This work identifies two transcription factors, MYC3 and MYC4, as targets of JAZ repressors and regulators of responses to jasmonate. It finds a specificity of transcription factor activity that could be a clue to understanding the diversity of JA-regulated responses.
Abstract
Jasmonates (JAs) trigger an important transcriptional reprogramming of plant cells to modulate both basal development and stress responses. In spite of the importance of transcriptional regulation, only one transcription factor (TF), the Arabidopsis thaliana basic helix-loop-helix MYC2, has been described so far as a direct target of JAZ repressors. By means of yeast two-hybrid screening and tandem affinity purification strategies, we identified two previously unknown targets of JAZ repressors, the TFs MYC3 and MYC4, phylogenetically closely related to MYC2. We show that MYC3 and MYC4 interact in vitro and in vivo with JAZ repressors and also form homo- and heterodimers with MYC2 and among themselves. They both are nuclear proteins that bind DNA with sequence specificity similar to that of MYC2. Loss-of-function mutations in any of these two TFs impair full responsiveness to JA and enhance the JA insensitivity of myc2 mutants. Moreover, the triple mutant myc2 myc3 myc4 is as impaired as coi1-1 in the activation of several, but not all, JA-mediated responses such as the defense against bacterial pathogens and insect herbivory. Our results show that MYC3 and MYC4 are activators of JA-regulated programs that act additively with MYC2 to regulate specifically different subsets of the JA-dependent transcriptional response.
INTRODUCTION
The plant hormones jasmonates (JAs) are fatty acid–derived oxylipins required for the regulation of multiple physiological aspects of plant growth, development, and defense (Wasternack, 2007; Kazan and Manners, 2008; Browse, 2009; Chung et al., 2009; Pauwels et al., 2009). Thus, JAs are widely recognized as regulators of plant responses to environmental stresses such as pathogen and pest attack, wounding, ozone exposure, and water deficit (Devoto et al., 2005; Browse and Howe, 2008). They are also important regulators of growth and developmental programs such as gamete development, the cell cycle, root growth, tendril coiling, and senescence in many plant species (Pauwels et al., 2008; Zhang and Turner, 2008; Reinbothe et al., 2009; Yoshida et al., 2009). JAs are being recognized as important integrators of developmental and stress signals to modulate the allocation of resources to grow or to defend (Moreno et al., 2009; Robson et al., 2010).
Transcription is a major regulatory step in the activation of these responses, and JAs trigger an important transcriptional reprogramming of the cells to switch the basal developmental programs into the necessary stress response program (Reymond et al., 2004; Devoto et al., 2005; Mandaokar et al., 2006; Pauwels et al., 2008). The signaling events that lead to transcriptional reprogramming are starting to be elucidated. Upon elicitation by exogenous or endogenous signals, the hormone (+)-7-iso-jasmonoyl-l-isoleucine [also known as (3R,7S)-jasmonoyl-l-isoleucine or JA-Ile] is synthesized by JAR1 (Fonseca et al., 2009b; Suza et al., 2010; Wasternack and Kombrink, 2010). JA-Ile is perceived by a receptor complex formed by the protein COI1 and the JAZ repressors (Xie et al., 1998; Thines et al., 2007; Katsir et al., 2008; Fonseca et al., 2009b; Sheard et al., 2010). COI1 is an F-box protein that participates in Skp1-Cul1-F-box protein (SCF)-type E3-ubiquitin ligase complex and is responsible for the recognition and recruitment of specific substrates. The hormone stimulates the specific binding of COI1 and JAZ proteins that leads to ubiquitination of JAZ by SCFCOI1 and subsequent degradation by the 26S proteasome (Chini et al., 2007; Maor et al., 2007; Thines et al., 2007; Yan et al., 2007; Saracco et al., 2009). In the absence of the hormone, JAZ repressors bind to transcription factors (TFs) and prevent their activity by recruiting the general corepressors TOPLESS (TPL) and TOPLESS-Related (TPR) proteins through an interaction with the adaptor protein NINJA (Pauwels et al., 2010). Upon hormone accumulation and perception, degradation of JAZ repressors liberates TFs from NINJA and TPL and initiates the transcriptional reprogramming of the cell and the activation of the JA responses.
However, the key signaling events identified so far still do not explain how the diversity of JA responses is specifically regulated. Identification of new TFs targeted by the JAZ repressors represents a necessary step to address this question, since MYC2/JIN1 is so far the only TF described as a direct JAZ target (Chini et al., 2007). MYC2 is a key bHLH TF regulating the expression of different subsets of JA-responsive genes (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007). However, MYC2 cannot be the only TF regulating JA responses since myc2/jin1 mutants do not show a complete loss of JA sensitivity. Besides MYC2, several TFs have been shown to be involved in specific aspects of JA-induced responses. These include TFs such as ERF1, WRKYs, and MYBs among others (Fonseca et al., 2009a). However, their interaction with JAZs has not been reported so far. It has been speculated that the specific interactions between JAZs and their respective (still unidentified) TF targets may be largely responsible for the diversity and specificity of JA responses to different stimuli. However, this hypothesis remains to be formally demonstrated.
Here, we show that at least two closely related bHLH TFs, MYC3 and MYC4, act additively with MYC2 in the activation of JA responses. Moreover, both TFs are required for full responsiveness to the hormone in several JA-regulated physiological processes, including gene expression, inhibition of root growth, and pathogen and insect resistance. Our results suggest that MYC2, MYC3, and MYC4 may form a cluster of TFs regulating specific JA-dependent responses.
RESULTS
Identification of New Targets of JAZ Repressors
Yeast two-hybrid screens using JAZ2 and JAZ3 as baits identified a total of 60 positive clones. Direct sequencing revealed that ~20% corresponded to MYC2, confirming previous reports (Chini et al., 2007, 2009; Melotto et al., 2008). Sequencing of the remaining clones allowed the identification of two previously unknown candidate JAZ targets: the TFs MYC3 and MYC4.
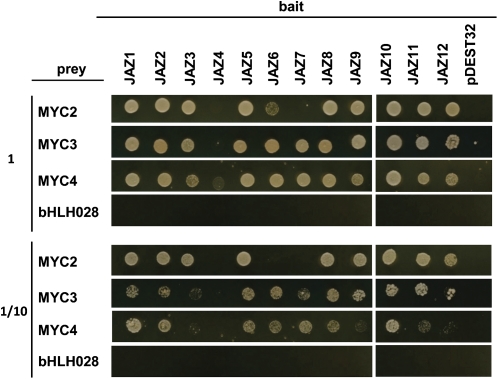
MYC3 and MYC4, together with At5g46830 (bHLH028), are the closest homologs of MYC2 in the Arabidopsis thaliana genome (see Supplemental Figure 1 online). To test if these MYC proteins interact with other JAZ proteins and if there is specificity in this interaction, we checked all possible combinations between all 12 JAZ proteins (as baits) and all four MYC TFs (as preys) by yeast two-hybrid assays (Figure 1). Consistent with previously reported data, MYC2 interacts with virtually all JAZ proteins with the exception of JAZ4 and JAZ7 (Figure 1; see Supplemental Figure 2 online; Melotto et al., 2008; Chini et al., 2009). Similarly, MYC3 interacts in yeast with all JAZ proteins except JAZ4, and MYC4 interacts with all JAZ, but very weakly with JAZ4. Thus, in contrast with MYC2, MYC3 and MYC4 interact with JAZ7, and MYC4 can weakly interact with JAZ4. In addition to this qualitative difference, quantitative differences in the intensities of the interactions were also present (Figure 1).
Figure 1.
MYC3 and MYC4 Interact with JAZ Repressors in Yeast Two-Hybrid Assays.
Yeast cells cotransformed with pDEST22-MYC2, pDEST22-MYC3, pDEST22-MYC4, or pDEST22-bHLH028 (preys) and pDEST32-JAZ1-12 (baits) were selected and subsequently grown on yeast synthetic dropout lacking Leu and Trp (−2) as a transformation control (shown in Supplemental Figure 2 online) or on selective media lacking Ade, His, Leu, and Trp (−4) to test protein interactions. The bottom panel shows one-tenth dilution yeast growth in −4 selective media. pDEST22-MYC2, pDEST22-MYC3, pDEST22-MYC4, and pDEST22-bHLH028 cotransformations with pDEST32 vector were included as controls.
[See online article for color version of this figure.]
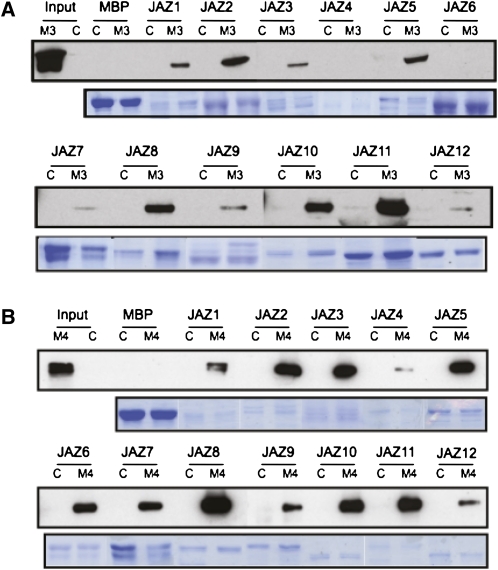
To support further that MYC3 and MYC4 are direct targets of JAZ proteins, we performed pull-down experiments using recombinant purified MBP-JAZ (maltose binding protein-JAZ) fusion proteins and extracts of transgenic plants expressing MYC3 or MYC4 derivatives (MYC3-HA or MYC4-GFP [for green fluorescent protein]). As shown in Figure 2, the results were consistent with those in yeast. MYC3 and MYC4 could be pulled down by most MBP-JAZ proteins, although again, qualitative and quantitative differences could be observed among different JAZ proteins and between MYC2, MYC3, and MYC4 (Figure 2; Chini et al., 2009).
