This work uses proteomics and functional analyses to identify the pepper absicic acid–responsive gene (ABR1) that is required for cell death and defense responses against microbial pathogens. Nuclear localization of ABR1 in plant cells is essential for cell death induction associated with ABA and SA antagonism.
Abstract
Abscisic acid (ABA) is a key regulator of plant growth and development, as well as plant defense responses. A high-throughput in planta proteome screen identified the pepper (Capsicum annuum) GRAM (for glucosyltransferases, Rab-like GTPase activators, and myotubularins) domain-containing ABA-RESPONSIVE1 (ABR1), which is highly induced by infection with avirulent Xanthomonas campestris pv vesicatoria and also by treatment with ABA. The GRAM domain is essential for the cell death response and for the nuclear localization of ABR1. ABR1 is required for priming cell death and reactive oxygen species production, as well as ABA-salicylic acid (SA) antagonism. Silencing of ABR1 significantly compromised the hypersensitive response but enhanced bacterial pathogen growth and ABA levels in pepper. High levels of ABA in ABR1-silenced plants antagonized the SA levels induced by pathogen infection. Heterologous transgenic expression of ABR1 in Arabidopsis thaliana conferred enhanced resistance to Pseudomonas syringae pv tomato and Hyaloperonospora arabidopsidis infection. The susceptibility of the Arabidopsis ABR1 putative ortholog mutant, abr1, to these pathogens also supports the involvement of ABR1 in disease resistance. Together, these results reveal ABR1 as a novel negative regulator of ABA signaling and suggest that the nuclear ABR1 pool is essential for the cell death induction associated with ABA-SA antagonism.
INTRODUCTION
Plants mount diverse defense responses to survive attack by microbial pathogens. One of these defense reactions to pathogen attack is the hypersensitive response (HR), a form of programmed cell death (PCD), which is characterized by the rapid death of plant cells at the site of pathogen infection (Lam, 2004). The HR eventually leads to the dramatic restriction of pathogen growth at infected sites (Dangl and Jones, 2001). In incompatible interactions between plants and pathogenic microorganisms, host plants recognize the avirulence gene products of individual pathogens using specific receptors, resistance (R) gene products. These interactions cause changes in ion fluxes, a burst of reactive oxygen species (ROS), activation of defense-related genes, salicylic acid (SA) or jasmonic acid accumulation, callose deposition, and eventually HR-like cell death (Yasuda et al., 2008; Zipfel and Robatzek, 2010). In susceptible plants, however, microbial pathogens secrete effector proteins inside plant cells, which target pathogen-associated molecular pathogen (PAMP) receptors or downstream components of PAMP-triggered immunity to achieve full virulence (Hann et al., 2010). In turn, plants have evolved new strategies to surmount a range of weapons from effector armories, which has led to effector-triggered immunity (Jones and Dangl, 2006).
The gram-negative bacterial pathogen Xanthomonas campestris pv vesicatoria (Xcv), the causal agent of bacterial spot disease in pepper (Capsicum annuum) plants, is a good model bacterial pathogen to provide insight into the molecular mechanisms underlying plant cell death and defense against microbial invasions. Pepper R genes, such as Bs2 (Leister et al., 2005) and Bs3 (Römer et al., 2007), recognize the pathogen-derived avirulence (avr) genes avrBs2 and avrBs3 of Xcv, respectively, to elicit PCD, also referred to as the HR. Some defense-related genes in pepper, such as BPR1 (basic PR-1) (Kim and Hwang, 2000), SAR82 (SAR8.2) (Lee and Hwang, 2003), DEF1 (defensin) (Do et al., 2004), PO2 (peroxidase) (Choi et al., 2007), AMP1 (antimicrobial protein) (Lee et al., 2008), MNR1 (menthone reductase) (Choi et al., 2008), and LOX1 (lipoxygenase) (Hwang and Hwang, 2010) are rapidly and differentially induced in pepper plants during their incompatible interactions with the avirulent strain Bv5-4a of Xcv carrying avrBsT (Kim et al., 2010). Infection with strain Bv5-4a produces a strong HR-like cell death in pepper leaves (Lee and Hwang, 2003, 2005). Cell death in pepper leaves is accompanied by strong induction of the SA-dependent gene BPR1 and the accumulation of H2O2 (Lee and Hwang, 2005; Choi et al., 2007). Agrobacterium tumefaciens–mediated transient expression of pepper defense-related genes PO2 (Choi et al., 2007), CaM1 (calmodulin) (Choi et al., 2009), LOX1 (Hwang and Hwang, 2010), HIR1 (hypersensitive induced reaction protein) (Choi et al., 2011), and MBL1 (mannose binding lectin) (Hwang and Hwang, 2011) induces the cell death response in pepper or Nicotiana benthamiana.
In Arabidopsis thaliana, genome-wide analysis of gene expression showed that more than 1300 genes are responsive to abscisic acid (ABA) (Hoth et al., 2002). Some ABA-responsive proteins are closely related to PR proteins. The two small peptide (17 and 18 kD) acidic proteins of pea (Pisum sativum), ABA-responsive proteins 17 and 18 (ABR17 and ABR18), have deduced amino acid sequences similar to those of other pea disease resistance-responsive proteins, PR proteins in other species, and the major birch (Betula verrucosa) pollen allergen Betv1 (Iturriaga et al., 1994). Colditz et al. (2005) analyzed the root proteome of Medicago truncatula in response to infection by the oomycete root pathogen Aphanomyces euteiches and found that the moderate susceptibility of the M. truncatula A17 line is mirrored by the abundance levels of one group of ABA-responsive proteins (ABR17) of the PR-10 class. Members of this gene family were demonstrated to be induced in several plant–pathogen interactions (Jwa et al., 2001; McGee et al., 2001). Thus, abundant expression of ABA-responsive genes in plants during pathogen infection indicated that ABA-mediated signaling is involved in PR protein induction for disease resistance.
The GRAM (for glucosyltransferases, Rab-like GTPase activators, and myotubularins) domain is ubiquitous in glucosyltransferases, myotubularins, and other membrane-associated proteins in eukaryotes (Doerks et al., 2000; Jiang et al., 2008). This domain, which is ~70 amino acids in length, is predicted to function in intracellular protein binding or lipid binding during membrane-associated processes (Doerks et al., 2000). However, functions of most of these family member proteins are poorly understood. Barley (Hordeum vulgare) aba45 was the first plant gene demonstrated to encode an ABA-inducible protein with the GRAM domain sequence (Liu et al., 1999). In Arabidopsis, VASCULAR ASSOCIATED DEATH1 (VAD1), which encodes a GRAM domain–containing protein, is expressed in response to pathogen infection and is involved in cell death and defense responses in vascular tissues (Lorrain et al., 2004). Several Arabidopsis GRAM domain genes are also regulated in response to abiotic or biotic stresses (Jiang et al., 2008). Furthermore, a few rice genes in the GRAM domain family are upregulated by drought, salinity, Xanthomonas oryzae pv oryzae infection, and ABA treatment (Jiang et al., 2008). ABA plays a role in response to biotic stresses, and many GRAM domain genes are regulated by abiotic and biotic stresses; however, the relationship between the GRAM domain and ABA-mediated plant responses to biotic stresses remains to be clarified.
Here, we analyzed the proteome of pepper leaves inoculated with the Xcv virulent strain Ds1 or the avirulent strain Bv5-4a to identify defense-related proteins using two-dimensional (2D) gel electrophoresis, matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry, and liquid chromatography–tandem mass spectrometry (LC/MS-MS). Most of the novel proteins induced in pepper leaves by Xcv infection were associated with disease, virulence, and defense. Among the defense-related proteins identified was ABR1, a GRAM domain–containing ABA-responsive protein. In this study, we isolated and functionally characterized full-length ABR1 from the pepper cDNA library. Virus-induced gene silencing (VIGS) and Agrobacterium-mediated transient expression of ABR1 were performed in pepper plants. Transgenic Arabidopsis plants overexpressing ABR1 were also generated to determine whether the gain of function of this gene is required for basal resistance to plant pathogens. Here, we demonstrate that expression of ABR1 confers enhanced resistance to pathogen infection in pepper and transgenic Arabidopsis plants, which is accompanied by cell death, callose deposition, and ABA-SA antagonism. Our data also suggest that the GRAM domain is required to initiate the cell death response and localize ABR1 to the nuclei of plant cells.
RESULTS
Proteomics Analysis of Pepper Leaves Infected by Xcv
To investigate alterations in protein expression induced by bacterial infection, total soluble proteins were identified in pepper leaves 15 h after inoculation with the virulent (compatible) strain Ds1 and the avirulent (incompatible) strain Bv5-4a of Xcv using proteomics techniques. The entire 2D gel images of total proteins from healthy and infected leaves are shown in Supplemental Figure 1 online. The protein spot numbers and volumes among 2D gel images were reproducibly detected and the protein profiles of healthy and bacterial-infected leaves were compared. Overall, the levels of 46 proteins were altered by Xcv infection. Thirty-six proteins were newly induced or upregulated, while 10 were downregulated. To identify these proteins, protein spots excised from the gels were analyzed by the LC/MS-MS or MALDI-TOF techniques. The genomic sequence data analysis of all the differentially expressed proteins identified only 19 proteins from pepper leaves (Table 1) and three Xcv pathogen proteins (see Supplemental Table 1 online). Homologs of some of these proteins were identified in other plant species, and the identification of pepper proteins was very low due to the incomplete status of the pepper genome database.
Table 1.
Proteins Differentially Expressed in Pepper Leaves Infected by Virulent (Compatible) Strain Ds1 and Avirulent (Incompatible) Strain Bv5-4a of Xcv, as Identified by MALDI-TOF MS and LC/MS-MS
| Spot No. | Accession No. | Experimental pl/MM (kD) | Theoretical pI/MM (kD) | SC (%) | MS Used for Identification | Spot Density |
|||
| Protein (Organism) | Healthy | Compatible | Incompatible | ||||||
| Novel protein | |||||||||
| I. RNA synthesis | |||||||||
| N1 | RNA binding protein precursor (Persea americana) | CAD18921 | 4.29/33 | 4.49/32.9 | 10 | LC-MS | 0a | 0.1045b | 0.0644b |
| II. Disease, virulence, and defense | |||||||||
| N2 | Pathogenesis-related protein 10 (C. annuum) | AAF63519 | 5.31/24 | 5.2/17.38 | 16 | LC-MS | 0a | 0.0932a | 1.2633b |
| N3 | ABA-responsive protein (C. annuum) | CA524559 | 6.29/38 | 8.96/13.01 | 30 | LC-MS | 0a | 0.0171a | 0.5689b |
| N4 | Precursor β-1,3-glucanase (Nicotiana plumbaginifolia) | CAA38540 | 7.96 / 32 | 7.79/39.96 | 10 | LC-MS | 0a | 0.8030b | 0.8050b |
| N5 | Precursor β-1,3-glucanase (N. plumbaginifolia) | CAA38540 | 8.19/32 | 7.79/39.96 | 10 | LC-MS | 0a | 0.3080c | 0.0543b |
| N6 | Glucan endoglucosidase (Nicotiana tabacum) | 1410344A | 7.34/32 | 6.6/39.13 | 13 | LC-MS | 0a | 0.0776b | 0.0568b |
| N7 | Peroxidase (C. annuum) | AAL35364 | 7.47/34 | 6.88/36.05 | 26 | LC-MS | 0a | 0.2630b | 0.2770b |
| N8 | Peroxidase (C. annuum) | AAL35364 | 7.46/33 | 6.88/36.05 | 29 | LC-MS | 0a | 0.0123a | 0.1020b |
| N9 | Endochitinase (C. annuum) | AAY90154 | 6.99/31 | 7.7/19.13 | 58 | LC-MS | 0a | 0.0199b | 0.0497c |
| N10 | Osmotin-like protein precursor (Pathogenesis-related protein PR-5d) (N. tabacum) | P25871 | 7.00/26 | 6.49/27.63 | 5 | LC-MS | 0a | 0.1150b | 0.0900b |
| N11 | TMV induced protein 1-2 (C. annuum) | AAO49266 | 7.00/20 | 5.5 / 17.87 | 51 | LC-MS | 0a | 0.0424b | 0.1130c |
| Upregulated protein | |||||||||
| I. C-compound and carbohydrate metabolism | |||||||||
| U1 | Acetyl-CoA carboxylase (Triticum aestivum) | AAC49275 | 6.48/21 | 6/252.86 | 6 | MALDI-TOF | 0.1580a | 0.4973b | 0.4288b |
| II. Photosynthesis | |||||||||
| U2 | Light-harvesting chlorophyll a/b binding protein (Nicotiana sylvestris) | BAA25388 | 4.95/29 | 5.2/28.29 | 13 | MALDI-TOF | 0.0494a | 0.1038b | 0.1642c |
| III. Extracellular/secretion protein | |||||||||
| U3 | Thioredoxin (C. annuum) | AAR83852 | 4.91/16 | 5.01/13.71 | 20 | LC-MS | 0.0821a | 0.1646c | 0.0119b |
| IV. Disease, virulence, and defense | |||||||||
| U4 | Putative pathogenesis-related protein (Capsicum chinensis) | CAI48023 | 5.22/21 | 5.22/17.27 | 18 | LC-MS | 0.0649a | 0.7988b | 0.7090b |
| V. Unclassified protein | |||||||||
| U5 | Chloroplast protease (C. annuum) | CAA09935 | 5.05/81 | 5.8/74.15 | 26 | MALDI-TOF | 0.0593a | 0.1270b | 0.1462b |
| U6 | Chloroplast protease (C. annuum) | CAA09935 | 5.29/81 | 5.8/74.15 | 39 | MALDI-TOF | 0.0732a | 0.2013b | 0.1617b |
| U7 | JH02012D09 JH-201 C. annuum cDNA 5′, mRNA sequence (C. annuum) | CO776410 | 5.03/37 | 9.65/16.29 | 20 | LC-MS | 0.2840a | 0.4007b | 0.5081b |
| Downregulated protein | |||||||||
| I. C-compound and carbohydrate metabolism | |||||||||
| D1 | Glyceraldehyde-3-phosphate dehydrogenase (C. annuum) | CAC80374 | 6.26/47 | 6.3/33.13 | 21 | MALDI-TOF | 0.1470a | 0.0493b | 0.0866c |
Spot density is expressed as the percentage volume to correct the variability due to silver staining, which is calculated by ImageMaster 2D Platinum 6.0 (Amersham Biosciences). Significant differences were determined by Student's t test (P ≤ 0.05, labeled a to c). SC, sequence coverage; MM, molecular mass.
