This work examines maize argonaute104, finding that ago104 mutants have apomixis-like phenotypes (unreduced gametes), defective chromatin condensation, alterations in methylation of repetitive DNA, and globally altered transcription in the ovary. AGO104 accumulation in somatic cells surrounding the female meiocyte suggests the action of a mobile signal repressing somatic fate in female germ cells.
Abstract
Apomixis is a form of asexual reproduction through seeds in angiosperms. Apomictic plants bypass meiosis and fertilization, developing offspring that are genetically identical to their mother. In a genetic screen for maize (Zea mays) mutants mimicking aspects of apomixis, we identified a dominant mutation resulting in the formation of functional unreduced gametes. The mutant shows defects in chromatin condensation during meiosis and subsequent failure to segregate chromosomes. The mutated locus codes for AGO104, a member of the ARGONAUTE family of proteins. AGO104 accumulates specifically in somatic cells surrounding the female meiocyte, suggesting a mobile signal rather than cell-autonomous control. AGO104 is necessary for non-CG methylation of centromeric and knob-repeat DNA. Digital gene expression tag profiling experiments using high-throughput sequencing show that AGO104 influences the transcription of many targets in the ovaries, with a strong effect on centromeric repeats. AGO104 is related to Arabidopsis thaliana AGO9, but while AGO9 acts to repress germ cell fate in somatic tissues, AGO104 acts to repress somatic fate in germ cells. Our findings show that female germ cell development in maize is dependent upon conserved small RNA pathways acting non-cell-autonomously in the ovule. Interfering with this repression leads to apomixis-like phenotypes in maize.
INTRODUCTION
Apomixis is an asexual mode of reproduction through seeds that is observed in more than 400 plant genera (reviewed in Grimanelli et al., 2001; Koltunow and Grossniklaus, 2003). The progeny of apomictic plants are genetic clones of the mother plants. This property of apomicts to fix any genotype over generations could be an important asset for plant breeding, in both propagation of hybrid seed varieties and fast-tracking plant breeding methods (Spillane et al., 2004). Apomixis, however, is absent from the major crops and found mostly in wild species. Harnessing its full potential would require inducing apomixis in crops. However, the molecular mechanisms of apomixis remain elusive.
Apomixis differs from sexual reproduction in two key steps of female gamete development: (1) the avoidance of meiosis, and (2) parthenogenesis (i.e. embryo development without fertilization of an egg). Apomixis can occur via different routes (reviewed in Koltunow and Grossniklaus, 2003). In the sporophytic type of apomixis (adventitious embryony), embryogenesis is initiated from maternal, sporophytic cells within the reproductive organs, totally avoiding gamete formation. In the gametophytic type, gametes are formed, but without meiosis. The mature gametes subsequently develop parthenogenetically into an embryo without fertilization. Diplospory is a type of gametophytic apomixis observed in several species, including Tripsacum, a wild relative of maize (Zea mays; Leblanc et al., 1995). In diplosporous apomixis, the precursor cell of the female gamete (called the megaspore mother cell [MMC]) differentiates from the nucellus and initiates meiosis. However, restitution of meiosis I takes place, with the meiocytes undergoing a mitotic division to produce unreduced spores. The resulting spore undergoes mitosis, similar to wild-type gametogenesis, which therefore results in unreduced female gametes (Grimanelli et al., 2003). Apospory is the second class of gametophytic apomixis, in which the development of the MMC is avoided altogether; the female spore directly differentiates, without preceding meiosis, from somatic tissues in the female reproductive organs and therefore is unreduced.
Cell-specific marker studies have shown that apomixis expression in different species results from a deregulation of the sexual process. As a result, either the temporal (gametophytic type) or spatial (sporophytic type) regulation of reproductive developmental is altered, resulting in heterochronic or ectopic expression of the core programs of sexual development (Grimanelli et al., 2003; Tuckey et al., 2003; Bicknell and Koltunow, 2004; Bradley et al., 2007; Sharbel et al., 2010). Genetic analyses of the inheritance of the apomixis trait in several species, including Tripsacum, have further shown that apomixis is a simple dominant trait. In several species, including species from the Pennisetum, Tripsacum, Bracchiaria, Panicum, and Paspalum genera, it is controlled by a single chromosomal segment often referred to as the apomixis locus; alternatively, in Hieracium and Taraxacum species, avoidance of meiosis and parthenogenesis segregate as two independent traits (reviewed in Ozias-Akins and van Dijk, 2007). All these examples point toward a relatively simple Mendelian inheritance. Mutagenesis, therefore, has been proposed as a strategy to dissect the mechanism of spontaneous forms of apomixis through the identification of mutants that either mimic apomixis in sexual plants or alter apomictic development toward sexual reproduction (Bicknell and Koltunow, 2004). This approach is supported by the availability of several mutants that phenocopy aspects of apomictic development, for example the elongate1 (el1) and ameiotic1 mutants of maize (Rhoades and Dempsey, 1966; Pawlowski et al., 2009). Mutagenesis has also been proposed as a way to engineer a de novo form of apomixis, potentially distinct from natural forms of apomixis, by combining mutations that affect the sexual pathway (Ravi et al., 2008; d’Erfurth et al., 2009).
Several lines of evidence also suggest that apomixis may be epigenetically regulated. First, it has been shown that the apomixis locus is a heterochromatic region enriched in retrotransposons and repetitive DNA (Calderini et al., 2006; Conner et al., 2008). Leblanc et al. (2009) further showed a parent-of-origin effect of meiosis on apomixis expression: in segregating populations, the apomixis locus was not inherited in a functional state (i.e., able to induce apomixis) when transmitted through a reduced female gamete, but it remained functional when transmitted via male meiosis. More evidence for a possible epigenetic basis of apomixis came recently from the analysis of Arabidopsis thaliana plants defective for the ARGONAUTE9 (AGO9) gene (Olmedo-Monfil et al., 2010). The ago9 mutants affect the specification of precursor cells of the gametes in the Arabidopsis ovule in a dominant way and display a multiple-spore phenotype that resembles apospory. In the same study, it was also shown that mutants of the RNA-DEPENDENT RNA POLYMERASE6 and SUPRESSOR OF GENE SILENCING3 genes, both key players in the trans-acting small interfering RNA pathway, cause a similar phenotype. Interestingly, AGO9 is also responsible for silencing of transposons in the female germ line, as ago9 mutants show transposon reactivation in the egg. We further showed recently (Garcia-Aguilar et al., 2010) that in maize, the inactivation of the DNA methyltransferases dmt102 and dmt103, which are expressed in the ovule, results in phenotypes that are strongly reminiscent of apomictic development, including the production of unreduced gametes and the formation of multiple embryo sacs. Presumably, DMT102 activity in the maize ovule negatively regulates the transcriptional competence of chromatin in the archesporial tissue, a condition that might be essential for proper sexual development (Garcia-Aguilar et al., 2010). Collectively, the mutant phenotypes of ago9 in Arabidopsis and dmt103 in maize suggest that the regulation of DNA methylation in the precursor cells of the female gametes plays a role in the differentiation between apomictic and sexual reproduction. However, extranumerary spores or embryo sacs in ago9 and dmt103 mutants do not form functional unreduced female gametes, and none of these mutants produces apomictic progeny.
To evaluate the possibility of inducing apomixis in maize, we set up a genetic screen to identify dominant mutants that mimic apomictic development by forming viable unreduced female gametes. Here, we report the characterization of one of the mutants identified through this screen, called Dominant non-reduction4 (Dnr4), which has a phenotype that strongly resembles diplospory. Dnr4 is defective in chromatin condensation during meiosis and results in a high frequency of unreduced but viable female gametes. Dnr4 codes for the AGO104 protein, which accumulates in somatic tissues surrounding the female meiocytes, thus acting non-cell-autonomously, a pattern similar to the related Arabidopsis gene AGO9. We showed that AGO104 controls non-CG DNA methylation at centromeric and knob heterochromatin. These results indicate that small RNA-mediated silencing during early plant reproduction is critical to sexual development and possibly to the differentiation between apomixis and sexuality.
RESULTS
Genetic Screen to Identify Mutants with Unreduced Gametes
During diplospory in Tripsacum, an apomictic wild relative of maize, the MMC initiates meiosis but subsequently switches to a mitotic division, leading to the production of unreduced gametes (Grimanelli et al., 2003). We designed a forward genetic screen to identify maize mutants producing unreduced female gametes in a manner mimicking that of Tripsacum. The screen was thus designed to increase the odds of recovering dominant mutations and to pick up only phenotypes with viable, unreduced female gamete formation (Figure 1A). The screen exploited the response of the maize endosperm to maternal:paternal genome ratio imbalance, which results in aborted seeds, as a phenotypic marker (Birchler, 1993; Figure 1B). Active Mutator (Mu) transposon lines were first crossed to an inbred line (CML216) to generate a population of plants heterozygous for Mu insertion sites. These F1 plants were subsequently pollinated with a tetraploid tester line. Plants that produced only normal reduced female gametes did not set seeds when crossed to a tetraploid due to paternal genomic excess in the endosperm. Plants with plump kernels, possibly resulting from the fertilization of unreduced (diploid) female gametes by a reduced (diploid) male gamete from the tetraploid tester, were recovered in the screen at a frequency of 6 out of 17,000 individual F1 plants. Given that the mutants arose in plants likely heterozygous for all mutagenic insertions, they were tentatively named Dnr1 to Dnr6. One of them, Dnr4, showed a ratio of plump-to-defective seeds higher than expected from a gametophytic mutant (87:42 observed against 1:1 expected for a gametophytic effect; χ2 test, P < 0.01; Figure 1B), suggesting rather a maternal sporophytic effect with incomplete penetrance. To test whether the plump seeds indeed arose from unreduced female gametes, we estimated ploidy levels in the resulting embryos by flow cytometry. Results showed that plump (n = 10) and shrunken (n = 10) seeds contained tetraploid and triploid embryos, respectively (Figure 1C), confirming the occurrence of unreduced gametes.
