (FAIMS),[60, 61] and T-wave Ion Mobility Spectrometers[62, 63], because the data obtained from such experiments cannot be used for first principle CCS calculations.
Table 1.
Ion-helium collision cross section values for single-charged peptides, proteins, and peptide and protein complexes.
| Analyte | m/z | n-mer | Average CCS (A2) | Average Ko (cm2V−1s−1) |
|---|---|---|---|---|
| Neuropeptide Y (1-24) | ||||
| monomer | 2657.9 | 1 | 417 ± 36 | 1.29 ± 0.096 |
| dimer | 5314.8 | 2 | 651 ± 34 | 0.83 ± 0.041 |
| trimer | 7971.7 | 3 | 771 ± 34 | 0.70 ± 0.028 |
| tetramer | 10628.6 | 4 | 932 ± 39 | 0.58 ± 0.026 |
| pentamer | 13285.5 | 5 | 1044 ± 46 | 0.51 ± 0.026 |
| hexamer | 15942.4 | 6 | 1159 ± 35 | 0.46 ± 0.012 |
| heptamer | 18599.3 | 7 | 1192 ± 105 | 0.48 ± 0.038 |
| octamer | 21256.2 | 8 | 1227 ± 103 | 0.46 ± 0.040 |
| Melittin | ||||
| monomer | 2847.5 | 1 | 469 ± 30 | 1.15 ± 0.057 |
| dimer | 5694 | 2 | 774 ± 35 | 0.69 ± 0.015 |
| trimer | 8540.5 | 3 | 881 ± 68 | 0.62 ± 0.038 |
| tetramer | 11387 | 4 | 1009 ± 92 | 0.56 ± 0.040 |
| Insulin β-chain | ||||
| monomer | 3496.9 | 1 | 501 ± 27 | 1.07 ± 0.043 |
| dimer | 6992.8 | 2 | 730 ± 30 | 0.74 ± 0.028 |
| trimer | 10488.7 | 3 | 872 ± 41 | 0.62 ± 0.030 |
| tetramer | 13984.6 | 4 | 1016 ± 40 | 0.53 ± 0.026 |
| pentamer | 17480.5 | 5 | 1167 ± 101 | 0.47 ± 0.039 |
| hexamer | 20976.4 | 6 | 1270 ± 103 | 0.45 ± 0.033 |
| heptamer | 24472.3 | 7 | 1370 ± 122 | 0.41 ± 0.035 |
| octamer | 27968.2 | 8 | 1436 ± 134 | 0.40 ± 0.038 |
| B-amyloid (1-40) | ||||
| monomer | 4330.9 | 1 | 560 ± 35 | 0.96 ± 0.067 |
| dimer | 8660.8 | 2 | 773 ± 46 | 0.70 ± 0.055 |
| trimer | 12990.7 | 3 | 950 ± 57 | 0.57 ± 0.044 |
| tetramer | 17320.6 | 4 | 1118 ± 60 | 0.48 ± 0.031 |
| pentamer | 21650.5 | 5 | 1300 ± 114 | 0.44 ± 0.041 |
| hexamer | 25980.4 | 6 | 1377 ± 115 | 0.41 ± 0.034 |
| Insulin | ||||
| monomer | 5734.6 | 1 | 654 ± 37 | 0.82 ± 0.044 |
| dimer | 11468.2 | 2 | 930 ± 41 | 0.58 ± 0.029 |
| trimer | 17201.8 | 3 | 1119 ± 52 | 0.48 ± 0.028 |
| tetramer | 22935.4 | 4 | 1356 ± 101 | 0.43 ± 0.033 |
| pentamer | 28669 | 5 | 1486 ± 110 | 0.40 ± 0.037 |
| hexamer | 34402.6 | 6 | 1592 ± 144 | 0.38 ± 0.050 |
| heptamer | 40136.2 | 7 | 1705 ± 141 | 0.35 ± 0.037 |
| Ubiquitin | ||||
| monomer | 8565.8 | 1 | 821 ± 45 | 0.66 ± 0.040 |
| dimer | 17130.6 | 2 | 1147 ± 57 | 0.47 ± 0.029 |
| trimer | 25695.4 | 3 | 1410 ± 129 | 0.41 ± 0.045 |
| tetramer | 34260.2 | 4 | 1580 ± 122 | 0.37 ± 0.041 |
| Cytochrome C | ||||
| monomer | 12232 | 1 | 983 ± 58 | 0.55 ± 0.030 |
| dimer | 24463 | 2 | 1303 ± 101 | 0.41 ± 0.031 |
| RNase A | ||||
| monomer | 13683.4 | 1 | 983 ± 52 | 0.55 ± 0.039 |
| dimer | 27365.8 | 2 | 1407 ± 107 | 0.41 ± 0.043 |
| trimer | 41048.2 | 3 | 1838 ± 132 | 0.34 ± 0.040 |
| Lysozyme | ||||
| monomer | 14306.1 | 1 | 1017 ± 56 | 0.53 ± 0.031 |
| dimer | 28611.2 | 2 | 1464 ± 106 | 0.39 ± 0.035 |
| trimer | 42916.3 | 3 | 1895 ± 107 | 0.33 ± 0.032 |
| Apomyoglobin | ||||
| monomer | 16952.5 | 1 | 1077 ± 77 | 0.50 ± 0.052 |
| dimer | 33904 | 2 | 1587 ± 144 | 0.37 ± 0.040 |
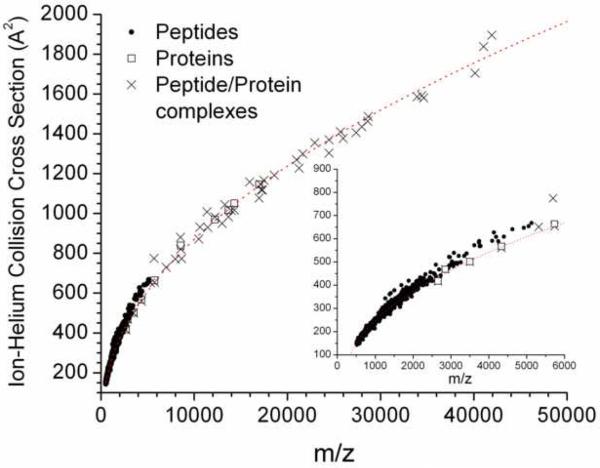
Figure 2 contains a plot of CCS versus m/z for a large number of single-charged, peptide and protein ions, as well as oligomeric peptide and protein ionic complexes. The dataset includes CCS values of previously reported single-charged peptide ions obtained by digesting nine standard proteins with three proteolytic enzymes (trypsin, chymotrypsin, and pepsin).[2] Inspection of Figure 2 shows that peptide ions and peptide/protein ionic complexes fall on the same trend line in the CCS versus m/z plot, and this average CCS trend line can be described as a square-root function of m/z: <CCS>= 8.8 × (m/z − 210)½. We have previously observed that in the case of peptide ions, a small number of ion signals deviate from the average CCS trend line, while other molecular classes fall on very different trend lines. [2, 64] For calibration purposes, we recommend the use of the CCS data reported in Table 1. The broad distribution of CCSs contained in Figure 2 illustrates the structural diversity of the peptide and peptide ion complexes while highlighting the utility of the IM-MS separation to identify a single molecular class. For example, the CCS for melittin [M + H]+ ions (see Table 1) lies above (by approximately 4%) the average peptide ion CCS trend line. The deviation from the expected value could be an indication of helical character for the melittin [M + H]+ ion, which in its native state is ~ 88% helical. [65-67] In addition, helical character was previously used to explain the results for gas-phase MS fragmentation reactions and MALDI hydrogen/deuterium (H/D) exchange,[19, 68] and formation of melittin oligomers with helical character has been also observed in basic or high ionic strength solutions using NMR studies.[69]