Pepper and Bacterial Proteins Differentially Expressed Following Xcv Infection
The SOLANACEAE EST Analysis System (http://sol.pdrc.re.kr/) was used to functionally categorize pepper proteins that were identified. Arabidopsis putative orthologs of the identified pepper proteins were used to functionally categorize the pepper proteins into six classes based on the Munich Information Center for Protein Sequences Arabidopsis thaliana genome database (MatDB, http://mips.gsf.de/proj/funcatDB/search_main_frame.html): C-compound and carbohydrate metabolism (5.5%), photosynthesis (5.5%), RNA synthesis (5.5%), extracellular/secretion (5.5%), disease, virulence, and defense (61%), and unclassified proteins (17%) (see Supplemental Figure 2 online).
Most of the novel proteins induced in pepper leaves during Xcv infection belong to the disease, virulence, and defense class (Table 1). These defense-related proteins include pathogenesis-related protein 10 (PR-10, N2; Shin et al., 2001), an ABA-responsive protein (N3), precursor β-1,3-glucanase (N4; Gheysen et al., 1990), glucan endoglucosidase (N6), peroxidase (N7; Do et al., 2003), endochitinase (N9), osmotin-like protein precursor (N10), and tobacco mosaic virus–induced protein 1-2 (N11) (see Supplemental Figures 3 and 4 online). In particular, PR protein 10 (N2) and the C. annuum ABA-responsive protein (N3) were strongly induced in pepper leaves during the incompatible interaction with Xcv. Among them, we selected this ABA-responsive protein for further analysis (see Supplemental Figures 5 and 6 online).
Some bacterial proteins were expressed in pepper leaves during Xcv infection. Three bacterial proteins were identified by MALDI-TOF in this study (see Supplemental Table 1 online). However, all bacterial proteins were found to be derived from other bacterial species.
Isolation and Sequence Analysis of Pathogen-Induced ABR1 cDNA
A protein newly induced by Xcv infection was identified as KS12040A12 KS12 C. annuum cDNA (N3 [EST clone], Table 1; see Supplemental Figure 5 online). We also previously isolated a full-length KS12040A12 KS12 C. annuum cDNA encoding the protein from a pepper cDNA library using a differential hybridization technique (Jung and Hwang, 2000). This full-length cDNA sequence has a GRAM domain also present in glucosyltransferases, myotubularins, and other putative membrane-associated proteins (see Supplemental Figure 6 online; Doerks et al., 2000), which are evolutionarily conserved in eukaryotes (Jiang et al., 2008). The isolated pepper GRAM domain–containing protein shares 51 to 66% sequence identities with uncharacterized grapevine (Vitis vinifera) protein products (accession numbers XP_002263365 and XP_002263309), and Arabidopsis (NP_196824), rice (Oryza sativa; ABA98234), and barley (AAD09343) ABA-responsive proteins (see Supplemental Figure 7A online). Therefore, we named this novel pathogen-induced gene the C. annuum ABA-responsive protein (ABR1) gene. A phylogenetic tree of pepper ABR1 and ABA-responsive proteins was constructed based on GRAM domain sequences derived from several plant species (see Supplemental Figure 7B and Supplemental Data Set 1 online). ABR1 is more similar to a group of grapevine GRAM domain–containing proteins than to rice GRAM domain–containing proteins (see Supplemental Figure 7A online). The two GRAM domain–containing proteins of grapevine are included in group II, although they are highly similar to ABR1, which falls into group 1.
Expression Pattern of ABR1
To identify the ABR1 protein spot separated on 2D gels, immunoblot analysis was performed with a specific antiserum raised against a peptide of the ABR1 protein (Figure 1A). The one-dimensional (1D) and 2D analyses identified the N3 spot as the ABR1 protein. However, other ABR1 isoforms were not detected on the 2D gels by immunoblotting.
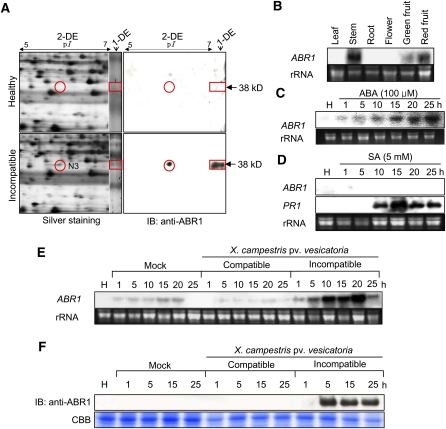
Figure 1.
RNA Gel Blot, 2D, and Immunoblot Analyses of the Expression of the ABR1 Protein and Gene in Pepper Leaves Infected by Xcv or Treated with ABA and SA.
(A) Identification of the ABR1 protein by 1D and 2D electrophoresis and immunoblot analysis. The red circles and rectangles indicate ABR1 expression. IB, imunnoblotting.
(B) Organ-specific expression of ABR1 in pepper plants. rRNA is used as loading control ([B] to [E]).
(C) Expression of ABR1 in leaves at various times after treatment with 100 μM ABA. H, healthy leaves.
(D) Expression of ABR1 and PR1 in leaves at various times after treatment with 5 mM SA. Pepper basic pathogenesis-related protein gene (PR1) was used as a comparable control. H, healthy leaves.
(E) Expression of the ABR1 gene in pepper leaves at various times after inoculation with the virulent (compatible) strain Ds-1 and the avirulent (incompatible) strain Bv5-4a of Xcv. H, healthy leaves.
(F) Immunoblot analysis of expression of ABR1 protein in leaves at various times after inoculation with the Ds-1 and Bv5-4a strains of Xcv. Immunoblotting used a specific antiserum raised against an ABR1 peptide. H, healthy leaves; IB, imunnoblotting; CBB, Coomassie blue.
[See online article for color version of this figure.]
RNA gel blot analysis of ABR1 expression in pepper plants showed that ABR1 transcripts were present in stems and green and red fruits, but not in leaves, roots, and flowers (Figure 1B). RNA gel blot analysis also revealed that ABR1 was induced in pepper leaves upon ABA treatment but not in untreated plants (Figure 1C). However, ABR1 was not induced by SA treatment, which induced the pepper PATHOGENESIS-RELATED PROTEIN1 (PR1) gene that was used as a comparable control (Figure 1D). As shown in Figure 1E, however, ABR1 was strongly expressed in leaves inoculated with the Xcv avirulent strain Bv5-4a. ABR1 transcripts were induced at an early infection time and increased until 20 h after inoculation. Immunoblot analysis demonstrated expression of ABR1 during avirulent Xcv infection but not during virulent Xcv infection (Figure 1F). These results indicate that ABR1 is induced in pepper plants during the HR to Xcv infection, as well as in response to ABA exposure.
Agrobacteruim-Mediated Transient Expression of ABR1 Induces Cell Death Response
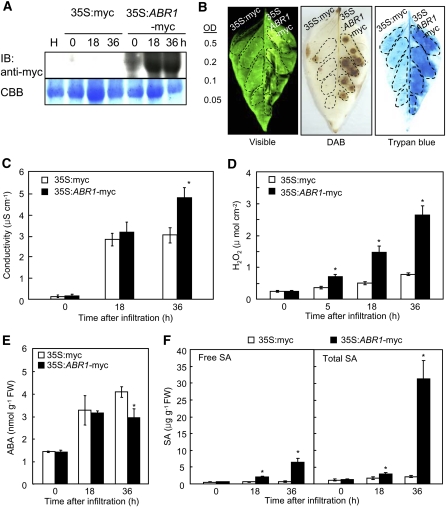
A transient expression experiment using an Agrobacterium infiltration system was conducted to define the roles of ABR1 in pepper leaves. Immunoblot analysis showed that the myc-tagged ABR1 protein was expressed in pepper leaves (Figure 2A). Transient expression of ABR1 in pepper triggered a rapid cell death response 36 h after agroinfiltration (Figure 2B). Cell death was measured by electrolyte leakage from leaf discs infiltrated with Agrobacterium-containing empty vector (35S:myc) or 35S:ABR1-myc constructs (OD600 = 0.2) (Figure 2C). Overexpression of ABR1 in pepper leaves enhanced electrolyte leakage, indicating that ABR1 is involved in the induction of hypersensitive cell death in plants. In addition, Agrobacterium-mediated transient expression of ABR1 in pepper leaves induced H2O2 production at the infiltrated site, as determined by diaminobenzidine (DAB) staining and a ferrous ammonium sulfate/xylenol orange assay (Figures 2B and 2D). H2O2 from oxidative bursts is known to drive programmed cell death at challenged sites (Levine et al., 1994; Torres et al., 2005). A significantly enhanced oxidative burst was induced by transient expression of ABR1; however, H2O2 accumulated to a much smaller extent in the empty vector control (Figure 2D). The endogenous levels of ABA and SA in pepper leaves after agroinfiltration with ABR1 were also analyzed. As expected, transient expression of ABR1 significantly suppressed ABA levels (Figure 2E) but enhanced SA levels (Figure 2F). Taken together, these results indicate that transient expression of ABR1 triggers cell death, accompanied by an oxidative burst and an antagonistic ABA and SA interaction.
Figure 2.
Effects of Agrobacterium-Mediated Transient Epression of ABR1 on the Cell Death Response of Pepper Leaves.
(A) Immunoblot analysis of transiently expressed myc-ABR1 in leaves at different time points after agroinfiltration. An anti-myc antibody was used to detect myc-ABR1 on the immunoblot. IB, imunnoblotting.
(B) Induction of the cell death response by transient expression of ABR1. The leaf areas between the lateral veins received the indicated Agrobacterium strain at the indicated OD600. Photographs were taken 36 h after agroinfiltration. The experiment was performed three times with similar results. Visible, visible light image; trypan blue, trypan blue staining.
(C) Electrolyte leakage from leaf discsagroinfiltrated with the empty vector control and the ABR1 transient expression construct (OD600 = 0.2).