Figure 1.
Genetic Screen to Identify Dnr Phenotypes.
(A) Crossing scheme. Predicted maternal-to-paternal genomic ratio in the endosperm (m:p) are indicated. 2m:1p and 4m:2p ratios result in normal endosperm development, while 2m:2p results in an abortive endosperm and defective kernels.
(B) Ear phenotypes of wild-type and mutant females. R, Seeds containing putatively reduced gametes; UR, seeds containing putatively unreduced gametes.
(C) Flow cytometry analysis of embryos from Dnr4 × tetraploid ears dissected from defective (left) and normally developed kernels (right; in green, with the embryo dissected from defective kernels in blue as a control). The defective kernels contained triploid embryos (3×), while the plump kernels contained tetraploid embryos (4×), most likely originating from unreduced female gametes.
(D) Flow cytometry analysis of pollen ploidy in Dnr4 plants. Identical results were obtained for four independent plants with a Dnr4 female phenotype.
To verify whether the mutant phenotype was transmitted to the next generation, we grew plants selected from plump kernels. Except for ploidy (all were confirmed tetraploids by flow cytometry), no obvious developmental phenotype was observed in the progeny of the Dnr4 mutant before gamete formation. Eight plants were tested for the production of unreduced gametes using tetraploid pollen donors (see Supplemental Figure 1 online). They produced a mosaic of plump and defective seeds, similar to what we observed in the original mutant, with a variable penetrance ranging from 20 to 80%. Flow cytometric analyses showed that the defective kernels carried hexaploid embryos (n = 10 per plant), indicating fertilization of unreduced (4×) female gametes by reduced (2×) male gametes, while the plump kernels carried tetraploid embryos (n = 10 per plant). This indicates that the phenotype of the original Dnr4 plant was transmitted to the next generation through unreduced gametes. To analyze the transmission of the mutation through meiosis, and eventually confirm that the Dnr4 mutation was dominant, we performed reciprocal crosses using a Dnr4-expressing plant and a wild-type tetraploid tester. We took advantage of the incomplete penetrance of the phenotype and selected the plump seeds (thus produced from meiotically derived male and female gametes) from both crosses and analyzed the expression of the Dnr4 ear phenotype in the resulting plants (see Supplemental Figure 1 online). Wild-type and Dnr4 plants were found in both reciprocal crosses, in a proportion not significantly different from a 1:1 segregation ratio (Fisher exact tests for the paternally transmitted allele, P = 0.25; for the paternally transmitted allele, P = 0.67). This indicates transmission of a single dominant allele in the mutant through both male and female meiosis. In addition, we tested whether the mutant produced spontaneous seeds in the absence of pollination. No kernels were produced upon no pollination (10 independent ears).
To test whether the mutation also affected the production of male gametes, we then compared ploidy levels in mature male gametophytes (pollen grains) collected from the tetraploid plants grown from the plump Dnr4 seeds and from wild-type autotetraploid maize lines (Figure 1D). We observed that the wild-type tetraploid stocks produced mostly reduced male gametes. By contrast, the Dnr4 plants produced an excess of gametes that were either unreduced or aneuploids with high ploidy levels (>50%). This indicates that the mutation affected the production of both the male and female gametes. The absence of aneuploid plants in the progeny of the reciprocal crosses suggests that aneuploid male gametophytes were probably counterselected.
Dnr4 Unreduced Gametes Correlate with Abnormal Chromatin Condensation during Meiosis
To further define the Dnr4 phenotype, we analyzed the development of the male and female meiocytes at different stages. We first looked at female reproductive development using whole-mount clearing of dissected ovules (see Supplemental Figure 2 online). We observed that a single MMC (the precursor of the female meiocyte) was formed in mutant ovules (n = 100 observations), similar to wild-type plants. Contrary to the wild type, however, we also noticed the frequent formation (31%; n = 100) of dyads rather than tetrads as a result of sporogenesis. Dyads of spores were also retained after the selection of the functional spore in 24% of the ovules (n = 100), while only a single spore was observed in wild-type plants (100%; n = 100). Later, during gametogenesis, we observed the formation of a single gametophyte per ovule in both the wild type and mutants (n = 50). This collectively indicates that unreduced gametophytes and gametes were most likely derived from aberrant meiotic or mitotic divisions of the MMC rather than accessory somatic cells in the ovule. Additional defects linked to chromosome behavior or cell divisions are not detectable using clearing procedures, and since the mutant produces both male and female unreduced gametes, we focused further cytological analyses on male meiocytes, which are more accessible.
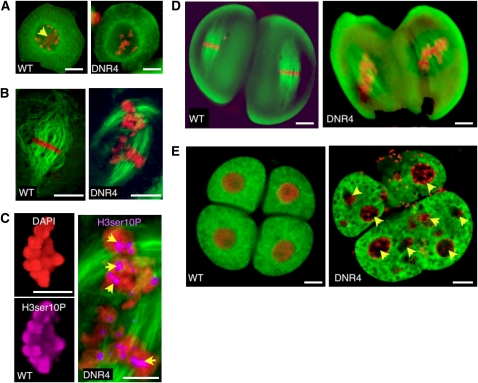
Using 4′,6-diamidino-2-phenylindole (DAPI) staining and immunofluorescence of male meiocytes, we observed that chromatin condensation, spindle formation, and chromosome segregation were severely affected in mutant plants compared with the wild type (Figure 2). The earliest defects were observed at diakinesis, with >70% of meiocytes (n = 500) presenting abnormal condensation (Figure 2A). Chromatin condensation failure was even more evident at metaphase I (>80% of the meiocytes; n = 500; Figures 2B and 2C). At that stage, chromatin remained in a partially decondensed state. Also, the cytoskeleton failed to form the bipolar spindle observed in the wild-type cells (Figures 2B and 2C). Similar defects were observed at metaphase II (n = 500), with dyads showing partial chromosome condensation (Figure 2D) and irregular spindle formation. Later (Figure 2E), we observed abnormal tetrads that included triads, dyads, and microspores with multiple nuclei (68%; n = 500). Since B73 is diploid and the mutant stock resulting from the screen was tetraploid, we further examined metaphase plates of B73, its autotetraploid, and the tetraploid tester line used in the screen using DAPI staining. All three stocks showed comparable levels of chromatin compaction (Figure 2B) at metaphase (n > 100 meiocytes each), clearly distinct from the mutant phenotype.
Figure 2.
Dnr4 Mutants Show Multiple Defects during Meiosis I and II.
Analysis of male meiosis in wild-type and mutant plants shows that Dnr4 mutant plants are deficient in chromatin condensation during both meiotic divisions. Chromatin was revealed using DAPI staining (red); an antibody against β-tubulin (green) was used to visualize the cytoskeleton. Representative images are from n > 50 observations for each stage. Bars = 25 μm.
(A) Dyakinesis-stage male meiocytes. This is the earliest stage at which chromosome condensation is noticeably altered. Note the conspicuous absence of the nucleolar organizing region (arrowhead) in the mutant line.
(B) Metaphase I. Poorly condensed chromosomes are clearly visible in the mutant, associated with a highly disorganized spindle.
(C) Staining with an antibody against phospho H3ser10P shows exclusively pericentromeric staining (arrowheads) during meiosis I, suggesting that the mutant turns meiosis I into a mitosis-like division (see Methods).
(D) Metaphase II. Poorly condensed chromosomes are visible in the mutant
(E) Abnormal tetrads are produced in the mutant, characterized by multiple micronuclei (arrowheads) and cell division failure.
A phenotype somewhat related to Dnr4 is observed in the el1 mutant in maize (Rhoades and Dempsey, 1966), which also affects chromosome condensation and results in viable unreduced male and female gametes, presumably by skipping meiosis II (Barrell and Grossniklaus, 2005). Very similar phenotypes are also observed during the development of unreduced male meiocytes in Tripsacum apomicts (Grimanelli et al., 2003), which skips meiosis I. Phosphorylation of histone H3 at Ser-10 (H3ser10P) is associated with sister chromatid cohesion in plants and is a good discriminating marker for meiosis I and meiosis II/mitosis (Kaszás and Cande, 2000). During meiotic metaphase I, H3ser10P is spread on the whole chromosome. However, during meiotic metaphase II or mitotic metaphase, H3ser10P is restricted to the pericentromeric regions of the chromosomes. To test if the meiotic defects in Dnr4 altered the H3ser10P localization pattern, immunolocalization was performed with antibodies against H3ser10P using both the diploid B73 and the tetraploid line as wild-type controls compared with Dnr4 (Figure 2C). Metaphase I chromosomes in Dnr4 mutant plants showed a pericentromeric localization of H3ser10P that resembled the pattern of mitotic metaphase. By contrast, both wild-type controls showed an identical whole-chromosome staining pattern. This suggests that meiocytes in Dnr4 plants underwent a mitotic rather than a meiotic division, consequently developing unreduced gametes.