Figure 2.
Plot of ion-helium collision cross sections vs m/z for single-charged peptide and peptide and protein ionic complexes. In the inset, CCS for single-charge peptide ions obtained by digesting nine standard proteins with three proteolytic enzymes (trypsin, chymotrypsin, and pepsin) is shown. [1, 2] The total fitting function is CCS = 8.8 × (m/z − 210)½
On the other hand, the [M + H]+ ion of neuropeptide Y 1-24 presents a large negative deviation in CCS (−4.3%), that could be related to the lower helical content (< 40 %) observed in native state studies.[70] As the molecular size increases, the influence of the helical/sheet content on the CCS may be overshadowed by other structural features, i.e., side chain and intramolecular interactions. The CCS for insulin [M + H]+ ions agrees very well with the expected value, whereas the CCS for apomyoglobin [M + H]+ ions has a large negative deviation (−5.4 %) from the average CCS. In their native states, these proteins contain ~46% and ~74% helical character, respectively.[56, 71, 72] Quite possibly the intramolecular disulfide bonding of insulin makes the total secondary structure spherical/globular, where the helical contribution to a more elongated form (and larger CCS) has been reduced by these side-chain interactions.[56] Similarly, for apomyoglobin the side-chain interaction with the heme group in the native state forces the [M + H]+ ion to adopt a globular/spherical compact configuration surrounding the heme group.[71, 72] Overall, side-chain and intramolecular interactions may also stabilize the secondary structure of the ionic complex in “non-native” states, while not conserving the original secondary structure and/or helical/sheet content. Only large differences in the secondary structure of native versus “non-native” states will significantly contribute to the packing density or CCS, making higher resolution IMS experiments necessary to successfully observe these effects for large peptides and proteins. The CCS values contained in Table 1 are in good agreement with reported data obtained using DMA analyzers,[73-75] where the electromobility particle diameter correspond to the diameter of a sphere if the molecular complex is assumed to have a globular shape.
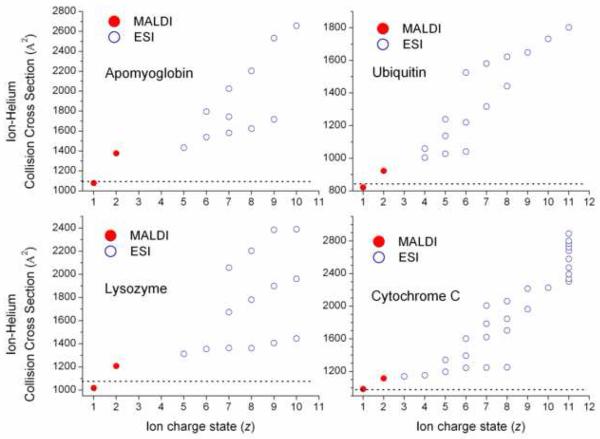
The CCS values reported in Table 1 are for single-charged, protonated molecules, [M+H]+, which are always the most abundant ions formed by MALDI. In some cases, double-charged ions, [M + 2H]2+ ions can be observed in MALDI-IM-MS experiments (e.g., results of McLean et al. and Blase et al.),[76, 77] and measured CCSs for some of the [M + 2H]2+ ions are compared with previously reported CCS values obtained by using ESI (Figure 3).[78, 79] Note that the collision cross section increases as the charge-state increases. Thus it appears that as the charge state increases Coulombic interactions also increase, forcing the molecular structure to separate the charge sites, which in turn increases the total conformational space and leads to larger CCS values. Comparison of the CCS values with reported Protein Data Bank structures obtained by using nuclear magnetic resonance (solution-phase structure) and X-ray diffraction (crystal structure) results shows that the best agreement is observed for the lower charge states CCS values obtained using a MALDI source (Figure 3).[71, 72, 80-86] Larger deviations for CCS for the higher charge states may also arise if higher energy configurations or “non-native” states are sampled. For example, denaturing of proteins in the gas-phase at high charge states has been previously observed in ESI-IM-MS experiments.[87, 88]
Figure 3.