(D) Accumulation of H2O2 in leaves transiently expressing ABR1.
(E) and (F) Quantification of ABA (E) and SA (F) levels in empty vector control leaves and in leaves transiently expressing ABR1 after agroinfiltration (OD600 =0 .5). FW, fresh weight. Data are the means ± sd (n = 3) from three independent experiments. Asterisks indicate a significant difference, as determined by the two-tailed t test (P < 0.05) ([C] to [F]).
[See online article for color version of this figure.]
The GRAM Domain Is Required to Initiate the Cell Death Response and to Localize ABR1 to the Nucleus
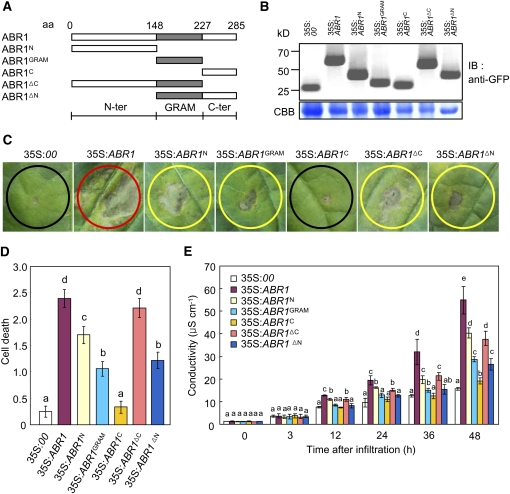
To further investigate whether the GRAM domain is crucial for PCD, we generated N-terminal and C-terminal deletion constructs of ABR1 under the 35S promoter (Figure 3A). Transient expression of these constructs was detected in pepper leaves by immunoblot analysis (Figure 3B). Agrobacterium-mediated transient expression of all deletion fragments induced a cell death response in pepper leaves, except for the ABR1C deletion mutant (residues 227 to 285) (Figures 3C and 3D). However, full-length ABR1 was the most effective in triggering cell death at the infiltrated site. All the deletion mutants containing the GRAM domain induced the cell death response. Cell death events were monitored over the subsequent 2 d, and visual evidence of PCD was substantiated by an electrolyte leakage analysis (Figure 3E). These results support our findings that the GRAM domain is required for the cell death response in pepper leaves.
Figure 3.
Deletion Analysis of the GRAM Domain.
(A) Schematic of ABR1 structures used in the cell death assay and for protein localization. aa, amino acids.
(B) Expression of the ABR1 variant deletion proteins in pepper leaves, as detected by immunoblotting using an anti-GFP antibody. IB, imunnoblotting; CBB, Coomassie blue.
(C) Development of cell death responses in pepper leaves caused by infiltrating Agrobacterium (OD600 = 0.5) strains carrying different ABR1 constructs. Agrobacterium carrying an empty vector (35S:00) was used as a control. Red, yellow, and black circles indicate full, partial, and no cell death, respectively.
(D) The extent of ABR1 construct-induced PCD is classified with the following scales: 0, no PCD (<10%); 1, weak PCD (10 to 30%); 2, partial PCD (30 to 80%); and 3, full PCD (80 to 100%). The experiments were performed three times with similar results.
(E) Electrolyte leakages from leaf discs infiltrated by Agrobacterium (OD600 = 0.5) strains carrying different ABR1 constructs.
Error bars represent ± sd (n = 3) from three independent experiments and different letters (a to e) indicate significant differences, as determined by Fisher's protected least significant difference (LSD) test (P < 0.05) ([D] and [E])
[See online article for color version of this figure.]
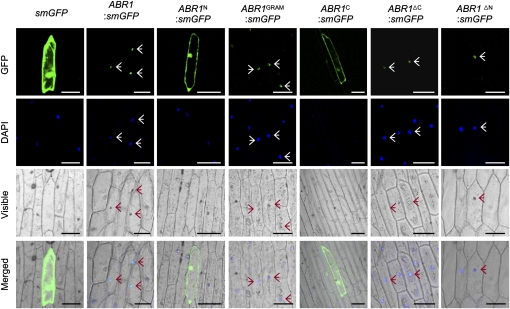
To determine the subcellular localization of ABR1, the soluble-modified green fluorescence protein (smGFP) was fused in frame to the C termini of several ABR1 deletion fragments. smGFP was used as a control. The transiently overexpressed ABR1:smGFP fusion protein localized exclusively to the nucleus in onion epidermal cells (Figure 4). Subsequent 4',6-diamidino-2-phenylindole (DAPI) staining confirmed nuclear localization. GRAM domain fusion constructs, including ABR1GRAM:smGFP, ABR1ΔC:smGFP, and ABR1ΔN:smGFP, were targeted to the nuclei; however, ABR1N:smGFP and ABR1C:smGFP, which lack the GRAM domain, were expressed throughout the cytoplasm. These results indicate that the GRAM domain is essential for the localization of ABR1 to plant nuclei.
Figure 4.
Subcellular Localization of ABR1.
GFP fusions of full- or partial-length ABR1 were transiently transformed into onion epidermal cells. The overall schematic structures of each construct are shown in Figure 3A with the addition of a GFP fusion motif at the 3′ termini. The plant nuclei were stained with DAPI. Images were taken using confocal microscopy (GFP fluorescence, green; DAPI fluorescence, blue; visible, visible light image; merged, merged images of above three images). Empty vector (smGFP) transformed cells are shown as a control. Arrows indicate ABR1-localized nuclei. Bar = 100 μm.
[See online article for color version of this figure.]
Nuclear Localization of ABR1 Is Essential for Cell Death Induction and Hormone Regulation
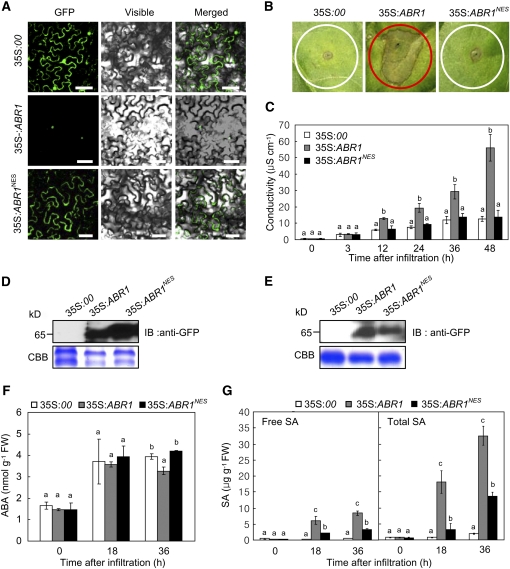
To determine the functional role of the nuclear ABR1 pool, we fused a nuclear export sequence (NES) to the C terminus of ABR1 (Wen et al., 1995). Transient expression of the 35S:ABR1NES-GFP construct revealed an absence of nuclear fluorescence signals in epidermal cells of N. benthamiana, despite clearly visible cytoplasmic GFP fluorescence (Figure 5A). Consistent with the nuclear localization in onion epidermal cells (Figure 4), ABR1 localized to the nuclei in N. benthamiana. Subsequent experiments of pepper transient expression show that ABR1 export from the nuclei inhibits the ABR1-induced cell death response (Figure 5B). Quantitative analysis of electrolyte leakage also supported drastic cell death reduction in ABR1NES-expressed pepper leaves (Figure 5C). Transient expression of both 35S:ABR1-GFP and 35S:ABR1NES-GFP was detected in leaves of N. benthamiana and pepper by immunoblot (Figures 5D and 5E). We next tested whether transient expression of ABR1NES affects the levels of plant hormones ABA and SA in pepper leaf tissues. When transiently expressed in pepper leaves, ABR1 and ABR1NES expression had a similar effect on ABA induction, except that ABA induction was lower in plants transiently expressing ABR1 36 h after agroinfiltration (Figure 5F). By contrast, ABR1 transient expression induced significantly higher free SA and total SA (free SA plus glycoside-conjugated SA) levels than the transient expression of ABR1NES (Figure 5G). Together, these data indicate that nuclear localization of ABR1 is required for cell death induction, which is associated with ABA and SA antagonism in plants.
Figure 5.
The Function of ABR1 Is Dependent on Nuclear Localization.
(A) Localization of ABR1- and ABR1NES-GFP fusion proteins in N. benthamiana leaves as visualized by confocal microscopy. GFP, GFP fluorescence; visible, visible light images; merged, merged images of GFP and visible light images. Bars = 50 μm.
(B) The ABR1- and ABR1NES-GFP–mediated cell death response in pepper leaves.
(C) Quantification of ABR1- and ABR1NES-GFP–mediated cell death by electrolyte leakage measurement from leaf discs.
(D) Transient expression of ABR1- and ABR1NES-GFP in N. benthamiana leaves as detected by immunoblotting.
(E) Transient expression of ABR1- and ABR1NES-GFP in pepper leaves as detected by immunoblotting.
(F) and (G) Quantification of ABA (F) and SA (G) levels in the empty vector control (35S:00) and pepper leaves transiently expressing ABR1- and ABR1NES-GFP after agroinfiltration (OD600 = 0.5). FW, fresh weight. Data are means ± sd (n = 3) from three independent experiments. Different letters indicate significant differences, as determined by Fisher's protected LSD test (P < 0.05) ([C], [F], and [G]).
[See online article for color version of this figure.]
VIGS of ABR1 in Pepper Plants
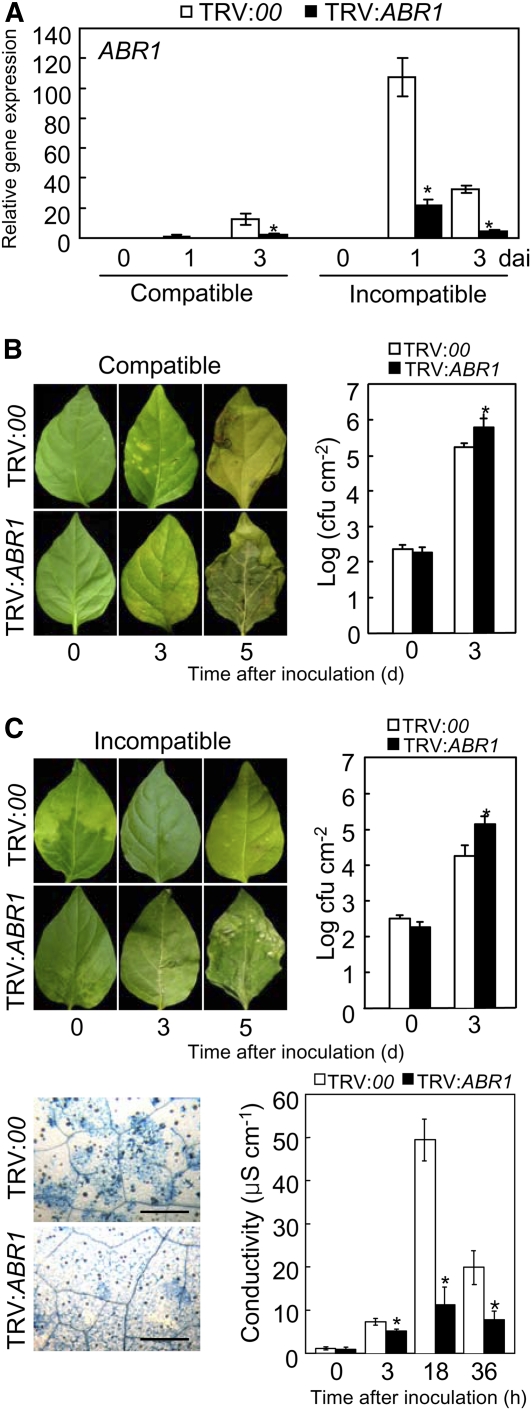
To determine the ABR1 loss-of-function phenotype in pepper leaves, VIGS was performed using recombinant tobacco rattle virus (TRV) silencing constructs (Liu et al., 2002) containing the full-length ABR1 open reading frame. A quantitative RT-PCR analysis showed that ABR1 transcripts were significantly reduced in ABR1-silenced pepper leaves during Xcv infection (Figure 6A), indicating that ABR1 was effectively silenced in pepper. Immunoblot analysis also confirmed that ABR1 silencing compromised ABR1 expression at the protein level in pepper leaves (see Supplemental Figure 8 online).