Dnr4 Encodes a Predicted AGO Protein
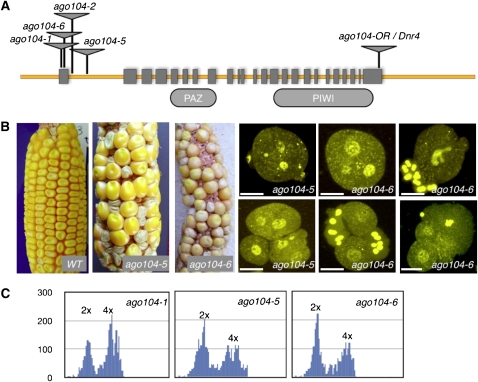
The Dnr4 phenotype was identified from a population of active transposable Mu elements. A putative lesion was identified by Mu tagging (see Methods). The Mu insertion is located in locus GRMZM2G141818, which is annotated as ago104 in the maize genome database. The Mu element is inserted in the last exon of the predicted gene (Figure 3A). We used reverse genetics to isolate four new Mu insertion alleles in the same predicted gene. The four alleles carry a Mu insertion near the 5′ end of the gene, either in the first exon (ago104-1 and -6) or in the first intron (ago104-2 and -5; Figure 3A). The four mutant alleles produced unreduced female gametes when assayed with a tetraploid tester as male (Table 1, Figure 3B). At least ago104-1, ago104-5, and ago104-6 also produced unreduced male gametes (Figure 3C; ago104-2 was not tested). They all exhibited defects during male meiosis I and II similar to Dnr4, leading to the formation of abnormal tetrads, including dyads, triads, and microspores with multiple nuclei. At early stages of development, a large proportion of meiocytes were arrested at stages from diakinesis to metaphase I in ago104-1 and ago104-5 homozygous mutants (Table 2). At the tetrad stage, we observed numerous defects in both mutants, including metaphase II arrest, dyads, and abnormal tetrads (28 and 30%, respectively; Table 3). We conclude that ago104 alleles phenocopy essential aspects of the Dnr4 mutant; accordingly, we renamed Dnr4 as ago104-or. Because the original allele was only available in a tetraploid background, all subsequent analyses were performed using alleles ago104-2, ago104-5, and ago104-6 in diploid backgrounds.
Figure 3.
Cloning of the Dnr4 Locus.
(A) Structure of the Dnr4 locus and of the mutant alleles. The triangles indicate the insertion sites of Mu elements. The gene contains the canonical PAZ and PIWI domains found in all AGO proteins. The original Dnr4 allele, located in the 3′ end of the sequence, was renamed ago104-or following the molecular identification of the locus.
(B) Phenotypes of ago104-5 and ago 104-6 plants when crossed to a tetraploid tester line used as male. Phenotypes very similar to the original allele were observed; ago104 ears exhibit semisterility and kernel abortion representative of unreduced female gametes. Defects in meiosis II, visualized using DAPI staining, lead to the production of abnormal tetrads, including triads and microspores with multiple nuclei, similar to Dnr4. Bar = 50 μm.
(C) Flow cytometry analysis of pollen ploidy in ago104 mutant plants. Identical results were obtained for four independent plants for each allele.
Table 1.
Production of Unreduced Female Gametes in ago104 Mutants
| Allele | Unreduced | Reduced | Total |
| ago104-1 | 62 | 93 | 155 |
| ago104-2 | 111 | 75 | 186 |
| ago104-5 | 117 | 52 | 169 |
| ago104-6 | 56 | 80 | 136 |
| Wild type | 0 | 200 | 200 |
Ploidy of the female gametes as inferred from seed morphology (defective seeds, meiotic spore; plump seeds, unreduced functional spores) with three independent ears, crossed to a tetraploid tester. Ploidy was verified by flow cytometry for a subsample of the seeds (see text).
Table 2.
Meiocyte Developmental Arrest in Anthers of ago104 Mutants
| Allele | Diak | MI | AI | TI | Dyad | MII | AII | TII | Tetrad |
| ago104-1 | 177 | 193 | 22 | 7 | 192 | 187 | 7 | 2 | 211 |
| ago104-5 | 137 | 80 | 17 | 15 | 69 | 92 | 12 | 36 | 52 |
| Wild type | 0 | 0 | 0 | 0 | 15 | 25 | 30 | 45 | 182 |
Diak, diakinesis; MI, meiosis I; MII, meiosis II; AI, anaphase I; AII, anaphase II; TI, telophase I; TII, telophase II.
Table 3.
Abnormal Tetrads in Anthers of ago104 Mutants
| Allele | Tetrads | Abnormal Tetrads | Dyads | MII |
| ago104-1 | 192 | 27 | 61 | 37 |
| ago104-5 | 139 | 38 | 41 | 38 |
| Wild type | 200 | 4 | 7 | 0 |
MII, meiosis II.
The AGO104 Protein Specifically Accumulates during Sporogenesis in Somatic Nucellar Cells of the Ovule Primordium
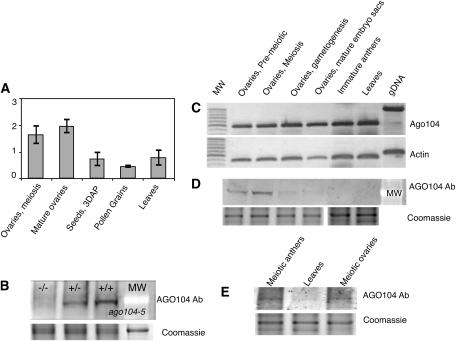
ago104 transcription was studied with quantitative RT-PCR at various stages of ovary and anther development and in leaves. The transcript was detected in all the tissues analyzed (Figure 4A). Highest expression was found in the ovaries, with a maximum in mature ovules prior to fertilization (Figure 4A). To analyze the accumulation of AGO104 protein, we generated an anti-AGO104 antibody against an AGO104 peptide. To test the specificity of the antibody, we probed meiosis-stage ovaries of mutant and wild-type plants. The antibody detected a signal of ~100 kD in ovaries of wild-type plants at meiosis, consistent with the predicted size of the protein. The signal could not be detected in the ovaries of ago104-5 homozygous mutant plants at the same stage, while it was reduced in the heterozygous mutants compared with the wild type (Figure 4B), suggesting that the antibody was highly specific for the AGO104 peptide. Immunoblot analyses revealed that AGO104 accumulates strongly in ovaries during both female and male sporogenesis but not during gametogenesis (when transcription is highest), as weak or no signal was detected (Figures 4C–4E). In addition, we observed no accumulation in young leaves. Collectively, this shows that the gene is likely broadly transcribed but that the AGO104 protein specifically accumulates in the ovaries and the anthers around the time of meiosis, possibly because of posttranscriptional regulation.
Figure 4.
ago104 Expression and Protein Accumulation.
(A) Quantitative RT-PCR profiles of ago104 expression. Profiles were determined relative to gpm120 expression. Error bars indicate se. Four replicates for two different samples were used for each data point. The y axis shows arbitrary units.
(B) Immunoblot. Total protein extracts from young (meiotic stage) ovules in ago104-5 mutant plants (homozygous, −/−; heterozygous, +/−) and wild-type plants (+/+) were assayed with an antibody raised against an AGO104 peptide.
(C) and (D) RT-PCR assay in reproductive tissues (C) and immunoblot with the same samples (D). The protein is detected mostly in premeiotic and meiotic ovaries. A very faint signal is detected during gametogenesis and barely detectable in anthers or leaves.
(E) Protein accumulation during anther development. Anthers and ovaries were staged using DAPI staining and whole-mount ovule clearing, respectively, before protein extraction.
To precisely localize the protein within ovaries, immunolocalization was performed with the AGO104 antibody in wild-type and ago104-5 homozygous plants. We observed that during early sporogenesis in all ovules examined, AGO104 was confined to the nucellar tissue of wild-type ovules and localized to the somatic cells surrounding, but strikingly not including, the precursors of the gametic cells (i.e., the MMC; Figure 5A; n > 50 ovules). This signal was highly specific, as no corresponding signal was observed in homozygous ago104-5 mutants (Figure 5A; n = 25 ovules). The protein, at our resolution, appears preferentially located in cytoplasmic foci (Figure 5B; n > 50 ovules). Later, at the mature gametophyte stage, AGO104 localization becomes very specific to the micropylar end of the female gametophyte, in the region where the egg cell and the synergids are located (Figure 5C; n > 50 ovules), a pattern not observed in ago104-5 mutant ovules (n = 15).
Figure 5.
Immunolocalization of AGO104 in Maize Ovules.
(A) Immunostaining of premeiotic ovules in wild-type and ago104 homozygous plants. An AGO104 signal is visible in the nucellar cells only. The arrows indicate the position of the MMC precursor cell. A differential interference contrast (DIC) image of a wild-type ovule is shown at right.
(B) Closeup of nucellar tissues shows that most signals are cytoplasmic and do not colocalize with DAPI-stained nuclei.
(C) Immunostaining of a mature gametophyte in wild-type and ago104 homozygous plants. The AGO104-specific signal is restricted to the micropylar pole of the embryo sac, where the egg apparatus (egg cell + synergids) is located. The exact location of the AGO104 signal in the egg apparatus is difficult to specify.
Bars = 50 μm.
AGO104 Clusters with the Arabidopsis AGO4/AGO6/AGO9 Clade and Is Likely a Functional Homolog of Arabidopsis AGO9
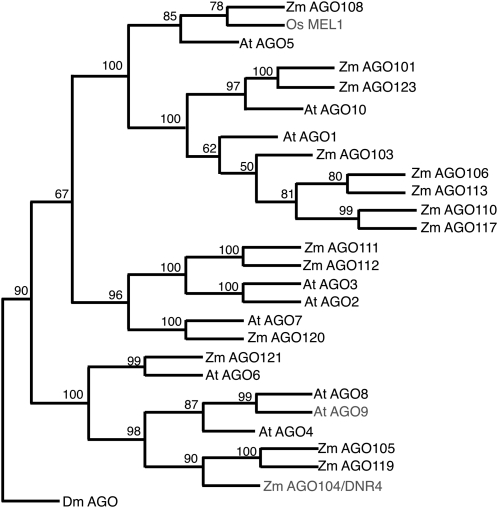
The ago104 cDNA is 3017 bp in length with an open reading frame of 2733 bp that encodes a protein of 910 amino acids. The AGO104 protein contains the characteristic PAZ and PIWI domains found in all AGO proteins and is highly homologous to all known AGO proteins. Phylogenetic analysis (Figure 6) of maize (numbered 101–119) and Arabidopsis (numbered 1–10) AGO proteins and MEIOCYTES ARRESTED AT LEPTOTENE1 (MEL1), an AGO protein from rice (Oryza sativa) that has also been shown to have a meiosis-related function (Nonomura et al., 2007), showed a division into three clades. AGO1, AGO10, and AGO5 from Arabidopsis fall in the same clade with a complex set of maize AGOs. AGO2, AGO3, and AGO7 constitute the second clade, while AGO4, AGO6, AGO8, and AGO9 form the third clade. AGO104 is classified in the latter clade. MEL1 is more closely related to AGO5 and AGO1 and is clearly distinct from AGO104. Two other maize AGO proteins, AGO105 and AGO119, are closely related to AGO104. They are more closely related to each other and may represent a duplication of the same gene. AGO8 in Arabidopsis is likely a pseudogene, without known expression. This reconstruction thus suggests that AGO104 is likely orthologous to either AGO9 or AGO4. AGO4 in Arabidopsis, however, is ubiquitously found in vegetative tissues, mostly nuclear localized (Li et al., 2006), and mutant alleles do not have any obvious developmental phenotypes. By contrast, Arabidopsis AGO9 is expressed specifically in reproductive tissues and similarly localizes to cytoplasm foci in somatic cells surrounding the MMC (Olmedo-Monfil et al., 2010). We thus propose that AGO104 and AGO9, rather than AGO4, are orthologs.