Ion-helium CCS variation as a function of the ion charge state, z. Single- and double-charged ions were produced using a MALDI source, and the higher order charge states were previously reported using a ESI source.[78, 79] The dashed lines represent the corresponding average CCS values of nuclear magnetic resonance (solution-phase structure) and X-ray diffraction (crystal structure) reported structures. [71, 72, 80-86]
Many of the peptides studied in the present work are also observed as oligomeric structures (see Table 1). The CCS of the protein complexes do not deviate significantly from the predicted trend line, and as discussed earlier can be used to infer structural information of the ionic protein complexes. That is, the CCS provides an experimental verification for the packing density of the oligomeric structures, which may ultimately be correlated with the peptide/protein aggregation mechanism, or as shown recently by Ruotolo et al. could be used to infer denaturation of peptide/protein ionic complexes. [87, 88]
In previous studies we have shown that IMS data are greatly enhanced when complemented with theoretical studies of candidate structures by direct comparison of the theoretical and experimental CCS values.[89-92] In this study we use a protein-protein structure algorithm generator to compare experimental and theoretical CCS values for insulin oligomers (n = 1 - 7). The spherical/globular shape of the insulin monomer makes it a good candidate to illustrate the utility of this method.
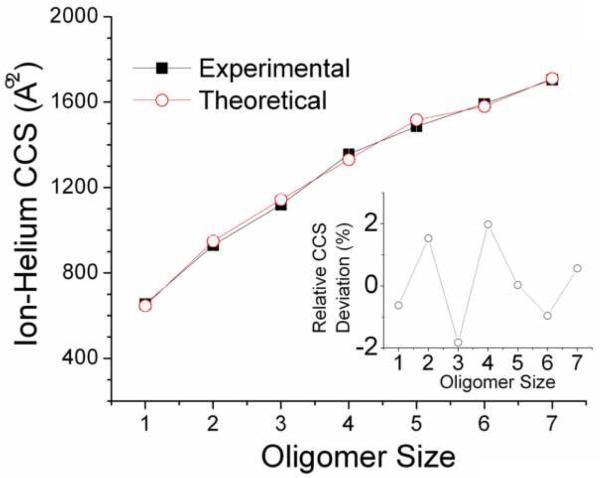
Figure 4 contains a comparison between theoretical and experimental CCS for the insulin oligomers. The protein-protein structure algorithm generator assumes that there are no significant conformational changes during complex formation. That is, the secondary structure of the insulin monomer does not significantly change as the individual units assemble. The binding orientations will ultimately define the total conformational state of the protein complex, which can be related to the gas-phase packing density.
Figure 4.
Theoretical and Experimental ion-helium CCS for the insulin oligomers. In the inset, relative CCS deviations to the average experimental CCS values are shown. Notice the negative deviations for the insulin trimer and hexamer. The average theoretical and experimental CCS values agree within 2%.
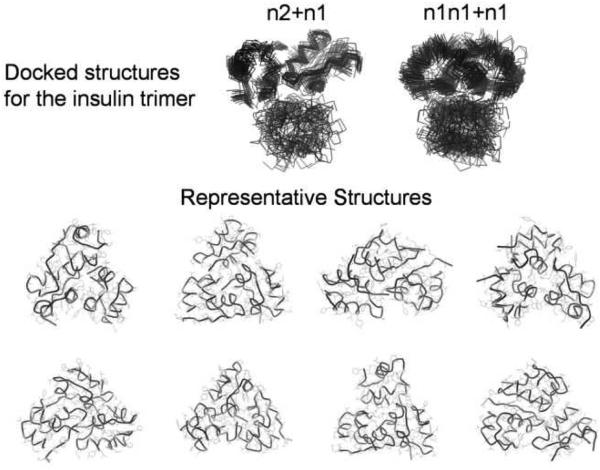
Theoretical CCSs are obtained after averaging the highest score structures obtained in the protein-protein structure algorithm generator. The docking parameters (e.g., angular step, docking resolution and added radius) of the protein-protein structure algorithm generator were verified by the observation of the n2 structure of the insulin dimer as one of the 100 highest scores for insulin monomer (n1+n1) combinations. Good agreement is observed between the predicted and experimentally observed CCS values (< 2 %). Close inspection reveals smaller CCSs for the n = 3 and n = 6 oligomers (see Figure 4 inset), which suggests a more compact structure for this species. Figure 5 contains the most abundant backbone orientation of the insulin trimer (n = 3) obtained from the protein-protein structure algorithm generator. The insulin trimer is obtained from a combination of the insulin dimer (n2) and docked structures of insulin monomers (n1+n1) with the insulin monomer (n1). Eight representative structures were obtained for the insulin trimer, but the main differences between these structures are related to the orientations of the docking sites. It appears that the spherical/globular shape of the insulin monomers makes the trimer structure more stable owing to a larger number of interactions. Each monomer unit in the trimer structure is attached to two other monomer units. For larger oligomers, multiple docking regions are observed, which in some cases promotes formation of a triangular pyramid.