Figure 6.
Enhanced Susceptibility of ABR1-Silenced Pepper Leaves to Xcv Infection.
(A) Relative expression of ABR1 transcript using real-time RT-PCR analysis. dai, days after inoculation.
(B) and (C) Disease symptoms induced on leaves of empty vector control (TRV:00) or ABR1-silenced (TRV:ABR1) pepper plants 0, 3, and 5 d after inoculation (dai) with the Xcv virulent (compatible) strain Ds1 ([B], left panel) and avirulent (incompatible) strain Bv5-4a ([C], left panel) (106 colony-forming units [cfu] mL−1). Bacterial growth in leaves inoculated with strain Ds1 ([B], right panel) and strain Bv5-4a ([C], right panel) (104 cfu mL−1). Trypan blue staining of leaves ([C], bottom left panel) and electrolyte leakage from leaf discs ([C], bottom right panel) of empty vector and ABR1-silenced plants inoculated with strain Bv5-4a (107 cfu mL−1). Error bars indicate sd (n = 3) from three independent experiments. Asterisks indicate a significant increase in bacterial growth and electrolyte leakage, as determined by the two-tailed t test (P < 0.05) ([A] to [C]). Bar = 500 μm.
[See online article for color version of this figure.]
Silencing of the ABR1 gene conferred enhanced susceptibility during compatible and incompatible interactions of pepper plants with Xcv (Figures 6B and 6C). Growth of the virulent strain Ds1 and the avirulent strain Bv5-4a of Xcv was significantly increased in ABR1-silenced plants compared with that in empty vector plants. Rapid cell death was compromised in ABR1-silenced leaves by avirulent Xcv Bv5-4a infection (Figure 6C, left). Electrolyte leakage from leaf discs of empty vector and ABR1-silenced pepper plants was measured to evaluate damage to plasma membranes caused by HR-like cell death (Figure 6C, right). During infection by avirulent Xcv Bv5-4a, electrolyte leakage levels from ABR1-silenced leaves was significantly lower than that from empty vector leaves, supporting the hypothesis that transient expression of ABR1 induces cell death in pepper leaves (Figure 3C). These data indicate that the ABR1 gene plays a crucial role in basal defense and the HR-associated resistance of pepper plants to Xcv infection.
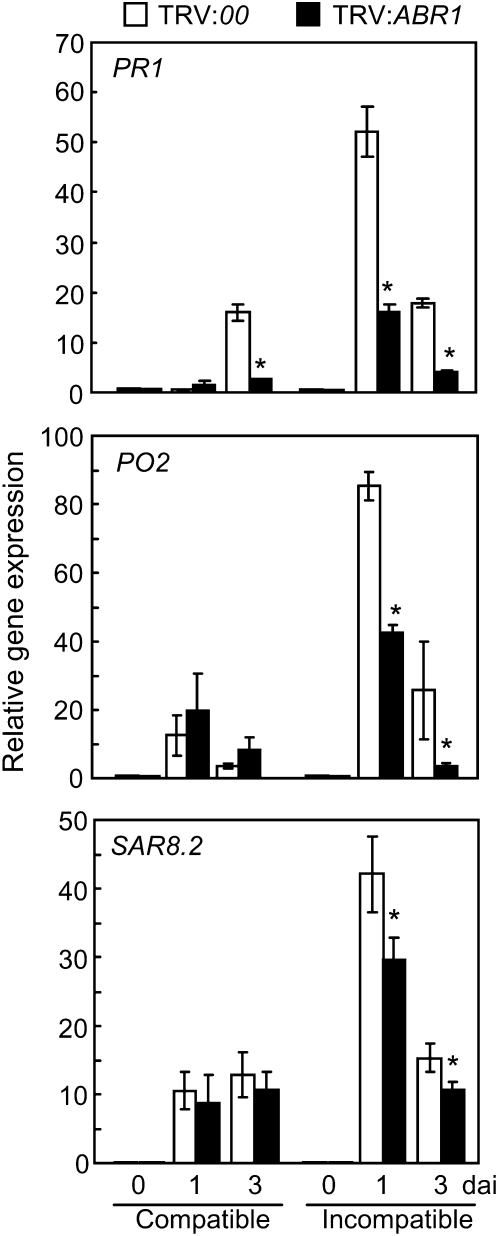
Quantitative RT-PCR was used to determine whether ABR1 silencing affects the expression of defense-related genes in pepper leaves during infection (Figure 7). The defense-related genes BPR1 (basic PR-1) (Kim and Hwang, 2000), SAR82 (SAR8.2) (Lee and Hwang, 2003), and PO2 (peroxidase) (Do et al., 2003) were significantly downregulated in ABR1-silenced leaves during avirulent Xcv infection. These results indicate that ABR1 silencing compromises the induction of these defense-related genes by Xcv infection, thus leading to disease susceptibility of pepper plants.
Figure 7.
Real-Time RT-PCR Analysis of Defense Marker Gene Expression in Empty Vector (TRV:00) and ABR1-Silenced (TRV:ABR1) Pepper Plants.
PR1, pepper basic PR-1; PO2, peroxidase; SAR82, SAR8.2. Data are the means ± sd from three independent experiments. Asterisks indicate significant differences in gene expression, as determined by the two-tailed t test (P < 0.05). dai, days after inoculation.
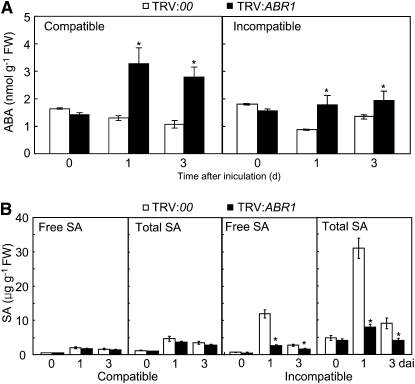
Interplay and antagonism between ABA and SA may be crucial for basal and induced resistance in plants (Flors et al., 2008; Spoel and Dong, 2008; de Torres-Zabala et al., 2009). To investigate the effect of endogenous ABA and SA on pepper disease resistance, we analyzed ABA and SA levels in leaves of empty vector and ABR1-silenced pepper plants infected with Xcv strains Ds1 and Bv5-4a (Figure 8). As shown in Figure 8A, ABA levels drastically increased in the ABR1-silenced leaves during Xcv infection. By contrast, silencing of ABR1 significantly reduced SA levels in pepper leaves during Xcv infection, especially in incompatible interactions (Figure 8B). These results suggest that the enhanced disease susceptibility in ABR1-silenced plants may be largely attributed to the decreased SA level but also to the elevated ABA level.
Figure 8.
Quantification of ABA and SA Levels in VIGS Plants.
Data are the means ± sd from three independent experiments. Asterisks indicate significant differences in ABA (A) and SA (B) levels, as determined by the two-tailed t test (P < 0.05). Total SA, free SA plus its glucoside (SAG); FW, fresh weight.
Enhanced Disease Resistance of ABR1-OX Transgenic Arabidopsis
Because it is difficult to transform pepper plants, we generated transgenic plants overexpressing pepper ABR1 in the Arabidopsis ecotype Columbia (Col-0) using the floral dipping method (Clough and Bent, 1998). Arabidopsis T3 progeny selected in the presence of kanamycin were not distinctly different in morphology and development from untransformed plants. The ABR1-OX (overexpression) lines #1, #2, and #3, which constitutively express the ABR1 protein (Figure 9A), were selected for further experiments. Notably, ABR1 overexpression induced a spontaneous cell death response in transgenic leaves (Figure 9B).
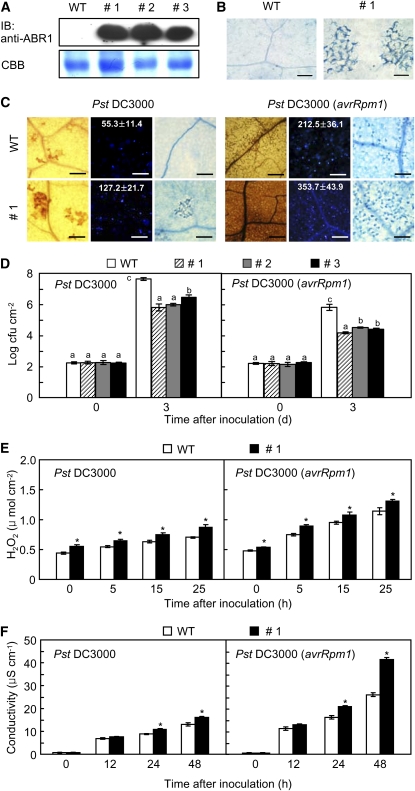
Figure 9.
Enhanced Resistance of ABR1-OX Transgenic Arabidopsis Plants to Pst Infection.
(A) Constitutive protein expression of ABR1 in three lines (#1, #2, and #3) of transgenic plants by immunoblotting (IB). WT, wild type.
(B) Spontaneous cell death response in the transgenic leaf tissue. Bars = 100 μm.
(C) DAB (1 dai, left), aniline blue (1 dai, middle), and trypan blue staining (2 dai, right) of leaves of wild-type and ABR1-OX plants inoculated with Pst DC3000 and Pst DC3000 (avrRpm1) (107 cfu mL−1). The number of calloses per mm2 is represented with the means ± sd (n = 3) in the box. Bars = 100 μm.
(D) Bacterial growth in leaves of wild-type and ABR1-OX plants inoculated with Pst DC3000 and Pst DC3000 (avrRpm1) (5 × 104 cfu mL−1). Data are the means ± sd (n = 3) from three independent experiments. Different letters and asterisks indicate a significant difference, as determined by Fisher’s protected LSD test (P < 0.05) and the two-tailed t test (P < 0.05), respectively ([D] to [F]). (E) Accumulation of H2O2 in ABR1-OX transgenic Arabidopsis.
(F) Electrolyte leakage from seven leaf discs (7 mm in diameter) of wild-type and ABR1-OX transgenic Arabidopsis plants.
[See online article for color version of this figure.]
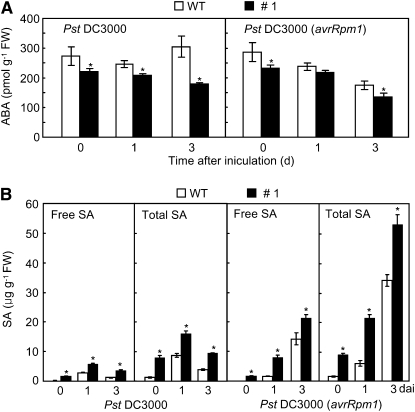
ABR1-OX transgenic Arabidopsis plants exhibited enhanced resistance to virulent or avirulent Pseudomonas syringae pv tomato (Pst) DC3000 infection (Figures 9C and 9D). Wild-type plants showed progressive susceptible chlorotic symptoms on inoculated leaves, whereas disease symptoms were not visible on the leaves of the tested transgenic lines 3 d after inoculation. The ABR1-OX transgenic lines #1, #2, and #3 exhibited a significant reduction in bacterial growth 3 d after inoculation with Pst DC3000 (Figure 9D). The growth of Pst DC3000 (avrRpm1) was also remarkably decreased in the leaf tissues of all tested transgenic lines. Infection with Pst DC3000 resulted in a small increase in H2O2 production, whereas Pst DC3000 (avrRpm1) infection drastically increased H2O2 accumulation in leaves of ABR1-OX plants (Figures 9C and 9E). We further tested whether expression of ABR1 induces cell death in ABR1-OX leaves. Cell death was gradually induced in ABR1-OX leaves infected by Pst DC3000. Strikingly, overexpression of ABR1 in Arabidopsis conferred enhanced cell death during avirulent Pst infection (Figures 9C and 9F). Avirulent Pst infection drastically induced electrolyte leakage from ABR1-OX leaves compared with that in wild-type leaves (Figure 9F). We also measured the levels of endogenous ABA in leaves of the two Arabidopsis genotypes (Figure 10A). During disease development, ABA levels significantly declined in leaves of ABR1-OX transgenic Arabidopsis infected by virulent and avirulent Pst strains compared with those in wild-type plants. By contrast, both virulent and avirulent Pst infection caused accumulation of significant amounts of free SA and total SA (free SA plus Glc-conjugated SA) in the leaves of ABR1-OX transgenic Arabidopsis (Figure 10B).