Figure 2.
MYC3 and MYC4 Interact with JAZ Repressors in Pull-Down Experiments.
Immunoblots with anti-HA antibody of recovered MYC3-HA (A) or with anti-GFP antibody of recovered MYC4-GFP (B) after pull-down reactions using crude protein extracts from 35S:MYC3-HA (M3), 35S:MYC4-GFP (M4), or Col-0 (C) Arabidopsis plants and resin-bound recombinant MBP or MBP-fused JAZ proteins (top). Input lanes show the level of expression of recombinant proteins in transgenic and control plants. Coomassie blue staining shows the amount of recombinant proteins used (bottom). Double bands correspond to degradation products of MBP-fused JAZ proteins.
[See online article for color version of this figure.]
In spite of being the closest phylogenetic homolog to MYC2, bHLH028 was not identified in our screens (neither in yeast two-hybrid nor in tandem affinity purification (TAP) tagging in cultured cells; see below). Consistent with this, direct testing of the interaction of (bHLH028) with the 12 JAZ proteins both in yeast two-hybrid and pull-down experiments did not render any positive results (Figure 1; see Supplemental Figure 2 online).
To confirm further their interaction with JAZ proteins in planta, we performed TAP tagging of protein complexes in cultured PSB-D Arabidopsis cells using MYC2, MYC3, and MYC4 as baits. As expected for a direct target of JAZ repressors (Chini et al., 2007), MYC2-TAP allowed the copurification of several JAZ proteins, including JAZ2, JAZ11, and JAZ12 (Table 1; see Supplemental Table 1 online). Moreover, MYC2-TAP also copurified NINJA, which has been described as an adaptor protein between JAZ repressors and the corepressors TPL and TPR proteins (Pauwels et al., 2010). Therefore, these results indicate that MYC2 can form complexes in vivo with JAZ repressors and NINJA, as previously proposed (Chini et al., 2007; Pauwels et al., 2010). More interestingly, MYC3-TAP and MYC4-TAP baits also identified NINJA and several JAZ proteins (JAZ2 and JAZ12 in the case of MYC3 and JAZ2, JAZ11, and JAZ12 in the case of MYC4; Table 1). These results strongly support that these two new MYC TFs, similarly to MYC2, can form complexes with JAZ repressors and NINJA in vivo and therefore participate in JA signaling modules. In addition to JAZs and NINJA, these TAP tagging screens also identified MYC3 as an interactor of MYC2 and MYC4, and MYC4 as an interactor of MYC3, suggesting that they can form heterodimers in vivo (Table 1; see Supplemental Table 1 online).
Table 1.
Interactors of MYC and JAZ Proteins in TAP Tagging Screens
| Identified Proteins | MYC4 | MYC3 | MYC2 | JAZ3 | JAZ5 |
| JAZ2 | 3 | 4 | 2 | 2 | |
| JAZ3 | 2 | ||||
| JAZ5 | 4 | ||||
| JAZ11 | 4 | 1 | |||
| JAZ12 | 4 | 4 | 3 | 4 | |
| MYC4 | 4 | 2 | |||
| MYC3 | 4 | 4 | 3 | 1 | 2 |
| MYC2 | 3 | ||||
| NINJA | 4 | 4 | 3 | 2 | 4 |
| TPR1/TPL | 1 | 2 | |||
| TPR2 | 1 | ||||
| TPR3 | 1 |
The left column shows identified proteins that copurified with TFs MYC2-, MYC3-, and MYC4-TAP and repressors JAZ3- and JAZ5-TAP expressed in Arabidopsis cells suspension cultures (PSB-D) as TAP-tagged fusion proteins. Numbers within the table indicate the number of positive results for each combination of prey/bait in four independent TAP experiments, except in the case of JAZ3-TAP, where only two experiments were available.
As an alternative method to identify new JAZ-interacting TFs, we performed TAP tagging screens using JAZ3 and JAZ5 as baits. Consistent with previous results (Chini et al., 2009; Pauwels et al., 2010), these baits identified NINJA, several JAZs, TPL, and TPRs, confirming that these two JAZ proteins (JAZ3 and JAZ5) can participate in JA signaling modules in vivo (Table 1; see Supplemental Table 1 online). Interestingly, both baits identified MYC3, further supporting that interactions with this TF occur in living cells (Table 1; see Supplemental Table 1 online).
It is noteworthy that peptides corresponding to MYC2 were also identified in several TAP tagging experiments (using JAZ3, JAZ5, MYC3, or MYC4 as baits). However, the statistical significance was below cutoff values, indicating that endogenous expression of MYC2 or accumulation of the MYC2 protein may be very low in PSB-D Arabidopsis cultured cells.
The JAZ Interaction Domain of MYC Proteins
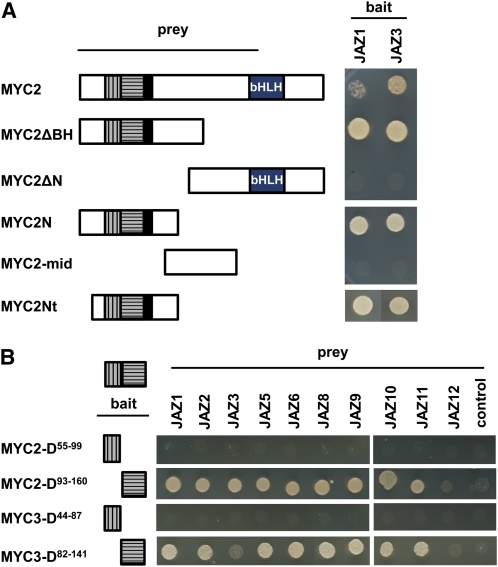
To characterize further the domain in MYC proteins responsible for the interaction with JAZ repressors, we performed yeast two-hybrid analysis using full-length MYC2 or several MYC2 derivatives (fused to the GAL4 activation domain) and JAZ1 or JAZ3 (fused to the GAL4 DNA binding domain). As shown in Figure 3A, the full-length MYC2 protein and all derivatives that included the N terminus were able to interact with both JAZ1 and JAZ3. However, derivatives lacking this N terminus did not interact with any of these JAZs. The smallest positive MYC2 fragment tested had three regions also conserved in MYC3 and MYC4: the MYC activation domain (in black) and two additional conserved regions represented with gray vertical or horizontal stripes in Figure 3 and Supplemental Figure 3 online. To delineate further the MYC2 interaction domain, we separated these two conserved domains and tested the corresponding protein derivatives (MYC2-D93-160 and MYC2-D55-99 fused to the GAL4 DNA binding domain; Figure 3B) in yeast two-hybrid assays against all MYC2-interacting JAZ proteins (fused to the GAL4 activation domain). The results showed that the MYC2-D93-160 region was sufficient for the interaction with most JAZ proteins, whereas the other conserved domains were dispensable. To verify that this new domain also mediates the interaction of additional MYC proteins with the JAZs, we tested the corresponding MYC3 derivative (MYC3-D82-141) and confirmed that this region was also sufficient for the interaction of MYC3 with most JAZ proteins (Figure 3B).
Figure 3.
Identification of the Domain in MYC TFs Interacting with JAZ Proteins.
(A) Full-length MYC2 or truncated derivatives were tested for interaction with JAZ1 and JAZ3. Yeast cells cotransformed with pGBKT7-JAZ1 or pGBKT7-JAZ3 (bait) and pGADT7-MYC2 or pGADT7-MYC2 derivatives (prey) were selected and subsequently grown on selective media lacking Ade, His, Leu, and Trp (−4) to test protein interactions. The different domains in MYC proteins are represented in (A) and (B): conserved domains among MYC proteins in gray with vertical and horizontal stripes and activation domain in black and bHLH motif.
(B) Different N-terminal fragments of MYC2 and MYC3 were assayed for interaction with MYC2-interacting JAZ proteins (JAZ1, 2, 3, 5, 6, 8, 9, 10, 11, and 12). Yeast cells cotransformed with pGBKT7-MYC2 derivatives or pGBKT7-MYC3 derivatives (bait) and pGADT7-JAZ proteins (prey) were selected and subsequently grown on selective media lacking Ade, His, Leu, and Trp (−4) to test protein interactions. pGBKT7-MYC2 or pGBKT7-MYC3 cotransformations with the pGADT7 vector were included as controls.
[See online article for color version of this figure.]