Figure 5.
Illustration of the docking procedure for the insulin trimer using the protein-protein structure algorithm generator. Top) Overlay of the backbone structures obtained from the combination of n2+n1 and n1n1+n1. Bottom) Backbone and side-chain representation of the representative structures obtained from the cluster analysis of the highest score structures.
The hexameric insulin ion also appears to favor a single structural motif, where each unit is coordinated with three insulin monomer units forming a compact structure that resembles two parallel, triangular trimer structures coupled together. This compact hexamer configuration shares some similarities with solution based studies, where the insulin hexamer counterparts are coordinated to Zn metal ions and each insulin monomer unit is coordinated to three other insulin monomer units.[93-95] A high degree of stabilization of the oligomeric complexes with the larger number of docking-site interactions can be directly correlated with negative CCS deviations if the monomer unit has a spherical/globular shape. When the monomer unit deviates from the spherical/globular shape, docking-site interactions between the oligomer counterparts may be sufficiently strong to stabilize the oligomer complex in site-oriented configurations, where their correlation with the CCS may not be straightforward.
Conclusions
A high confidence level peptide and peptide/protein ionic complex ion-helium CCS reference database has been compiled, which significantly extends the conformational space of single-charged ions up to ~50 kDa. This database provides essential data for future theoretical studies and contains calibration standards for other gas-phase separation techniques where direct CCS measurements cannot be obtained.
We also describe the use of a protein-protein structure algorithm generator to estimate CCSs for protein ions as well as protein ionic complexes. On the basis of these studies it appears that intramolecular interactions stabilize the secondary structure of the ionic complexes in native and/or “non-native” states. The end result is that only large differences in secondary structure of native versus “non-native” states will significantly contribute to the measured CCS, thus higher resolution IMS experiments will be needed to observe these effects for large molecule systems. We anticipate that a 3 - 4 fold increase in the IMS resolution will permit the study of docking-site orientations on the peptide and protein complex assembly, disassembly and exchange of subunits between protein complexes.
The advantages of a protein-protein structure algorithm generator were illustrated by the comparison of theoretical and experimental CCS of insulin oligomers, where deviations in the CCS values were associated with structural motifs of the protein complex. This work provides a step forward for the development of theoretical algorithms to efficiently sample the protein complex conformational space. Future efforts will be focused on the implementation of higher-level docking strategies and the study of protein complexes where side-chain groups and/or “structural pockets” have been modified to induce specific molecular attachment as a method to study drug delivery mechanisms and their effect on the biological function of protein complexes.
Acknowledgments
This work was supported by the Robert A. Welch Foundation (A-1176), the U.S. Department of Energy, Division of Chemical Sciences, BES (DE-FG-04ER-15520) and the NSF-MRI (DSI0821700) grants. We would like to thank Dr. Lisa M. Perez from the Laboratory for Molecular Simulations at Texas A&M University for the helpful discussions and support during the development of the computational methods.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Valentine SJ, Counterman AE, Clemmer DE. A database of 660 peptide ion cross sections: use of intrinsic size parameters for bona fide predictions of cross sections. J. Am. Soc. Mass Spectrom. 1999;10:1188–1211. doi: 10.1016/S1044-0305(99)00079-3. [DOI] [PubMed] [Google Scholar]
- 2.Tao L, McLean JR, McLean JA, Russell DH. A Collision Cross-Section Database of Singly-Charged Peptide Ions. J. Am. Soc. Mass Spectrom. 2007;18:1232–1238. doi: 10.1016/j.jasms.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Palma PN, Krippahl L, Wampler JE, Moura JJG. BiGGER: A new (soft) docking algorithm for predicting protein interactions. Proteins: Structure, Function, and Genetics. 2000;39:372–384. [PubMed] [Google Scholar]
- 4.Cox TF, Cox MAA. Multidimensional scaling. CRC Press; London: 1994. [Google Scholar]