Figure 10.
Alteration of ABA and SA Levels in ABR1-OX Transgenic Arabidopsis Plants.
Endogenous ABA (A) and SA (B) levels in leaves of wild-type (WT) and ABR1-OX (line #1) plants inoculated with Pst DC3000 and Pst DC3000 (avrRpm1) (5× 104 cfu mL−1). Data are the means ± sd from three independent experiments. Asterisks indicate significant differences in ABA and SA levels, as determined by the two-tailed t test (P < 0.05). Total SA, free SA plus its glucoside (SAG); FW, fresh weight.
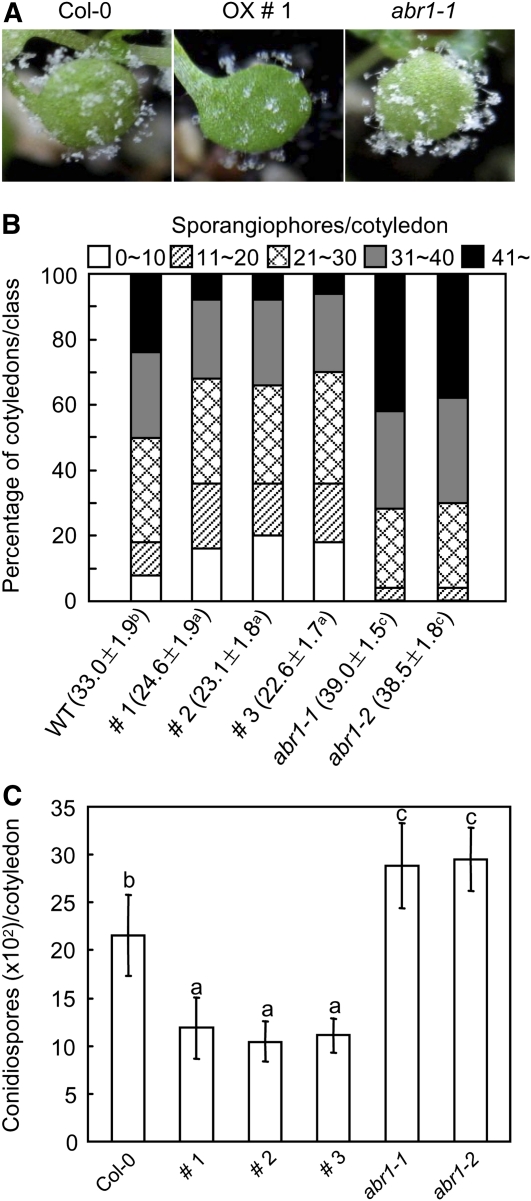
To determine whether transgenic Arabidopsis plants expressing ABR1 are resistant to biotrophic oomycete pathogens, we inoculated wild-type (Col-0) and ABR1-OX transgenic seedlings with Hyaloperonospora arabidopsidis isolate Noco2. Five days after inoculation, the production of sporangiophores on transgenic seedlings was greatly reduced compared with that on wild-type Arabidopsis (Figure 11A). To assess disease severity, we scored the numbers of sporangiophores and conidiospores per cotyledon 7 d after inoculation (Figures 11B and 11C). These levels were significantly lower in transgenic plants than in wild-type plants. These data indicate that ABR1 overexpression is sufficient to enhance the resistance of Arabidopsis to downy mildew disease.
Figure 11.
Responses of Arabidopsis ABR1-OX Plants and ABA-Responsive Protein-Like abr1 Mutants to Infection with H. arabidopsidis Isolate Noco2.
(A) Visual images of diseased cotyledons 7 d after inoculation.
(B) Quantification of asexual sporangiophore formation on cotyledons 7 d after inoculation. The numbers at the bottom indicate the mean sporagiophores/cotyledon ± sd from three independent experiments. Different letters above sd indicate significant difference, as determined by Fisher’s protected LSD test (P < 0.05). The number of sporagiophores per cotyledon was determined and cotyledons were classified into five scales: 0 to 10, 11 to 20, 21 to 30, 31 to 40, and >40. WT, wild type.
(C) Numbers of conidiospores produced on >50 cotyledons 7 d after inoculation. Statistical analyses were performed using the LSD test. Different letters above the bars indicate significantly different means (P < 0.05).
[See online article for color version of this figure.]
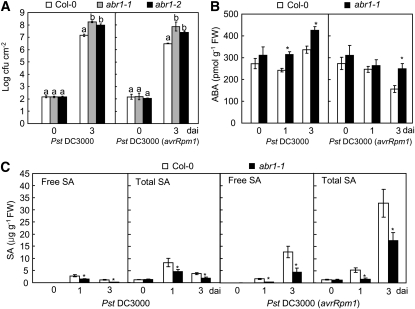
Enhanced Disease Susceptibility of the Arabidopsis T-DNA Insertion Mutant abr1
To determine whether the Arabidopsis putative orthologs of ABR1 contribute to susceptibility to Pst and H. arabidopsidis infection, we tested the Arabidopsis ABA-responsive–related protein gene (At ABR1, NP_196824), which shares 63% sequence identity with Ca ABR1 (see Supplemental Figure 7 online). The T-DNAs inserted into the homozygous Arabidopsis mutant abr1 are located in the first exon (abr1-1, GK-783H10) and in the 3′ untranslated region near the termination codon (abr1-2, SALK_017675) (see Supplemental Figure 9A online). RT-PCR was used to determine the effect of T-DNA insertion on the mRNA levels of Arabidopsis ABR1. At ABR1 was expressed in wild-type Col-0 plants but not in abr1 homozygous lines (see Supplemental Figure 9B online). We first examined bacterial growth in leaves of wild-type and abr1 loss-of-function mutant lines during Pst infection (Figure 12A). As expected, the cotyledons of abr1 mutants were more susceptible to Pst DC3000 or DC3000 (avrRPM1) infection than were wild-type cotyledons. As shown in Figure 12B, enhanced ABA levels were detected in abr1 mutants upon Pst challenge, indicating that a high level of ABA is required for Pst infection in abr1 mutants. In contrast with the high ABA levels in abr1 plants, SA significantly declined in abr1 plants upon Pst infection, compared with that in wild-type plants (Figure 12C). These data support the proposed antagonism between ABA and SA in disease resistance against bacterial pathogens.
Figure 12.
Responses of Arabidopsis ABA-Responsive Protein-Like abr1 Mutants to Pst Infection.
Bacterial growth (A) and endogenous ABA (B) and SA (C) levels in leaves of wild-type and abr1 plants inoculated with Pst DC3000 and Pst DC3000 (avrRpm1) (5× 104 cfu mL−1). Error bars indicate ± sd (n = 3) from three independent experiments. Different letters above the bars indicate significant difference, as determined by Fisher's protected LSD test (P < 0.05). dai, days after inoculation.
To investigate the response of abr1 mutants to a biotrophic oomycete pathogen, we inoculated abr1 mutant seedlings with H. arabidopsidis isolate Noco2 (Figure 11). While ABR1-OX transgenic Arabidopsis plants were more resistant than wild-type plants, abr1 mutants produced higher numbers of sporangiophores. Measurement of the numbers of sporangiophores and conidiospores per cotyledon 7 d after inoculation (Figures 11B and 11C) showed that the levels of these spores were significantly higher in abr1 mutants than in wild-type plants. These results indicate that the Arabidopsis putative ortholog of the pepper ABR1 gene, ABR1, may contribute to disease resistance.
DISCUSSION
Proteomics Analysis and Identification of Pepper Defense-Related Proteins
In this work, several pathogen-inducible proteins that were differentially expressed in pepper leaves infected by avirulent Xcv were identified using a proteomics approach. Despite a number of distinct proteins on 2D gels, the incomplete genome sequence limited the identification of pepper proteins. Therefore, protein spots of interest were identified on 2D gels using LC/MS-MS as well as MALDI-TOF. In general, the matching rate for LC/MS-MS data is higher than that for MALDI-TOF data (van Wijk, 2001). As expected, most of the novel proteins induced in pepper leaves by Xcv infection were involved in disease, virulence, and defense. The proteins that were specifically expressed in pepper leaves infected by Xcv are likely to be valuable as defense marker proteins.
The Pepper ABA-Responsive Protein ABR1 Contains the GRAM Domain Required for Nuclear Localization and the Cell Death Response
Among the proteins identified in this proteomics study, we analyzed the biological function of a pepper ABA-responsive protein that is encoded by ABR1. ABR1 was strongly induced in pepper leaves by avirulent Xcv infection as well as by treatment with ABA. Expression analysis supports the notion that many GRAM domain family genes function as responders to various environmental factors (Liu et al., 1999; Jiang et al., 2008). ABR1 has high sequence similarity to the ABA-responsive proteins of Arabidopsis and rice that contain the GRAM domain conserved in eukaryotes (Jiang et al., 2008). The GRAM domain is suggested to have an important function during membrane-associated metabolic processes (Doerks et al., 2000). However, the functions of most GRAM proteins remain to be clarified. Many GRAM proteins have an additional domain named C2, thus leading to a C2-GRAM or C2-C2-GRAM structural organization. In mice, Drosophila, and yeasts, GRAM proteins contain 20 other domains, except for the C2 domain (Jiang et al., 2008). Like the ABA-responsive proteins of Arabidopsis and rice, ABR1 has only one GRAM domain, but no other known motifs or domains. Comparative sequence analyses suggest that pepper may have differentiated from common ancestors of the GRAM domain–containing protein family for Arabidopsis and rice.
We first expected that ABR1 may be localized to the plasma membrane, since some membrane-associated proteins contain a GRAM domain (Doerks et al., 2000). However, subcellular localization experiments in onion epidermal cells demonstrated that ABR1 localizes to the nucleus. In addition, three deletion mutants containing the GRAM domain were also expressed in nuclei, which provides compelling evidence that the GRAM domain of ABR1 is sufficient for nuclear localization. Deletion of the GRAM domain abrogates the nuclear accumulation of ABR1. Thus, we suggest that the GRAM domain functions in the nuclear import of ABR1 through a nuclear membrane-dependent process.
Another possible function of the GRAM domain of ABR1 is to induce HR-like cell death. Transient expression of the GRAM domain construct of ABR1 effectively induced the cell death response in pepper leaves. We reasoned that the nuclear accumulation of ABR1 does not fully correlate with HR-like cell death because the N-terminal fragment without the GRAM domain induces more severe cell death than does the GRAM domain alone. However, ABR1 export from inside nuclei by ABR1NES expression provides evidence that the nuclear ABR1 pool is essential for the cell death induction associated with ABA and SA antagonism. More recently, barley MLA10 (intracellular mildew A10) R protein (Shen et al., 2007) and Arabidopsis nucleocytoplasmic protein EDS1 (for Enhanced Disease Susceptibility1) (García et al., 2010) have been demonstrated to function in the nucleus to confer resistance against pathogen attack. A phosphatase type 2C (PP2C) protein, ABI1, is involved in the ABA-regulated process, and the nuclear compartmentalization of ABI1 is important for the ABA response (Moes et al., 2008). The complex functions of the GRAM domain family were recently proposed to reflect its roles as a regulator of environmental and hormonal signaling (Jiang et al., 2008). Together, these results suggest that one of the pepper GRAM domain members, ABR1, plays specialized roles in nuclear localization and HR-like cell death as well as ABA and SA antagonism.
ABR1 Plays a Role in Cell Death and Defense Responses in Pepper and Arabidopsis
Several members of the plant GRAM domain family have been identified, and a few of them have been shown to be involved in disease responses. In Arabidopsis, VAD1, a gene encoding a GRAM domain–containing protein, is induced in response to pathogen infection (Lorrain et al., 2004). Avirulent Xcv infection and transient expression of ABR1 by agroinfiltration induced exponential cell death and concomitantly enhanced expression of the ABR1 protein in pepper leaves. Cell death was distinctly compromised in ABR1-silenced pepper leaves, which exhibited an enhanced susceptibility to Xcv infection. Collectively, these results suggest that ABR1 functions as a positive regulator in HR-like cell death in pepper leaves. An important feature of cell death is that ABR1-dependent cell death may require ROS production at the infection site.