The identified JAZ interaction domain is conserved among MYC proteins from several plant species, and the degree of conservation is very high among MYC2, MYC3, and MYC4 (see Supplemental Figure 3 online). Using this conserved domain in a BLAST search, we identified additional MYC proteins bearing it and therefore representing new candidate JAZ targets (see Supplemental Figure 3 online).
Homo- and Heterodimerization between MYC TFs
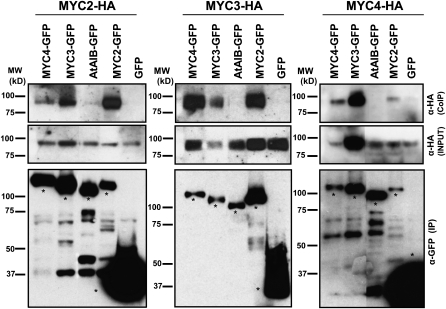
The TAP tagging results suggested that MYC2, MYC3, and MYC4 TFs could dimerize in vivo. To test this hypothesis further, we transiently expressed these proteins in Nicotiana benthamiana leaves and checked all possible combinations by coimmunoprecipitation experiments. Consistent with TAP tagging results, MYC2, MYC3, and MYC4 could form homo- and heterodimers, whereas none of them interact with the closely related MYC TF At AIB (Figure 4). MYC4/MYC4 and MYC4/MYC2 signals are weak, suggesting that the strength of the interactions may be different for different MYC/MYC combinations.
Figure 4.
Homo- and Heterodimerization among MYC Proteins in Planta.
Immunoblots of coimmunoprecipitated MYC2-HA, MYC3-HA, and MYC4-HA (CoIP; top panel), immunoprecipitated MYC2-GFP, MYC3-GFP, and MYC4-GFP (IP; bottom panel), and crude extracts (input HA; middle panel) from transiently expressed proteins in N. benthamiana leaves. Immunoprecipitation was performed using anti-GFP matrix, and coimmunoprecipitated proteins were detected using anti-HA antibody. The expression levels of the input HA-fused proteins were assessed by anti-HA of crude extracts (middle panel). The closely related bHLH At AIB was used as a negative control for interaction and GFP vector as a background control for interaction. Asterisks in the bottom panel mark the full-length protein band. Protein molecular mass (MW) ladder is shown on the left side of each blot. In all three panels, the lanes corresponding to MYC3 and MYC4 were originally separated from the other three lanes by one additional lane, which has been removed in the figure.
DNA Binding Specificity of MYC3 and MYC4
The interaction of MYC3 and MYC4 with JAZ repressors in vitro and in vivo and their heterodimerization with MYC2 suggests that these three TFs may share redundant functions. To test this idea, we first confirmed their predicted nuclear localization in vivo using GFP fusions of both MYC3 and MYC4 proteins. As for MYC2 (Lorenzo et al., 2004), both MYC3 and MYC4 are nuclear proteins (see Supplemental Figure 4 online).
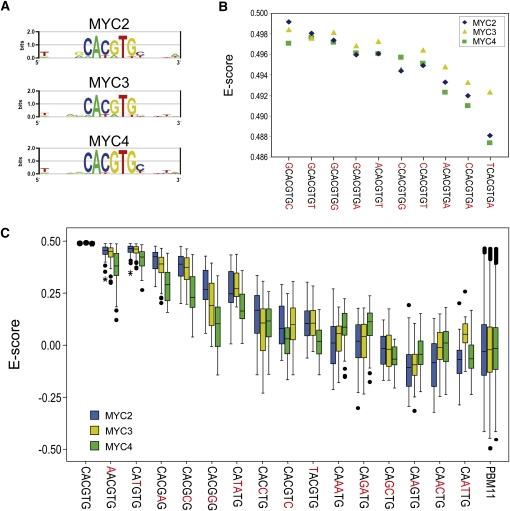
Next, we characterized the DNA binding specificities of MYC3 and MYC4 and compared them with that of MYC2 (recently characterized by Godoy et al., 2011). We determined the consensus DNA binding site of MYC3 and MYC4 using a protein binding microarray developed in our laboratory (PBM11; Godoy et al., 2011). The PBM11 contains all possible combinations of double-stranded 11mers (~4.2 million sequences) combined in ~240,000 oligonucleotides. The PBM11 is hybridized against the TF fused to MBP, and the result of specific binding of the TF to its DNA binding sites revealed by an antibody against MBP. This PMB11 has been successfully used for the determination of the consensus DNA binding site of MYC2 and other TFs (Godoy et al., 2011). As shown in Figure 5, the consensus binding sites obtained for MYC3 and MYC4 are strikingly similar to that of MYC2 (the G-box), including their preferences for 5′- and 3′-end nucleotides (Figures 5A and 5B). In addition, we also analyzed E-scores (reflecting binding affinities) of all three MYC proteins for all the possible G-related variants generated by replacing each nucleotide in the canonical G-box by the three remaining bases. In this analysis, we observed the highest E-scores for the elements G, T/G, G-like, G/A, and G/C, indicating that the MYC proteins tested showed similar binding affinities. However, we observed that whereas MYC2 and MYC3 were undistinguishable, MYC4 showed lower affinity for the G-box variants (Figure 5C). These results indicate that MYC2 and MYC3 have almost identical DNA binding specificities and, therefore, likely recognize similar targets in vivo. By contrast, MYC4, although showing a very similar binding affinity to the other two MYC proteins, may recognize a slightly different subset of target genes in vivo, at least in its homodimeric conformation.
Figure 5.
Identification of MYC2, MYC3, and MYC4 DNA Binding Motifs in Vitro.
(A) Position weight matrix representation of the top-scoring 8-mers corresponding to MYC2, MYC3, and MYC4. All three proteins showed highest binding affinity to a canonical G-box (CACGTG).
(B) Enrichment scores (E-scores) of all the possible G-box–containing 8-mers for the three MYC proteins tested, showing similar binding preferences of the three proteins for nucleotides at the 5′- and 3′-ends of the G-box 6-mer.
(C) Box plot of E-scores of G-box variants, including both single-site mutations and E-boxes (CANNTG). Boxes represent quartiles 25 to 75%, and black line within represents the median of the distribution (quartile 50%). Bars indicate quartiles 1 to 25% (above) and 75 to 100% (below), and dots denote outliers of the distribution. Boxes in blue correspond to MYC2, yellow boxes represent MYC3, and green ones correspond to data from MYC4.
Expression Patterns of MYC3 and MYC4
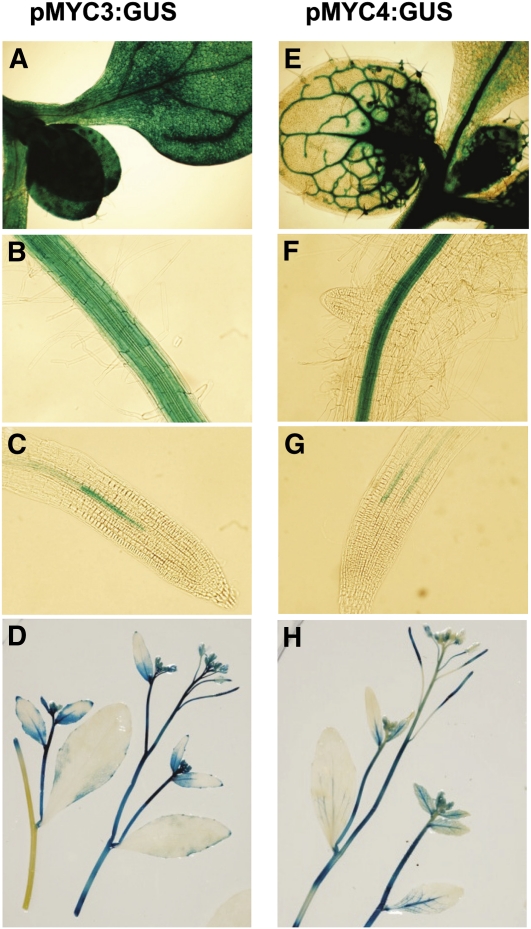
To gain further insight into MYC3 and MYC4 function, we analyzed their expression patterns using fusions of their promoters to the β-glucuronidase (GUS) reporter in stable transgenic plants. Both MYC3 and MYC4 showed strong expression in aerial parts of young seedlings (Figures 6A and 6E). MYC3 expression was observed in all tissues of the hypocotyls, cotyledons, and leaves, whereas MYC4 was preferentially expressed in the vasculature. Similarly, both were expressed in developed roots (Figures 6B and 6F), but MYC4 expression was restricted to vascular tissues. In young roots (Figures 6C and 6G), expression of both of them was very weak. In adult plants (Figures 6D and 6H), GUS staining was observed in most organs of the plant, including stems, siliques, flowers, and young leaves. No evident induction or changes in gene expression patterns could be detected after JA treatment (50 μM JA treatment for 1, 3, 8, 12, or 24 h). Quantitative PCR analysis of MYC3 and MYC4 gene expression confirmed that MYC3 and MYC4 are only very weakly induced by JA treatment (see Supplemental Figure 5 online). These expression patterns are consistent with available microarray data (www.genevestigator.com; see Supplemental Figure 6 online).
Figure 6.
Tissue Expression Patterns of MYC3 and MYC4.
