The common oxidative DNA lesion 8-oxoguanine is highly mutagenic. Here, we show that, in Arabidopsis, DNA polymerase λ plays a crucial role in the error-free bypass of this lesion, together with proliferating cell nuclear antigen 2. Expression of DNA polymerase λ is induced by UV in vivo, and altered levels of DNA polymerase λ affect the growth of transgenic plants.
Abstract
The oxidized base 7,8-oxoguanine (8-oxo-G) is the most common DNA lesion generated by reactive oxygen species. This lesion is highly mutagenic due to the frequent misincorporation of A opposite 8-oxo-G during DNA replication. In mammalian cells, the DNA polymerase (pol) family X enzyme DNA pol λ catalyzes the correct incorporation of C opposite 8-oxo-G, together with the auxiliary factor proliferating cell nuclear antigen (PCNA). Here, we show that Arabidopsis thaliana DNA pol λ, the only member of the X family in plants, is as efficient in performing error-free translesion synthesis past 8-oxo-G as its mammalian homolog. Arabidopsis, in contrast with animal cells, possesses two genes for PCNA. Using in vitro and in vivo approaches, we observed that PCNA2, but not PCNA1, physically interacts with DNA pol λ, enhancing its fidelity and efficiency in translesion synthesis. The levels of DNA pol λ in transgenic plantlets characterized by overexpression or silencing of Arabidopsis POLL correlate with the ability of cell extracts to perform error-free translesion synthesis. The important role of DNA pol λ is corroborated by the observation that the promoter of POLL is activated by UV and that both overexpressing and silenced plants show altered growth phenotypes.
INTRODUCTION
The DNA of all living organisms is subjected to damage by physical and chemical environmental agents (UV and ionizing radiations, chemical mutagens, etc.) and by free radicals or alkylating agents endogenously generated by metabolism (Britt, 1999). DNA is also damaged because of errors during its replication. The DNA lesions produced by these damaging agents may result in base change, base loss, base mismatch, base deletion or insertion, linked pyrimidines, strand breaks, and intra- and interstrand cross-links (Bray and West, 2005). These DNA lesions can be both genotoxic and cytotoxic. Plants are particularly affected by the UV-B radiation of sunlight, which penetrates cells and damages their genome by inducing DNA–protein and DNA–DNA cross-links, thymidine dimers, and oxidative damage through the generation of reactive oxygen species (ROS) (Collins, 1999). ROS are produced not only through the action of exogenous agents, but also during normal cell metabolism. When ROS react with DNA, the most frequently generated lesion (103 to 104 per cell/per day in human cells) is 7,8-dihydro-8-oxoguanine (8-oxo-G), which is potentially mutagenic (Kamiya, 2003, 2004). In fact, the presence of 8-oxo-G in the replicating strand can lead to frequent misincorporation of A opposite the lesion by the replicative DNA polymerases (DNA pols) α, δ, and ε, resulting in an error-prone synthesis (Maga et al., 2009).
Removal of A:8-oxo-G mismatches arising from the activity of replicative DNA pols requires a two-step mechanism. First, the mismatch is recognized by the glycosylase MutY, which removes the incorrectly paired A, leaving a 1-nucleotide gap on the DNA with the 8-oxo-G as the template base. At this point, a DNA pol is required that incorporates dCTP opposite the lesion to reconstitute a C:8-oxo-G base pair; this is subsequently recognized by a second glycosylase, Ogg1, which removes the oxidized base. Thus, the presence of a specialized translesion DNA pol able to efficiently incorporate C opposite 8-oxo-G is of paramount importance in the mechanism of tolerance toward oxidative DNA damage. In human cells, we have recently shown that, after removal of the erroneously incorporated A opposite 8-oxo-G by the glycosylase MutYH, the subsequent error-free bypass of the lesion requires the specialized DNA pol λ, along with the auxiliary proteins proliferating cell nuclear antigen (PCNA) and Replication Protein A (RP-A) (Maga et al., 2007, 2008), to catalyze the correct incorporation of C opposite 8-oxo-G during the resynthesis step, reconstituting a C:8-oxo-G base pair that could subsequently be repaired by the base excision repair mechanism (Macpherson et al., 2005).
In plants, the general knowledge about DNA pol λ structure and functions is still limited. Analysis of the Arabidopsis thaliana genome shows that this enzyme, encoded by the gene At1g10520, is the only member of the X polymerase family. Likewise, a single gene member of the X-family encoding DNA pol λ (POLL) was identified in the rice (Oryza sativa) genome. The Os DNA pol λ protein consists of 552 amino acid residues with a molecular mass of 60.9 kD and shares high sequence identity with Arabidopsis DNA pol λ. Both Os DNA pol λ and At DNA pol λ show a strongly conserved PIP box (PCNA binding domain; Warbrick, 1998). At DNA pol λ possesses the amino acidic stretch QKLGLKYF, common to the DNA pol λ of two other dicots, Populus trichocarpa and Vitis vinifera. Conversely, Os DNA pol λ has a PIP box (QRIGLKFF) found to be conserved in the monocot Sorghum bicolor. Both amino acid sequences are different from the animal DNA pol λ PIP-box (i.e., QAIGLKHY in mammals), thus reflecting a different phylogenetic origin.
A second PCNA-interacting box (VPVLELF) has been identified in human (Homo sapiens) DNA pol λ (Maga et al., 2004). At DNA pol λ also contains a similar domain (VRTISLF). The Os DNA pol λ contains a BRCT domain in the N-terminal region and a pol X domain in the C-terminal domain, appearing similar to mammalian DNA pol λ (Uchiyama et al., 2004). The comparison between the Os DNA pol λ amino acid sequence and those from other known eukaryotic DNA polymerases, belonging to the X family, revealed that it shares 29.6% identity with Hs DNA pol λ and 60.5% with At DNA pol λ. Sequence conservation between Os DNA pol λ and Hs DNA pol λ was higher (37%) in the C-terminal domain. Moreover, the N-terminal BCRT domain, which proved to be an important mediator of the protein–protein interactions involved in the DNA repair mechanisms, showed a lower degree of identity with Hs DNA pol λ (Uchiyama et al., 2004). From a biochemical and functional point of view, it was found that the Os DNA pol λ catalysis is metal dependent, and Mn2+ appeared to be an activator more efficient than Mg2+. Os DNA pol λ catalyzes DNA synthesis with high fidelity, preferentially using poly(dA)/oligo(dT) as a template, and these observations were similar to those reported for Hs DNA pol λ.
As previously mentioned, Hs DNA pol λ exerts a deoxyribonucleotide-terminal transferase activity and directly interacts with human PCNA, which was shown to increase its processivity. PCNA is an essential component of the DNA replication and repair machinery of all eukaryotic organisms (Umar et al., 1996; Gary et al., 1999; Maga and Hubscher, 2003). The primary sequence of PCNA is highly conserved in all eukaryotes, and PCNA homologs have been described in many plants (Krishna et al., 1994a, 1994b). PCNA genes have been duplicated in Arabidopsis: PCNA1 (At1g07370) is on chromosome 1 in a region duplicated from a chromosome-2 segment encompassing PCNA2 (At2g29570) (Blanc et al., 2003). Both Arabidopsis PCNA proteins (Shultz et al., 2007) have a nuclear location.
RP-A is a heterotrimeric protein conserved in all eukaryotes. It is the major single-stranded DNA (ssDNA) binding protein and stabilizes ssDNA during DNA replication, repair, and transcription (Iftode et al., 1999; Fanning et al., 2006). In plants, RP-A participates also in the repair and replication of plastid DNA (Ishibashi et al., 2006). In Arabidopsis, the largest subunits of RP-A (RP-A1) has the ability to bind ssDNA, while the two small subunits (RP-A2 and RP-A3) stabilize the complex and the interactions with the replicative machinery (Zou et al., 2006). Plants are the only known eukaryotes with multiple copies of the RP-A genes. Arabidopsis possesses five putative genes for RP-A1 and two genes each for RP-A2 and RP-A3 (Shultz et al., 2007). Given the conservation in plants of all the essential components of the 8-oxo-G tolerance pathway identified in human cells, we investigated their functional relationships in carrying out the bypass of this highly mutagenic lesion. In addition, we examined the effects of human PCNA and RP-A on the activity of At DNA pol λ synthesis opposite the 8-oxo-G lesion. Our results show that a highly efficient mechanism for error-free bypass of the 8-oxo-G lesion operates also in plant cells. We identified important differences in the functional interaction among DNA pol λ and the two PCNA proteins present in Arabidopsis cells, as well as a high degree of functional conservation between the plant and human machineries. Moreover, the use of transgenic plants characterized by an altered expression of DNA pol λ allowed us to establish a correlation between this enzyme and translesion synthesis (TLS).
RESULTS
Purification and Characterization of Recombinant Arabidopsis DNA Pol λ
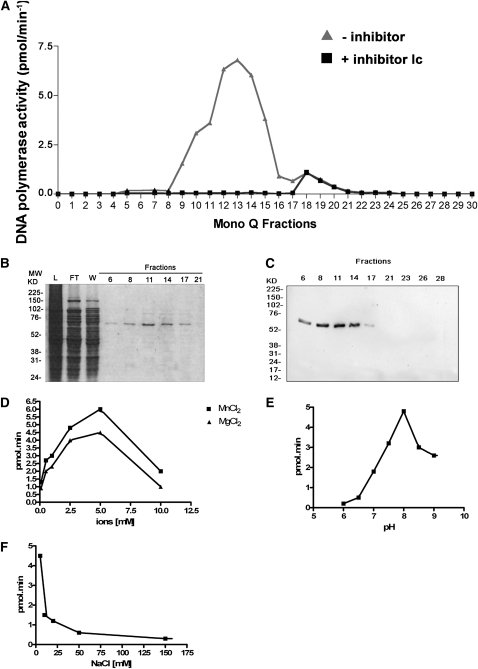
To characterize the biochemical features of DNA pol λ, the corresponding coding sequence (CDS) was expressed in Escherichia coli, and the recombinant His-tagged protein was purified by fast protein liquid chromatography (FPLC). Figure 1A shows the elution profile of polymerase activity from the last chromatographic step (Mono Q). To distinguish DNA pol λ activity from that of any interfering bacterial DNA pol possibly present in the eluted fractions, the assay was performed in parallel in the presence of Ic (N-9-fluorenylmethoxycarbonyl-aminoalkyl-triphosphate), a selective inhibitor of eukaryotic DNA pol λ (Crespan et al., 2005). The single peak of activity detected in fractions 8 to 14 was inhibited by Ic, thus confirming that the enzymatic activity measured was actually that of DNA pol λ. A small residual pol activity in fractions 18 to 21, which was not sensitive to Ic, was due to E. coli DNA pol I contamination and was discarded. SDS-PAGE analysis of the same fractions showed the presence of a single polypeptide of the expected molecular mass, indicating a degree of homogeneity of the recombinant protein higher than 95% (Figure 1Β). Protein identity was further confirmed by protein gel blot analysis using anti-Hs DNA pol λ polyclonal antibodies (Figure 1C). With respect to enzymological parameters, DNA pol λ, as anticipated, required the presence of bivalent metal ions for the catalysis, with a preference for Mn2+ over Mg2+ (Figure 1D) and displayed optimal activity at pH 8.0 (Figure 1E). Moreover, the catalytic efficiency was very sensitive to salt concentration, displaying almost complete inhibition at NaCl concentration higher that 50 mM (Figure 1F).