ABA accumulation is directly correlated with the growth of virulent bacterial pathogens in Arabidopsis (de Torres-Zabala et al., 2009). It has recently been demonstrated that ABA suppresses inducible defense responses by downregulating SA biosynthesis and SA-mediated defenses (de Torres-Zabala et al., 2009). In our study, ABR1-OX transgenic Arabidopsis plants were resistant to Pst and H. arabidopsidis infection, suggesting that ABR1 overexpression may trigger disease resistance by downregulating ABA levels in Arabidopsis. In particular, the T-DNA insertion mutant abr1, which lacks the Arabidopsis putative ortholog of ABR1, was susceptible to these pathogens. Taken together, our results support the idea that ABR1 is involved in disease resistance of pepper and Arabidopsis.
Silencing of ABR1 in pepper plants through VIGS compromised the defense response by enhancing the growth of Xcv, which was accompanied by an increased level of endogenous ABA in pepper leaves. These results raise the possibility that expression of ABR1 may reduce ABA levels in pepper leaves, which is associated with basal defense or resistant responses to Xcv infection. This finding is also consistent with previous reports that the increase in ABA levels is correlated with disease susceptibility in Arabidopsis (Anderson et al., 2004; Kariola et al., 2006). Expression of ABR1 was rapidly and constantly induced in pepper by exogenous ABA treatment. In general, exogenous application of ABA has been proposed to enhance disease susceptibility by inhibiting SA/jasmonic acid/ethylene-mediated defense signaling (Anderson et al., 2004; Mauch-Mani and Mauch, 2005; Flors et al., 2008); however, there are a few examples suggesting that ABA has positive roles in disease resistance (Flors et al., 2005; Melotto et al., 2006; Adie et al., 2007).
Crosstalk between ABA- and SA-Mediated Signaling in Pepper
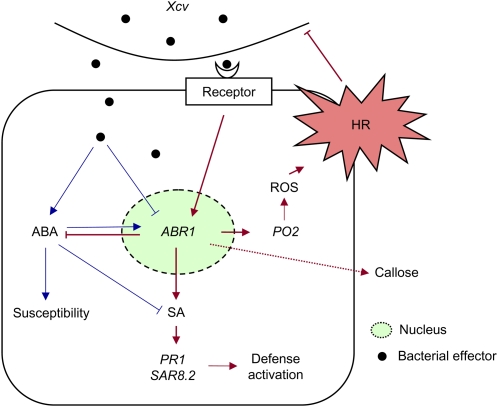
Comprehensive evidence supports an antagonistic relationship between ABA and SA in host-pathogen interactions (Flors et al., 2008; Yasuda et al., 2008; de Torres-Zabala et al., 2009). ABA accumulation is proposed to suppress SA-dependent defense signaling mechanisms, ultimately leading to basal susceptibility to bacterial and fungal pathogens in plants (Audenaert et al., 2002; Mohr and Cahill, 2007). The relationship between SA and ABA reflects early host-pathogen conflict and modulates plant defense responses (de Torres-Zabala et al., 2009). Our data revealed that ABA and SA have antagonistic functions in Xcv–pepper interactions. Increased ABA levels, together with SA reduction, were apparently observed in ABR1-silenced pepper leaves. To resist bacterial pathogen attack, ABR1 expression may regulate and fine-tune endogenous ABA levels. Our findings that ABR1 is rapidly and strongly induced by ABA treatment but not SA treatment raise the possibility that ABR1 expression may enhance SA biosynthesis to trigger defense responses in pepper. Importantly, endogenous ABA levels enhanced by ABR1 silencing suppressed the SA-dependent marker gene PR-1 in pepper leaves infected with an avirulent strain of Xcv. Ectopic expression of ABR1 in Arabidopsis supported the role of ABR1 in basal and HR-like resistant responses to bacterial challenge. These results also gained further support from our studies of the Arabidopsis abr1 mutants. Taken together, we propose a working model that strong induction of ABR1 suppresses ABA biosynthesis but upregulates SA and ROS production, ultimately leading to cell death and disease resistance in pepper leaves (Figure 13).
Figure 13.
Proposed Model of ABA-SA Antagonism in the Xcv–Pepper Interaction.
The pepper ABA-responsive protein, ABR1, which localizes to the nucleus, negatively regulates ABA signaling in an SA-dependent manner to resist pathogen attack. Arrows indicate positive regulation and blunt ends denote negative regulation. Blue and red lines show compatible and incompatible interaction events, respectively.
[See online article for color version of this figure.]
Collectively, our data provide information regarding the alteration of the pepper proteome upon Xcv infection and also evidence for interplay between various defense signaling pathways in pepper during the HR against Xcv. The pepper ABA-responsive protein, ABR1, may contribute to HR-like cell death and defense responses associated with the crosstalk between ABA and SA. Notably, the GRAM domain of ABR1 is likely to be essential for the nuclear localization and cell death response. Proteomics and functional analyses of Xcv–pepper interactions allowed further definition of the role of defense signaling-related proteins, such as ABR1, in cell death and defense responses against microbial pathogens.
METHODS
Plant Growth and Pathogen Inoculation
Pepper (Capsicum annuum cv Nockwang) plants were grown in a steam-sterilized soil mix (peat moss, perlite, and vermiculite, 5:3:2, v/v/v), sand, and loam soil (1:1:1, v/v/v) at 25 ± 2°C under fluorescent light (70 μmol photons m−2 s−1) for 16 h d−1. The virulent (compatible) strain Ds1 and the avirulent (incompatible) strain Bv5-4a of Xanthomonas campestris pv vesicatoria were cultured overnight in yeast nutrient broth (YN; 5 mg yeast extract and 8 mg nutrient broth mL−1) at 28°C. Pepper leaves were infiltrated with the bacterial suspension, and inoculated pepper plants were incubated in a moist chamber for 15 h.
Arabidopsis thaliana ecotype Col-0 and the T-DNA insertion mutants abr1-1 (GK-783H10) and abr1-2 (SALK_017675) from the Nottingham Arabidopsis Stock Centre and the ABRC, respectively, were used in this study. The leaves of wild-type (Col-0), ABR1-OX, and abr1 plants were infiltrated with Pseudomonas syringae pv tomato DC3000 and DC3000 (avrRpm1). Bacterial populations in leaf tissues from wild-type and ABR1-OX plants were monitored on King’s B agar medium containing 100 μg mL−1 rifampicin.
Hyaloperonospora arabidopsidis isolate Noco2 was maintained on the susceptible Arabidopsis ecotype Col-0. Inoculation was performed by spraying 10-d-old seedlings of wild-type, ABR1-OX, and aba1 plants with a suspension of 5 × 104 conidiosporangia mL−1 in distilled tap water containing 0.05% Tween 20. Seedlings were covered with a transparent dome to maintain high humidity and were grown at 17°C with a 14-h photoperiod. Asexual sporulation was assessed 7 d after inoculation by counting spores per cotyledon.
2D Electrophoresis and Protein Staining
For 2D-PAGE, pepper proteins were prepared following a modified protocol of Damerval et al. (1986). Frozen leaf tissues were ground in liquid nitrogen to a fine powder and homogenized in extraction buffer (cold acetone [−20°C], 10% [w/v] trichloroacetic acid, and 0.07% [w/v] DTT). The first dimensional gel separation was performed following the manufacturer's protocol with modifications (Bio-Rad). Immobilized pH gradient (IPG) strips (24 cm, pH 3 to 10 nonlinear; Bio-Rad) were rehydrated with the protein samples (100 μg for silver staining or 800 μg for Coomassie Brilliant Blue staining) diluted in 450 μL sample buffer (9 M urea, 100 mM DTT, 4% [w/v] CHAPS, 0.5% [v/v] Bio-lyte lyte 3-10 carrier ampholytes, and 0.002% bromophenol blue). For first dimension isoelectric focusing, strips were loaded onto a PROTEAN IEF Cell (Bio-Rad). IPG strips were rehydrated at 50 V for 24 h and focused at gradient steps of 250 V for 1 h, 500 V for 1 h, 1000 V for 2 h, and 10,000 V for 4 h, and a final step of 10,000 V toward a total of 90 kVh. IPG strips were incubated in equilibration buffer (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% [v/v] glycerol, and 2% [w/v] SDS) containing 1% (w/v) DTT for 15 min as the first equilibration step and then in 4% (w/v) iodoacetamide as the second step. For 2D-PAGE, IPG strips were fixed on 12.5% acrylamide gels. 2D-PAGE was performed using the Ettan DALTsix electrophoresis unit (Amersham Biosciences) at 5 W per gel for 1 h and then 15 W per gel for 6 h, until the bromophenol blue dye front had reached the bottom of the gels.
Silver staining was performed to detect proteins. Gels were incubated in fixing solution (50% [v/v] ethanol and 12% [v/v] acetic acid) for at least 2 h and washed twice with 50% (v/v) ethanol for 20 min. Gels were sensitized with 0.02% (w/v) sodium thiosulfate for 2 min and washed with water three times for 3 min each. After being stained with silver staining solution (0.2% [w/v] silver nitrate and 0.02% [v/v] formaldehyde) for 20 min, gels were developed with developer (6% [w/v] sodium carbonate, 0.05% [v/v] formaldehyde, and 0.0004% [w/v] sodium thiosulfate) prechilled on ice for 30 min.
Coomassie Brilliant Blue G 250 staining was performed according to the protocol of Giavalisco et al. (2005) with modifications. After 2D-PAGE, gels were fixed in a 40% (v/v) methanol and 7% (v/v) acetic acid solution for 2 h and stained in a solution (0.1% [w/v] Coomassie Brilliant Blue G 250, 34% [v/v] methanol, 3% [v/v] phosphoric acid, and 17% [w/v] ammonium sulfate).
2D gels were scanned using a UMAX PowerLook 1100XL scanner, and the images were analyzed using ImageMaster 2D Platinum 6.0 (Amersham Biosciences). To determine protein abundance, protein spots of interest were quantified as relative percentage volume, where the individual spot volumes are divided by the total spot volume of the whole image.
Identification of Proteins by LC/MS-MS or MALDI-TOF MS
Proteins differentially expressed in healthy and infected plants were identified by peptide sequencing using LC/MS-MS or MALDI-TOF. Protein spots of interest from Coomassie Brilliant Blue–stained gels were excised and digested with modified sequencing grade trypsin (Promega), as previously described (Bahk et al., 2004). Tryptic peptides remaining in the gel matrix were extracted for 40 min at 30°C with 20 μL 50% (v/v) aqueous acetonitrile containing 0.1% (v/v) formic acid for MS analysis.
The resulting tryptic peptides were separated and analyzed using a reversed phase capillary HPLC directly coupled to a Finnigan LCQ ion trap mass spectrometer (LC/MS-MS) (Zuo et al., 2001), with a slight modification. For MS-MS, the full mass scan range mode was m/z = 450 to 2000 D. After determination of the charge states of an ion on zoom scans, product ion spectra were acquired in the MS-MS mode with a relative collision energy of 55%. The individual MS/MS spectra were processed using TurboSEQUEST software (Thermo Quest). The generated peak list files were used to query either the MSDB database or National Center for Biotechnology Information (NCBI) using the MASCOT program (http://www.matrixscience.com). Protein identification was also performed using Ettan MALDI-TOF (Amersham Biosciences). The resulting peptide mass, pI, and molecular mass were used to search the NCBI or SWISS-PROT and TrEMBL databases with Profound (http://prowl.rockefeller.edu/prowl-cgi/profound.exe) for peptide mass fingerprinting.
Isolation and Sequence Analysis of Pathogen-Induced ABR1 cDNA
The full-length cDNA clone of the ABR1 gene was previously obtained from a pepper cDNA library using a differential hybridization technique (Jung and Hwang, 2000). The ABR1 cDNA clone hybridized strongly and specifically to cDNA probes from leaves infected by the avirulent Xcv strain Bv5-4a. The ABR1 clone was sequenced with an ABI 310 DNA sequencer (Applied Biosystems) using the PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Biosystems).
Database Search and Sequence Alignment
Domain search was performed using Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de). DNA sequences aligned in this study were searched from the NCBI (www.ncbi.nlm.nih.gov/blast) database. Amino acid alignment was processed using ClustalW (www.ebi.ac.uk/clustalw) and manually adjusted to optimize alignments.