Histochemical GUS activity of 6-d-old Arabidopsis transgenic seedlings ([A] to [C] and [E] to [G]) or 4-week-old Arabidopsis plants ([D] and [H]) expressing the GUS reporter gene under the control of the promoter of MYC3 (pMYC3:GUS) or MYC4 (pMYC4:GUS). GUS activity was detected between 3 and 12 h after staining.
Phenotypic Characterization of myc3 and myc4 Mutants
Our results suggest a role of MYC3 and MYC4 in the regulation of JA responses. To test this hypothesis, we obtained T-DNA insertional mutants (from Gabi-Kat; www.gabi-kat.de) for both genes, selected homozygous plants, and analyzed the response to JA of each single, double (of both and with myc2), and triple mutant. We analyzed typical JA-regulated responses, such as JA-dependent gene expression, root growth inhibition, and defense responses to insects and bacterial pathogens. Mutations in myc3 or myc4 affected to different degrees all tested responses to the hormone, as described below for each phenotypic analysis.
JA-Dependent Gene Expression
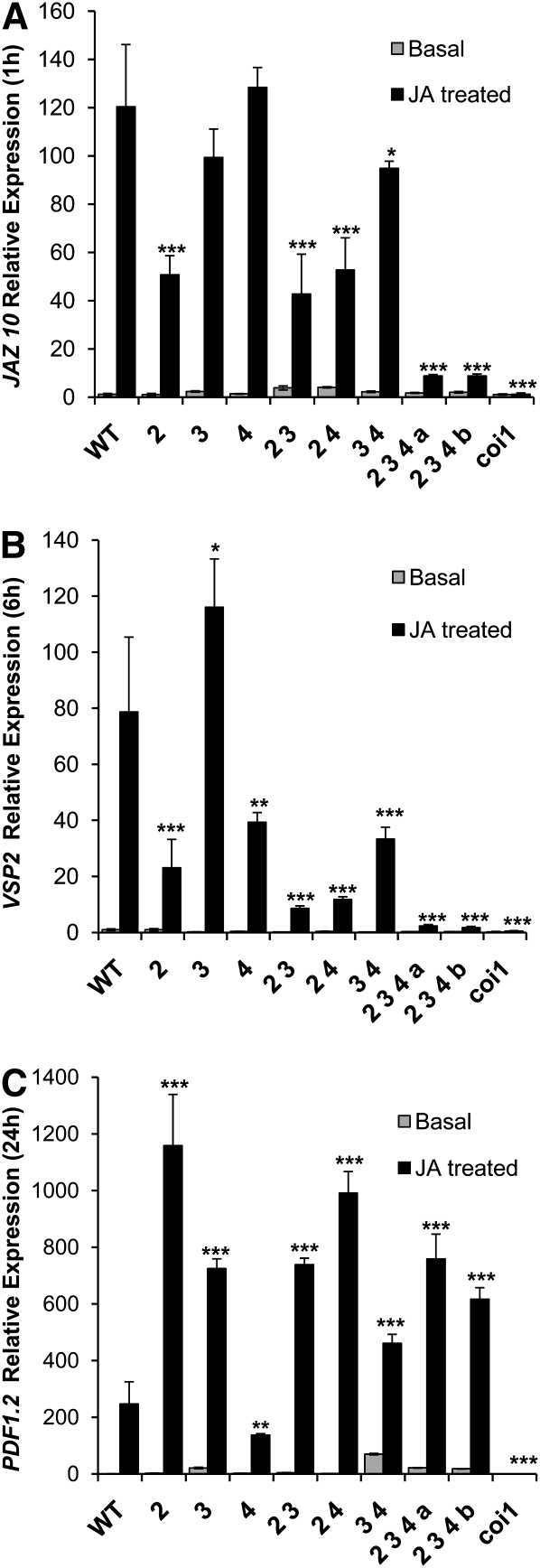
We analyzed induction of JA marker gene expression in 8-d-old wild-type and mutant seedlings. As JA markers, we chose genes induced at different times after JA treatment (i.e., immediate early [JAZ10], medium [VSP2], and late [PDF1.2] expressed genes). As shown in Figure 7, both myc3 and myc4 single mutants exhibit minor effects on JA induction of JAZ10,VSP2, and PDF1.2. Interestingly, analysis of double and triple mutants revealed, however, that each MYC TF has a striking additive effect on JA inducibility of JAZ10 and VSP2, rendering the triple mutants almost as impaired in JA-dependent expression of these genes as coi1-1, a JA receptor loss-of-function mutant that is completely insensitive to JA. In contrast, JA induction of PDF1.2 does not seem affected in the triple mutant. Therefore, these results indicate that MYC3 and MYC4 contribute to the activation of gene expression in response to JA, particularly of genes positively regulated by MYC2 (Lorenzo et al., 2004).
Figure 7.
Effect of myc Mutants on Induction of JA Marker Genes.
Quantitative RT-PCR of JAZ10 (1-h treatments; [A]), VSP2 (6-h treatments; [B]), and PDF1.2 (24-h treatments; [C]) expression in mutant and wild-type (WT) plants. The measurements (three technical replicates) represent the expression level between mock (basal) and treated (50 μM JA) plants relative to wild-type basal expression. The level of each gene is relative to that of ACTIN8. Error bars represent sd. Asterisks indicate statistically significant differences compared with Col-0 (Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001). WT:Col-0, 2:myc2, 3:myc3, 4:myc4, 2 3:myc2 myc3, 2 4:myc2 myc4, 3 4:myc3 myc4, 2 3 4 a: myc2 myc3 myc4 a, 2 3 4 b:myc2 myc3 myc4 b, myc2 myc3 myc4 a, and myc2 myc3 myc4 b represent two different triple mutants obtained from independent crosses but using the same alleles.
It is noteworthy that the single mutant myc3 showed an enhancement of VSP2 and PDF1.2 induction by JA, which is reminiscent of the negative effect of MYC2 on PDF1.2 expression (Figure 7; Lorenzo et al., 2004). This effect could suggest that MYC2 and MYC3 behave as repressors of PDF1.2 and VSP2, respectively. However, the analysis of triple mutants discards this possibility since myc2 myc3 myc4 shows a lower expression of VSP2, for instance, than does the myc2 myc4 double mutant. Gene expression analysis using quantitative RT-PCR of the three MYC genes showed a very low variation of MYC expression in each mutant background, indicating that the enhancement of PDF1.2 and VSP2 in myc2 and myc3 mutants is unlikely to be caused by a compensatory enhancement of the expression of the other MYC genes (see Supplemental Figure 7 online). However, whether this effect could be due to compensatory activation of the other two MYCs (e.g., by favoring particular dimeric combinations) or to enhanced expression of other unidentified MYCs needs further study.
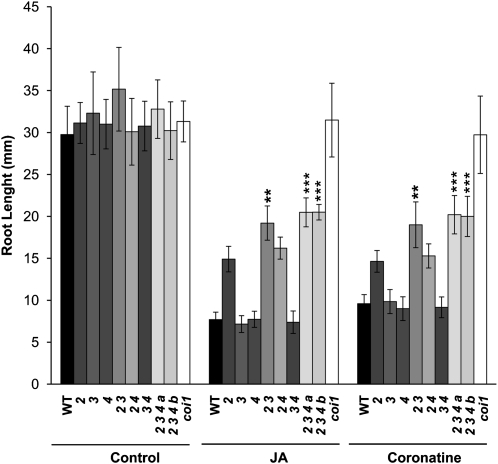
Root Growth Inhibition by JA
Consistent with the low expression of MYC3 and MYC4 in young roots (Figure 6; see Supplemental Figure 6 online), but in contrast with the striking effect on JA-dependent gene expression in whole seedlings, myc3 or myc4 single mutants or the double myc3 myc4 mutant did not show any alteration in the inhibition of root growth by JA compared with the wild type (Figure 8). In spite of this, the double myc2 myc3 mutant and the triple myc2 myc3 myc4 mutant showed a lower reduction of root growth induced by JA than did the single myc2 mutant, indicating that MYC3 and MYC4 also contribute to this JA-dependent phenotype. However, this contribution is weak since the triple mutant is substantially more sensitive to JA than coi1-1 is. These results indicate that MYC2 has a major role in the activation of JA responses in the root, whereas MYC3 and MYC4 have only a minor contribution in this tissue. This is consistent with the preferential expression of MYC2 in roots (see Supplemental Figure 6 online). Therefore, other TFs are expected to participate in the regulation of JA responses in the root. In this context, we already identified several MYC TFs bearing the conserved JAZ interaction domain, and these are good candidates to regulate these responses (see Supplemental Figure 3 online).
Figure 8.
Effect of myc Mutants on Root Growth Inhibition by JA.
Root growth inhibition assay of 8-d-old Arabidopsis seedlings from the wild type (WT), coi1-1, myc2, myc3, and myc4 single, double, and triple mutants grown in 50 μM JA, 0.5 μM coronatine, or mock media. Results shown are the mean ± sd of measurements from 30 seedlings. Asterisks indicate statistically significant differences between double or triple mutants and myc2 (one-way analysis of variance, *P < 0.05, **P < 0.01, and ***P < 0.001). Numbering is as in Figure 7.