Figure 6.
Phylogenetic Relationships among Maize and Arabidopsis AGO Proteins.
Consensus tree following 1000 bootstraps. The numbers on the branches indicate the number of times the partition into the two sets that are separated by that branch occurred among the trees, out of 100 trees. Drosophila melanogaster AGO (Dm AGO) was used as an outgroup. Os MEL1 (rice MEL1), an AGO protein with a meiotic role (Nonomura et al., 2007), was also included.
Centromeric and Knob-Repeat DNA Is Hypomethylated at Non-CG Sites in ago104 Mutants
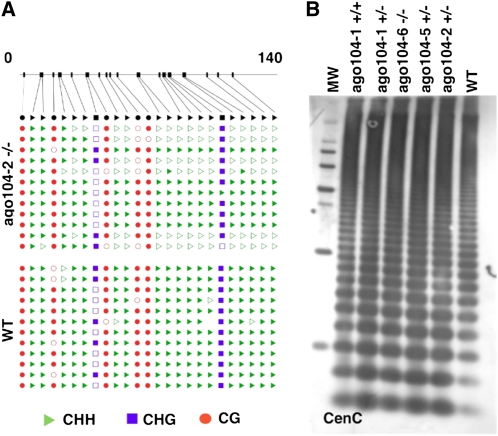
Both AGO4 and AGO9 in Arabidopsis are important for heterochromatic silencing. AGO4, at least, and possibly AGO9 acts via DNA methylation. We thus investigated DNA methylation in ago104 mutants in various heterochromatic contexts. Chromosomal knobs are subtelomeric heterochromatic regions of the maize chromosomes that can form neocentromeres (Dawe and Hiatt, 2004). Heterochromatin in the knobs is composed of a 180-bp tandem repeat and a 350-bp tandem repeat (TR-1; Ananiev et al., 1998). DNA methylation at 180-bp knob repeats was analyzed by bisulfite sequencing using homozygous ago104-2 mutant plants and compared with the W23 inbred line as a control. The ago104-2 mutant allele showed a strong reduction in asymmetric CHH methylation compared with W23 (Figure 7A). By contrast, CG methylation in the mutant was similar to the wild type, suggesting that ago104 might target non-CG methylation. CHG sites were poorly represented in the repeat and thus could not be reliably assayed. To further test the effect of ago104 mutations on non-CG heterochromatin, we tested methylation at centromeric repeats CentC using DNA gel blot analysis and methylation-sensitive enzymes. We observed that ago104 mutants have reduced DNA methylation in the CentC repeats compared with the two wild-type inbred lines W23 and B73, as detected by the MspI restriction enzyme, which is sensitive to CNG methylation (Figure 7B). Among the mutants, homozygous plants showed more hypomethylation compared with the heterozygous plants, which were themselves hypomethylated compared with wild-type plants. When a 5S ribosomal DNA repeat was used as a probe, however, CNG methylation was not affected in the ago104 mutants. Collectively, these data suggest that ago104 is involved in establishing or maintaining DNA methylation at non-CG sites at least in heterochromatin, where most methylated non-CG sites are found.
Figure 7.
DNA Methylation Analysis of Repeat Sequences in ago104 and Wild-Type Plants.
(A) Bisulfite sequencing of knob-repeat DNA of wild-type (B73) and homozygous mutant (ago104-2) immature ears. Positions of the 25 cytosines found within the 140-bp repeat are indicated on the top line. CHH, CHG, and CG contexts are indicated by green triangles, blue squares, and red circles, respectively, and are shown as filled for methylated sites and unfilled for nonmethylated sites. The results suggest a requirement for methylation at the CHH site.
(B) DNA was digested with the methyl-sensitive enzyme MspI and probed with centromeric tandem repeat sequence CentC. The MspI enzyme will cut at most once within the CentC sequence (forming a ladder from the tandem repeat) and will not cut if the sequences are methylated. Thus, a greater intensity of bands at lower molecular weights indicates reduced methylation of the CentC repeat. Homozygous ago104 mutants showed reduction in CNG methylation compared with mutant-derived wild-type homozygous plants or wild-type control W23 (similar results were obtained for B73). Results were replicated with four separate blots prepared from DNA samples.
Loss of Function in ago104 Results in a Large Alteration of Transcription in the Ovaries, Affecting Both Genes and Repeats
To evaluate the impact of ago104-dependent DNA methylation on transcription in the ovule, we performed a comparative profiling experiment using sporogenesis-stage ovaries dissected from wild-type (B73) and ago104-deficient plants. We used digital gene expression tag profiling, an open-ended approach based on large-scale sequencing of mRNA transcripts. Using Illumina/Solexa technology, we generated 20,652,000 and 21,852,000 short sequences for the wild-type and mutant samples, respectively. Following base calling and quality filtering, we identified 6.59 and 7.51 million 17-bp tags, corresponding to 363,000 and 404,000 unique sequences in wild-type and mutant libraries, respectively (see Methods).
To evaluate the effect of ago104 loss of function on the transcription of repeats, including transposable elements, we first compared the number of tags originating from low-copy genic regions and repetitive DNA in both samples (Figure 8A). A total of 3.54 and 3.32 million tags (wild-type and mutant libraries, respectively) mapped to single-copy locations, while 3.05 and 4.19 million tags mapped to repeats. Thus, the number of tags originating from repeats was 46 and 56% of the total in the wild-type and mutant samples, respectively. This suggests a significant increase of transcriptional activity in repeats. To further define the repeats that might be specifically affected in the ago104 mutant, we mapped individual tags to specific families of repetitive elements (Figure 8B). Transcription of class I retrotransposons (long terminal repeat [LTR] and non-LTR) was somehow affected in the mutant, their expression being 141% of the wild-type level (n = 156,120 unique tags, >10,000 loci). This difference, estimated over a large number of different retrotransposons (>50), is highly significant (Fisher exact test, P < e-05). Further distinction among LTR retrotransposons does not indicate a clear target for AGO104 activity. Interestingly, although certainly biased by the Mu-active background used to generate the ago104 mutant, a much larger effect was observed for class II transposons (401% of the wild-type expression level; n = 86,257, >100 loci), including, but not restricted to, Mu-like sequences. Nevertheless, the most striking difference was observed for the centromeric repeats (491% of the wild-type expression level; n = 25,264), which showed a massive transcriptional activity in the mutant as compared with the wild type. This is consistent with the results of the DNA methylation assays of centromeric repeats and further suggests a possible centromeric role for AGO104. Also consistent with the DNA gel blot, we did not detect any change in the transcription of nuclear ribosomal repeats, including 5S.
Figure 8.
RNA Profiling in Wide-Type and Mutant Plants.
Digital gene expression analysis comparing transcripts in ovaries from ago104-5 and B73.
(A) Targeting of repeats and predicted protein-coding loci in the maize genome in wild-type and mutant ovaries. Percentage values refer to the total tag counts. CDS, coding sequences.
(B) Expression differences among different repeat classes, expressed as percentages of the wild-type level. Note that the mutant line was derived from a Mu-active line.
(C) Differential expression among predicted protein-coding loci, a subset of differentially expressed genes (2982 out of 15,130 loci with more than six tags; 19.7%). Differences were mostly qualitative (present/absent). Among differentially expressed genes found in both samples, 531 were found with a ratio of relative expression r > 10 (987 with r > 5).
(D) Proportion of loci expressed in both samples showing upregulation and downregulation of gene expression when comparing RNA from wild-type and mutant plants (for r > 10). Percentage values refer to the number of loci.
To analyze the effect of the mutation on protein-coding gene expression in the ovule, we selected tags that mapped to nonambiguous, unique loci. We identified tags for 22,690 and 26,058 (number of tags > 1) predicted transcripts in wild-type and mutant libraries, respectively. Using a cutoff of six tags per locus (eventually as a cumulative number when a given locus was identified by several tags), this resulted in expression estimates for 15,130 loci. We found that knocking out ago104 resulted in large changes in transcription levels for more than 3000 predicted loci in ovaries (Figure 8C). The changes were evenly distributed between genes upregulated and downregulated in the mutant (Figure 8D; see Supplemental Data Set 2 online). Among differentially expressed genes, 1356 were not detected in the wild type but expressed in the mutant, and reciprocally, 1095 were expressed in the wild type but absent in the mutant. Another 531 were expressed in both backgrounds but quantitatively upregulated (n = 251) or downregulated (n = 280) in the mutant, considering an arbitrary fold change value of r > 10 (595 and 456 with r > 5). Classifying the differentially expressed loci by biological functions (see Supplemental Data Set 3 online) did not indicate any obvious targets of the AGO104-dependent pathway. Rather, the scale of differential expression, widely spread among diverse biological functions, illustrates a massive disruption of the transcriptional landscape in ovaries when comparing the mutant with B73.