Figure 1.
Purification of Recombinant At DNA Pol λ.
(A) Elution profile of DNA pol λ from the MonoQ column. Polymerase activity (expressed as pmols of nucleotides incorporated/min) in all MonoQ fractions was tested in the absence (triangles) or in the presence (squares) of the selective DNA pol λ inhibitor Ic.
(B) Coomassie blue–stained SDS-PAGE of the Mono Q column fractions. Fraction numbers are indicated on top of each lane. Molecular weight marker (MW) positions are indicated at the left of the panel. L, loading; FT, flow through (unbound proteins); W, wash.
(C) Protein gel blot analysis of the MonoQ fractions with anti-HsDNA pol λ antibodies. Fraction numbers are indicated at the top of each lane. Molecular weight marker positions are indicated at the left of the panel.
(D) Variation of DNA pol λ activity in the presence of varying concentrations of MnCl2 (squares) or MgCl2 (triangles).
(E) Variation of DNA pol λ activity at different pH values (from 6.0 to 9.0).
(F) Variation of DNA pol λ activity in the presence of increasing NaCl concentrations (from 5 to 150 mM).
Expression and Purification of Recombinant Arabidopsis PCNA1 and PCNA2
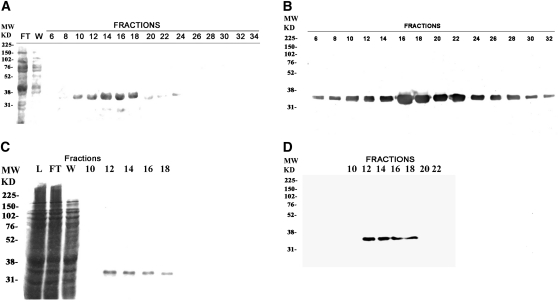
Contrary to animal cells, which possess only one PCNA gene, Arabidopsis has two PCNA genes. To obtain recombinant PCNA1 and PCNA2, the corresponding CDSs were cloned and expressed as His-tagged proteins in E. coli cells. The recombinant proteins were purified by FPLC and the eluted fractions analyzed by SDS-PAGE and protein blots, using an antibody against human PCNA that cross-reacts with both plant proteins. In the case of PCNA1, a single polypetide of the expected molecular mass could be detected, with a peak in fractions 10 to 24 (Figures 2A and 2B). PCNA2 was similarly produced, as shown in Figure 2C (fractions 12 to 18). Protein gel blot analysis with anti-Hs PCNA antibodies again confirmed its identity (Figure 2D). Thus, both PCNA1 and PCNA2 were successfully expressed and purified as His-tagged proteins.
Figure 2.
Purification of Recombinant Arabidopsis PCNA1 and PCNA2.
(A) Coomassie blue staining after SDS-PAGE of the fractions containing PCNA1. Fraction numbers are indicated on top of each lane. Molecular weight marker (MW) positions are indicated at the left of the panel. L, loading; FT, flow through (unbound proteins); W, wash.
(B) Protein gel blot analysis of the PCNA1-containing fractions with anti-PCNA antibody. Fraction numbers are indicated on top of each lane. Molecular weight marker (MW) positions are indicated at the left of the panel.
(C) As in (A), but with the fractions containing PCNA2.
(D) As in (B), but with the fractions containing PCNA2.
DNA Pol λ Recognizes 8-Oxo-G on a Template as a Normal Guanine and Preferentially Incorporates dCTP over dATP Opposite This Lesion
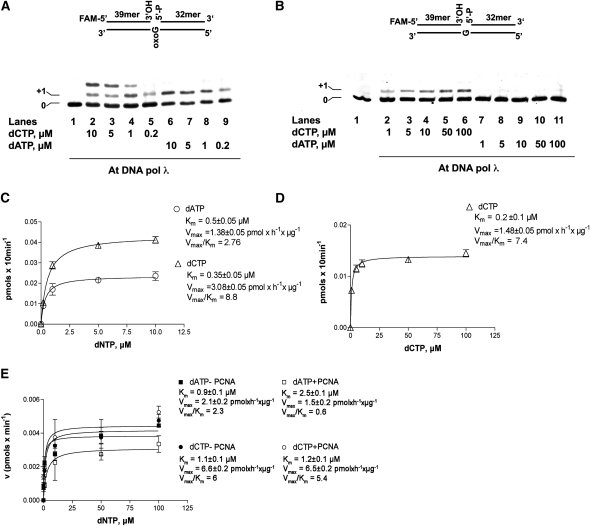
The mutagenic potential of 8-oxo-G when present on the template strand is due to its ability to direct the preferential incorporation of dATP by some DNA pols opposite the lesion. We have shown that Hs DNA pol λ, on the contrary, preferentially incorporates the correct dCTP opposite 8-oxo-G (Maga et al., 2007). Thus, the next step was to test the efficiency of nucleotide incorporation by DNA pol λ opposite an 8-oxo-G lesion present into a 1-nucleotide gap substrate. As shown in Figure 3A, both dCTP (lanes 2 to 5) and dATP (lanes 6 to 9) were incorporated opposite the lesion. At high dCTP concentrations, a limited (1 nucleotide) strand displacement synthesis could be observed (lanes 2 to 4) as revealed by the incorporation of a second dCTP residue opposite to the template G immediately downstream of the lesion. This feature was observed also for the human enzyme (Singhal and Wilson, 1993). When the same experiment was repeated using a control substrate containing an unmodified guanine, only dCTP was incorporated (Figure 3B, lanes 2 to 6), confirming that the incorporation of dATP was strictly dependent on the presence of an 8-oxo-G and was not due to an intrinsic infidelity of DNA pol λ. Quantification of the products allowed us to determine the kinetic parameters for these reactions. As shown in Figure 3C, the catalytic efficiency (Vmax/Km) for dATP incorporation opposite 8-oxo-G was 3-fold higher than for dATP, thus confirming a preferential dCTP incorporation opposite the lesion. Remarkably, the Vmax/Km value for dCTP incorporation opposite 8-oxo-G (8.8 pmol × μM−1 × h−1 × μg−1) was nearly identical to the one displayed opposite a normal G (7.4 pmol × μM−1 × h−1 × μg−1) (Figure 3D), suggesting that DNA pol λ recognized an 8-oxo-G lesion on the template as a normal guanine.
Figure 3.
PCNA2 Favors Error-Free Translesion Synthesis by DNA Pol λ Opposite the 8-Oxo-G Lesion.
(A) Incorporation was monitored with varying concentrations of dCTP (lanes 2 to 5) or dATP (lanes 6 to 9). Lane 1, control in the absence of nucleotides. The structure of the substrate is shown on top of the panel. The position of the band corresponding to the incorporation opposite the 8-oxo-G lesion (+1) is indicated at the left of the panel.
(B) Incorporation of dATP and dCTP opposite to a normal G by DNA pol λ was monitored with increasing concentration of dCTP (lanes 2 to 6) or dATP (lanes 7 to 11). Lane 1, control in the absence of nucleotides. The structure of the substrate is shown on top of the panel. The position of the band corresponding to the incorporation opposite the undamaged base (+1) is indicated on the left of the panel.
(C) Quantification of dCTP (triangles) and dATP (circles) incorporation opposite 8-oxo-G. The Km and Vmax values are reported for both nucleotides.
(D) Quantification of dCTP incorporation (triangles) opposite normal G. The corresponding Km and Vmax values are indicated. Values are the mean of three independent experiments. Error bars are ± sd.
(E) Variation of the initial velocity (v), induced by DNA pol λ on the 1-nucleotide gapped 8-oxo-G template for dCTP incorporation and in the presence (white circles) or in the absence (black circles) of At PCNA2 or for dATP incorporation in the presence (white squares) or in the absence (black circles) of At PCNA2 as a function of nucleotide substrate concentration. Values are the mean of three biological replicates. The corresponding Km and Vmax values are indicated. Error bars are ± sd.
Arabidopsis PCNA2 but Not PCNA1 Increases the Selectivity for dCTP Incorporation Opposite 8-Oxo-G by DNA Pol λ
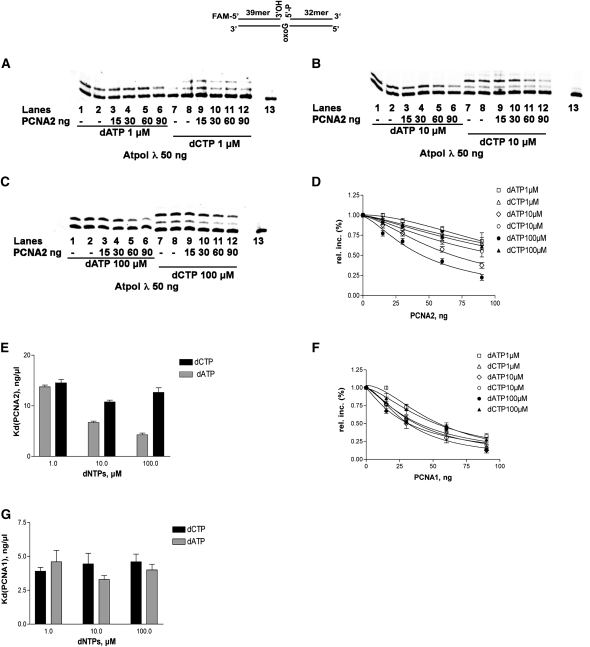
Hs PCNA was shown to increase the processivity of Hs DNA pol λ and its fidelity during 8-oxo-G bypass. We produced both PCNA recombinant proteins from Arabidopsis and sought to verify whether they had the same effect on At DNA pol λ activity. To investigate whether PCNA2 contributed also to translesion synthesis, increasing concentrations of dATP or dCTP were incubated with a 1-nucleotide gapped substrate containing the 8-oxo-G lesion in the presence or in the absence of PCNA2. Quantification of the data showed that the incorporation efficiency, defined as the Vmax/Km ratio, for dATP incorporation opposite to the 8-oxo-G damage was decreased 4-fold, while the incorporation of dCTP was not affected (Figure 3E). To better investigate the effects of PCNA2 on 8-oxo-G bypass, similar experiments were performed at dATP or dCTP concentrations of 1 μM (Figure 4A), 10 μM (Figure 4B), or 100 μM (Figure 4C). Quantification of the data allowed us to determine dose–response curves for At PCNA2 inhibition at different nucleotide concentrations. As shown in Figure 4D, while the effect on dCTP incorporation was modest (10 to 25% inhibition), a strong inhibition of dATP incorporation was observed, which increased with the nucleotide concentration, showing a 75% reduction of incorporation at 100 μM dATP. This was reflected by a decrease of the apparent equilibrium dissociation constant (Kd) for PCNA2 as a function of increasing dATP concentrations (Figure 4E). Thus, PCNA2 acted as an a-competitive inhibitor of dATP incorporation by DNA pol λ, with respect to the nucleotide substrate. These data suggest that PCNA2 binds to the ternary complex of DNA pol λ and its substrates nucleic acid and nucleotides. Similar assays were performed with PCNA1. Dose–response curves showed a clear inhibition of both dATP and dCTP incorporation opposite 8-oxo-G (Figure 4F), the extent of which, however, was not influenced by the nucleotide concentration, as shown by the similar values obtained for the Kd of PCNA1 (Figure 4G).