Analysis of JA-Dependent Defense Responses
The contrast between the strong effect of myc3 and myc4 mutants in gene expression analyses and the weak effect on root growth inhibition was consistent with the low expression of MYC3 and MYC4 in young roots. Therefore, we studied JA-regulated defense responses that occur in aerial tissues, where MYC3 and MYC4 are strongly expressed. As an example of JA-dependent resistance, we studied herbivory by the generalist herbivore Spodoptera littoralis, and as an example of JA-mediated susceptibility, we analyzed the infection by Pseudomonas syringae pv tomato DC3000.
In spite of numerous replicate experiments, myc2 mutant plants always showed a relatively modest increase in susceptibility to S. littoralis larvae (Figure 9), which is consistent with its low expression in aerial tissues (see Supplemental Figure 6 online). We thus hypothesized that MYC2 homologs might play a role in insect resistance. Indeed, single myc3 and myc4 mutants showed compromised resistance, each of them having a stronger effect than that of myc2 mutants (Figure 9). Interestingly, the analysis of myc3 myc4 double and triple mutants showed an additive phenotype, in the case of the triple myc2 myc3 myc4 mutant rendering plants as susceptible as coi1-1. These results are fully consistent with the gene expression analysis and indicate that each one of the three MYC TFs contribute to the activation of JA-dependent defenses against S. littoralis, with a prominent role for MYC3 and MYC4.
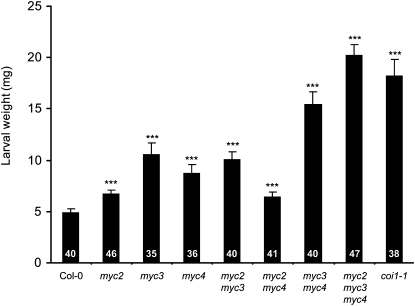
Figure 9.
Susceptibility of myc Mutants to a Generalist Herbivore.
Freshly hatched S. littoralis larvae were placed simultaneously on each genotype, and larval weight (mean ± se) was measured after 7 d of feeding. The number of larvae used in each experiment is shown within the bars. Asterisks indicate statistically significant differences compared with Col-0 (Student’s t test, ***P < 0.001). Similar results were observed in two other independent replicate experiments. The missing additive effect in myc2 myc3 and myc2 myc4 double mutants suggests a minor role of MYC2 in insect defense, which is consistent with its low expression in leaves.
We also analyzed the response of the mutants to infection by the hemibiotrophic pathogen P. syringae pv tomato DC3000. Contrary to insects that activate the JA pathway and trigger a strong defense response, this bacterial pathogen activates the JA pathway to promote susceptibility (Feys et al., 1994; Kloek et al., 2001; Laurie-Berry et al., 2006). Consistent with previous reports, the JA-insensitive mutants coi1-1 and myc2 showed increased resistance (both in terms of bacterial growth and leaf symptoms) that correlated with their levels of JA insensitivity (Figures 10A and 10B). Single myc3 and myc4 mutants showed an enhanced resistance (reduction of bacterial growth and leaf symptoms) to similar levels to those of myc2. As in the case of S. littoralis, analysis of double and triple mutants demonstrated an additive effect of all three genes rendering myc2 myc3 myc4 mutant plants almost as resistant to Pto DC3000 as coi1-1 was (Figures 10A and 10B).
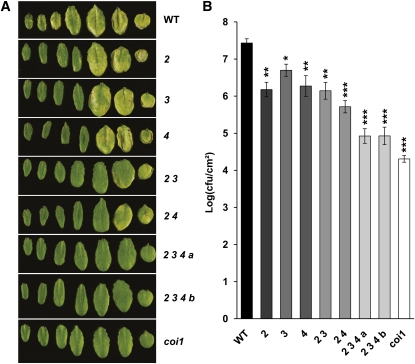
Figure 10.
Resistance of myc Mutants to the Bacterial Hemibiotrophic Pathogen Pto DC3000.
(A) Disease symptoms on Col-0 (WT), myc2 (2), myc3 (3), myc4 (4), myc2 myc3 (2;3), myc2 myc4 (2;4), myc2 myc3 myc4 a (2;3;4 a), myc2 myc3 myc4 b (2;3;4 b), and coi1-1 plants after spray inoculation with Pto DC3000 bacteria at 108 colony-forming units mL−1 (cfu/mL). Pictures were taken 3 d after inoculation. Leaves show representative symptoms of three independent experiments.
(B) Growth of Pto DC3000 on wild-type (WT) and mutant Arabidopsis plants 2 d after spray inoculation as in (A). Bacterial counts are expressed as log (cfu/cm−2). Error bars indicate se. The results are representative of three independent experiments. Asterisks indicate statistically significant differences compared with Col-0 (Student’s t test, *P < 0.05, **P < 0.01, and ***P < 0.001).
These results indicate that MYC3 and MYC4 have an additive effect with MYC2 in regulating JA-dependent responses mainly in aerial tissues.
DISCUSSION
Activation of JA responses requires a profound transcriptional reprogramming of cellular genetic programs that involves a complex interplay between positive and negative regulators (i.e., TFs and JAZ repressors). Several TFs activating JA responses have been described already, but MYC2 was the only direct target of the JAZ repressors identified so far (Fonseca et al., 2009a). Nonetheless, MYC2 cannot be the sole JAZ target for a number of reasons. First, loss-of-function mutations in this TF do not affect all JA-dependent phenotypes. For instance, myc2/jin1 mutants are fully fertile, suggesting that other TFs should regulate this developmental process. Second, most JA-insensitive phenotypes associated with myc2/jin1 are weaker than those of coi1-1 (Feys et al., 1994; Lorenzo et al., 2004; Katsir et al., 2008; Fonseca et al., 2009b; Sheard et al., 2010). Third, MYC2 negatively regulates the expression of some genes that are positively activated by JA, such as PDF1.2 (Lorenzo et al., 2004; Dombrecht et al., 2007). Therefore, other direct targets of JAZ are expected to exist and the identification of such TFs has become a major task in the field.
Using several JAZ proteins as bait, we identified two TFs, MYC3 and MYC4, that are phylogenetically closely related to MYC2, and show that both are direct targets of JAZ repressors. In addition, we show that they act additively with MYC2 as activators of JA-regulated programs. Supporting these conclusions, we show that MYC3 and MYC4 are nuclear proteins that bind DNA with similar specificity to that of MYC2. They interact in vitro and in vivo with several JAZ repressors and can also form homo- and heterodimers among themselves and with MYC2. Loss-of-function mutations in MYC3 and MYC4 impair full responsiveness to JA and enhance the JA insensitivity of myc2/jin1 mutants. Moreover, the triple mutant myc2 myc3 myc4 is as impaired as coi1-1 in the activation of several JA-mediated responses tested, such as the susceptibility to hemibiotrophic pathogens (P. syringae) and the resistance to insect herbivory (S. littoralis), as well as the induction of JA-dependent gene expression (JAZ10 and VSP2). However, since other JA-regulated responses are not completely blocked (i.e., inhibition of root growth) or are not altered at all (i.e., fertility) in the triple mutant, still more TFs may be expected to act redundantly with MYC2, MYC3, and MYC4 to achieve full responsiveness to JA, at least in particular tissues or developmental processes.
A genetic screen for mutants altered in Trp metabolism identified atr2D, a dominant mutant in MYC3 (Smolen et al., 2002). This mutant showed an enhanced expression of stress-related genes such PDF1.2 and Trp-related genes that participate in the biosynthesis of indole-glucosinolates (Smolen et al., 2002). These atr2D phenotypes are consistent with our results and compatible with the activation of JA-mediated defense responses by a constitutively active form of MYC3. Interestingly, the mutation in atr2D affects a conserved amino acid (D94N) within the JAZ interaction domain of MYC3. Whether this mutation prevents the interaction with JAZ proteins remains to be determined. However, this could be a plausible explanation for the constitutive MYC3 activity in the dominant atr2D mutant. Supporting the importance of the JAZ interaction region, At5g46830, the closest homolog of MYC2, does not interact with any JAZ proteins at all and bears a nonconservative amino acid change (Gly to Lys) in this conserved region (equivalent to Gly-91 in MYC3) (see Supplemental Figure 3 online).
In addition to the identification of new TFs regulating JA responses, one of the current major questions in the JA signaling field is how the specificity and diversity of JA responses is determined throughout the plant. In the case of auxins, this diversity seems to be a consequence of a combination of abundance and biochemical differences among TIR1/AFB auxin receptors, local concentration of bioactive auxins, the rate of degradation of AUX/IAA repressors, and the tissue-specific expression patterns of the components of the auxin signaling modules (TIR/AFB receptors, AUX/IAA repressors, and ARF TFs; Dreher et al., 2006; Leyser, 2006; Muto et al., 2007; Mockaitis and Estelle, 2008; Parry et al., 2009; Vanneste and Friml, 2009). In the case of JA, where only one receptor and one bioactive hormone has been described so far (Katsir et al., 2008; Fonseca et al., 2009b; Sheard et al., 2010), the rate of JAZ degradation and the differences in JAZ–MYC interactions, together with the tissue specificity of their expression patterns, may determine how specificity is achieved in the regulation of the diversity of JA-regulated processes. Supporting this hypothesis, JAZ spliced isoforms are more stable than full-length JAZ proteins, which could potentially contribute to the strength and specificity of responses (Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2010). Moreover, in this work, we provide evidence showing that part of this diversity and specificity can be explained by differences in MYC TF function. MYC3 and MYC4 share functions with MYC2, but each of them seems to have a predominant role in particular processes. Whereas MYC2 has a major role in root growth inhibition, MYC3 and MYC4 seem to be, for instance, more important than MYC2 in the regulation of responses to herbivory. This suggests that MYC2, MYC3, and MYC4 are not fully redundant, but rather have evolved some specificity on their functions. This specificity is likely due to the differences observed in their tissue expression patterns. However, we cannot discard that the small variations in DNA binding specificity and the differences observed in their affinities for JAZ repressors and among themselves (homo- and heteromeric interactions) may also contribute to the specificity of MYC function.