- 5.Karp DA, Gittis AG, Stahley MR, Fitch CA, Stites WE, Garcia-Moreno E B. High Apparent Dielectric Constant Inside a Protein Reflects Structural Reorganization Coupled to the Ionization of an Internal Asp. Biophys. J. 2007;92:2041–2053. doi: 10.1529/biophysj.106.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitch CA, Whitten ST, Hilser VJ, García-Moreno E B. Molecular mechanisms of pH-driven conformational transitions of proteins: Insights from continuum electrostatics calculations of acid unfolding. Proteins: Structure, Function, and Bioinformatics. 2006;63:113–126. doi: 10.1002/prot.20797. [DOI] [PubMed] [Google Scholar]
- 7.Bone S. Dielectric properties of biomacromolecules: some aspects of relevance to biological systems. Journal of Bioelectricity. 1985;4:389–418. [Google Scholar]
- 8.Wang Y, Zhang H, Li W, Scott RA. Discriminating compact nonnative structures from the native structure of globular proteins. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:709–713. doi: 10.1073/pnas.92.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss WFIV, Hodgdon TK, Kaler EW, Lenhoff AM, Roberts CJ. Nonnative Protein Polymers: Structure, Morphology, and Relation to Nucleation and Growth. Biophys. J. 2007;93:4392–4403. doi: 10.1529/biophysj.107.112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karas M, Bachmann D, Hillenkamp F. Influence of the wavelength in high-irradiance ultraviolet laser desorption mass spectrometry of organic molecules. Anal. Chem. 1985;57:2935–2939. [Google Scholar]
- 11.Hillenkamp F, Karas M. Matrix-assisted laser desorption/ionisation, an experience. Int. J. Mass Spectrom. 2000;200:71–77. [Google Scholar]
- 12.Whitehouse CM, Dreyer RN, Yamashita M, Fenn JB. Electrospray interface for liquid chromatographs and mass spectrometers. Anal. Chem. 1985;57:675–679. doi: 10.1021/ac00280a023. [DOI] [PubMed] [Google Scholar]
- 13.Cole RB. Electrospray Ionization Mass Spectrometry. New York; John Wiley & Sons: 1997. [Google Scholar]
- 14.Miranker A, Robinson CV, Radford SE, Aplin RT, Dobson CM. Detection of transient protein folding populations by mass spectrometry. Science. 1993;262:896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- 15.Valentine SJ, Clemmer DE. Temperature-dependent H/D exchange of compact and elongated cytochrome c ions in the gas phase. J. Am. Soc. Mass Spectrom. 2002;13:506–517. doi: 10.1016/S1044-0305(02)00372-0. [DOI] [PubMed] [Google Scholar]
- 16.Resing KA, Hoofnagle AN, Ahn NG. Modeling deuterium exchange behavior of ERK2 using pepsin mapping to probe secondary structure. J. Am. Soc. Mass Spectrom. 1999;10:685–702. doi: 10.1016/S1044-0305(99)00037-9. [DOI] [PubMed] [Google Scholar]
- 17.Deng Y, Zhang Z, Smith DL. Comparison of continuous and pulsed labeling amide hydrogen exchange/mass spectrometry for studies of protein dynamics. J. Am. Soc. Mass Spectrom. 1999;10:675–684. doi: 10.1016/S1044-0305(99)00038-0. [DOI] [PubMed] [Google Scholar]
- 18.Nemirovskiy O, Giblin DE, Gross ML. Electrospray ionization mass spectrometry and hydrogen/deuterium exchange for probing the interaction of calmodulin with calcium. J. Am. Soc. Mass Spectrom. 1999;10:711–718. doi: 10.1016/S1044-0305(99)00036-7. [DOI] [PubMed] [Google Scholar]
- 19.Figueroa ID, Russell DH. Matrix-assisted laser desorption ionization hydrogen/deuterium exchange studies to probe peptide conformational changes. J. Am. Soc. Mass Spectrom. 1999;10:719–731. doi: 10.1016/S1044-0305(99)00040-9. [DOI] [PubMed] [Google Scholar]
- 20.Jaime AS, Monia G, Oleg VB, Thomas RR. Conformation-specific infrared and ultraviolet spectroscopy of tyrosine-based protonated dipeptides. J. Chem. Phys. 2007;127:154322. doi: 10.1063/1.2798111. [DOI] [PubMed] [Google Scholar]
- 21.Hunig I, Kleinermanns K. Conformers of the peptides glycine-tryptophan, tryptophan-glycine and tryptophan-glycine-glycine as revealed by double resonance laser spectroscopy. Physical Chemistry Chemical Physics. 2004;6:2650–2658. [Google Scholar]
- 22.Jarrold MF. Peptides and Proteins in the Vapor Phase. Annual Review of Physical Chemistry. 2000;51:179–207. doi: 10.1146/annurev.physchem.51.1.179. [DOI] [PubMed] [Google Scholar]
- 23.Ruotolo BT, McLean JA, Gillig KJ, Russell DH. The influence and utility of varying field strength for the separation of tryptic peptides by ion mobility-mass spectrometry. J. Am. Soc. Mass Spectrom. 2005;16:158–165. doi: 10.1016/j.jasms.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Kanu AB, Hill HH., Jr Identity confirmation of drugs and explosives in ion mobility spectrometry using a secondary drift gas. Talanta. 2007;73:692–699. doi: 10.1016/j.talanta.2007.04.058. [DOI] [PubMed] [Google Scholar]
- 25.Zilch LW, Kaleta DT, Kohtani M, Krishnan R, Jarrold MF. Folding and Unfolding of Helix-Turn-Helix Motifs in the Gas Phase. J. Am. Soc. Mass Spectrom. 2007;18:1239–1248. doi: 10.1016/j.jasms.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez-Lima FA, Wei H, Gao YQ, Russell DH. On the structure elucidation using IMS and Molecular Dynamics. J. Phys. Chem. A. 2009;113:8221–8234. doi: 10.1021/jp811150q. [DOI] [PubMed] [Google Scholar]