DISCUSSION
AGO104 Is Required for Proper Chromatin Condensation during Meiosis
AGOs and the related PIWI proteins form a large family with diverse functions during development. AGOs are found in plants, animals, and fungi. They act in transcriptional gene silencing, posttranscriptional gene silencing, or through modification of chromatin by RNA interference (reviewed in Peters and Meister, 2007; Hutvagner and Simard, 2008; Vaucheret, 2008). AGOs are ~100-kD proteins that contain characteristic PAZ and PIWI domains. The PAZ domain binds the small RNA, while the PIWI domain is purported to have RNaseH-like endonuclease activity, which is required to cleave bound RNA targets for AGOs that have a slicer function. The germ line–specific PIWI proteins, by contrast, have only been described and characterized in animals, where they are evolutionarily conserved (reviewed in Thomson and Lin, 2009). PIWI proteins are essential for germ line specification and germ line stem cell maintenance, meiosis, gametogenesis, and transposon silencing in the gametes. These diverse functions involve PIWI-interacting small RNAs (piRNAs), 24 to 32 nucleotides long, which have been found in mammals, zebrafish, and Drosophila. Neither piRNA-like RNAs nor the PIWI family are found in plants.
We have identified an AGO gene in maize, ago104, whose function is required for the production of meiotically derived male and female spores. We primarily characterized meiosis in male meiocytes, owing to the difficulty of accessing female meiocytes. Whole-mount clearing of mutant ovules together with the genetic evidence, however, suggest that the mechanisms of unreduced gamete formation are likely similar in both male and female reproductive organs. In male meiocytes, mutations in ago104 result in defective chromatin condensation, misalignment of chromosomes on the metaphase plate, and failure in spindle formation at male meiosis I and II. Condensation during prometaphase I was highly defective, and the chromosomes never condensed to the wild-type levels. During meiosis I, metaphase meiocytes showed H3ser10 phosphorylation patterns, indicating an equational division instead of the expected reductional division. Whether condensation defects per se are responsible for the substitution of meiosis I by a mitosis- like division is not clear. However, is it well established that proper chromosome compaction is necessary for correct segregation of chromosomes during both meiosis I and II; in either yeast or animals, depletion of the conserved condensin complexes, which mediate chromosome condensation, results in poor compaction, sister chromatid cohesion defects, and missegregation of chromosomes (Hagstrom et al., 2002; Chan et al., 2004; Yu and Koshland, 2005; Hartl et al., 2008). Data are scant in plants, but both the el1 mutant in maize (Rhoades and Dempsey, 1966) and natural apomicts in Tripsacum (Grimanelli et al., 2003) exhibit condensation defects and high production of unreduced gametes, suggesting a similar relationship.
Unreduced gamete formation in ago104 might not be restricted to meiosis I defects. During metaphase II, we also observed a population of cells arrested at metaphase or undergoing irregular anaphase, suggesting restitution of meiosis II. Both these phenomena have the potential to give rise to unreduced gametes, although only restitution of meiosis I will produce unreduced gametes that are exact genetic clones of the mother plant.
In plants, including maize, transcriptionally silent heterochromatin is highly methylated at non-CG sites (Vaillant and Paszkowski, 2007; Wang et al., 2009). Here, we observed that in ago104 mutants, centromeric and/or pericentromeric repeats are hypomethylated in a non-CG context. These repeats are also transcribed at a much higher level than in wild-type plants. Additionally, knob heterochromatin repeats in the mutants are also hypomethylated in a non-CG context. Furthermore, profiling indicates a significant effect of the loss of ago104 function in the transcription of repetitive DNA, which constitutes the bulk of heterochromatin. This collectively suggests that ago104 might have a broad function in establishing or maintaining heterochromatin in the meiocytes, likely explaining the decondensation phenotype of the mutant.
AGO-dependent centromeric and pericentromeric heterochromatin organization is essential for centromere function and cell division in diverse organisms. In Drosophila, mutants of AGO2 lead to defective mitotic divisions during early embryogenesis due to defects in centromeric heterochromatin formation (Deshpande et al., 2005), affecting centromere formation, chromosome condensation, nuclear kinesis, and assembly of the spindle apparatus. AGO2 is also required for centromeric localization of a centromere-specific histone 3 variant. In Caenorhabditis elegans, the AGO protein CSR-1 is required for proper segregation of the holocentric chromosomes through a chromatin-mediated mechanism (Claycomb et al., 2009). Interestingly, csr-1 mutants show cell division phenotypes very similar to ago104 mutants. At metaphase, csr-1 mutants are defective for the alignment of chromosomes on the metaphase plate, leading to missegregation. These mutants are also defective in the pattern of loading of centromeric H3 variant HCP3 and cohesion-related proteins. Our data suggest a specific function for ago104 in meiotic centromere organization in maize. Whether the centromeric function of ago104, its broader role on heterochromatin, or both are sufficient to explain the nonreduction phenotype remains to be determined.
Localization of AGO104 Suggests a Non-Cell-Autonomous, Ovule-Specific Mechanism for Chromatin Modification, Targeting Both Genes and Repeats
In animals, PIWI proteins play a crucial role in germ line determination, germ line stem cell maintenance, meiosis, and transposon silencing in the germ line (Aravin et al., 1997; Girard et al., 2006; Lau et al., 2006; Megosh et al., 2006; Vagin et al., 2006; Houwing et al., 2007). Plants do not have germ lines as such, and neither PIWI-like proteins nor small RNAs corresponding to piRNAs have been identified. Recently, however, two AGOs have been identified in rice and Arabidopsis with specific functions during sporogenesis. MEL1 in rice was the first sporogenesis-related AGO characterized in plants (Nonomura et al., 2007). mel1 null mutants have defective premeiotic and meiotic development. More recently, Arabidopsis AGO9 has been shown to act in the specification of MMC identity and in silencing transposons in the female gametes (Olmedo-Monfil et al., 2010), a role played by PIWIs in animals.
While the reproductive phenotype of ago104 mutants is clearly meiotic, we found that the AGO104 protein does not localize to the meiocytes but instead accumulates in somatic cells from the maternal tissues surrounding the female germ cell in premeiotic and meiotic ovaries. A very similar expression pattern was detected for the AGO9 protein in Arabidopsis (Olmedo-Monfil et al., 2010). Incidentally, ago9 loss-of-function alleles result in multiple spores in the ovule tissue adjacent to the L1 layer, where AGO9 protein is expressed. This, together with the phylogenetic relationship between both proteins, suggests that AGO9 and AGO104 are functionally related proteins and that AGO104 in maize, similar to AGO9 in Arabidopsis, acts in a conserved non-cell-autonomous manner to affect MMC development. Interestingly, however, the mutant phenotypes indicate that the Arabidopsis and maize genes likely have opposite functions: while AGO9 acts to repress germ cell fate in somatic tissues, ago104 acts to repress somatic fate in germ cells. The basis for this opposition is unclear. Note, however, that our interpretation here is based on relatively limited data regarding changes in cell identity and development in the mutant ovules in both Arabidopsis and maize. Critically, it is currently unknown whether female meiosis in ago9 is identical to that in the wild type. Similarly, lack of appropriate markers hampers the identification of more subtle changes in cell identity in the ago104-deficient maize ovule beyond cytologically obvious changes. Further analyses in both mutants will be critical to refine our working models. Note also that while their expression domains suggest non-cell-autonomous action (from the somatic cells to the precursor of the germ cells) for both proteins, they are in fact not expressed in the same somatic tissues. AGO9 is found in the L1 layer of cells surrounding the (diminutive) Arabidopsis nucellus. By contrast, AGO104 is detected in parts of the maize nucellus itself. Possibly, the much larger maize nucellus is already divided into domains prior to germ cell differentiation, adding an additional layer of complexity to cell identity in the maize ovule that is absent in Arabidopsis.
It is not known if AGO9 targets chromatin in a manner similar to that observed for AGO104. However, AGO9 is involved in the suppression of transposon activity in the female gametes (Olmedo-Monfil et al., 2010), a process that in somatic tissues at least is dependent on DNA methylation at non-CG sites (reviewed in Vaillant and Paszkowski, 2007). Furthermore, small interfering RNAs originating from centromeric repeats and transposons are highly represented in AGO9-interacting small RNAs (Havecker et al., 2010). It is thus probable that both AGO104 and AGO9 act similarly on non-CG methylation in the ovule. Interestingly, the results also suggest that the ago104 pathway differs from the canonical RNA-directed DNA methylation (RdDM) pathway. RdDM mutants in Arabidopsis affect non-CG methylation of 5S repeats but have no effect on the methylation or transcription of centromeric repeats. We observed the opposite in ago104. We further detected increased transcription in the ovule for both retrotransposons and DNA transposons, with the strongest effect on DNA transposons. Our results, therefore, suggest a pathway, targeting plant germ cells, in which the conserved AGO9/104 proteins have likely been recruited to perform functions similar to the animal PIWIs. We further extend their role, at least in maize, to the control of chromatin structure during meiosis, a role also demonstrated for zebrafish PIWIs (Houwing et al., 2008).
At the same time, the ago104 mutant can affect the expression pattern of thousands of gene-coding loci, which is intriguing. Because the ago104 mutants used in our experiments were derived from Mu-active stocks, we cannot discard the possibility that the differences observed in the transcriptomic data relate to Mu activity. Strong transcriptional effects of Mu activity, which can reprogram the transcriptome of developing maize anthers, have been well documented (Skibbe et al., 2009). Further experiments are thus required to define more precisely the contribution of an ago104-dependent pathway to transcriptional control in the ovule. Interestingly, however, similar changes have been reported recently in the shoot apical meristem for the modifier of paramutation1 (mop1) mutant of maize, which encodes a homolog of Arabidopsis RNA-DEPENDENT RNA POLYMERASE II, a member of the RdDM pathway (Jia et al., 2009). This might indicate that both genes belong to the same pathway, acting in reproductive and meristematic tissues. However, expression patterns for mop1 in reproductive cells or ago104 in meristems remain to be determined.
ago104 Loss of Function Induces Diplospory-Like Development
Our findings demonstrate a crucial role for an ago104-dependent pathway in regulating sexual development in maize. These findings also suggest that this pathway may be an important component of diplosporic development. First, we found that ago104 loss of function results in an apomeiosis-like phenotype. Many individual meiotic mutants have been identified in Arabidopsis or maize that produce unreduced spores (Curtis and Doyle, 1992; Mercier et al., 2001; Agashe et al., 2002; Golubovskaya et al., 2006; Ravi et al., 2008). However, these mutations are recessive (contrary to diplosporous development), result in almost complete male and female sterility, and produce viable unreduced gametes, if any, at extremely low frequency. Only the accumulation of multiple recessive mutations allows for the formation of such gametes at high frequency (d’Erfurth et al., 2009). An important aspect of the ago104 mutants, clearly, is that abnormalities during meiosis result in the formation of functional unreduced gametes. The only other known meiotic mutant with a related phenotype is el1 in maize, which also affects chromosome condensation and results in viable unreduced male and female gametes. While the molecular function of el1 is unknown, it has been mapped to chromosome 8L, while ago104 is located on chromosome 6L. Both are thus different loci. Therefore, linking meiotic failure to gametogenesis initiation seems much more frequent in ago104 mutants or in el1 than in meiotic mutants in maize or Arabidopsis.