Analyses of JA marker gene expression in the single, double, and triple mutants revealed that all three MYC proteins are required for full responsiveness to JA. In the case of immediate-early and medium targets, such as JAZ10 and VSP2, triple mutants greatly reduce induction by JA. By contrast, in the case of late-responsive genes, such as PDF1.2, their expression is only weakly affected in the triple mutant. This suggests that MYC3 and MYC4 cooperate with MYC2 in the positive regulation of only a subset of JA-regulated genes. It is noteworthy that single mutants myc2 and myc3 enhance the response to the hormone of PDF1.2 and VSP2. This would indicate that MYC2 and MYC3 act as repressors of PDF1.2 and VSP2 expression. However, the analyses of double and triple mutants clearly show that at least in the case of VSP2 its overinduction in myc3 mutants is an indirect effect rather than a consequence of a direct repressive function of MYC3. This opens new perspectives on the understanding of MYC regulatory function and suggests that MYC TFs are subjected to a complex regulatory network of interactions wherein mutations in MYC2 or MYC3 promote unbalanced compensatory effects leading to this overinduction of PDF1.2 and VSP2. The molecular mechanism explaining such compensatory effect awaits further investigation.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana Columbia (Col-0) is the genetic background of wild-type and transgenic lines used throughout the work. Plants were growth in Johnson’s media at 21°C under a 16-h-light/8-h-dark cycle as previously described (Lorenzo et al, 2003).
The knockout lines myc3 (GK445B11) and myc4 (GK 491E10) were obtained from the Nottingham Arabidopsis Stock Centre, myc2/jin1-2 was previously described (Lorenzo et al., 2003), and coi1 was provided by J. Turner. The 35S:MYC2-GFP was previously described (Chini et al., 2009).
Double myc2 myc3, myc2 myc4, and myc3 myc4, and triple myc2 myc3 myc4 (a and b) mutants were generated by crossing the corresponding parental single or double (in the case of triple mutants) homozygous lines. F2 segregating progenies of these crosses were genotyped for plants homozygous for each gene.
To generate transgenic plants expressing MYC3 or MYC4 in Col-0 background, full-length MYC3 and MYC4 coding sequences carrying the stop codon or not were amplified with Expand High Fidelity polymerase (Roche) using Gateway-compatible primers (see Supplemental Table 2 online). PCR products were cloned into pDONR207 with a Gateway BP II kit (Invitrogen) and sequence verified. These plasmids, a Gateway LR II kit (Invitrogen), and the pGWB5 and pGWB14 (Mita et al., 1995) destination vectors were used to generate 35S:MYC3-GFP, 35S:MCY3-HA, 35S:MYC4-GFP, and 35S:MYC4-HA. These constructs were transferred to Agrobacterium tumefaciens strain C58C1 by freeze thawing and then transformed in Col-0 plants by floral dipping method (Clough and Bent, 1998). Kanamycin- and hygromycin-resistant plants were selected and their T2 progenies propagated for subsequent analysis.
Root Measurements
For root growth inhibition assays, root length of 20 to 30 seedlings was measured 8 d after germination in presence or absence of 50 μM JA (Sigma-Aldrich) or 0.5 μM coronatine (Sigma-Aldrich). Three independent replicates (20 to 30 seedlings each) were measured for each sample. Values represent mean ± sd. Comparisons between double and triple mutants of myc2, myc3, and myc4 versus myc2 were done by one-way analysis of variance. Comparisons between OE lines and the wild type (Col-0) were done by Student’s t test.
Yeast Two-Hybrid Screen
JAZ2 and JAZ3 sequences were PCR amplified with the Expand High Fidelity PCR system (Roche) from plasmid templates provided by The Arabidopsis Information Resource (TAIR) as described. All primers are listed in Supplemental Table 2 online. PCR products were digested with EcoRI and PstI and cloned into EcoRI-PstI–digested pGBKT7 (with GAL4 DNA binding domain; Clontech), and the constructs were sequence verified. The pGBKT7-JAZ2 and pGBKT7-JAZ3 plasmids were used as baits and transformed into yeast strain Y187. The prey cDNA library, from Arabidopsis seedlings 12 d old, grown on Pi-starved medium, was prepared in the plasmid pGADT7 and in yeast mating strain AH109 following BD Matchmaker Library Construction (version PR32047; Clontech). Bait (Y187) and prey (AH109) were mated by growing 50 mL of bait and 500 μL of prey overnight on 2× YPDA medium at 30°C. Yeast diploids were selected by plating at 30°C for 4 d on minimal medium SD lacking His, Leu, Trp, and adenine and supplemented with 20 mM 3-aminotriazole. For JAZ2 and JAZ3 baits, 67 and 38 clones containing putative interacting preys were selected, respectively. These clones were then sequenced and confirmed.
Yeast Two-Hybrid Assays
Full-length MYC3, MYC4, and BHLH028 coding sequences carrying a stop codon and truncated derivatives of MYC2 and MYC3 were amplified with Expand High Fidelity polymerase (Roche) using Gateway-compatible primers (see Supplemental Table 2 online). PCR products were cloned into pDONR207 with a Gateway BP II kit (Invitrogen) and sequence verified. MYC3, MYC4, and BHLH028 constructs were used in Gateway (Invitrogen) LR reactions, in combination with the destination low-copy yeast expression vectors pDEST22 (Gal4 AD) and pDEST32 (Gal4 BD), and were then checked by sequencing. Truncated derivatives of MYC2 and MYC3 in pDONR207 were used in Gateway (Invitrogen) LR reactions, in combination with the destination high-copy yeast expression vectors pGADT7gateway (Gal4 AD) and pGBKT7gateway (Gal4 BD) (Chini et al., 2007). JAZ1 to JAZ12 construct in pDEST32 and pGADT7gateway and MYC2 truncated derivatives MYC2ΔBH (equivalent to MYC2ΔC) and MYC2ΔN were previously described (Chini et al., 2007, 2009).
To assess protein interactions, the corresponding plasmids were cotransformed into Saccharomyces cerevisiae AH109 cells following standard heat shock protocols (Chini et al., 2009). Successfully transformed colonies were identified on yeast synthetic dropout lacking Leu and Trp. At 3 d after transformation, yeast colonies were grown in selective –Leu, –Trp liquid media for 6 h, and the cell density was adjusted to 3 × 107 cells mL−1 (OD600 = 1). A 3-μL sample of the cell suspensions was plated out on yeast synthetic dropout lacking Ade, His, Leu, and Trp and supplemented with 3 mM 3-aminotriazole to test protein interaction. Plates were incubated at 28°C for 2 to 4 d. The empty vectors pDEST22 or pDEST32 or pGADT7gateway or pGBKT7gateway were also cotransformed as negative controls.
Protein Extracts and Pull-Down Assays
MBP-JAZ fusion proteins were generated as previously described (Chini et al., 2009). Ten-day-old Arabidopsis wild-type seedlings and lines expressing 35S:MYC3-HA or 35S:MYC4-GFP were ground in liquid nitrogen and homogenized in extraction buffer containing 50 mM Tris-HCl, pH 7.4, 80 mM NaCl, 10% glycerol, 0.1% Tween 20, 1 mM DTT, 1 mM phenylmethylsulphonyl fluoride, 50 μM MG132 (Sigma-Aldrich), and complete protease inhibitor (Roche). After centrifugation (16,000g at 4°C), the supernatant was collected. For in vivo pull-down experiments, 6 μg of resin-bound MBP fusion protein was added to 1 mg of total protein extract and incubated for 1 h at 4°C with rotation. After washing, samples were denaturalized, loaded on 8% SDS-PAGE gels, transferred to nitrocellulose membranes, and incubated with anti-HA-horseradish peroxidase (Roche) or anti-GFP-horseradish peroxidase antibody (Milteny Biotec).
A 3-μL aliquot of MBP-fused protein of each sample was run into SDS-PAGE gels and stained with Coomassie Brilliant Blue to confirm equal protein loading.