Figure 4.
The Inhibition of dATP Incorporation by PCNA2, but Not by PCNA1, Is Dependent on the Nucleotide Concentration.
(A) DNA pol λ (50ng) was incubated in the presence of the 1-nucleotide gapped 8-oxo-G template and with 1 μM of either dATP (lanes 1 to 6) or dCTP (lanes 7 to 12) in the absence (lanes 1, 2, 7, and 8) or in the presence (lanes 3 to 6 and 9 to 12) of increasing amounts of PCNA2 (15, 30, 60, and 90 ng). Lane 13, control reaction in the absence of DNA pol λ.
(B) As in (A), but in the presence of 10 μM dATP or dCTP.
(C) As in (A), but in the presence of 100 μM dATP or dCTP.
(D) Effects of increasing amounts of PCNA2 on the relative incorporation of different concentrations (1, 10, and 100 μM) of dATP and dCTP opposite to 8-oxo-G by DNA pol λ. Values are the mean of three independent biological replicates. Error bars represent ± sd values.
(E) Variations of the apparent dissociation constant of PCNA2 (Kd) for DNA pol λ as a function of increasing concentrations of dATP (light-gray bars) or dCTP (dark-gray bars) on a 1-nucleotide gapped 8-oxo-G template. Values are the mean of three independent biological replicates. Error bars represent ± sd values.
(F) Effects of increasing amounts of PCNA1 on the relative incorporation of different concentrations (1, 10, and 100 μM) of dATP and dCTP opposite 8-oxo-G by DNA pol λ. Values are the mean of three independent biological replicates. Error bars represent ± sd values.
(G) Variations of the apparent dissociation constant of PCNA1 (Kd) for DNA pol λ as a function of increasing concentrations of dATP (light-gray bars) or dCTP (dark-gray bars) on a 1-nucleotide gapped 8-oxo-G template. Values are the mean of three independent biological replicates. Error bars represent ± sd values.
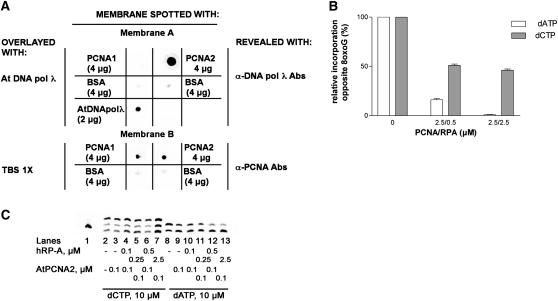
Arabidopsis PCNA2, but Not PCNA1, Physically Interacts with DNA Pol λ
The results described above indicated a different effect of PCNA1 PCNA2 on the bypass activity of DNA pol λ. In particular, they suggested a physical interaction between PCNA2 and the translesion polymerase. Since both PCNA and DNA pol λ had a His-tag, pull-down experiments could not be performed. Coimmunoprecipitation experiments were attempted, but were not conclusive, due to the high tendency of DNA pol λ to unspecifically bind to Protein A or Protein G Sepharose (data not shown). Thus, a protein dot blot experiment was devised, where both PCNA proteins were separately spotted onto a nitrocellulose membrane and overlaid with DNA pol λ. After extensive washing of the membrane, the presence of DNA pol λ bound to PCNA spotted on the filter was revealed by anti-Hs DNA pol λ polyclonal antibodies. As shown in Figure 5A, DNA pol λ could bind only to PCNA2 (membrane A). As a control, an identical membrane was incubated with buffer without DNA pol λ (membrane B) and revealed by anti-Hs PCNA antibodies to visualize the presence of the two proteins. The interaction appeared to be specific, since no signal was detected in positions corresponding to equivalent amounts of BSA, spotted as negative controls (membrane A). Similarly, no cross-reaction between anti-Hs DNA pol λ antibodies and PCNA1 was noticed, as indicated by the absence of a signal corresponding to the spotted area, whereas antibodies recognized the recombinant At DNA pol λ spotted on the membrane as a positive control (membrane A). These results confirm that the functional interaction between DNA pol λ and PCNA2 during translesion synthesis, which allows bypassing of the 8-oxo-G lesion with high fidelity, is due to their direct physical interaction. Moreover, this physical interaction was selective, since no binding was observed with PCNA1.
Figure 5.
Functional Interaction of At PCNA2, Hs RP-A, and Hs PCNA with At DNA Pol λ during the Synthesis Opposite to 8-Oxo-G Damage.
(A) Protein gel blot analysis was performed using recombinant At PCNA2, At PCNA1, and At DNA pol λ proteins. Membrane A was spotted with PCNA1, PCNA2 (4 μg), or DNA pol λ (2μg); then membranes were overlaid with TBS buffer containing 0.1 mg/mL recombinant DNA pol λ and whose presence was revealed by anti-HsDNA pol λ antibodies. Membrane B was spotted with PCNA1, PCNA2, or BSA and then washed with TBS buffer and finally revealed by anti-Hs PCNA antibodies, as control experiment. The indicated amounts of BSA were spotted on both membranes as negative controls.
(B) Relative levels of dATP (white bars) and dCTP (gray bars) incorporation (expressed as percentage relative to the control in the absence of RP-A and PCNA) opposite to 8-oxo-G in the presence of different PCNA/RP-A combinations. Values are the mean of three independent biological replicates. Error bars represent ± sd values.
(C) Incorporation of dCTP (lanes 2 to 7) or dATP (lanes 8 to 13) in the absence (lanes 1 and 8) or in the presence (lanes 3 to 7 and 9 to 13) of 0.1 μM PCNA2 alone or in combination (lanes 4 to 7 and 10 to 13) with increasing amounts of Hs RP-A.
Human PCNA and RP-A Can Substitute for the Plant Proteins to Favor the Error-Free Bypass Synthesis by At DNA Pol λ Opposite 8-Oxo-G
Hs RP-A and Hs PCNA have previously been shown in our laboratory to be able to increase the efficiency of translesion synthesis by Hs DNA pol λ in animal cells. Due to the high level of conservation between At DNA pol λ, At PCNA2, and their respective human orthologs, we analyzed the effect of the human proteins Hs RP-A and Hs PCNA on the ability of At DNA pol λ to bypass the 8-oxo-G damage. As shown in Figure 5B, there was a significant inhibitory effect of Hs RP-A and Hs PCNA proteins on At DNA pol λ efficiency for dATP incorporation opposite 8-oxo-G damage, which was higher than in the case of dCTP. Indeed, dATP incorporation in the presence of the highest tested amounts of Hs PCNA and Hs RP-A (2.5 μM) was almost undetectable, whereas under the same conditions, total dCTP incorporation was reduced by 50%.
Human RP-A Cooperates with At PCNA2 to Increase the Fidelity of 8-Oxo-G Bypass by At DNA Pol λ
Since Hs RP-A and Hs PCNA could synergistically enhance the fidelity of 8-oxo-G bypass by At DNA pol λ, we tested whether At PCNA2 displayed the same effect when combined with Hs RP-A. As shown in Figure 5C, addition of At PCNA2 alone or in combination with Hs RP-A did not affect dCTP incorporation opposite 8-oxo-G (lanes 2 to 7). On the contrary, At PCNA2, which alone was effective in reducing dATP incorporation (lane 9), in combination with Hs RP-A resulted in a strongly enhanced inhibition of dATP insertion (lanes 10 to 13). Overall, these data demonstrate that the functional interaction of RP-A with At DNA pol λ and PCNA in 8-oxo-G bypass is highly conserved, as it occurs also between human and plant proteins. Such a high level of evolutionary conservation highlights the importance of this pathway in preventing the incorrect dATP incorporation opposite the 8-oxo-G damage, thus reducing its mutagenic potential.
Both PCNA1 and PCNA2 Are Localized in the Nucleus of Plant Cells, but Only PCNA2 Interacts with DNA Pol λ in Vivo
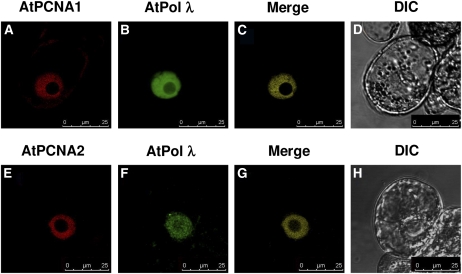
Since in vitro experiments indicated a functional cooperation between DNA pol λ and PCNA2, we confirmed the physical interaction between these two proteins in vivo. Transient expression assays using BY-2 tobacco (Nicotiana tabacum) protoplasts with constructs encoding At PCNA1, At PCNA2, and At DNA pol λ N-terminally fused to green fluorescent protein (GFP) and yellow fluorescent protein (YFP), under the control of cauliflower mosaic virus 35S promoter (Guilley et al., 1982), showed an in vivo nuclear localization of all three proteins (see Supplemental Figure 1 online). Additional assays also showed that nuclear localization was not affected by the N- or C-terminal position of the tag (data not shown). As shown in Figures 6A and 6E, both PCNA1 and PCNA2 localize in the nucleus, but not in the nucleolus. On the contrary, DNA pol λ accumulates in the whole nucleus even though GFP labeling of the nucleolus was weak (Figures 6B and 6F). The in vivo colocalization of DNA pol λ with PCNA1 and PCNA2 was tested by cotransfecting tobacco BY-2 protoplasts with two different constructs encoding the fluorescent fusion proteins GFP-At DNA pol λ and AtPCNA1-RFP (for red fluorescent protein), or GFP-At DNA pol λ and At PCNA2-RFP. Digital merging of red and green single-channel fluorescent images confirmed the nuclear colocalization (Figures 6C and 6G).