Phylogenetic Analysis
All protein sequences used in phylogenetic analysis are shown in Supplemental Data Set 1 online. An unrooted tree was visualized using the MEGALIGN program (DNASTAR) with default setting. Statistical support was evaluated with 100 bootstrap values.
Bombardment Assays of Onion Epidermal Cells with GFP Constructs
Particle bombardment assays were performed using the Bio-Rad He/1000 particle delivery system. For the construction of 35S:ABR1:smGFP, the full- and partial-length coding regions of ABR1 without its stop codon were amplified with forward and reverse primers (see Supplemental Table 3 online) carrying XbaI and BamHI restriction sites at their 5′ ends, respectively, and cloned into the vector pCR2.1-TOPO. After digestion with XbaI and BamHI, insert fragments were subcloned on the 5′ of the smGFP gene in the vector p326-GFP. Transfection into onion epidermal cells was performed using gold particles (1.0 μm) and a Biolistic PDS-1000/He particle delivery system (Bio-Rad) with 1300 p.s.i. helium pressure. After bombardment, the onion layers were incubated in the dark for 18 h at 24°C. The cell layers were mounted in staining buffer containing 0.1% DAPI in 5% DMSO and 1% Tween 20. GFP fluorescence was imaged in an LSM 5 Exciter confocal laser scanning microscope (Carl Zeiss) with an excitation wavelength of 488 nm and a 505- to 530-nm band-pass emission filter. DAPI fluorescence was also imaged using an excitation wavelength of 405 nm and a 435- to 480-nm band-pass emission filter.
RNA Isolation, RNA Gel Blot Analysis, and Quantitative Real-Time RT-PCR
Total RNA was isolated from pepper plants as previously described (Chomczynski and Sacchi, 1987). To generate a gene-specific probe, the coding region of the ABR1 gene was amplified using the primers 5′-ATGACAGGCACAACAGAAG-3′(forward) and 5′-AATAAGTTATGACAGAGCCATT-3′ (reverse). The amplified PCR product was 32P-labeled using a random priming kit (Boehringer Mannheim). Agarose gel electrophoresis, RNA transfers, and hybridization with the ABR1 fragment were performed following standard procedures.
First-strand cDNA synthesis was performed using reaction mixtures containing 1 μL (200 units) Moloney Murine Leukemia Virus Reverse Transcriptase (Enzynomics), 1.0 μg total RNA, 10 pmol oligo (dT)20 primer, 1× reaction buffer, 0.25 mM deoxynucleotide triphosphate mixture, and 1 unit of ribonuclease inhibitor for 1 h at 42°C and was stopped by incubation at 70°C for 10 min. cDNA was diluted 1:10 with distilled water and used for real-time RT-PCR with SYBR green (Bio-Rad) using iCycler iQ (Bio-Rad). 18S rRNA was used as an internal control to normalize transcript levels. Each experiment was repeated twice. Specific primers are listed in Supplemental Table 2 online.
Immunoblot Analysis
After gel electrophoresis (1D or 2D), proteins were electrotransferred to polyvinylidene difluoride membranes (GE Healthcare Biosciences). Membranes were incubated with a specific antiserum raised against a ABR1 peptide (Young In Frontier) at 1:10,000 dilution or a rabbit anti-c-Myc or anti-GFP antibody (Sigma-Aldrich) at 1:2000 dilution. Antigen-antibody complexes were detected using peroxidase-conjugated goat anti-rabbit IgG (Sigma-Aldrich).
VIGS in Pepper
The pTRV vector and Agrobacterium tumefaciens for VIGS were prepared as described by Senthil-Kumar et al. (2007). The amplified full-length ABR1 coding region was inserted into the vector pTRV2 to yield pTRV2:ABR1. The pTRV1 vector and the pTRV2 vector with or without ABR1 were transformed into Agrobacterium strain GV3101. A 5-mL culture of each strain was grown overnight at 28°C in YEP (yeast extract/ bactopeptone) broth (10 mg mL−1 yeast extract, 10 mg mL−1 peptone, and 5 mg mL−1 NaCl) with appropriate antibiotics (50 mg mL−1 kanamycin and 50 mg mL−1 rifampicin). The cells were resuspended into Agrobacterium infiltration buffer (10 mM MgCl and 10 mM MES, pH 5.7), and adjusted to OD600 = 0.4. Cultures were then exposed to 150 μM acetosyringone at room temperature with shaking for 3 h. Agrobacterium strains containing the pTRV1 vector and pTRV2:00 or pTRV2:ABR1 were mixed at a 1:1 ratio and coinfiltrated into the cotyledons of pepper seedlings. Agrobacterium-infiltrated pepper plants were grown at 25°C with a 16-h-light/8-h-dark photoperiod cycle and were used after 6 weeks of VIGS treatment.
Chemical Treatment
For ABA and SA treatment, 100 μM (±)-cis, trans-ABA (Sigma-Aldrich), and 5 mM SA (Sigma-Aldrich) solution was sprayed onto the 4-week-old empty vector and ABR1-silenced plants to ensure total coverage of the foliage area. Plants treated with ABA were incubated at room temperature under a 16-h-light/8-h-dark condition. In parallel experiments, water was sprayed as a control. All experiments were repeated at least three times, and results from a representative experiment are shown.
Arabidopsis Transformation
Transgenic Arabidopsis plants overexpressing the ABR1 gene were generated using the floral dipping method (Clough and Bent, 1998). The ABR1 coding region was amplified and inserted into the binary vector pBIN35S under the control of the cauliflower mosaic virus 35S promoter. A pBIN35S:ABR1 construct was introduced into Agrobacterium strain GV3101 through electroporation. Eight lines of putative transgenic Arabidopsis plants harboring the 35S:ABR1 construct were selected by planting seeds on Murashige and Skoog (Duchefa) medium containing 50 mg L−1 kanamycin.
Agrobacterium-Mediated Transient Expression
For Agrobacterium-mediated transient expression of ABR1 deletion mutants, the full-length (ABR1), partial coding region for N terminus (ABR1N), GRAM domain (ABR1GRAM), or C terminus (ABR1C) and deletion mutants without N- (ABR1ΔN) or C terminus (ABR1ΔC) of ABR1 were amplified via PCR using the full-length ABR1 gene as a template. To determine the functional role of the nuclear ABR1 pool, we also fused an NES (LALKLAGLDI) to the C terminus of ABR1 (Wen et al., 1995). The PCR products were directly subcloned into pTOP TA V2 vector (Enzynomics). All fragments digested with BamHI and XbaI were combined into a binary vector pBIN35S-GFP and transferred to Agrobacterium strain GV3101 through electroporation.
Agrobacterium strain GV3101 harboring the myc- or GFP-tagged constructs was grown overnight in YEP medium containing appropriate antibiotics. Cells were suspended in infiltration buffer (10 mM MgCl2, 10 mM MES, and 200 uM acetosyringone, pH 5.7). Pepper leaves were infiltrated with Agrobacterium cells (OD600 = 0.05 to ~0.5). For detection of GFP fusion protein localization, N. benthamiana leaves were infiltrated with Agrobacterium cultures. The images were taken using a LSM 5 Exciter confocal laser scanning microscope (Carl-Zeiss) 36 h after agroinfiltration.
Measurement of Endogenous ABA and SA Levels
The extraction and measurement of endogenous ABA in pepper and Arabidopsis leaves were performed as described by Kang et al. (2008). Leaves were harvested 0, 1, and 3 d after inoculation and were immediately frozen in liquid nitrogen. The frozen leaves were ground in liquid nitrogen and were extracted overnight at 4°C on a rotary shaker with 100% methanol containing 0.5 g L−1 citric acid monohydrate and 0.1 g L−1 butylated hydroxytoluene (Sigma-Aldrich) as antioxidants. ABA in the supernatants was further purified using Sep-Pak C18 cartridges (Waters). Endogenous ABA levels were quantified with the Phytodetekt ABA Enzyme Immunoassay Test Kit (Agdia) in an ELISA reader according to the manufacturer’s instructions.
SA was extracted from leaves excised from Arabidopsis and pepper plants and quantified by HPLC as previously described (Mohr and Cahill, 2007) with minor modifications. Briefly, the leaf powder was ground in liquid nitrogen and was suspended in 90% (v/v) methanol. As an internal standard for SA, 3-hydroxybenzoic acid (Sigma-Aldrich) was added at a mass ratio of 50 mg g−1 fresh weight. SA extracts were analyzed automatically by reversed-phase HPLC (Waters). A C18 analytical column (Xbridge C18 5 μm, 4.6 × 250 mm; Waters) fitted with a guard column (Xbridge C18 5 μm, 4.6 × 20 mm; Waters) was used for chromatography. SA was detected by passage through a fluorescence detector (excitation at 305 nm and emission at 405 nm; 2998 Photo Diode Array Detector; Waters).
DAB Staining and H2O2 Measurement
For H2O2 detection, healthy or infected leaves were incubated in 1 mg mL−1 DAB-HCl, pH 3.8 (Sigma-Aldrich), in the dark for 8 h (Thordal-Christensen et al., 1997). The leaves were then cleared by boiling in alcoholic lactophenol (95% ethanol:lactophenol, 2:1 [v/v]) for 20 min. The reddish color of the leaves as evidence of H2O2 was visualized by light microscopy.
H2O2 was quantified with a ferrous ammonium sulfate/xylenol orange assay (Galletti et al., 2008) with modifications. The assay mixture contained 250 μM ferrous ammonium sulfate and 100 μM sorbitol in 25 mM H2SO4. Excised leaf discs were soaked in the mixture, followed by centrifugation at 5000g for 10 min. The supernatant was added to 100 μM xylenol orange reagent and incubated for 30 min.
Cell Death Assay
Dead cell staining with trypan blue was performed as described by Koch and Slusarenko (1990). To visualize cell death, leaves were stained by boiling in lactophenol-trypan blue (10 mL lactic acid, 10 mL glycerol, 10 g phenol, and 10 mg trypan blue, dissolved in 10 mL distilled water), followed by destaining with chloral hydrate (2.5 g mL−1). Electrolyte leakage was measured from dying and dead cells as described (Mackey et al., 2002). Seven discs (7 mm in diameter) were prepared from leaves at different time points after inoculation with bacteria and incubated in 10 mL distilled water. Water conductance was measured with a sensION7 electrical conductivity meter (Hach).
Aniline Blue Staining
Aniline blue was used to stain papillary callose deposits as described (Dietrich et al., 1994). Leaves were cleared with alcoholic lactophenol and stained for 30 min at room temperature in a solution containing 0.01% (w/v) aniline blue in 0.15 M K2HPO4 and examined using an UV epifluorescence microscope.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: C. annuum ABR1 (GQ373000), Vitis vinifera hypothetical proteins (XP_002263365 and XP_002263309), Arabidopsis ABR (At5g13200), Oryza sativa ABR (ABA98234), Hordeum vulgare ABR (AAD09343), Nicotiana sylvestris Cab (BBA25388), Nicotiana tabacum (P27493), Lycopersicon esculentum Cab (P07370), Solanum tuberosum Cab (AAA80593), C. annuum BPR1 (AF053343), C. annuum SAR82A (AF313766), and C. annumm PO2 (DQ489711).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. 2D Gel Images of Proteins in Pepper Leaves Uninfected and Infected by Xcv.
Supplemental Figure 2. The Proportion of Proteins that are Differentially Expressed in Pepper Leaves Infected by the DS1 and Bv5-4a Strains of Xcv.
Supplemental Figure 3. Relative Protein Expression Changes in Pepper Leaves Infected by Xcv.
Supplemental Figure 4. Differentially Expressed Protein Spots.
Supplemental Figure 8. Identification of ABR1 by LC/MS-MS.
Supplemental Figure 6. Nucleotide and Deduced Amino Acid Sequences of the Pepper (Capsicum annuum) ABA-Responsive Protein (ABR1) Gene cDNA.
Supplemental Figure 7. Comparison of ABR1 with Other ABA-Responsive Proteins.
Supplemental Figure 8. Protein Gel Blot Analysis of Expression of ABR1 in Leaves of Empty Vector and ABR1-Silenced Pepper Plants.