TAP
Cloning of transgenes encoding tag fusions under control of the constitutive cauliflower tobacco mosaic virus 35S promoter and transformation of Arabidopsis cell suspension cultures were performed as previously described (Van Leene et al., 2007; see Supplemental Table 2 online). TAP of protein complexes was done using the GS tag (Bürckstümmer et al., 2006) followed by protein precipitation and separation, according to Van Leene et al. (2008). For the protocols of proteolysis and peptide isolation, acquisition of mass spectra by a 4800 Proteomics Analyzer (Applied Biosystems) and MS-based protein homology identification based on the TAIR genomic database, refer to Van Leene et al. (2010). Experimental background proteins were subtracted based on ~40 TAP experiments on wild-type cultures and cultures expressing TAP-tagged mock proteins GUS, red fluorescent protein, and GFP (Van Leene et al., 2010).
Coimmunoprecipitation
Nicotiana benthamiana leaves were infiltrated with Agrobacterium harboring MYC3 and MYC4 proteins fused to GFP or HA tags. Briefly, Agrobacterium cultures OD 0.6 were applied with a syringe to the undersides of three leaves of 4-week-old plants grown at 16 h light/8 h dark and 21°C. Empty pGWB vectors were used for the expression of GFP and HA proteins as negative controls. After 2 d from agroinfiltration, 0.6 g of agroinfiltrated leaves were collected and homogenized in 2 mL of coimmunoprecipitation buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2mM DTT, 0.1% Tween 20, 1mM phenylmethylsulphonyl fluoride, 50 μM MG132, and complete protease inhibitor cocktail (Roche) and were centrifuged twice at 16,000g at 4°C. The supernatant (1.5 mg of total protein) was incubated for 2 h (4°C, with rotation) with the agarose-conjugated anti-GFP matrix (MBL) and was washed three times with 1 mL of immunoprecipitation buffer. After denaturation in 90 μL of Laemmli SDS-PAGE loading buffer, samples were loaded into 8% SDS-PAGE gels, transferred to polyvinylidene fluoride membranes (Millipore), and incubated with anti-HA-horseradish peroxidase (Roche) and anti-GFP-horseradish peroxidase antibodies (Milteny Biotec). A 30-μL aliquot of total protein extract was also used for immunoblot with the same antibodies to confirm equal amount of recombinant proteins in each sample.
Protein Purification and Determination of MYC3 and MYC4 DNA Binding Motifs
Translational fusions of MYC3 and MYC4 fused to MBP were obtained by cloning their corresponding cDNAs into the pMAL-c2 vector (New England Biolabs) using Gateway technology. Donor templates were obtained through PCR amplification of pMYC3 and pMYC4 with oligonucleotides MYC3Fwgateway; MYC3Rvstopgateway (MYC3) and MYC4Fwgateway; and MYC4Rvstopgateway (MYC4) (see Supplemental Table 2 online). Recombinant inserts were verified by sequencing and plasmids introduced into BL-21 strain. Expression and purification of recombinant proteins were as described for the pMAL purification system (New England Biolabs).
MYC3 and MYC4 DNA binding specificities were determined using protein binding microarrays (PBM11) as described by Godoy et al. (2011). Briefly, recombinant protein (1 μg) was incubated for 2.5 h at room temperature with a double-stranded DNA microarray containing all 11-bp sequences (~4.2 million) compacted in ~240,000 spots. After washes, mircroarrays were incubated with a primary antibody against MBP and a secondary antibody coupled with DyLight 549 fluorophore. Finally, slides were scanned at 5 μm in a GenePix 4000B scanner (Axon Instruments) and signal intensities quantified in the GenePix Pro 5.1 software. All the steps (synthesis in situ of double-stranded DNA, incubation and washes of recombinant proteins and antibodies, scanning, quantification, and determination of DNA motifs) were performed as described (Godoy et al., 2011).
GUS Staining
To generate transgenic plants expressing GUS protein under the regulation of MYC3 and MYC4 promoter regions, 2028 and 1549 bp, respectively, upstream of ATG (including the first 30 nucleotides of the coding sequence of each gene) were amplified with Expand High Fidelity polymerase (Roche) using appropriate primers (see Supplemental Table 2 online). PCR products were cloned into pENTR/D-TOPO (Invitrogen) and sequence verified. These clones, a Gateway LR II kit (Invitrogen), and pGWB3 (Mita et al., 1995) destination vector were used to generate pMYC3:GUS and pMYC4:GUS. These constructs were transferred to Agrobacterium strain C58C1 by freeze thawing and then transformed in Col-0 plants by the floral dipping method (Clough and Bent, 1998). Six-day-old seedlings or adult plant tissues from several T2 transgenic lines were stained for GUS activity. Samples were placed in staining solution 50 mM phosphate buffer, pH 7, 0.1% (v/v) Triton (Sigma-Aldrich), 2 mM X-Gluc (Glycosynth), 1 mM K-ferrocyanide (Sigma-Aldrich), and 1 mM K ferricyanide (Sigma-Aldrich) and incubated at 37°C overnight. After staining, the tissue was soaked in several changes of 75% ethanol and kept in 5% glycerol until being photographed with a Leica DMR UV/VIS microscope (seedlings) or with a digital NIKON D1-x camera (adult plants). In the case of the roots, a root destaining protocol was applied (Malamy and Benfey, 1997): 15 min incubation at 57°C in 0.24 n HCl and 20% methanol and 15 min incubation at room temperature with shaking in 7% NaOH and 60% ethanol. After these two incubations, tissue was rehydrated by washing in decreasing ethanol series and vacuum treated in 5% ethanol and 25% glycerol.
Quantitative RT-PCR
Quantitative RT-PCR experiments were performed with RNA extracted from seedlings that were treated with 50 μM JA for 1, 6, or 24 h or mock treated (with DMF). For each experiment, three biological replicates, consisting of tissue pooled from 15 to 20 plants from different plates, were taken. RNA extraction and cleanup was done using Trizol reagent (Invitrogen) followed by RNeasy mini kit (Qiagen) and DNase digestion to remove genomic DNA contamination. cDNA was synthesized from 0.5 to 1 μg of total RNA with the high-capacity cDNA reverse transcription kit (Applied Biosystems). Five microliters from one-tenth diluted cDNA was used to amplify JAZ10 (1 h treatment), VSP2 (6 h treatment), PDF1.2 (24 h treatment), and the housekeeping gene ACTIN8 using Power SYBR Green (Applied Biosystems). Primer sequences are in Supplemental Table 2 online. Quantitative PCR was performed in 96-well optical plates in a 7300 Real Time PCR system (Applied Biosystems). Thermocycler conditions comprised an initial holding at 50°C for 120 s and then 95°C for 10 min. This step was followed by a two-step SYBR PCR program consisting of 95°C for 15 s and 60°C for 60 s for 40 cycles. Data analysis shown was done using three technical replicates from one biological sample; similar results were obtained with two others biological replicates.
Insect Bioassays
Arabidopsis (Col-0) and the mutants were vernalized in water for 4 d at 4°C. myc2 myc3 myc4 mutants were vernalized in water containing 0.1 mM gibberellic acid to stimulate germination. Seeds were then transferred to pots containing potting compost. coi1-1 seeds were germinated on Murashige and Skoog medium (Sigma-Aldrich) containing 3% sucrose and 30 μM JA and incubated under light (150 μmol m−2 s−1) for 7 d in a growth chamber. Homozygous coi1-1 mutants showing normal greening of leaves and no inhibition of root growth (Feys et al., 1994) were transferred to pots. Plants were grown for 3 weeks in a growth chamber as previously described (Reymond et al., 2000).
Three-week-old plants were placed in transparent plastic boxes in a growth chamber (20°C, 65% relative humidity, 100 μmol m−2 s−1, 10/14-h photoperiod). Forty to fifty newly hatched Spodoptera littoralis larvae were placed on 70 plants for 7 d of feeding. Surviving larvae were then collected and weighed. Data were analyzed on log-transformed values by Student’s t test. The experiment was repeated three times independently.
Bacterial Assays on Arabidopsis
Pseudomonas syringae pv tomato (Pto) DC3000 growth assays in Arabidopsis were performed by spray inoculation. Briefly, overnight bacterial cultures were pelleted and resuspended in sterile 10 mM MgCl2. Three- to four week-old plants were sprayed with a bacterial suspension containing 108 (colony-forming units)/mL bacteria (OD600 = 0.2) with 0.04% Silwet L-77. Leaf discs were harvested after 2 d and ground in 10 mM MgCl2. Population counts were performed at 2 d after infiltration. In both cases, serial dilutions of leaf extracts were plated on LB agar with appropriate antibiotics. Each data point represents the average of four replicates, each containing two leaf discs from different plants. Error bars indicate se. These experiments were repeated three times with similar results, and representative results are shown. Pictures of disease symptoms 3 d after inoculation on analyzed genotypes (Col-0, myc2, myc3, myc4, myc2 myc3, myc2 myc4, myc2 myc3 myc4 a, myc2 myc3 myc4 b, and coi1-1) were taken with a digital NIKON D1-x.
Confocal Microscopy
GFP of Arabidopsis 35S:MYC3-GFP and 35S:MYC4-GFP transgenic seedlings grown in Johnson’s media was visualized by a confocal microscope at 495 to 610 nm (Leica). Photographs of cells expressing GFP were taken under UV (GFP expression) and visible light (root structure).