Figure 6.
Arabidopsis DNA Pol λ, PCNA1, and PCNA2 Colocalize in the Nucleus of Plant Cells.
Images of tobacco BY-2 protoplasts 24 h after cotransfection with constructs encoding GFP-AtPCNA1 and RFP-AtPCNA2 ([A] to [D]) or GFP-AtDNA pol λ and RFP-AtPCNA2 ([E] to [G]) constructs. Both DNA pol λ ([B] and [F]) and PCNA1 (A) and PCNA2 (E) are targeted to the nucleus, as previously reported (Raynaud et al., 2006). Digital merging of images reveals the colocalization of DNA pol λ both with PCNA1 (C) and with PCNA2 (G). Respective bright-field differential interference contrast (DIC) images are shown ([D] and [H]). Bars = 25 μm.
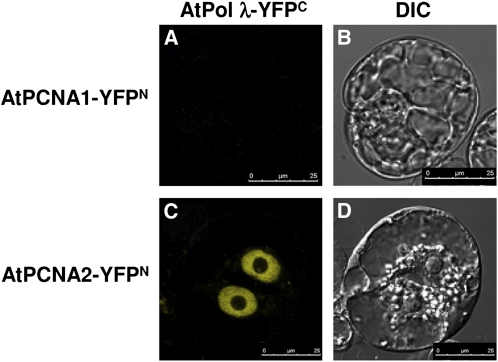
To prove the physical interaction between DNA pol λ and PCNA1 or PCNA2, we performed a bimolecular fluorescence complementation assay (Figure 7). BY-2 protoplasts were cotransfected with a construct encoding a fusion protein between At DNA pol λ and YFP C-terminal region (amino acids 156 to 238) (At POLL-YFPC) and with those encoding either At PCNA1 or At PCNA2 fused to YFP N-terminal region (amino acids 1 to 155) (At PCNA1-YFPN or At PCNA2-YFPN). Both YFP moieties were C-terminally fused. Fluorescence was observed only with PCNA2 as a result of molecular complementation of YFPN and YFPC due to in vivo physical interaction between DNA pol λ and PCNA2 (Figure 7C), while no fluorescence was observed with PCNA1 (Figure 7A). The construct At PCNA1-YFPN was previously proven to encode a functional fusion protein able to interact with ATXR5,6 proteins (Raynaud et al., 2006).
Figure 7.
Arabidopsis DNA Pol λ and PCNA2, but Not PCNA1, Interact in the Nucleus.
Images of BY-2 protoplasts 24 h after cotransfection with plasmids encoding the fusion proteins AtDNA pol λ-YFPC and AtPCNA1-YFPN or with AtDNA pol λ-YFPC and AtPCNA2-YFPN constructs. DNA pol λ interacts in vivo with PCNA2 (A) but not with PCNA1 (C). Respective bright-field differential interference contrast (DIC) images are shown ([B] and [D]). Bars = 25 μm.
Arabidopsis DNA Pol λ Is Responsible for Error-Free Bypass of 8-Oxo-G Lesions in Planta
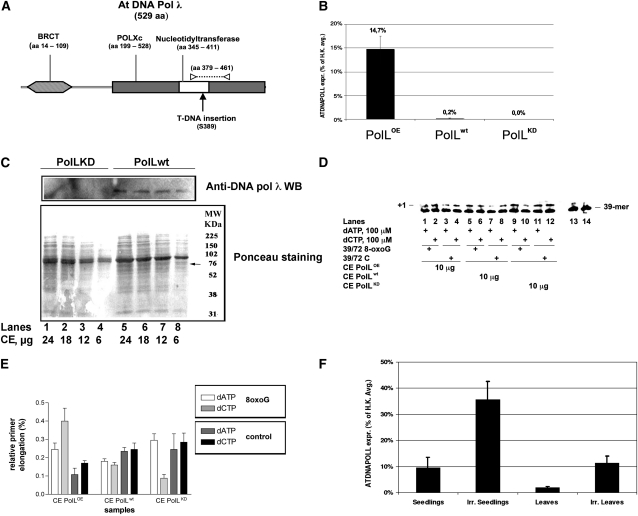
POLL overexpressing plants (POLOE) were produced by Agrobacterium tumefaciens–mediated transformation of Columbia (Col-0) flowers using the vector pGBIN1-At POLL (see Supplemental Figure 2 online) where the CDS encoding DNA pol λ is under the control of the constitutive cauliflower mosaic virus 35S promoter. Three-week-old kanamycin-resistant T1 lines were analyzed by quantitative RT-PCR (qRT-PCR) with respect to the level of POLL transcripts. Out of more than 20 overexpressors, a line characterized by a 70-fold increased content of POLL transcripts was chosen to evaluate its behavior in a translesion assay (Figure 8B). The protein level of overexpressing lines was assessed by protein gel blot analysis (see Supplemental Figure 3 online). The relative increase in protein level with respect to wild-type plants was 2 ± 0.5-fold, which was lower than the increase of the relevant transcript level. This suggests that DNA pol λ is subjected to a tight posttranslational regulation that controls protein accumulation, as already demonstrated in the case of mammalian cells (Wimmer et al., 2008).
Figure 8.
Arabidopsis DNA Pol λ Is Responsible for Error-Free Bypass of 8-Oxo-G Lesion in Plant Cells.
(A) Schematic drawing of the protein domains encoded by DNA pol λ gene. Functional domains are reported. BRCT, BRCA-1 C-terminal domain; POLXc, DNA polymerase family X catalytic domain. The Ser-389 residue corresponding to T-DNA insertion is indicated by a black arrow. The portion of catalytic domain (amino acids 379 to 461) corresponding to amplicon produced by qRT-PCR with primers POLL FW and POLL RV is indicated by the dotted line. Primers position is reported as white triangles.
(B) Relative expression level of POLL in 10-d-old seedlings of different lines. qRT-PCR results show that POLL transcript levels in wild-type (POLWT), knockdown (POLKD), and overexpressing (POLOE) lines. Transcript levels are expressed as percentage of the average expression level of two housekeeping genes (H.K.): EF1α translation elongation factor (At5g60390) and glyceraldehyde-3-phosphate dehydrogenase C2 (At1g13440). POLL transcripts are ~14.7% of H.K in the overexpressing line and only 0.2% in Col-0. No expression was detected in knockdown line. Error bars represent ± sd of two technical replicates
(C) Protein gel blot analysis of the expression of DNA pol λ in wild-type (lanes 5 to 8) and KD (lanes 1 to 4) cell extracts (CE). Top panel: protein blot with anti-Hs DNA pol λ antibodies. Bottom panel: Ponceau staining of the same membrane. The migration position of DNA pol λ is indicated with an arrow.
(D) Reactions were performed with cell extracts under the conditions specified in Methods. POLLOE (lanes 1 to 4), POLLwt (lanes 5 to 8), or POLLKD (lanes 9 to 12) crude extracts (10 μg total proteins) were incubated in the presence of 100 μM dATP (lanes 1, 3, 5, 7, 9, and 11) or 100 μM dCTP (lanes 2, 4, 6, 8, 10, and 12) with the 39/72 8-oxo-G DNA substrate or with the 39/72 control DNA substrate. Lanes 13 (with 8-oxo-G substrate) and 14 (with control substrate) are control reactions in the absence of extracts.
(E) Relative activity, expressed as percentage of elongated primer ends, for dATP (light white bars) and dCTP (light-gray bars) in the 39/72 8-oxo-G DNA substrate and for dATP (dark-gray bars) and dCTP (black bars) in the 39/72 8-oxo-G DNA control substrate. Values are the mean of two independent biological replicates. Error bars are ± sd.
(F) qRT-PCR on 10-d-old seedlings and on mature leaves (30 d old) show a higher transcript level in young tissues, 9.5% (±4.1%) of H.K. versus 1.8% (±0.5%). After 30 min of UV irradiation (0.3 mW/cm2, corresponding to 2.9 J/m2/s, at 243-nm wavelength), transcript level was ~3.7-fold higher in seedlings and >6-fold higher in mature leaves, 35.5% (±7%) versus 11.3% (±2.7%). Error bars represent ± sd values among three independent biological replicates.
The only available Arabidopsis line with an insertion in the POLL gene (At1g10520) was the SALK_075391C line, in which the T-DNA (4.5 kb) is located in the intron between the 9th and 10th exons, corresponding in the spliced mRNA to an exon-exon junction at 1167 bp starting from ATG, within the codon for S389 of the translated peptide (Figure 8A). Since the active site of DNA pol λ spans from amino acid 346 to 411, it was anticipated that the insertion could prevent the production of a functional protein. Following further selection on kanamycin-containing medium, the analysis by PCR using genomic DNA (see Supplemental Figure 3 online) showed that SALK_075391C knockdown line was actually homozygous for the T-DNA insertion. Moreover, qRT-PCR (Figure 8B) and protein gel blot analysis (Figure 8C) failed to detect POLL transcripts or protein, thus proving the actual knocking down of the POLL gene. Both types of plants, POLL overexpressors and knockdown, are characterized by an altered phenotype. Overexpressors show a retarded growth, while the knockdown features early flowering (see Supplemental Figure 4 online). The reasons of these phenotypes are currently under investigation; however, taking into account the modest increase in protein levels observed in POLLOE plants (see Supplemental Figure 3 online), these alterations reflect the importance of the maintenance of a balanced level of DNA pol λ in the living plant organism.
To further investigate the role of DNA pol λ in 8-oxo-G lesion bypass, we prepared cell extracts from 10-d-old seedlings of overexpressing POLL (POLLOE) and knockdown (POLLKD) lines. The extracts were tested in in vitro translesion assays in comparison with extracts coming from Col-0 plants (POLLwt) (see Figure 8D for a representative experiment). The error-free (dCTP) incorporation opposite the 8-oxo-G lesion (Figure 8E) was increased ~2-fold in the POLLOE extracts, while it was reduced in the POLLKD extracts with respect to POLLwt. The level of error-prone (dATP) bypass was basically unaffected. These data strongly suggest that DNA pol λ is an essential component of the machinery responsible for error-free 8-oxo-G bypass in plants.