- 27.Gao YQ, Yang LJ. On the enhanced sampling over energy barriers in molecular dynamics simulations. J. Chem. Phys. 2006;125:114103. doi: 10.1063/1.2244548. [DOI] [PubMed] [Google Scholar]
- 28.Gao YQ. An integrate-over-temperature approach for enhanced sampling. J. Chem. Phys. 2008;128:064105. doi: 10.1063/1.2825614. [DOI] [PubMed] [Google Scholar]
- 29.Gao YQ. Self-adaptive enhanced sampling in the energy and trajectory spaces: Accelerated thermodynamics and kinetic calculations. J. Chem. Phys. 2008;128:134111. doi: 10.1063/1.2901037. [DOI] [PubMed] [Google Scholar]
- 30.Bartels C, Karplus M. Probability Distributions for Complex Systems: Adaptive Umbrella Sampling of the Potential Energy. J. Phys. Chem. B. 1998;102:865–880. [Google Scholar]
- 31.Darve E, Pohorille A. Calculating free energies using average force. J. Chem. Phys. 2001;115:9169. [Google Scholar]
- 32.Bussi G, Laio A, Parrinello M. Equilibrium Free Energies from Nonequilibrium Metadynamics. Phys. Rev. Lett. 2006;96:090601. doi: 10.1103/PhysRevLett.96.090601. [DOI] [PubMed] [Google Scholar]
- 33.Hamelberg D, McCammon JA. Fast Peptidyl cis–trans Isomerization within the Flexible Gly-Rich Flaps of HIV-1 Protease. J. Am. Chem. Soc. 2005;127:13778–13779. doi: 10.1021/ja054338a. [DOI] [PubMed] [Google Scholar]
- 34.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: A promising and efficient simulation method for biomolecules. J. Chem. Phys. 2004;120:11919. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 35.Berg BA, Neuhaus T. Multicanonical algorithms for first order phase transitions. Phys. Lett. B. 1991;267:249–253. doi: 10.1103/PhysRevLett.68.9. [DOI] [PubMed] [Google Scholar]
- 36.Mitsutake A, Okamoto Y. Replica-exchange simulated tempering method for simulations of frustrated systems. Chem. Phys. Lett. 2000;332:131–138. [Google Scholar]
- 37.Sugita Y, Kitao A, Okamoto Y. Multidimensional replica-exchange method for free-energy calculations. J. Chem. Phys. 2000;113:6042. [Google Scholar]
- 38.Hansmann UHE. Parallel tempering algorithm for conformational studies of biological molecules. Chem. Phys. Lett. 1997;281:140–150. [Google Scholar]
- 39.Damsbo M, Kinnear BS, Hartings MR, Ruhoff PT, Jarrold MF, Ratner MA. Application of evolutionary algorithm methods to polypeptide folding: Comparison with experimental results for unsolvated Ac-(Ala-Gly-Gly)5-LysH+ Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7215–7222. doi: 10.1073/pnas.0401659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ehrlich LP, Wade RC. Protein-protein docking. Reviews in Computational Chemistry. 2001;17:61–97. [Google Scholar]
- 41.Marshall AG, Vakser IA. Protein-protein docking methods. Protein Reviews. 2005;3:115–146. [Google Scholar]
- 42.Lee K, Lee J-W. Computational approaches to protein-protein docking. Current proteomics. 2008;5:10–19. [Google Scholar]
- 43.Ritchie DW. Recent progress and future directions in protein-protein docking. Current Protein and Peptide Science. 2008;9:1–15. doi: 10.2174/138920308783565741. [DOI] [PubMed] [Google Scholar]
- 44.Gillig KJ, Ruotolo B, Stone EG, Russell DH, Fuhrer K, Gonin M, Schultz AJ. Coupling High-Pressure MALDI with Ion Mobility/Orthogonal Time-of-Flight Mass Spectrometry. Anal. Chem. 2000;72:3965–3971. doi: 10.1021/ac0005619. [DOI] [PubMed] [Google Scholar]
- 45.Stone EG, Gillig KJ, Ruotolo BT, Russell DH. Optimization of a matrix-assisted laser desorption ionization-ion mobility-surface-induced dissociation-orthogonal-time-of-flight mass spectrometer: simultaneous acquisition of multiple correlated MS1 and MS2 spectra. Int. J. Mass Spectrom. 2001;212:519–533. [Google Scholar]
- 46.Ruotolo BT, Gillig KJ, Stone EG, Russell DH, Fuhrer K, Gonin M, Schultz JA. Analysis of protein mixtures by matrix-assisted laser desorption ionization-ion mobility-orthogonal-time-of-flight mass spectrometry. Int. J. Mass Spectrom. 2002;219:253–267. [Google Scholar]
- 47.Gillig KJ, Ruotolo BT, Stone EG, Russell DH. An electrostatic focusing ion guide for ion mobility-mass spectrometry. Int. J. Mass Spectrom. 2004;239:43–49. [Google Scholar]
- 48.Watts P, Wilders A. On the resolution obtainable in practical ion mobility systems. Int. J. Mass Spectrom. Ion Processes. 1992;112:179–190. [Google Scholar]
- 49.McDaniel EW, Mason EA. Mobility and diffusion of ions in gases. John Wiley and Sons, Inc., New York; New York: 1973. [Google Scholar]
- 50.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. Structural Information from Ion Mobility Measurements: Effects of the Long-Range Potential. J. Phys. Chem. 1996;100:16082–16086. [Google Scholar]
- 51.Shvartsburg AA, Jarrold MF. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chemical Physics Letters. 1996;261:86–91. [Google Scholar]