Molecular Phylogenetic Analyses
Alignment of the N-terminal JAZ interacting region of MYC2, MYC3, MYC4, and (BHLH028) from Arabidopsis and MYC2-closest relatives from rice (Oryza sativa), Populus, and Physcomitrella patens and sequences of the JAZ interaction domain of MYC2, MYC3, MYC4, (BHLH028), (GL3), (EGL3), (TT8), (BHLH013), (At AIB), and (BHLH003) was performed using a multiple alignment method with ClustalW algorithm (T-Coffee Web server, http://tcoffee.vital-it.ch, default parameters, blosum matrix). Similarly, full-length protein sequences of 20 MYC2 bHLH-related proteins from Arabidopsis, Populus, rice, and P. patens were used to generate a multiple alignment with ClustalW algorithm (T-Coffee Web server, http://tcoffee.vital-it.ch, default parameters, blosum matrix). The alignment is available as Supplemental Data Set 1 online. A phylogenetic tree of these 20 proteins was generated by MEGA4.0.2 using Maxima Parsimonia method (10,000 bootstrap trials).
Accession Numbers
Sequence data from this article can be found in TAIR or GenBank/EMBL databases under the following accession numbers: MYC2 (locus At1G32640, GenBank NM_102998), MYC3 (locus AT5G46760, GenBank NM_124046.1), MYC4 (locus AT4G17880, GenBank NM_117897.3), AIB (locus AT2G46510, GenBank NM_130216.2), BHLH028 (locus AT5G46830, GenBank NM_124054.1), GL3 (locus At5G41315, GenBank NM_148067), EGL3 (locus At1G63650, GenBank NM_202351), TT8 (locus At4G09820, GenBank NM_117050), BHLH013 (locus AT1G01260, GenBanK NM_100009), BHLH003 (locus AT4G16430, GenBank NM_117738), BHLH014 (locus AT4G00870, GenBank NM_116313), BHLH020 (locus AT2G22770, GenBank NM_179700), BHLH019 (locus AT2G22760, GenBank NM_127841), BHLH025 (locus AT4G37850, GenBank NM_119946), BHLH092 (locus AT5G43650, GenBank NM_123731), BHLH027 (locus AT4G29930, GenBank NM_119139), ALCATRAZ (locus AT5G67110, GenBank NM_126111), PIF4 (locus AT2G43010, GenBank NM_129862), JAZ3 (locus AT3G17860, GenBank NM_112667), JAZ5 (locus AT1G17380, GenBank NM_101599), JAZ10 (locus AT5G13220, GenBank NM_001161241), PDF1.2 (locus AT5G44420, GenBank NM_123809), and VSP2 (locus AT5G24770, GenBank NM_001036860). Accession numbers of MYC2 closest relatives from other species are as follows: O. sativa (locus Os10g0575000, GenBank NM_001072010), Populus trichocarpa (GenBank XM_002329476.1), and P. patens (GenBank XM_001765109).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Tree of MYC2-Related Proteins from Arabidopsis and Other Plants.
Supplemental Figure 2. Control Growth of Yeast Cells Transformed with MYC3 or MYC4 and with JAZ Repressors in Yeast Two-Hybrid Assays.
Supplemental Figure 3. Alignment of the JAZ Interaction Domain of MYC2-Related Proteins.
Supplemental Figure 4. MYC3 and MYC4 Are Nuclear-Localized bHLH Proteins.
Supplemental Figure 5. JA-Dependent Induction of MYC3 and MYC4 Gene Expression.
Supplemental Figure 6. MYC2, MYC3, and MYC4 Tissue Expression Patterns.
Supplemental Figure 7. Expression of MYC Genes in myc Mutant Backgrounds.
Supplemental Table 1. MALDI-TOF/TOF-MS Identification of MYC2, MYC3, MYC4, JAZ3, and JAZ5 Interactors.
Supplemental Table 2. List of Primers Used.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Phylogenetic Tree in Supplemental Figure 1.
Acknowledgments
We thank Sylvia Gutiérrez for assistance with the confocal imaging and members of the Solano lab for critical reading of the manuscript. This work was financed by grants to R.S. from the Spanish Ministerio de Ciencia y Tecnología (BIO2007-66935, CSD2007-00057, and EUI2008-03666). P.F.-C. was supported by a predoctoral fellowship from Comunidad de Madrid (FPI-CAM). A.C. was supported by a “Juan de la Cierva” Fellowship from the Spanish Ministerio de Educación y Ciencia and an “EMBO long-term” Fellowship. J.-M.C. was supported by a postdoctoral contract from the Spanish Ministerio de Educación y Ciencia (CSD2007-00057-B).
References
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Browse J., Howe G.A. (2008). New weapons and a rapid response against insect attack. Plant Physiol. 146: 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer T., Bennett K.L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. (2006). An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Chico J.M., Fernández-Calvo P., Solano R. (2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Cooke T.F., Depew C.L., Patel L.C., Ogawa N., Kobayashi Y., Howe G.A. (2010). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Niu Y., Browse J., Howe G.A. (2009). Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry 70: 1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Ellis C., Magusin A., Chang H.-S., Chilcott C., Zhu T., Turner J.G. (2005). Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol. Biol. 58: 497–513 [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K.A., Brown J., Saw R.E., Callis J. (2006). The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J.F., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R. (2009a). The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009b). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Godoy M., Franco-Zorrilla J.M., Pérez-Pérez J., Oliveros J.C., Lorenzo O., Solano R. (2011). Improved Protein-Binding-Microarrays for the identification of DNA-binding specificities of transcription factors. Plant J., http://dx.doi.org/10.1111/j.1365-313X.2011.04519.x [DOI] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2008). Jasmonate signaling: Toward an integrated view. Plant Physiol. 146: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloek A.P., Verbsky M.L., Sharma S.B., Schoelz J.E., Vogel J., Klessig D.F., Kunkel B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26: 509–522 [DOI] [PubMed] [Google Scholar]
- Laurie-Berry N., Joardar V., Street I.H., Kunkel B.N. (2006). The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol. Plant Microbe Interact. 19: 789–800 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2006). Dynamic integration of auxin transport and signalling. Curr. Biol. 16: R424–R433 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J.J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J.E., Benfey P.N. (1997). Analysis of SCARECROW expression using a rapid system for assessing transgene expression in Arabidopsis roots. Plant J. 12: 957–963 [DOI] [PubMed] [Google Scholar]
- Mandaokar A., Thines B., Shin B., Lange B.M., Choi G., Koo Y.J., Yoo Y.J., Choi Y.D., Choi G., Browse J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46: 984–1008 [DOI] [PubMed] [Google Scholar]
- Maor R., Jones A., Nühse T.S., Studholme D.J., Peck S.C., Shirasu K. (2007). Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol. Cell. Proteomics 6: 601–610 [DOI] [PubMed] [Google Scholar]
- Melotto M., Mecey C., Niu Y., Chung H.S., Katsir L., Yao J., Zeng W., Thines B., Staswick P., Browse J., Howe G.A., He S.Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S., Suzuki-Fujii K., Nakamura K. (1995). Sugar-inducible expression of a gene for beta-amylase in Arabidopsis thaliana. Plant Physiol. 107: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K., Estelle M. (2008). Auxin receptors and plant development: A new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Moreno J.E., Tao Y., Chory J., Ballaré C.L. (2009). Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc. Natl. Acad. Sci. USA 106: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H., Watahiki M.K., Nakamoto D., Kinjo M., Yamamoto K.T. (2007). Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol. 144: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Calderon-Villalobos L.I., Prigge M., Peret B., Dharmasiri S., Itoh H., Lechner E., Gray W.M., Bennett M., Estelle M. (2009). Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. USA 106: 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Inzé D., Goossens A. (2009). Jasmonate-inducible gene: What does it mean? Trends Plant Sci. 14: 87–91 [DOI] [PubMed] [Google Scholar]
- Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inzé D., Goossens A. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe C., Springer A., Samol I., Reinbothe S. (2009). Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 276: 4666–4681 [DOI] [PubMed] [Google Scholar]
- Reymond P., Bodenhausen N., Van Poecke R.M.P., Krishnamurthy V., Dicke M., Farmer E.E. (2004). A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P., Weber H., Damond M., Farmer E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Okamoto H., Patrick E., Harris S.R., Wasternack C., Brearley C., Turner J.G. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco S.A., Hansson M., Scalf M., Walker J.M., Smith L.M., Vierstra R.D. (2009). Tandem affinity purification and mass spectrometric analysis of ubiquitylated proteins in Arabidopsis. Plant J. 59: 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen G.A., Pawlowski L., Wilensky S.E., Bender J. (2002). Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics 161: 1235–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suza W.P., Rowe M.L., Hamberg M., Staswick P.E. (2010). A tomato enzyme synthesizes (+)-7-iso-jasmonoyl-L-isoleucine in wounded leaves. Planta 231: 717–728 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Van Leene J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Van Leene J., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., Witters E., Inzé D., De Jaeger G. (2008). Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci. 13: 517–520 [DOI] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009). Auxin: A trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C., Kombrink E. (2010). Jasmonates: Structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem. Biol. 5: 63–77 [DOI] [PubMed] [Google Scholar]
- Xie D.-X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Sano R., Wada T., Takabayashi J., Okada K. (2009). Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136: 1039–1048 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]