Arabidopsis POLL Expression Is Higher in Young Tissues and Is Induced by UV Irradiation
On the basis of annotated coding sequences of Arabidopsis available at The Arabidopsis Information Resource website (www.Arabidopsis.org), the length of the intergenic region upstream of the coding sequence of POLL (At1g10520) is only 177 bp long because of the proximity of the upstream gene (At1g10510). The analysis of the putative promoter by PlantPAN (Plant Promoter Analysis Navigator; http://PlantPAN.mbc.nctu.edu.tw), which takes into account 197 Arabidopsis transcription factors from PLACE, TRANSFAC, AGRIS, and JASPAR databases (Chang et al., 2008), revealed the presence of T-box and SORLIP5 sites, which are overrepresented in promoters of light-activated genes (Chan et al., 2001; Hudson and Quail, 2003), along with two distinct Myb4 binding sites. The Myb family of transcription factors is known to mediate biotic and abiotic stresses in plants (Chen et al., 2002). In particular, the Myb4 subgroup was found to repress the transcription of UV response genes, since the related mutants were found to be more tolerant to UV. Moreover, it was observed that MYB4 transcription is downregulated after UV-B irradiation (Jin et al., 2000). Thus, the presence of two Myb4 binding sites in the putative POLL promoter implicates UV as a possible regulator of POLL expression. To prove this hypothesis, we compared the relative level of 10-d-old At POLL transcripts between control and UV-irradiated seedlings and 30-d-old leaves by qRT-PCR. Three independent biological replicates were performed for each group of samples. The amount of POLL transcripts found in untreated seedling and leaves was ~9.5 and 1.8% of the geometric average of two independent housekeeping genes, respectively (Figure 8F). The fact that in seedlings the level of POLL transcripts was higher than in mature leaves is consistent with the observation that the major DNA repair pathway in rice mature leaves is due to photoreactivation performed by cyclobutane pyrimidine dimers photolyase and (6-4) photolyase (Kimura et al., 2004). On the other hand, the higher expression level in seedlings may reflect a higher rate of cell proliferation and DNA replication due the presence of meristematic tissues. In fact, the horthologous rice POLL was shown to be expressed at a higher level in proliferating cells (Uchiyama et al., 2004).
Following UV irradiation, a remarkable increase of POLL expression was observed both in 10-d-old seedlings and in leaves. In particular, irradiated seedlings showed an increase of over 3.7-fold from ~9.5 to >35.5% with respect to the expression of housekeeping genes (Figure 8F). In leaves, the expression level raised from 1.8 to ~11.3% (6-fold) following irradiation (Figure 8F). To confirm the increased POLL expression caused by UV, we produced transgenic plants containing a chimeric reporter uidA gene driven by the POLL promoter. Results of histochemical staining showed that β-glucuronidase activity was detectable only in irradiated plants, thus indicating a very low basal expression of POLL under normal conditions (see Supplemental Figure 5 online). These data clearly show that POLL transcription is induced by UV.
DISCUSSION
DNA pol λ is the only member of DNA family X present in higher plants. Arabidopsis DNA pol λ shows 30% similarity with its human homolog. On the other hand, while human cells possess only one gene for each of the three subunits (p70, p32, and p14) of the ssDNA binding protein RP-A and one for the auxiliary factor PCNA, Arabidopsis contains two genes for PCNA, five genes for the p70, and two each for p32 and p14 subunits of RP-A (Kimura and Sakaguchi, 2006). Whether this fact simply constitutes an example of extreme functional redundancy or reflects a diversification of functions occurring in plant cells with respect to animal cells is not known. Our recent results have defined the role of human PCNA and RP-A in increasing the fidelity of synthesis of human DNA pol λ in bypassing the 8-oxo-G lesion (Maga et al., 2008). To investigate whether the mechanism operating in animal cells was also involved in increasing the tolerance to oxidative DNA damage in plants, we produced recombinant Arabidopsis DNA pol λ, PCNA1, and PCNA2. These proteins were then tested in vitro in specific translesion synthesis assays in the presence of an 8-oxo-G lesion.
The results presented here, while confirming that DNA pol λ is likely playing a major role in the error-free bypass of 8-oxo-G lesions in plants, highlight some important differences with respect to animal cells. The most intriguing one is the diversification of functions of the two plant PCNA proteins. Our analysis showed that both PCNA1 and PCNA2 are expressed and localized in the nucleus, together with DNA pol λ. However, only PCNA2 physically interacts with DNA pol λ both in vitro and in vivo. This differential behavior is also reflected at the enzymatic level. PCNA2 enhances the fidelity of DNA pol λ by reducing its ability to incorrectly incorporate dATP opposite 8-oxo-G, whereas PCNA1 shows a general inhibition of translesion synthesis by DNA pol λ. Interestingly, the effect of PCNA2 is dependent on nucleotide concentration and exerts its maximal efficiency in preventing error-prone bypass of the 8-oxo-G lesion by DNA pol λ precisely when it is most needed, that is, in the presence of high concentrations of dATP, that should give the highest probability for misincorporation. At PCNA1 and At PCNA2 are nearly identical (99% similarity) and show a very high similarity (85%) to Hs PCNA. In particular, all the conserved boxes that were found to be involved in the interaction between Hs DNA pol λ and Hs PCNA (SHV motif, amino acids 43 to 45; QLGI motif amino acids 125 to 128; LAPK motif, amino acids 251 to 254) are conserved also in At PCNA1 and AtPCNA2. However, PCNA1 and PCNA2 show two and three extra amino acids, respectively, at their C terminus compared with the human protein.
The recent resolution of the crystal structure of At PCNA1 and At PCNA2 in complex with a peptide derived from the human p21 protein, a known interactor of Hs PCNA, further confirmed the functional equivalence of both plant proteins with the Hs PCNA (Strzalka et al., 2009). PCNA1 and PCNA2 have been also proposed to be able to form heterotrimers, an observation that is particularly relevant in the light of the different functional effects of these two proteins revealed by our study.
Thus, it is possible to speculate that PCNA2 might act as a specialized DNA repair or translesion auxiliary factor, promoting the switch from highly error-prone replicative and/or repair plant DNA pols to the more faithful DNA pol λ. On the other hand, PCNA1 might be required for other functions (for example during DNA replication or certain types of DNA repair) not requiring the action of DNA pol λ. Heterotrimers composed of mixed PCNA1 and PCNA2 monomers might also display different properties with respect to the corresponding homotrimers, possibly adding a further level of regulation to plant PCNA functions. Further mutagenesis and in vivo expression studies will help to clarify these issues.
DNA pol η, a Y-family TLS polymerase of Arabidopsis, is able to rescue a yeast UV-sensitive phenotype only when cotransformed with the PCNA2 gene but not with PCNA1 (Anderson et al., 2008; Kunz, 2008), further emphasizing the specific role of PCNA2 protein as an auxiliary factor in translesion synthesis. PCNA1 and PCNA2 differ by eight amino acidic residues, but only Asn-201, present in PCNA2, was proven to be critical for the interaction with DNA pol η in yeast. In fact, when Asn was substituted with Lys (Lys-201 as is the case of PCNA1), PCNA2 failed to complement the RAD18 mutant.
Since monoubiquitylation of PCNA Lys-164, close to residue 201 in the three-dimensional structure, is required for the functionality of DNA pol η (van der Kemp et al., 2009), it was proposed that the amino acidic substitution might have some differential effect on PCNA1 and PCNA2 monoubiquitylation, thus influencing their functional interaction with DNA pol η (Kunz, 2008).
Recent data point to a role of ubiquitylation of PCNA in TLS (Chen et al., 2010; Das-Bradoo et al., 2010) suggesting a major role for posttranslational modulation of PCNA function rather than transcriptional regulation. It is tempting to speculate that PCNA is always present in the nuclear environment, and its interaction with translesion DNA pols is prevented unless DNA damage induces a rapid activation through ubiquitylation of PCNA, thus allowing interaction with TLS pols, the fidelity of which is lower compared with that of replicative ones.
However, PCNA monoubiquitylation in plants is still to be confirmed, and Arabidopsis has no clear homolog of RAD18, the enzyme that can perform PCNA monoubiquitylation in humans and yeast (Potuschak et al., 1998; Kraft et al., 2005). Nevertheless, it is not possible to exclude that monoubiquitylation of PCNA2 might further enhance DNA pol λ fidelity.
We previously demonstrated that, in animal cells, DNA pol λ is the specialized enzyme required for error-free bypass of 8-oxo-G lesions, and it is recruited to the damage site thanks to the coordinated action of two auxiliary proteins: PCNA and RP-A. Here, we show that At DNA pol λ, the plant homolog of Hs DNA pol λ, fulfills a similar function in coordination with At PCNA2. In addition, we show that the human proteins RP-A and PCNA can cooperate with a plant DNA pol λ. Such a high degree of evolutionary conservation is a characteristic of essential biochemical processes, resembling the interaction of replicative DNA pols with the auxiliary factors PCNA and Replication Factor C, which is conserved from yeast to man. Thus, our results further support an essential role of the DNA pol λ–dependent pathway for oxidative DNA damage tolerance in all eukaryotic organisms.
Sequence annotation of the genomes of Arabidopsis and rice has shown the presence in plants of conserved genes encoding components of repair pathways, including those for specialized DNA pols necessary for TLS (Kimura and Sakaguchi, 2006). Some of these have been characterized. For instance, it was shown that a cDNA encoding DNA pol η of Arabidopsis is able to complement a UV-sensitive yeast strain deficient in this polymerase (Santiago et al., 2006). Increased UV sensitivity was also reported for a Rev1 mutant (Takahashi et al., 2005). In the case of Arabidopsis DNA pol κ, deletion studies of POLK showed that the ability to bypass 8-oxo-G increases following the removal of the C-terminal region (not involved in catalytic activity), thus indicating a possible regulatory role of this region (García-Ortiz et al., 2007).
Also, Arabidopsis DNA pol ζ, which belongs to the B family, appears to be involved in TLS since deletion of its Rev3 subunits is responsible for a drastic increase in UV sensitivity (Sakamoto et al., 2003).
A reverse-genetic analysis was used to evaluate the role of genes encoding At DNA pol η (At POLH) and At DNA pol ζ (At POLZ) in the tolerance of roots to UV radiation, and it was shown that these pols contribute to the integrity of stem cells of the root meristem (Curtis and Hays, 2007). To evaluate the role played by At DNA pol λ in translesion synthesis in vivo, we analyzed transgenic plants characterized by either overexpression or silencing of POLL. Results of TLS assays using cell extracts of these plants showed that dCTP incorporation due to error-free bypass of 8-oxo-G lesion was increased ~2-fold in POLLOE extracts, while it was reduced in the POLLKD cell extracts with respect to control.