- 52.Wang Y, Zhang H, Li W, Scott RA. Discriminating compact nonnative structures from the native structure of globular proteins. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:709–713. doi: 10.1073/pnas.92.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Zhang H, Scott RA. A new computational model for protein folding based on atomic solvation. Protein Science. 1995;4:1402–1411. doi: 10.1002/pro.5560040714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Accelrys Cerius2 manual. 4.9 ed. Accelrys Inc; 2003. [Google Scholar]
- 55.Dinur U, Hagler AT. In: Reviews in Computational Chemistry. Lipkowitz KB, Boyd DB, editors. Vol. 2. VCH; New York: 1991. pp. 99–164. [Google Scholar]
- 56.Nanao MH, Sheldrick GM, Ravelli RB. Improving radiation-damage substructures for RIP. Acta Crystallogr., Sect.D. 2005;61:1227–1237. doi: 10.1107/S0907444905019360. [DOI] [PubMed] [Google Scholar]
- 57.Smith J. 1997 http://structbio.vanderbilt.edu/~jsmith/
- 58.Seo J, Shneiderman B. Interactively Exploring Hierarchical Clustering Results. IEEE Computer. 2002;35:80–86. [Google Scholar]
- 59.Schnier PD, Price WD, Jockusch RA, Williams ER. Blackbody Infrared Radiative Dissociation of Bradykinin and Its Analogues: Energetics, Dynamics, and Evidence for Salt-Bridge Structures in the Gas Phase. Journal of the American Chemical Society. 1996;118:7178–7189. doi: 10.1021/ja9609157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shvartsburg AA, Tang K, Smith RD. Understanding and Designing Field Asymmetric Waveform Ion Mobility Spectrometry Separations in Gas Mixtures. Analytical Chemistry. 2004;76:7366–7374. doi: 10.1021/ac049299k. [DOI] [PubMed] [Google Scholar]
- 61.Shvartsburg AA, Li F, Tang K, Smith RD. Distortion of Ion Structures by Field Asymmetric Waveform Ion Mobility Spectrometry. Analytical Chemistry. 2007;79:1523–1528. doi: 10.1021/ac061306c. [DOI] [PubMed] [Google Scholar]
- 62.Shvartsburg AA, Smith RD. Fundamentals of Traveling Wave Ion Mobility Spectrometry. Analytical Chemistry. 2008;80:9689–9699. doi: 10.1021/ac8016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thalassinos K, Grabenauer M, Slade SE, Hilton GR, Bowers MT, Scrivens JH. Characterization of Phosphorylated Peptides Using Traveling Wave-Based and Drift Cell Ion Mobility Mass Spectrometry. Analytical Chemistry. 2009;81:248–254. doi: 10.1021/ac801916h. [DOI] [PubMed] [Google Scholar]
- 64.McLean JA, Ruotolo BT, Gillig KJ, Russell DH. Ion mobility-mass spectrometry: a new paradigm for proteomics. Int. J. Mass Spectrom. 2005;240:301–315. [Google Scholar]
- 65.Terwilliger TC, Eisenberg D. The Structure of Melittin. I. Structure Determination and Partial Refinement. J. Biol. Chem. 1982;257:6010. doi: 10.2210/pdb1mlt/pdb. [DOI] [PubMed] [Google Scholar]
- 66.Terwilliger TC, Eisenberg D. The Structure of Melittin. II. Interpretation of the Structure. J. Biol. Chem. 1982;257:6016. [PubMed] [Google Scholar]
- 67.Anderson D, Terwilliger TC, Wickner W, Eisenberg D. Melittin Forms Crystals which are Suitable for High Resolution X-Ray Structural Analysis and which Reveal a Molecular 2-Fold Axis of Symmetry. J. Biol. Chem. 1980;255:2578. [PubMed] [Google Scholar]
- 68.Anderegg RJ, Wagner DS, Stevenson CL, Borchardt RT. The mass spectrometry of helical unfolding in petides. J. Am. Soc. Mass Spectrom. 1994;5:425–433. doi: 10.1016/1044-0305(94)85058-5. [DOI] [PubMed] [Google Scholar]
- 69.Buckley JS, Edison AS, Kemple MD, Prendergast FG. 13 Calpha-NMR assignments of melittin in methanol and chemical shift correlations with secondary structure. J. Biomol. NMR. 1993;3:639–652. doi: 10.1007/BF00198369. [DOI] [PubMed] [Google Scholar]
- 70.Monks SA, Karagianis G, Howlett GJ, Norton RS. Solution structure of human neuropeptide Y. J. Biomol. NMR. 1996;8:379–390. doi: 10.1007/BF00228141. [DOI] [PubMed] [Google Scholar]
- 71.Copeland DM, West AH, Richter-Addo GB. Crystal structures of ferrous horse heart myoglobin complexed with nitric oxide and nitrosoethane. Proteins: Struct., Funct., Genet. 2003;53:182–192. doi: 10.1002/prot.10495. [DOI] [PubMed] [Google Scholar]
- 72.Maurus R, Overall CM, Bogumil R, Luo Y, Mauk AG, Smith M, Brayer GD. A myoglobin variant with a polar substitution in a conserved hydrophobic cluster in the heme binding pocket. Biochim. Biophys. Acta. 1997;1341:1–13. doi: 10.1016/s0167-4838(97)00064-2. [DOI] [PubMed] [Google Scholar]
- 73.Kaufman SL, Skogen JW, Dorman FD, Zarrin F, Lewis KC. Macromolecule Analysis Based on Electrophoretic Mobility In Air: Globular Proteins. Anal. Chem. 1996;68:3703–3703. doi: 10.1021/ac951128f. [DOI] [PubMed] [Google Scholar]
- 74.Kaufman SL. Analysis of biomolecules using electrospray and nanoparticle methods: the gas-phase electrophoretic mobility molecular analyzer (gemma) J. Aerosol Sci. 1998;29:537–552. [Google Scholar]