Supplemental Figure 9. T-DNA Insertion Sites and Expression of Arabidopsis ABA-Responsive Protein-like At ABR1.
Supplemental Table 1. Pathogen Proteins Newly Expressed in Pepper Leaves Infected by the Ds1 and the Bv5-4a Strains of Xcv.
Supplemental Table 2. Gene-Specific Primers for qRT-PCR Used in This Study.
Supplemental Table 3. Primers for Generation of GFP Fusion Constructs Used in This Study.
Supplemental Data Set 1. Sequences Used to Generate the Phylogeny Presented in Supplemental Figure 7B online.
Supplementary Material
Acknowledgments
We thank S.P. Dinesh-Kumar (Yale University) for the pTRV1 and pTRV2 vectors and U. Bonas (Martin-Luther-Universitaet) for the Agrobacterium strain GV3101. This work was supported by a grant (CG1133) from the Crop Functional Genomics Center of the 21st Century, Frontier Research Program, funded by the Ministry of Education, Science, and Technology, Korea, a grant (K0821813) from Korea University, and a grant (2010-0007194) from the Basic Science Research Program, the National Research Foundation of Korea, funded by the Ministry of Education, Science, and Technology, Korea.
References
- Adie B.A.T., Pérez-Pérez J., Pérez-Pérez M.M., Godoy M., Sánchez-Serrano J.-J., Schmelz E.A., Solano R. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.P., Badruzsaufari E., Schenk P.M., Manners J.M., Desmond O.J., Ehlert C., Maclean D.J., Ebert P.R., Kazan K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert K., De Meyer G.B., Höfte M.M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahk Y.Y., Kim S.A., Kim J.S., Euh H.J., Bai G.H., Cho S.N., Kim Y.S. (2004). Antigens secreted from Mycobacterium tuberculosis: Identification by proteomics approach and test for diagnostic marker. Proteomics 4: 3299–3307 [DOI] [PubMed] [Google Scholar]
- Choi H.W., Kim Y.J., Hwang B.K. (2011). The hypersensitive induced reaction and leucine-rich repeat proteins regulate plant cell death associated with disease and plant immunity. Mol. Plant Microbe Interact. 24: 68–78 [DOI] [PubMed] [Google Scholar]
- Choi H.W., Kim Y.J., Lee S.C., Hong J.K., Hwang B.K. (2007). Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 145: 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.W., Lee B.G., Kim N.H., Park Y., Lim C.W., Song H.K., Hwang B.K. (2008). A role for a menthone reductase in resistance against microbial pathogens in plants. Plant Physiol. 148: 383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.W., Lee D.H., Hwang B.K. (2009). The pepper calmodulin gene CaCaM1 is involved in reactive oxygen species and nitric oxide generation required for cell death and the defense response. Mol. Plant Microbe Interact. 22: 1389–1400 [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colditz F., Braun H.P., Jacquet C., Niehaus K., Krajinski F. (2005). Proteomic profiling unravels insights into the molecular background underlying increased Aphanomyces euteiches-tolerance of Medicago truncatula. Plant Mol. Biol. 59: 387–406 [DOI] [PubMed] [Google Scholar]
- Damerval C., de Vienne D., Zivy M., Thiellement H. (1986). Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis 7: 52–54 [Google Scholar]
- Dangl J.L., Jones J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M., Bennett M.H., Truman W.H., Grant M.R. (2009). Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 59: 375–386 [DOI] [PubMed] [Google Scholar]
- Dietrich R.A., Delaney T.P., Uknes S.J., Ward E.R., Ryals J.A., Dangl J.L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77: 565–577 [DOI] [PubMed] [Google Scholar]
- Do H.M., Hong J.K., Jung H.W., Kim S.H., Ham J.H., Hwang B.K. (2003). Expression of peroxidase-like genes, H2O2 production, and peroxidase activity during the hypersensitive response to Xanthomonas campestris pv. vesicatoria in Capsicum annuum. Mol. Plant Microbe Interact. 16: 196–205 [DOI] [PubMed] [Google Scholar]
- Do H.M., Lee S.C., Jung H.W., Sohn K.H., Hwang B.K. (2004). Differential expression and in situ localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Plant Sci. 166: 1297–1305 [Google Scholar]
- Doerks T., Strauss M., Brendel M., Bork P. (2000). GRAM, a novel domain in glucosyltransferases, myotubularins and other putative membrane-associated proteins. Trends Biochem. Sci. 25: 483–485 [DOI] [PubMed] [Google Scholar]
- Flors V., Ton J., Jakab G., Mauch-Mani B. (2005). Abscisic acid and callose: Team players in defence against pathogens? J. Phytopathol. 153: 377–383 [Google Scholar]
- Flors V., Ton J., van Doorn R., Jakab G., García-Agustín P., Mauch-Mani B. (2008). Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 54: 81–92 [DOI] [PubMed] [Google Scholar]
- Galletti R., Denoux C., Gambetta S., Dewdney J., Ausubel F.M., De Lorenzo G., Ferrari S. (2008). The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 148: 1695–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A.V., Blanvillain-Baufumé S., Huibers R.P., Wiermer M., Li G., Gobbato E., Rietz S., Parker J.E. (2010). Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 6: e1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen G., Inzé D., Soetaert P., Van Montagu M., Castresana C. (1990). Sequence of a Nicotiana plumbaginifolia beta(1,3)-glucanase gene encoding a vacuolar isoform. Nucleic Acids Res. 18: 6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco P., Nordhoff E., Kreitler T., Klöppel K., Lehrach H., Klose J., Gobom J. (2005). Proteome analysis of Arabidopsis thaliana by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation-time of flight mass spectrometry. Proteomics 5: 1902–1913 [DOI] [PubMed] [Google Scholar]
- Hann D.R., Gimenez-Ibanez S., Rathjen J.P. (2010). Bacterial virulence effectors and their activities. Curr. Opin. Plant Biol. 13: 388–393 [DOI] [PubMed] [Google Scholar]
- Hoth S., Morgante M., Sanchez J.-P., Hanafey M.K., Tingey S.V., Chua N.-H. (2002). Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Hwang I.S., Hwang B.K. (2010). The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 152: 948–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I.S., Hwang B.K. (2011). The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol. 155: 447–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriaga E.A., Leech M.J., Barratt D.H.P., Wang T.L. (1994). Two ABA-responsive proteins from pea (Pisum sativum L.) are closely related to intracellular pathogenesis-related proteins. Plant Mol. Biol. 24: 235–240 [DOI] [PubMed] [Google Scholar]
- Jiang S.Y., Ramamoorthy R., Ramachandran S. (2008). Comparative transcriptional profiling and evolutionary analysis of the GRAM domain family in eukaryotes. Dev. Biol. 314: 418–432 [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jung H.W., Hwang B.K. (2000). Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Mol. Plant Microbe Interact. 13: 136–142 [DOI] [PubMed] [Google Scholar]
- Jwa N.S., Kumar Agrawal G., Rakwal R., Park C.H., Prasad Agrawal V. (2001). Molecular cloning and characterization of a novel Jasmonate inducible pathogenesis-related class 10 protein gene, JIOsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem. Biophys. Res. Commun. 286: 973–983 [DOI] [PubMed] [Google Scholar]
- Kang L., Wang Y-S, Uppalapati S.R., Wang K., Tnag Y., Vadapalli V., Venables B.J., Chapman K.D., Blancaflor E.B., Mysore K.S. (2008). Overexpression of a fatty acid amid hydrolase compromises innate immunity in Arabidopsis. Plant J. 56: 336–349 [DOI] [PubMed] [Google Scholar]
- Kariola T., Brader G., Helenius E., Li J., Heino P., Palva E.T. (2006). EARLY RESPONSIVE TO DEHYDRATION 15, a negative regulator of abscisic acid responses in Arabidopsis. Plant Physiol. 142: 1559–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.H., Choi H.W., Hwang B.K. (2010). Xanthomonas campestris pv. vesicatoria effector AvrBsT induces cell death in pepper, but suppresses defense responses in tomato. Mol. Plant Microbe Interact. 23: 1069–1082 [DOI] [PubMed] [Google Scholar]
- Kim Y.J., Hwang B.K. (2000). Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol. Plant. 108: 51–60 [Google Scholar]
- Koch E., Slusarenko A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E. (2004). Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 5: 305–315 [DOI] [PubMed] [Google Scholar]
- Lee S.C., Hwang B.K. (2003). Identification of the pepper SAR8.2 gene as a molecular marker for pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Planta 216: 387–396 [DOI] [PubMed] [Google Scholar]
- Lee S.C., Hwang B.K. (2005). Induction of some defense-related genes and oxidative burst is required for the establishment of systemic acquired resistance in Capsicum annuum. Planta 221: 790–800 [DOI] [PubMed] [Google Scholar]
- Lee S.C., Hwang I.S., Choi H.W., Hwang B.K. (2008). Involvement of the pepper antimicrobial protein CaAMP1 gene in broad spectrum disease resistance. Plant Physiol. 148: 1004–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister R.T., Dahlbeck D., Day B., Li Y., Chesnokova O., Staskawicz B.J. (2005). Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17: 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A., Tenhaken R., Dixon R., Lamb C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Liu J.-H., Luo M., Cheng K.J., Mohapatra S.S., Hill R.D. (1999). Identification and characterization of a novel barley gene that is ABA-inducible and expressed specifically in embryo and aleurone. J. Exp. Bot. 50: 727–728 [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Lorrain S., Lin B., Auriac M.C., Kroj T., Saindrenan P., Nicole M., Balagué C., Roby D. (2004). Vascular associated death1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 16: 2217–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D., Holt B.F., III, Wiig A., Dangl J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B., Mauch F. (2005). The role of abscisic acid in plant-pathogen interactions. Curr. Opin. Plant Biol. 8: 409–414 [DOI] [PubMed] [Google Scholar]
- McGee J.D., Hamer J.E., Hodges T.K. (2001). Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea. Mol. Plant Microbe Interact. 14: 877–886 [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Moes D., Himmelbach A., Korte A., Haberer G., Grill E. (2008). Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J. 54: 806–819 [DOI] [PubMed] [Google Scholar]
- Mohr P.G., Cahill D.M. (2007). Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. tomato. Funct. Integr. Genomics 7: 181–191 [DOI] [PubMed] [Google Scholar]
- Römer P., Hahn S., Jordan T., Strauss T., Bonas U., Lahaye T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318: 645–648 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M., Hema R., Anand A., Kang L., Udayakumar M., Mysore K.S. (2007). A systematic study to determine the extent of gene silencing in Nicotiana benthamiana and other Solanaceae species when heterologous gene sequences are used for virus-induced gene silencing. New Phytol. 176: 782–791 [DOI] [PubMed] [Google Scholar]
- Shen Q.H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ülker B., Somssich I.E., Schulze-Lefert P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shin R., Lee G.J., Park C.J., Kim T.Y., You J.S., Nam Y.W., Paek K.H. (2001). Isolation of pepper mRNAs differentially expressed during the hypersensitive response to tobacco mosaic virus and characterization of a proteinase inhibitor gene. Plant Sci. 161: 727–737 [Google Scholar]
- Spoel S.H., Dong X. (2008). Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3: 348–351 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z., Wei Y., Collinge D.B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11: 1187–1194 [Google Scholar]
- Torres M.A., Jones J.D., Dangl J.L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- van Wijk K.J. (2001). Challenges and prospects of plant proteomics. Plant Physiol. 126: 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Meinkoth J.L., Tsien R.Y., Taylor S.S. (1995). Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473 [DOI] [PubMed] [Google Scholar]
- Yasuda M., Ishikawa A., Jikumaru Y., Seki M., Umezawa T., Asami T., Maruyama-Nakashita A., Kudo T., Shinozaki K., Yoshida S., Nakashita H. (2008). Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20: 1678–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S. (2010). Pathogen-associated molecular pattern-triggered immunity: veni, vidi…? Plant Physiol. 154: 551–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X., Echan L., Hembach P., Tang H.Y., Speicher K.D., Santoli D., Speicher D.W. (2001). Towards global analysis of mammalian proteomes using sample prefractionation prior to narrow pH range two-dimensional gels and using one-dimensional gels for insoluble and large proteins. Electrophoresis 22: 1603–1615 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.