Additional evidence pointing to a major role of At POLL in TLS comes from expression studies following UV irradiation of young plants. It is well known that UV, besides inducing the formation of cyclobutane pyrimidine dimers and photoproducts, contributes to ROS production. Thus, the observation that UV irradiation causes an increase of POLL transcripts both in seedlings and mature leaves is consistent with its role in the response to oxidative DNA damage.
METHODS
Chemicals
Deoxynucleotides were purchased from Gene Spin. All the other reagents were of analytical grade and purchased from Fluka or Merck. The 39-, 32-, and 72-oligonucleotides, either unlabeled or 5′-labeled, were purchased from MWG.
DNA Substrates
All oligonucleotides were purified from polyacrylamide denaturing gels. The sequences were as follows: 72-mer template, 3′-ATGTTGGTTCTCGTATGCTGCCGGTCACGGCTTAAGTGTGGCGGCCGCGGGTTGGAGGGCTTATAGATTATG- 5′; 39-mer primer, 5′-TACAACCAAGAGCATACGACGGCCAGTGCCGAATTCACA-3′; 32-mer primer, 5′-CGCCGGCGCCCAACCTCCCGAATATCTAATAC-3′. The 72-mer, either undamaged or containing the 8-oxo-G lesion (8-oxo-dG-CE Phosphoramide from Glen Research), and the corresponding primer were chemically synthesized and purified on a 12% (w/v) polyacrylamide, 7 M urea, and 10% formamide gel. After elution and ethanol precipitation, their concentrations were determined spectrophotometrically. The bold letter corresponds to the position of the 8-oxo-G lesion. The 39-mer primer was 5′-labeled with Blue FAM Fluorescence group by the Eurofin Company. Each labeled primer was mixed to the complementary template oligonucleotide at 1:1:1 (M/M) ratio in the presence of 25 mM TrisHCl, pH 8.0, and 50 mM KCl, heated at 75°C for 10 min, and then slowly cooled down at room temperature.
Antibodies and Proteins
Monoclonal antibody against PCNA (clone PC10, mouse) was purchased from Santa Cruz Biotechnology. Antibodies against human DNA pol λ (polyclonal rabbit) were a gift from U. Hübscher, University of Zürich-Irchel. Recombinant human PCNA and RPA were expressed and purified in the laboratory of U. Hübscher as described (Maga et al., 2008).
Buffers
The following buffers were used: lysis buffer, 0.1 M NaPO4, pH 8, 0.01 M TrisHCl, pH 8, 0.01% Nonidet NP-40; equilibrium Ni-NTA buffer, 50 mM TrisHCl, pH 8, 50 mM NaCl, 5% (v/v) glycerol, and 5 mM imidazole-HCl; elution Ni-NTA buffer, 50 mM TrisHCl, pH 8, 50 mM NaCl, 5% (v/v) glycerol, and 500 mM imidazole-HCl; Equilibrium Mono Q buffer, 20 mM TrisHCl, pH 8, 8 mM NaCl, 5% (v/v) glycerol, and 1 mM EDTA, pH 8; Elution Mono Q buffer, 20 mM TrisHCl, pH 8, 1 M NaCl, 5% (v/v) glycerol, and 1 mM EDTA, pH 8; Dialysis Mono Q buffer, 50 mM TrisHCl, pH 7.5, and 20% (v/v) glycerol; TDB buffer, 40 mM Tris-HCl, pH 7.5, 1 mM DTT, and 0.2 mg/mL BSA.
Cloning, Expression, and Purification of His-At DNA Pol λ
Arabidopsis thaliana MM1 suspension cells were maintained as described (Menges and Murray, 2002). Total RNA was extracted from actively proliferating cells 2 d after transfer to fresh medium using an RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. One microgram of total RNA was reverse transcribed with the ImProm-II reverse transcription system (Promega).
The coding sequences of AtDNA pol λ were amplified using the HotStart HiFidelity Polymerase Kit with the following primers: attB1-PolL, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGCGGCAAAGCGAGGGAGAAA-3′; attB2-PolL, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTA[TTA]TTAGAGATTCCTCTCGTGTGG-3′. The recognition sequences for BP Recombinase II (Invitrogen) are underlined. For C-terminal fusion proteins, the stop codon (brackets) was omitted.
The PCR product was purified and cloned by site-specific recombination, performed with Gateway BP Clonase II enzyme mix (Invitrogen), following the manufacturer’s instructions, into the entry vector pDONR221 (Invitrogen) and sequenced (Macrogen).
Then, the CDS of At DNA pol λ was subcloned with a Gateway LR recombinase into the expression vector pEXP1-DEST (Invitrogen), carrying a His-tag in frame with the CDS at the N-terminal end.
For expression and purification of His-tagged At DNA pol λ, pEXP1-DEST-pol λ was used to transform Escherichia coli DH5α bacteria grown in 1.5 liters of Luria-Bertani medium containing ampicillin (50 μg/mL) to reach an OD600 of 0.4 at 37°C. At DNA pol λ expression was induced for 4 h at 37°C by adding isopropyl β-thiogalactoside to a 1 mM final concentration.
Cells were centrifuged at 3000g for 1 h, and bacterial pellets were resuspended in a 10 μL lysis buffer for 1 h on ice with lysozyme (1 mg/mL). Next, they were briefly sonicated to destroy cell membranes and reduce viscosity and subjected to a mechanical lysis by a glass Dounce homogenizer followed by incubation for 10 min in ice. The insoluble material was removed by centrifugation for 90 min at 100,000g and the supernatants loaded onto 1 mL FPLC-Ni-NTA His Bind Resin column (Pharmacia) equilibrated with equilibrium Ni-NTA buffer. The column was washed with 8 mL equilibrium buffer, and His-At DNA pol λ was then eluted with an 8-mL gradient from 5 to 500 mM imidazole in elution buffer. The presence and the purity of protein were determined by SDS-PAGE, protein blotting, Coomassie Brilliant Blue staining, and in vitro polymerase activity assay. To eliminate imidazole, fractions were pooled and dialyzed for 4 h at 4°C against the Equilibrium Mono Q buffer. The pool was loaded onto a Mono Q column (Pharmacia) equilibrated in the same buffer. The column was eluted with a linear gradient from 8 mM to 1 M NaCl. Fractions were tested by protein blotting, Coomassie Brilliant Blue staining, and with an in vitro polymerase assay. For SDS-PAGE and protein blotting, aliquots of the fractions that contained proteins of interest were electrophoresed in a 12% SDS-polyacrylamide gel and transferred by standard procedures to a nitrocellulose filter. The membrane was blocked for 3 h with milk-TBS and then incubated for 3 h with the primary anti-pol λ antibodies. After washing with TBS containing 0.1% Tween 20, the membrane was incubated with anti-rabbit (Pierce) IgG conjugated to peroxidase. Immunodetection was performed using a light-enhanced chemiluminescence (ECL) detection system according to Amersham’s instructions.
Cloning, Expression, and Purification of His-At PCNA1 and 2
The CDSs of Arabidopsis PCNA1 and PCNA2 were amplified as previously described in the case of DNA pol λ using the the following primers: attB1-PCNA1, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTTGGAGCTACGTCTT-3′; attB2-PCNA1 NS, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTA[TTA]GGGATTAGTGTCTTC-3′; attB1-PCNA2, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGTTGGAGCTTCGTTTA-3′; attB2-PCNA2 NS, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTA[TTA]TTCTGGTTTGGTGTC-3′.
The recognition sequences for BP Recombinase II (Invitrogen) are underlined; for C-terminal fusion proteins, the stop codons (in brackets) were omitted. The PCR products were purified, cloned by site-specific Gateway recombination into the entry vector pDONR221 (Invitrogen), and sequenced (Macrogen). The CDSs of At PCNA1 and At PCNA2 were then subcloned into the expression vector pEXP1-DEST as previously described.
The plasmids pEXP1-DEST-PCNA1 and pEXP-DEST-PCNA2 were used to transform E. coli DH5α (VWR). Protein expression was induced as in the case of DNA pol λ. The two proteins were purified by a Ni-NTA His Bind Resin column (Pharmacia) and a Mono Q column (Pharmacia) as described for DNA pol λ purification. Fractions were tested by protein gel blotting and Coomassie Brilliant Blue staining.
In Vitro Polymerase Assays
The reaction mixtures contained the enzymatic fraction of DNA pol λ, PCNA1, or PCNA2 from chromatography profile in a 10-μL final volume containing Buffer SCA (50 mM Tris-HCl, pH 7.5, 1 mM DTT, 0.20 mg/mL BSA, and 2% glycerol), 0.01 mM [3H-TTP] (1490 cpm/pmol), deoxynucleotide triphosphate (1.5 Ci/mmol), 10 mM MgCl2, and 0.5 μg poly(dA)/oligo(dT) 5:1 in the presence of different concentrations of purified enzyme and with fixed doses of Ic, a specific DNA pol λ polymerase inhibitor, previously developed in our laboratory (Crespan et al., 2005). All the reaction mixtures were incubated for 20 min at 37°C, and subsequently 10 μL from each sample was spotted onto glass GF/C fiber filters (Printed Filtermat A; WALLAC). Filters were washed three times with 5% TCA and once with ethanol for 5 min and then dried; finally, EcoLume Scintillation cocktail (ICN, Research Products Division) was added to detect the acid-precipitable radioactivity by the Perkin-Elmer Trilux MicroBeta 1450 counter.
The effect of metal ions on the DNA pol λ activity was assayed under the conditions described above in the presence of increasing concentrations of MnCl2 or MgCl2. The pH influence on the DNA pol λ activity was assayed under the conditions described above in the presence of 25 mM Tris-HCl at different pH values.
In Vitro Translesion Synthesis Assays
For denaturing gel analysis of the DNA products, the reaction mixtures (10 μL) contained 25 nM 5′-(6-carboxyfluorescein)- labeled dsDNA substrate (8-oxo-G damaged and control), 50 nM At DNA pol λ, 1 mM Mg2+, and different concentrations of deoxynucleotide triphosphate and purified Hs PCNA, Hs RPA, At PCNA1, and At PCNA2 as indicated in the figure legends. Reaction mixtures were incubated for 25 min at 37°C and then stopped by addition of standard denaturing gel loading buffer (95% formamide, 10 mM EDTA, xylene cyanol, and bromophenol blue), heated at 95°C for 5 min and loaded on a 7 M urea 12% polyacrylamide gel. The reaction products were analyzed using the Molecular Dynamics PhosphorImager (Typhoon Trio GE Healthcare) and quantified by Image Quant and GraphPad Prism programs.