- 75.Bacher G, Szymanski WW, Kaufman SL, Zöllner P, Blaas D, Allmaier G. Charge-reduced nano electrospray ionization combined with differential mobility analysis of peptides, proteins, glycoproteins, noncovalent protein complexes and viruses. J. Mass Spectrom. 2001;36:1038–1052. doi: 10.1002/jms.208. [DOI] [PubMed] [Google Scholar]
- 76.McLean JA, McLean JR, Russell DH. Investigation of anhydrous protein structures: the influence of post-translational modifications on tertiary and quaternary structure; 19th Annual Symposium of the Protein Society; Boston. August 2005. [Google Scholar]
- 77.Blase RC, Fernandez-Lima FA, Becker C, Perez LM, Russell DH. Structure of Protein Aggregates in the gas-phase using MALDI-IM-MS; Denver; 56th ASMS Conference on Mass Spectrometry and Allied Topics. 2008. [Google Scholar]
- 78.Valentine SJ, Counterman AE, Clemmer DE. Conformer-Dependent Proton-Transfer Reactions of Ubiquitin Ions. J. Am. Soc. Mass Spectrom. 1997;8:954–961. [Google Scholar]
- 79.Valentine SJ, Anderson JG, Ellington AD, Clemmer DE. J. Phys. Chem. B. 1997;101:3891–3900. [Google Scholar]
- 80.Evrard C, Fastrez J, Declercq JP. Crystal structure of the lysozyme from bacteriophage lambda and its relationship with V and C-type lysozymes. J.Mol.Biol. 1998;276:151–164. doi: 10.1006/jmbi.1997.1499. [DOI] [PubMed] [Google Scholar]
- 81.Refaee M, Tezuka T, Akasaka K, Williamson M. Pressure-Dependent Changes in the Solution Structure of Hen Egg-White Lysozyme. J.Mol.Biol. 2003;327:857. doi: 10.1016/s0022-2836(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 82.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J.Mol.Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 83.Cornilescu G, Marquardt JL, Ottiger M, Bax A. Validation of Protein Structure from Anisotropic Carbonyl Chemical Shifts in a Dilute Liquid Crystalline Phase. J.Am.Chem.Soc. 1998;120:6836–6837. [Google Scholar]
- 84.Benini S, Gonzalez A, Rypniewski WR, Wilson KS, Van Beeumen JJ, Ciurli S. Crystal structure of oxidized Bacillus pasteurii cytochrome c553 at 0.97-A resolution. Biochemistry. 2000;39:13115–13126. doi: 10.1021/bi000402j. [DOI] [PubMed] [Google Scholar]
- 85.Hasegawa J, Yoshida T, Yamazaki T, Sambongi Y, Yu Y, Igarashi Y, Kodama T, Yamazaki K, Kyogoku Y, Kobayashi Y. Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy. Biochemistry. 1998;37:9641–9649. doi: 10.1021/bi9803067. [DOI] [PubMed] [Google Scholar]
- 86.Osapay K, Theriault Y, Wright PE, Case DA. Solution structure of carbonmonoxy myoglobin determined from nuclear magnetic resonance distance and chemical shift constraints. J.Mol.Biol. 1994;244:183–197. doi: 10.1006/jmbi.1994.1718. [DOI] [PubMed] [Google Scholar]
- 87.Hyung S-J, Robinson CV, Ruotolo BT. Gas-Phase Unfolding and Disassembly Reveals Stability Differences in Ligand-Bound Multiprotein Complexes. Chemistry & Biology. 2009;16:382–390. doi: 10.1016/j.chembiol.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 88.Badman ER, Myung S, Clemmer DE. Evidence for Unfolding and Refolding of Gas-Phase Cytochrome c Ions in a Paul Trap. J. Am. Soc. Mass Spectrom. 2005;16:1493–1497. doi: 10.1016/j.jasms.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 89.Ruotolo BT, Tate CC, Russell DH. Ion mobility-mass spectrometry applied to cyclic peptide analysis: conformational preferences of gramicidin S and linear analogs in the gas phase. J. Am. Soc. Mass Spectrom. 2004;15:870–878. doi: 10.1016/j.jasms.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 90.Sawyer HA, Marini JT, Stone EG, Ruotolo BT, Gillig KJ, Russell DH. The Structure of Gas-Phase Bradykinin Fragment 1-5 (RPPGF) Ions: An Ion Mobility Spectrometry and H/D Exchange Ion-Molecule Reaction Chemistry Study. J. Am. Soc. Mass Spectrom. 2005;16:893–905. doi: 10.1016/j.jasms.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 91.Fernandez-Lima FA, Becker C, Gillig K, Russell WK, Nascimento MAC, Russell DH. Experimental and Theoretical Studies of (CsI)nCs+ Cluster Ions Produced by 355 nm Laser Desorption Ionization. J. Phys. Chem. A. 2008;112:11061–11066. doi: 10.1021/jp8047086. [DOI] [PubMed] [Google Scholar]
- 92.Fernandez-Lima FA, Wei H, Gao YQ, Russell DH. On the structure elucidation using IMS and Molecular Dynamics. J. Phys. Chem. A. 2009 doi: 10.1021/jp811150q. submitted. [DOI] [PubMed] [Google Scholar]
- 93.ÒDonoghue SI, Chang X, Abseher R, Nilges M, Led JJ. Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure. J. Biomol. NMR. 2000;16:93–108. doi: 10.1023/a:1008323819099. [DOI] [PubMed] [Google Scholar]
- 94.Chang X, Jorgensen AM, Bardrum P, Led JJ. Solution structures of the R6 human insulin hexamer. Biochemistry. 1997;36:9409–9422. doi: 10.1021/bi9631069. [DOI] [PubMed] [Google Scholar]
- 95.Smith GD, Pangborn WA, Blessing RH. The structure of T6 bovine insulin, Acta Crystallogr., Sect.D. 2005;61:1476–1482. doi: 10.1107/S0907444905025771. [DOI] [PubMed] [Google Scholar]