Our data show that AGO104 modifies chromatin through DNA methylation. This modification may induce developmental plasticity and possibly facilitate a shift from meiotic to mitotic state, with profound changes for the transcriptional landscape in the germ cells. Such global transcriptional reprogramming has been hypothesized in another apomictic genus, Boechera (Sharbel et al., 2010). Also, the defects observed during male meiosis in ago104 mutants are very similar to those observed in apomictic Tripsacum (Grimanelli et al., 2003). Interestingly, the ago104 locus maps to a region on chromosome 6 of maize that is syntenic to the Tripsacum apomixis locus–containing region (Grimanelli et al., 1998). Thus, understanding ago104 function might provide a uniquely interesting route to characterize apomixis, and efforts are under way to functionally link AGO104 activity to apomixis expression in Tripsacum. Note that while ago104 mutants recapitulate diplosporous development, we never found any apomictic progeny in the mutants, all of which lacked parthenogenesis. Clearly, loss of ago104 function is not sufficient to induce complete apomictic development, and additional components still need to be determined.
Interestingly, we recently showed that sexual development in maize ovules and apomixis in the ovules of maize–Tripsacum hybrid plants differ for the transcription of a limited number of chromatin-modifying enzymes, most of which function (or are predicted to function) in DNA methylation pathways (Garcia-Aguilar et al., 2010). Loss of function for two of them, dmt102 (homologous to Arabidopsis CHROMOMETHYLASE3 [CMT3]) and dmt103 (homologous to Arabidopsis DOMAIN REARRANGED METHYLTRANSFERASE2 [DRM2]), results in apomixis-like phenotypes. Similar to AGO104, CMT3 and DRM2 affect non-CG DNA methylation patterns. Furthermore, dmt102 and dmt103 transcription domains in maize ovules are restricted to germ cells and nucellar cells and possibly overlap with AGO104 expression. The presence of multiple embryo sacs in mutant plants suggests that at least dmt103 functions to enforce a single gametophyte within each ovule. The phenotype is thus reminiscent of aposporous-like ago9 mutations (Olmedo-Monfil et al., 2010). Whether dmt103, dmt102, and ago104 act as part of a single pathway is currently unknown and needs to be biochemically tested.
An important aspect of the ago9 phenotype was the formation of unreduced spores (but not unreduced gametes), reminiscent of apospory. It has thus been proposed that ago9 might shed light on the mechanisms underlying aposporous apomictic development in plants. Conversely, we did not find any effect of the ago104 mutation on cell identity in the ovule, but loss of function of AGO104, which is likely a functional AGO9 ortholog in maize, mimics apomeiosis of the diplosporous type, as functional unreduced spores are formed after substitution of meiosis by a mitosis-like division. This suggests that in spite of fundamental differences in the apomeiotic process, both types of gametophytic apomixis might rely on similar regulatory pathways.
METHODS
Plant Materials
Seeds of the Mu stocks for the DNR mutant screen were kindly provided by M. Freeling (University of California, Berkeley). Inbred CML216 was obtained from the Centro Internacional de Mejoramiento de Mais y Trigo (CIMMYT) gene bank. Two tetraploid tester stocks (Zm4xR and Zm4xB) were created from a highly heterogenous population of tetraploid maize (Zea mays) plants available at CIMMYT by successive generations of selfing and selection for adaptation to our growth conditions. The autotetraploid of B73 (stock number N108A) was kindly provided by the Maize Genetics Stock Center. Additional Mu-insertion alleles for the ago104 gene were obtained from the Pioneer Hi-Bred Trait Utility System for Maize reverse genetics population. The mutant alleles were provided as F2 seeds and recurrently backcrossed to wild-type inbred lines, either W23 or B73, selecting at each generation for ploidy and the transmission of the defective alleles. Phenotypic analyses were performed by comparing plants carrying the mutant alleles after three generations of backcrossing to the corresponding wild-type background. The profiling experiments compared B73 as the wild type with mutant lines backcrossed three times to the reference line. Mutant plants were genotyped using a Mu-specific primer (5′-AGAGAAGCCAACGCCAWSGCCTCYATTTCGTC-3′) and an ago104-specific primer (5′-TGTCTCCTGTATCAACGGGGTGGTC-3′). For PCR, we used 10-μL reactions containing 100 ng of DNA, 2 μL of ReadyMix Taq PCR Reaction Mix (Sigma-Aldrich), and 10 μM of each primer. The PCR cycling program included 35 cycles of 94°C for 30s, 58°C for 30s, and 72°C for 30s, followed by an additional cycle of 2 min at 72°C.
Identification of the Dnr4 Locus
The Dnr4 allele originated from an active stock containing multiple Mu elements. We had previously observed (D. Grimanelli, unpublished data) that new Mu insertions in a gene often result in the production of chimeric transcripts containing sequence from the gene and the terminal inverted repeats (TIRs) of the Mu element, and originating either from the TIRs or as run-through transcription of the original transcription unit. To identify putative insertions responsible for the lesion, we thus used thermal asymmetric interlaced-PCR (Settles et al., 2004) and searched for such chimeric transcripts in male meiocytes of plants expressing the Dnr4 phenotype not found in either of the two original parental lines. In five independent assays, thermal asymmetric interlaced-PCR generated two reproducible fragments that were cloned and used as probes for cosegregation analyses with the Dnr4 phenotype. To generate the segregating population, one plant with a clear Dnr4 female phenotype was used as pollen donor to a wild-type plant (Zm4xB), and 56 plants from the progeny were selected for analysis. DNA from the 56 progeny derived from the original Dnr4 mutant were digested with EcoRI and HindIII, and DNA gel blots were hybridized with the two probes. One of the probes generated a band cosegregating perfectly with the Dnr4 phenotype. Sequencing of the probe and BLAST analysis showed that it contained 450 bp of genomic DNA with a unique match in the 3′ end of the GRMZM2G141818 locus. We then used the GRMZM2G141818 sequence to design a set of primers covering the predicted genomic sequence of the gene to screen for novel alleles in the Pioneer Trait Utility System for Maize population, as described (Bensen et al., 1995).
RT-PCR Assays
RNA extractions from ovary, anther, and leaf tissue were performed with TRIZOL reagent (Invitrogen) followed by DNase treatment (DNase I; Invitrogen). RNA was reverse transcribed using the SuperScript III RT-PCR kit (Invitrogen). All procedures were done following the manufacturer’s instructions. PCR was performed in 10-μL reactions containing 1 μL of cDNA, 2 μL of ReadyMix Taq PCR Reaction Mix (Sigma-Aldrich), and 10 μM of each primer (118-mf1 and 118-mb1). The PCR cycling program included 35 cycles of 94°C for 30s, 58°C for 30s, and 72°C for 30s, followed by an additional cycle of 2 min at 72°C. Primers were designed to include an intron, when possible, in order to identify genomic DNA contaminations. Primer sequences are as follows: MuTIR, 5′-AGAGAAGCCAACGCCAWSGCCTCYATTTCGTC-3′; ago104 (genotyping), 5′-TGTCTCCTGTATCAACGGGGTGGTC-3′; ago104 (RT), 5′-TGGTGGTCTGAACTCGTTGCTG-3′ (forward) and 5′-AACATTGTCTGGTGATGCCGTC-3′ (reverse). Maize actin1 was used as an amplification standard for all tissues using the following primer sequences: 5′-CCTGAAGATCACCCTGTGCT-3′ (forward) and 5′-GCAGTCTCCAGCTCCTGTTC-3′ (reverse).
Quantitative RT-PCR Assays
RNA was isolated as above, quantified with a NanoDrop ND-1000 (NanoDrop Technologies), and verified for integrity by electrophoresis on a 1% (w/v) agarose gel. Real-time PCR was performed in four replicates for two different samples at each stage, each synthesized from 1 μg of total RNA using SuperScript III (Invitrogen) and oligo(dT) primers. cDNAs were diluted 100-fold, and 5 mL was used in 20-mL PCRs using the Brilliant II SYBR Green QPCR (Stratagene) master mix following the manufacturer’s instructions. Primers were designed using the Beacon Designer software (Stratagene) and validated by analyzing the dissociation curve of each reaction. PCR efficiency and data analyses were performed using the qbasePLUS software (www.biogazelle.com). To normalize the expression of ago104 genes, we used gpm120, previously determined to be a good control gene in maize reproductive tissues (Garcia-Aguilar et al., 2010; here, the expression stability value of the reference gene < 0.2 and the coefficient of variation of the normalized reference gene < 0.05). Primer sequences for ago104 quantitative PCR were 5′-ATACTGAGGCAGCATTCCGCAG-3′ (forward) and 5′-TCACACCACCACCCAAGTCAAC-3′ (reverse).
Antibodies
A polyclonal antibody was raised in rabbits against the predicted AGO104 protein using peptide sequence H2N-SERICKEQTFPLRQR-CONH2. Antibodies against α- and β-tubulin were from Sigma-Aldrich (catalog numbers T6199 and T5201). The phospho H3ser10 antibody was from Upstate (now Millipore; catalog number 05-1336).