Steady State Kinetic Analysis
Reactions were performed as described above. Quantification was done by scanning densitometry with a PhosphorImager (Typhoon Trio; GE Healthcare). The initial velocity of the reaction was calculated (Image Quant and GraphPad Prism 3.0) from the values of integrated gel band intensities as follows:
where T is the target site, the template position of interest; I*T is the sum of the integrated intensities at positions T, T +1, . . ., T + n.
All of the intensity values were normalized to the total intensity of the corresponding lane to correct for differences in gel loading. The apparent Km and Vmax values were calculated by plotting the initial velocities in dependence of the nucleotide [deoxynucleotide triphosphate] or primer [3′-OH] substrate concentrations and fitting the data according to Michaelis- Menten equation as follows:
where [E]0 was the input enzyme concentration and [S] was the variable substrate. Substrate incorporation efficiencies were defined as the Vmax/Km ratio. Reactions were performed as describe above.
Protein Dot Blotting
DNA pol λ or both the recombinant PCNA were spotted on two identical nitrocellulose membranes (Hybond; Amersham), as indicated in the figure legends, and the membranes were blocked by incubation with skimmed milk. Then, membrane A was overlaid with DNA pol λ and revealed by anti-Hs DNA pol λ antibody. BSA protein was also spotted as a control. Membrane B, loaded with both the PCNA1 and PCNA2, was treated as described above but was not incubated with DNA pol λ and revealed using specific anti-HsPCNA antibody. After washing with TBS containing Tween 20, the membranes were incubated with the appropriate IgG conjugated to peroxidase. Immunodetection was performed using a light-enhanced chemiluminescence detection system (ECL; GE Healthcare) according to the manufacturer’s instructions. The images were then analyzed by the QuantyOne software.
Transgenes Constructs
The pGHC-XR, pGHC-GX, and pGHC-XG plasmids were obtained by subcloning into the pFF19 plasmid (Timmermans et al., 1990) the Gateway fluorescent cassette excised from binary vectors pK7RWG2, pK7FWG2, and pK7WGF2 (Karimi, 2005), respectively.
The sequences encoding DNA pol λ, PCNA1, and PCNA2 cloned in the pDONR221 vector were subcloned into the suitable vectors with a Gateway LR recombinase, as previously described. pDONR221-AtPCNA1 and pDONR221-AtPCNA2 were recombined with pGHC-RX and pGHC-GX, and pDONR221-AtDNA pol λ with pGHC-GX and pGHC-XG.
For bimolecular fluorescence complementation assays, we used pUC-SPYNE and pUC-SPYCE (Walter, 2004), carrying a cassette containing each a single half of YFP coding sequence, adjacent to the MCS; pUC-SPYNE carries the N-terminal half of YFP (YFPN, amino acids 1 to 155) and pUC-SPYCE the C-terminal half of YFP (YFPC, amino acids 156 to 239). Cloning of CDSs of interest into the MCS resulted in a C-terminal fusion protein with YFPN (pUC-SPYNE) or YFPC (pUC-SPYCE).
DNA pol λ coding sequence was amplified with the following primers: 5′-GAGTCTAGAATGGCGGCAAAGCGAGGGAG-3′ and 5′-CTCGGTACCGAGATTCCTCTCGTGTGGCTC-3′. The restriction sites are underlined. The PCR product was purified, digested with XbaI and KpnI, cloned into the pUC-SPYCE polylinker, and sequenced. PCNA1 and PCNA2 coding sequences cloned in pUC-SPYNE were kindly provided by C. Raynaud (Institut de Biotechnologie des Plantes).
Protoplast Isolation and Transformation
Nicotiana tabacum BY-2 suspension cells were grown in the dark at 23°C in MS medium supplemented with 0.5 mg L−1 2,4-D (Duchefa), 0.05 mg L−1 kinetin (Duchefa), and 30 g L−1 sucrose. Actively proliferating cells 2 d after transfer to fresh medium were centrifuged and the pellet resuspended in 50 mL of MGM (MS medium supplemented with 170 mM glucose and 170 mM mannitol) containing 1% (w/v) Cellulase Onozuka R10 (Duchefa) and 0.1% (w/v) Pectolyase Y-23 (Duchefa).
Protoplasts were filtered through a 100-μm nylon mesh, washed with 50 mL of MGM medium, resuspended in 4 mL of MSS medium (MS medium supplemented with 28 M sucrose), counted, and diluted to 106/mL.
For each transfection, 40 μg of plasmid was mixed with 150 μL of protoplasts and 450 μL of polyethylene glycol solution [25% (w/v) polyethylene glycol 6000, 0.45 M mannitol, and 0.1 M Ca(NO3)2, pH 9].
After 30 min of incubation in the dark, 3 mL of Ca(NO3)2 0.275 M was added, and then protoplasts were washed with MGM medium and incubated in the dark for 24 h before observation.
Light and Confocal Microscopy
Transfected protoplasts were observed with a Leica SP2 inverted confocal microscopy station. To observe GFP, excitation was 488 nm, and the spectral detector was set between 500 and 540 nm. To observe YFP, excitation was at 514 nm, and the spectral detector was set between 520 and 550 nm. To observe RFP, excitation was at 543 nm, and the spectral detector was set between 580 and 640 nm.
Images were analyzed with ImageJ (http://rsbweb.nih.gov/ij/). Colocalization was revealed using the colocalization threshold plugin (http://www.macbiophotonics.ca/imagej/colour_analysis.htm#6.3%20Colocalisation%20Threshold).
Plant Transformation, Growth, and UV Irradiation
All Arabidopsis transgenic lines used in this study were generated in the Col ecotype (Col-0) by the floral dip method (Clough and Bent, 1998) using the Agrobacterium tumefaciens GV3101/pMP90 strain. Progeny seeds were selected on half-strength MS medium without sucrose and containing kanamycin (50 mg/L). Kanamycin-resistant selection was performed as reported by Harrison et al. (2006).
Plants were then grown for 3 weeks in a growth chamber in short-day conditions (10 h light and 14 h dark), transferred to soil, and grown to maturity in long-day conditions (16 h light and 8 h dark) at 70% relative humidity and 23 ± 3°C. Insertion mutant information was obtained from the SIGnAL website at http://signal.salk.edu.
UV irradiation was performed for 30 min at 0.3 mW/cm2, corresponding to 2.9 J/m2/s, at 243 nm of wavelength. For determination of At POLL transcription level in untreated and UV-irradiated tissues with qRT-PCR, biological triplicates were performed.
RNA Extraction and Retrotranscription
RNA was extracted from 500 mg of fresh tissue. Samples frozen with liquid nitrogen were grinded and then processed using the Aurum RNA Fatty and Fibrous kit (Bio-Rad). Nine microliters of the extracted RNA were retrotranscribed using the ImProm-II reverse transcription system (Promega) with oligo(dT) as primer, following the manufacturer’s instructions.
Real-Time qPCR
Real-time qPCR was performed with GoTaq qPCR Master Mix (Promega) following the manufacturer's instruction in a Rotorgene 2000 thermalcycler (Corbett) using two-step cycling condition (95°C for 2 min, followed by 40 cycles of 95°C for 20 s, 60°C for 20 s, and 72°C for 20 s).
Three independent biological replicates were performed for each group of samples. Two reactions were set up in parallel, using 0.5 μL of cDNA diluted 1:4 and 0.5 μL of each primer (0.5 μM final concentration) in 20 μL of final volume. For each pair of primers, an NTC (no template control) reaction was set up, in double.
Raw data exported from the Rotorgene 2000 were analyzed using LinRegPCR 11.0 computer software. Primer efficiencies were calculated using an assumption-free method (Ruijter et al., 2009) that uses a linear regression of the fluorescence curve to estimate the efficiency for each run, with the following procedure. After individual baseline correction, reactions were grouped by gene, and a common window of linearity (three to five cycles) was determined to calculate mean efficiency for each gene through linear regression. Then, the fluorescence value at one cycle before the upper limit of window of linearity was taken as common threshold (Nt) for each gene and used, together with the relevant efficiency (Eg) and the fractional number of cycles needed to reach the threshold (Ct), to calculate the starting concentration (No) of each individual reaction using the formula No = Nt/(Eg^Ct) (Ruijter et al., 2009), where No is expressed in arbitrary fluorescence units. POLL transcription level was normalized on the geometric average (Hellemans et al., 2007) of two housekeeping genes, the EF1α translation elongation factor (At5g60390) and glyceraldehyde-3-phosphate dehydrogenase C2 (At1g13440), whose expression levels were previously reported to be constant in different tissues (Czechowski et al., 2005). Primers used were as follows: EF1α FW, 5′-TGAGCACGCTCTTCTTGCTTTCA-3′; EF1α RV, 5′-GGTGGTGGCATCCATCTTGTTACA-3′; G3P FW, 5′-TTGGTGACAACAGGTCAAGCA-3′; 3P RV, 5′-AAACTTGTCGCTCAATGCAATC-3′; POLL FW, 5′-TATCGTTGTCACCCATCCTG-3′; POLL RV, 5′-CTGTCCAGGCTATGAGTCCA-3′.
POLL FW and POLL RV anneal along the CDS at 1137 to 1156 bp and 1365 to 1384 bp from ATG, respectively, thus producing an amplicon of 247 bp that spans the insertion point of T-DNA, located at 1158 bp starting from ATG.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database or the Arabidopsis Genome Initiative database under the following accession numbers: At1g10520 (POLL), At1g07370 (PCNA1), At2g29570 (PCNA2), At5g60390 (EF1α), At1g13440 (glyceraldehyde-3-phosphate dehydrogenase C2), and At4g38620 (Myb4). The SALK ID number for the T-DNA line used in this article is SALK_075391C.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Images of BY-2 Protoplasts Transfected with Plasmids Encoding the Fusion Proteins GFP-At DNA pol λ, GFP-At PCNA1, or GFP-At PCNA2.
Supplemental Figure 2. Map of pGBIN1-At POLL Binary Vector Plasmid Used for Agrobacterium-Assisted Production of ATDNAPOLLOE Plants.
Supplemental Figure 3. Genotyping of Overexpressing and Knockdown Lines.
Supplemental Figure 4. Phenotypes of Col-0, At POLLOE, and At pollKD 30-d-Old Plants Grown in Short-Day Conditions.
Supplemental Figure 5. GUS Histochemical Staining of Arabidopsis Plantlets Following UV Irradiation.
Acknowledgments
This work was partially supported by Associazione Italiana per la Ricerca sul Cancro Grant IG4538 2008-2010 to G.M. E.C. is the recipient of a Fondazione Italiana per la Ricerca sul Cancro fellowship and Fondo Agevolazioni alla Ricerca.
References
- Anderson H.J., et al. (2008). Arabidopsis thaliana Y-family DNA polymerase eta catalyses translesion synthesis and interacts functionally with PCNA2. Plant J. 55: 895–908 [DOI] [PubMed] [Google Scholar]
- Blanc G., Hokamp K., Wolfe K.H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray C.M., West C.E. (2005). DNA repair mechanisms in plants: Crucial sensors and effectors for the maintenance of genome integrity. New Phytol. 168: 511–528 [DOI] [PubMed] [Google Scholar]
- Britt A.B. (1999). Molecular genetics of DNA repair in higher plants. Trends Plant Sci. 4: 20–25 [DOI] [PubMed] [Google Scholar]
- Chan C.S., Guo L., Shih M.C. (2001). Promoter analysis of the nuclear gene encoding the chloroplast glyceraldehyde-3-phosphate dehydrogenase B subunit of Arabidopsis thaliana. Plant Mol. Biol. 46: 131–141 [DOI] [PubMed] [Google Scholar]
- Chang W.C., Lee T.Y., Huang H.D., Huang H.Y., Pan R.L. (2008). PlantPAN: Plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genomics 9: 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Ai Y., Wang J., Haracska L., Zhuang Z. (2010). Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat. Chem. Biol. 6: 270–272 [DOI] [PubMed] [Google Scholar]
- Chen W., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collins A.R. (1999). Oxidative DNA damage, antioxidants, and cancer. Bioessays 21: 238–246 [DOI] [PubMed] [Google Scholar]
- Crespan E., Zanoli S., Khandazhinskaya A., Shevelev I., Jasko M., Alexandrova L., Kukhanova M., Blanca G., Villani G., Hübscher U., Spadari S., Maga G. (2005). Incorporation of non-nucleoside triphosphate analogues opposite to an abasic site by human DNA polymerases beta and lambda. Nucleic Acids Res. 33: 4117–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.J., Hays J.B. (2007). Tolerance of dividing cells to replication stress in UVB-irradiated Arabidopsis roots: Requirements for DNA translesion polymerases η and ζ. DNA Repair (Amst.) 6: 1341–1358 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das-Bradoo S., Nguyen H.D., Bielinsky A.K. (2010). Damage-specific modification of PCNA. Cell Cycle 9: 3674–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E., Klimovich V., Nager A.R. (2006). A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34: 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ortiz M.V., Roldán-Arjona T., Ariza R.R. (2007). The noncatalytic C-terminus of AtPOLK Y-family DNA polymerase affects synthesis fidelity, mismatch extension and translesion replication. FEBS J. 274: 3340–3350 [DOI] [PubMed] [Google Scholar]
- Gary R., Kim K., Cornelius H.L., Park M.S., Matsumoto Y. (1999). Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem. 274: 4354–4363 [DOI] [PubMed] [Google Scholar]
- Guilley H., Dudley R.K., Jonard G., Balàzs E., Richards K.E. (1982). Transcription of Cauliflower mosaic virus DNA: Detection of promoter sequences, and characterization of transcripts. Cell 30: 763–773 [DOI] [PubMed] [Google Scholar]
- Harrison S.J., Mott E.K., Parsley K., Aspinall S., Gray J.C., Cottage A. (2006). A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M.E., Quail P.H. (2003). Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol. 133: 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftode C., Daniely Y., Borowiec J.A. (1999). Replication protein A (RPA): The eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34: 141–180 [DOI] [PubMed] [Google Scholar]
- Ishibashi K., Suzuki K., Ando Y., Takakura C., Inoue H. (2006). Nonhomologous chromosomal integration of foreign DNA is completely dependent on MUS-53 (human Lig4 homolog) in Neurospora. Proc. Natl. Acad. Sci. USA 103: 14871–14876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weisshaar B., Martin C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya H. (2003). Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: Approaches using synthetic oligonucleotides and nucleotides: Survey and summary. Nucleic Acids Res. 31: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya H. (2004). Mutagenicities of 8-hydroxyguanine and 2-hydroxyadenine produced by reactive oxygen species. Biol. Pharm. Bull. 27: 475–479 [DOI] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kimura S., Sakaguchi K. (2006). DNA repair in plants. Chem. Rev. 106: 753–766 [DOI] [PubMed] [Google Scholar]
- Kimura S., Tahira Y., Ishibashi T., Mori Y., Mori T., Hashimoto J., Sakaguchi K. (2004). DNA repair in higher plants; photoreactivation is the major DNA repair pathway in non-proliferating cells while excision repair (nucleotide excision repair and base excision repair) is active in proliferating cells. Nucleic Acids Res. 32: 2760–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.S., Deng X.W., Callis J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna T.S., Fenyö D., Kong X.P., Gary S., Chait B.T., Burgers P., Kuriyan J. (1994b). Crystallization of proliferating cell nuclear antigen (PCNA) from Saccharomyces cerevisiae. J. Mol. Biol. 241: 265–268 [DOI] [PubMed] [Google Scholar]
- Krishna T.S., Kong X.P., Gary S., Burgers P.M., Kuriyan J. (1994a). Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79: 1233–1243 [DOI] [PubMed] [Google Scholar]
- Kunz B.A. (2008). A role for PCNA2 in translesion synthesis by Arabidopsis thaliana DNA polymerase eta. Plant Signal. Behav. 3: 897–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson P., Barone F., Maga G., Mazzei F., Karran P., Bignami M. (2005). 8-Oxoguanine incorporation into DNA repeats in vitro and mismatch recognition by MutSalpha. Nucleic Acids Res. 33: 5094–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G., Blanca G., Shevelev I., Frouin I., Ramadan K., Spadari S., Villani G., Hübscher U. (2004). The human DNA polymerase lambda interacts with PCNA through a domain important for DNA primer binding and the interaction is inhibited by p21/WAF1/CIP1. FASEB J. 18: 1743–1745 [DOI] [PubMed] [Google Scholar]
- Maga G., Crespan E., Wimmer U., van Loon B., Amoroso A., Mondello C., Belgiovine C., Ferrari E., Locatelli G., Villani G., Hübscher U. (2008). Replication protein A and proliferating cell nuclear antigen coordinate DNA polymerase selection in 8-oxo-guanine repair. Proc. Natl. Acad. Sci. USA 105: 20689–20694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G., Hubscher U. (2003). Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J. Cell Sci. 116: 3051–3060 [DOI] [PubMed] [Google Scholar]
- Maga G., van Loon B., Crespan E., Villani G., Hübscher U. (2009). The block of DNA polymerase delta strand displacement activity by an abasic site can be rescued by the concerted action of DNA polymerase beta and Flap endonuclease 1. J. Biol. Chem. 284: 14267–14275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G., Villani G., Crespan E., Wimmer U., Ferrari E., Bertocci B., Hübscher U. (2007). 8-Oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature 447: 606–608 [DOI] [PubMed] [Google Scholar]
- Menges M., Murray J.A.H. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Potuschak T., Stary S., Schlögelhofer P., Becker F., Nejinskaia V., Bachmair A. (1998). PRT1 of Arabidopsis thaliana encodes a component of the plant N-end rule pathway. Proc. Natl. Acad. Sci. USA 95: 7904–7908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Sozzani R., Glab N., Domenichini S., Perennes C., Cella R., Kondorosi E., Bergounioux C. (2006). Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen (PCNA) in Arabidopsis. Plant J. 47: 395–407 [DOI] [PubMed] [Google Scholar]
- Ruijter J.M., Ramakers C., Hoogaars W.M., Karlen Y., Bakker O., van den Hoff M.J., Moorman A.F. (2009). Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A., Lan V.T., Hase Y., Shikazono N., Matsunaga T., Tanaka A. (2003). Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and gamma-rays in Arabidopsis: Implication of the presence of a translesion synthesis mechanism in plants. Plant Cell 15: 2042–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago M.J., Alejandre-Durán E., Ruiz-Rubio M. (2006). Analysis of UV-induced mutation spectra in Escherichia coli by DNA polymerase eta from Arabidopsis thaliana. Mutat. Res. 601: 51–60 [DOI] [PubMed] [Google Scholar]
- Shultz R.W., Tatineni V.M., Hanley-Bowdoin L., Thompson W.F. (2007). Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol. 144: 1697–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal R.K., Wilson S.H. (1993). Short gap-filling synthesis by DNA polymerase beta is processive. J. Biol. Chem. 268: 15906–15911 [PubMed] [Google Scholar]
- Strzalka W., Oyama T., Tori K., Morikawa K. (2009). Crystal structures of the Arabidopsis thaliana proliferating cell nuclear antigen 1 and 2 proteins complexed with the human p21 C-terminal segment. Protein Sci. 18: 1072–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Sakamoto A., Sato S., Kato T., Tabata S., Tanaka A. (2005). Roles of Arabidopsis AtREV1 and AtREV7 in translesion synthesis. Plant Physiol. 138: 870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans M.C., Maliga P., Vieira J., Messing J. (1990). The pFF plasmids: Cassettes utilising CaMV sequences for expression of foreign genes in plants. J. Biotechnol. 14: 333–344 [DOI] [PubMed] [Google Scholar]
- Uchiyama Y., Kimura S., Yamamoto T., Ishibashi T., Sakaguchi K. (2004). Plant DNA polymerase lambda, a DNA repair enzyme that functions in plant meristematic and meiotic tissues. Eur. J. Biochem. 271: 2799–2807 [DOI] [PubMed] [Google Scholar]
- Umar A., Buermeyer A.B., Simon J.A., Thomas D.C., Clark A.B., Liskay R.M., Kunkel T.A. (1996). Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87: 65–73 [DOI] [PubMed] [Google Scholar]
- van der Kemp P.A., de Padula M., Burguiere-Slezak G., Ulrich H.D., Boiteux S. (2009). PCNA monoubiquitylation and DNA polymerase eta ubiquitin-binding domain are required to prevent 8-oxoguanine-induced mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 37: 2549–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Warbrick E. (1998). PCNA binding through a conserved motif. Bioessays 20: 195–199 [DOI] [PubMed] [Google Scholar]
- Wimmer U., Ferrari E., Hunziker P., Hübscher U. (2008). Control of DNA polymerase lambda stability by phosphorylation and ubiquitination during the cell cycle. EMBO Rep. 9: 1027–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Zhang S., Zhang W., Li G., Chen Z., Zhai W., Zhao X., Pan X., Xie Q., Zhu L. (2006). The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 48: 687–698 [DOI] [PubMed] [Google Scholar]