Cytology and Immunochemistry
Male meiocytes were processed following the method previously described by Bass et al. (1997). Briefly, anthers were first stage-characterized by squashing in acetocarmine. Anthers from appropriate stages were fixed for 1 h in fixative (4% paraformaldehyde, 1× PBS, and 2% Triton) and washed twice in 1× PBS. Meiocytes were squeezed from anthers and mounted on a microscope slide in a layer of acrylamide as described (Bass et al., 1997). Immunocytochemistry was performed as described (Grimanelli et al., 2003). For ovule samples, freshly harvested ears at the appropriate stage were sliced with a vibratome (LeicaVT1000E). Sections (~200 μm) were fixed for 3 h in the same fixative as anthers and washed twice in 1× PBS. Samples were digested in an enzymatic solution (1% driselase, 0.5% cellulase, 1% pectolyase, and 1% BSA; all from Sigma-Aldrich) for 15 to 45 min at 37°C, depending on the tissue and developmental stage, rinsed three times in 1× PBS, and permeabilized for 1 to 2 h in 1× PBS, 2% Triton, and 1% BSA. The samples were then incubated overnight at 4°C with a 1:200 dilution of the primary antibodies (TUB, phH3S10, or AGO104 depending upon the experiment). The sections were then washed in 1× PBS and 0.2% Triton for 8 h and incubated overnight with a 1:400 dilution of secondary antibodies (either fluorescein isothiocyanate conjugate or Cy3 conjugate, both from Sigma-Aldrich, or AlexaFluor488 conjugate from Molecular Probes). After washing in 1× PBS and 0.2% Triton for a minimum of 6 h, the sections were incubated with DAPI (1 μg/mL in 1× PBS) for 1 h, washed for 1 h in 1× PBS, and mounted in PROLONG medium (Molecular Probes). Three-dimensional ovule images were captured on a laser scanning confocal microscope (Leica SP2) equipped for 405-nm (DAPI), 488-nm (fluorescein isothiocyanate/Alexa488), or 525-nm (Cy3/Alexa568) excitation and 20×, 40×, or 63× objective. Maximum-intensity projections of selected optical sections were generated for this report and then edited using Graphic Converter (LemkeSOFT).
DNA Gel Blot Analysis
Total genomic DNA was extracted using standard protocols, and 15 μg of DNA was digested with MspI and blotted to Hybond-N membrane (GE Healthcare). To generate probes, centromeric repeat CentC and 5S rDNA fragments were amplified from the maize genomic DNA, cloned, and sequenced. Probes were PCR-labeled with digoxigenin-dUTP (Roche) in a 50-μL PCR mix with 100 ng of template DNA, 10 μL of ReadyMix Taq PCR Reaction Mix (Sigma-Aldrich), 25 μM of each forward and reverse primer, and 1 μL of digoxigenin-dUTP. Hybridization, washing, and detection were performed using DIG Easy Hyb buffer and DIG Luminescent Detection kit (Roche) following the manufacturer’s guidelines.
Methylation Analysis
A total of 500 ng of genomic DNA was converted by bisulfite treatment using the MethylCode Bisulfite Conversion kit (Invitrogen) according to the instructions provided. PCR amplifications were performed from treated DNA with 180-bp knob-repeat primers (Papa et al., 2001). By using the Qiaquick gel extraction kit (Qiagen), the specific amplicon was purified and cloned in pCR2.1 (Invitrogen TA cloning kit). Twelve to 15 clones of each treatment were sequenced using BigDye Terminator version 3.1 (Applied Biosystems) and analyzed on an ABI PRISM 3730xl (Applied Biosystems). Sequence analysis for cytosine conversion was performed using the CyMATE software (Hetzl et al., 2007).
Phylogenetic Reconstruction
Sequence information for the maize and Arabidopsis thaliana AGOs and the MEL1 rice (Oryza sativa) protein were obtained from the CHROMDB database and aligned using a BLOSUM30 matrix and ClustalW, with an open gap penalty of 10 and an extend gap penalty of 0.1 in pairwise alignments, an extend gap penalty of 0.05, and delay divergent setting of 40% in the multiple alignment (for alignment, see Supplemental Data Set 1 online). The resulting alignment was hand-corrected and used for phylogenetic reconstruction using PHYLIP (Felsenstein, 1989). One thousand bootstrap replicates were performed to obtain a consensus tree. We used the Drosophila melanogaster AGO gene (Dm AGO) as an outgroup.
Protein Analysis
Protein concentrations were determined using the Bradford method with BSA as a standard. Samples were resolved by SDS-PAGE in duplicate and stained with Coomassie Brilliant Blue G 250 to check the quality of proteins. The samples from the second gel were electrotransferred onto nitrocellulose membranes (GE Healthcare). Following protein transfer, membranes were probed with a 1:400 dilution of the AGO104 antibody. The blotted membranes were developed with a 1:10,000 dilution of Immunopure Antibody goat anti-rabbit IgG (Pierce) and revealed by chemiluminescence substrate with ECL Plus Western Blotting detection reagents (GE Healthcare).
Digital Gene Expression Tag Profiling
Total RNA from ovaries during sporogenesis was sampled from three wild-type and three mutant plants, individually quantified, and then bulked in identical proportions. Illumina libraries were constructed for the wild-type and mutant samples following Illumina/Solexa’s standard protocol for Digital Gene Expression Tag Profiling library preparation. Adapter was as follows: 5′-CAAGCAGAAGACGGCATACGATCTAACGATGTACGCAGCAG-3′, corresponding to DpnII-derived tags. The resulting libraries were sequenced on a Solexa Genome Analyzer at the Montpellier GenomiX platform (Institut de Génomique Fonctionnelle, Montpellier, France). The Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) accession number for the data used in the paper is GSE26859. All sequences were filtered for adaptors, presence of the restriction site, quality scores, and against the presence of N in the first 20 bp. For mapping, we used the B73 RefGen_v2 (March 2010; http://www.maizesequence.org) reference sequence. Only reads that could be mapped to the genome without mismatches across 17 bases (irrespective of the number of mapping locations) were used for subsequent analyses (N not allowed). For analyzing gene expression, we used the “filtered gene set” collection of gene models and derived a pseudomolecule containing all predicted tag sites and the corresponding loci information. The short reads were then aligned to the pseudomolecule using Illumina's eland alignment tool (version 1.3), accepting only reads with unique nonambiguous locations. Total read counts per locus were obtained by adding all reads mapping to a given predicted transcription unit. Because the expression data sets do not include replicates, the statistics for identifying individual genes with differential expression are weak but collectively provide a broad image of transcriptional activity in our samples. We used two different methods for comparing both data sets and detecting putative differential expression. The first method is fully described (Jia et al., 2009). The second one is according to Lal et al. (1999). Both methods yielded essentially similar outcomes, and the results reported here correspond to the latter method. To analyze the transcription of repetitive DNA, we mapped all reads to the annotated transposon and retrotransposon families and subfamilies using BOWTIE. Each read was allowed to be mapped to multiple sites (BOWTIE parameters -n, number of mismatches = 1, and -k, reportable alignments = 50). We further tested the sensitivity of the results of the BOWTIE parameters by varying the number of mismatches (-n 0 to 2) and reportable alignments (-k 50, 100, and «best»). While the absolute number of reportable alignments varied widely depending on the parameters, the relative expression levels when comparing wild-type and mutant plants were relatively stable with zero or one mismatch. Allowing two mismatches resulted in an excess of possible mapping sites and was discarded. Without replicates for detecting transposons with differential transcription, we performed the analyses by comparing families of transposons rather than individual loci, taking each locus as a replicate within the family. Thus, the number of replicates varies greatly between families, but only families with at least five clearly identified loci were taken into account.
Accession Numbers
Sequence data from this article can be found in the GenBank data library under accession numbers BT066893 (AGO104 mRNA) and ACN33790 (AGO104 protein) and in the maize genome sequence database under accession number GRMZM2G141818. The Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) accession number for the data used in the paper is GSE26859.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of the Dnr4 Mutant Line.
Supplemental Figure 2.Whole-Mount Ovule Clearing of Dnr4 Ovules.
Supplemental Data Set 1. Text File of the Alignment of Maize, Arabidopsis, and Rice MEL1 AGO Proteins Used for Phylogenetic Reconstructions.
Supplemental Data Set 2. Genes with Putative Differential Expression.
Supplemental Data Set 3. Results of BLAST for the Differentially Expressed Genes.
Acknowledgments
Many thanks to Jose Galvez and the staff at CIMMYT Tlaltizapan station for field assistance; Bruno Bedora, Marc Senouque, and the Syngenta staff at Garlin for providing access to and assistance in their nurseries; M. Freeling for the Mu stocks; N. Lautrédou for help with laser confocal microscope optimization; and the members of the Apomixis group at the Institut de Recherche pour le Développement, Montpellier, for critical comments and suggestions on the work and manuscript. The work was funded by an Apomixis Consortium grant from Pioneer Hi-Bred, Syngenta Seeds, Group Limagrain and the Institut de Recherche pour le Développement, and by the Agence Nationale de la Recherche (Grant ANR-07-BLAN-0120).
References
- Agashe B., Prasad C.K., Siddiqi I. (2002). Identification and analysis of DYAD: A gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development 129: 3935–3943 [DOI] [PubMed] [Google Scholar]
- Ananiev E.V., Phillips R.L., Rines H.W. (1998). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95: 13073–13078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M.L., Landgraf P., Iovino N., Bass H.W., Marshall W.F., Sedat J.W., Agard D.A., Cande W.Z. (1997). Telomeres cluster de novo before the initiation of synapsis: A three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J. Cell Biol. 137: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell P.J., Grossniklaus U. (2005). Confocal microscopy of whole ovules for analysis of reproductive development: The elongate 1 mutant affects meiosis II. Plant J. 43: 309–320 [DOI] [PubMed] [Google Scholar]
- Bass H.W., Marshall W.F., Sedat J.W., Agard D.A., Cande W.Z. (1997). Telomeres cluster de novo before the initiation of synapsis: A three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J. Cell Biol. 137: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen R.J., Johal G.S., Crane V.C., Tossberg J.T., Schnable P.S., Meeley R.B., Briggs S.P. (1995). Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell R.A., Koltunow A.M. (2004). Understanding apomixis: Recent advances and remaining conundrums. Plant Cell 16 (suppl): S228–S245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A. (1993). Dosage analysis of maize endosperm development. Annu. Rev. Genet. 27: 181–204 [DOI] [PubMed] [Google Scholar]
- Bradley J.E., Carman G.C., Jamison M.S., Naumova T.N. (2007). Heterochronic features of the female germline among several sexual diploid Tripsacum L. (Andropogoneae, Poaceae). Sex. Plant Reprod. 20: 9–17 [Google Scholar]
- Calderini O., Chang S.B., de Jong H., Busti A., Paolocci F., Arcioni S., de Vries S.C., Abma-Henkens M.H., Lankhorst R.M., Donnison I.S., Pupilli F. (2006). Molecular cytogenetics and DNA sequence analysis of an apomixis-linked BAC in Paspalum simplex reveal a non pericentromere location and partial microcolinearity with rice. Theor. Appl. Genet. 112: 1179–1191 [DOI] [PubMed] [Google Scholar]
- Chan R.C., Severson A.F., Meyer B.J. (2004). Condensin restructures chromosomes in preparation for meiotic divisions. J. Cell Biol. 167: 613–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J.M., Batista P.J., Pang K.M., Gu W., Vasale J.J., van Wolfswinkel J.C., Chaves D.A., Shirayama M., Mitani S., Ketting R.F., Conte D., Jr, Mello C.C. (2009). The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner J.A., et al. (2008). Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol. 147: 1396–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C.A., Doyle G.G. (1992). Production of aneuploid, diploid eggs by meiotic mutants of maize. J. Hered. 83: 335–341 [Google Scholar]
- Dawe R.K., Hiatt E.N. (2004). Plant neocentromeres: Fast, focused, and driven. Chromosome Res. 12: 655–669 [DOI] [PubMed] [Google Scholar]
- d’Erfurth I., Jolivet S., Froger N., Catrice O., Novatchkova M., Mercier R. (2009). Turning meiosis into mitosis. PLoS Biol. 7: e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G., Calhoun G., Schedl P. (2005). Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 19: 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1989). PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Garcia-Aguilar M., Michaud C., Leblanc O., Grimanelli D. (2010). Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22: 3249–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A., Sachidanam R., Hannon G.J., Carmell M.A. (2006). A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Golubovskaya I.N., Hamant O., Timofejeva L., Wang C.J., Braun D., Meeley R., Cande W.Z. (2006). Alleles of afd1 dissect REC8 functions during meiotic prophase I. J. Cell Sci. 119: 3306–3315 [DOI] [PubMed] [Google Scholar]
- Grimanelli D., Garcia M., Kaszas E., Perotti E., Leblanc O. (2003). Heterochronic expression of sexual reproductive programs during apomictic development in Tripsacum. Genetics 165: 1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D., Leblanc O., Espinosa E., Perotti E., González-de-León D., Savidan Y. (1998). Mapping diplosporous apomixis in tetraploid Tripsacum: One gene or several genes? Heredity 80: 33–39 [DOI] [PubMed] [Google Scholar]
- Grimanelli D., Leblanc O., Perotti E., Grossniklaus U. (2001). Developmental genetics of gametophytic apomixis. Trends Genet. 17: 597–604 [DOI] [PubMed] [Google Scholar]
- Hagstrom K.A., Holmes V.F., Cozzarelli N.R., Meyer B.J. (2002). C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16: 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl T.A., Sweeney S.J., Knepler P.J., Bosco G. (2008). Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet. 4: e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker E.R., Wallbridge L.M., Hardcastle T.J., Bush M.S., Kelly K.A., Dunn R.M., Schwach F., Doonan J.H., Baulcombe D.C. (2010). The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzl J., Foerster A.M., Raidl G., Mittelsten Scheid O. (2007). CyMATE: A new tool for methylation analysis of plant genomic DNA after bisulfite sequencing. Plant J. 51: 526–536 [DOI] [PubMed] [Google Scholar]
- Houwing S., Berezikov E., Ketting R.F. (2008). Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 27: 2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S., et al. (2007). A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Hutvagner G., Simard M.J. (2008). Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Jia Y., Lisch D.R., Ohtsu K., Scanlon M.J., Nettleton D., Schnable P.S. (2009). Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes and 24-nt small RNAs. PLoS Genet. 5: e1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaszás E., Cande W.Z. (2000). Phosphorylation of histone H3 is correlated with changes in the maintenance of sister chromatid cohesion during meiosis in maize, rather than the condensation of the chromatin. J. Cell Sci. 113: 3217–3226 [DOI] [PubMed] [Google Scholar]
- Koltunow A.M., Grossniklaus U. (2003). Apomixis: A developmental perspective. Annu. Rev. Plant Biol. 54: 547–574 [DOI] [PubMed] [Google Scholar]
- Lal A., et al. (1999). A public database for gene expression in human cancers. Cancer Res. 59: 5403–5407 [PubMed] [Google Scholar]
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E. (2006). Characterization of the piRNA complex from rat testes. Science 313: 363–367 [DOI] [PubMed] [Google Scholar]
- Leblanc O., Grimanelli D., Hernandez-Rodriguez M., Galindo P.A., Soriano-Martinez A.M., Perotti E. (2009). Seed development and inheritance studies in apomictic maize-Tripsacum hybrids reveal barriers for the transfer of apomixis into sexual crops. Int. J. Dev. Biol. 53: 585–596 [DOI] [PubMed] [Google Scholar]
- Leblanc O., Peel M.D., Carman J.G., Savidan Y. (1995). Megasporogenesis and megagametogenesis in several Tripsacum species (Poaceae). Am. J. Bot. 82: 57–63 [Google Scholar]
- Li C.F., Pontes O., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W., Lagrange T., Pikaard C.S., Jacobsen S.E. (2006). An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126: 93–106 [DOI] [PubMed] [Google Scholar]
- Megosh H.B., Cox D.N., Campbell C., Lin H. (2006). The role of PIWI, the miRNA machinery in Drosophila germline determination. Curr. Biol. 16: 1884–1894 [DOI] [PubMed] [Google Scholar]
- Mercier R., Vezon D., Bullier E., Motamayor J.C., Sellier A., Lefèvre F., Pelletier G., Horlow C. (2001). SWITCH1 (SWI1): A novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev. 15: 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K.I., Morohoshi A., Nakano M., Eiguchi M., Miyao A., Hirochika H., Kurata N. (2007). A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V., Durán-Figueroa N., Arteaga-Vázquez M., Demesa-Arévalo E., Autran D., Grimanelli D., Slotkin R.K., Martienssen R.A., Vielle-Calzada J.P. (2010). Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozias-Akins P., van Dijk P.J. (2007). Mendelian genetics of apomixis in plants. Annu. Rev. Genet. 41: 509–537 [DOI] [PubMed] [Google Scholar]
- Papa C.M., Springer N.M., Muszynski M.G., Meeley R., Kaeppler S.M. (2001). Maize chromomethylase Zea methyltransfer-ase2 is required for CpNpG methylation. Plant Cell 13: 1919–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski W.P., Wang C.J., Golubovskaya I.N., Szymaniak J.M., Shi L., Hamant O., Zhu T., Harper L., Sheridan W.F., Cande W.Z. (2009). Maize AMEIOTIC1 is essential for multiple early meiotic processes and likely required for the initiation of meiosis. Proc. Natl. Acad. Sci. USA 106: 3603–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters L., Meister G. (2007). Argonaute proteins: Mediators of RNA silencing. Mol. Cell 26: 611–623 [DOI] [PubMed] [Google Scholar]
- Ravi M., Marimuthu M.P., Siddiqi I. (2008). Gamete formation without meiosis in Arabidopsis. Nature 451: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Rhoades M.M., Dempsey E. (1966). Induction of chromosome doubling at meiosis by the elongate gene in maize. Genetics 54: 505–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles A.M., Latshaw S., McCarty D.R. (2004). Molecular analysis of high-copy insertion sites in maize. Nucleic Acids Res. 32: e54–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharbel T.F., Voigt M.L., Corral J.M., Galla G., Kumlehn J., Klukas C., Schreiber F., Vogel H., Rotter B. (2010). Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. Plant Cell 22: 655–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe D.S., Fernandes J.F., Medzihradszky K.F., Burlingame A.L., Walbot V. (2009). Mutator transposon activity reprograms the transcriptomes and proteomes of developing maize anthers. Plant J. 59: 622–633 [DOI] [PubMed] [Google Scholar]
- Spillane C., Curtis M.D., Grossniklaus U. (2004). Apomixis technology development—Virgin births in farmers’ fields? Nat. Biotechnol. 22: 687–691 [DOI] [PubMed] [Google Scholar]
- Thomson T., Lin H. (2009). The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect. Annu. Rev. Cell Dev. Biol. 25: 355–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckey M.R., Araujo A.C., Paech N.A., Hecht V., Schmidt D.L., Rossell J.B., de Vries S.C., Koltunow M.G. (2003). Sexual and apomictic reproduction in Hieracium subgenus Pilosella are closely interrelated developmental pathways. Plant Cell 15: 1524–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vaillant I., Paszkowski J. (2007). Role of histone and DNA-methylation in gene regulation. Curr. Opin. Plant Biol. 10: 528–533 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Wang X., Elling A.A., Li X., Li N., Peng Z., He G., Sun H., Qi Y., Liu X.S., Deng X.W. (2009). Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.G., Koshland D. (2005). Chromosome morphogenesis: Condensin-dependent cohesin removal during meiosis. Cell 123: 397–407 [DOI] [PubMed] [Google Scholar]