The authors show that mutation of the rice gene encoding the kinesin-like protein BC12/GDD1 causes a significant reduction in the endogenous GA level and in cell elongation. This is a result of direct regulation of the expression of KO2, a key gene in GA biosynthesis, by GDD1 binding to the KO2 promoter.
Abstract
The kinesins are a family of microtubule-based motor proteins that move directionally along microtubules and are involved in many crucial cellular processes, including cell elongation in plants. Less is known about kinesins directly regulating gene transcription to affect cellular physiological processes. Here, we describe a rice (Oryza sativa) mutant, gibberellin-deficient dwarf1 (gdd1), that has a phenotype of greatly reduced length of root, stems, spikes, and seeds. This reduced length is due to decreased cell elongation and can be rescued by exogenous gibberellic acid (GA3) treatment. GDD1 was cloned by a map-based approach, was expressed constitutively, and was found to encode the kinesin-like protein BRITTLE CULM12 (BC12). Microtubule cosedimentation assays revealed that BC12/GDD1 bound to microtubules in an ATP-dependent manner. Whole-genome microarray analysis revealed the expression of ent-kaurene oxidase (KO2), which encodes an enzyme involved in GA biosynthesis, was downregulated in gdd1. Electrophoretic mobility shift and chromatin immunoprecipitation assays revealed that GDD1 bound to the element ACCAACTTGAA in the KO2 promoter. In addition, GDD1 was shown to have transactivation activity. The level of endogenous GAs was reduced in gdd1, and the reorganization of cortical microtubules was altered. Therefore, BC12/GDD1, a kinesin-like protein with transcription regulation activity, mediates cell elongation by regulating the GA biosynthesis pathway in rice.
INTRODUCTION
The yield of crops, including rice (Oryza sativa), is a concern around the world. Dwarf plants with high yield potential, for the Green Revolution, are an attractive phenotype for crop breeding. More than 60 rice dwarf mutants have been found (Matsuo et al., 1997). Numerous dwarf mutants are deficient in the biosynthesis or perception of gibberellic acid (GA). One of the GA 20-oxidase (GA20ox) genes in rice, GA20ox2 (SD1), is a well-known gene studied in Green Revolution rice varieties (Sasaki et al., 2002). GIBBERELLIN INSENSITIVE DWARF1 (GID1) in rice encodes a soluble receptor that mediates GA signaling (Ueguchi-Tanaka et al., 2005). GID2 encodes an F-box protein important in protein degradation (Sasaki et al., 2003). Mutations in these loci cause semidwarfism.
GAs with biological activity are hormones that regulate plant development processes, such as shoot and stem elongation (Hedden and Phillips, 2000; Olszewski et al., 2002). A tentative GA metabolism pathway has been found in plants (Hedden and Phillips, 2000; Yamaguchi and Kamiya, 2000). GAs are biosynthesized from geranylgranyl diphosphate, which is converted to ent-kaurene by 2 terpene syntheses, ent-copalyl diphosphate synthase, and ent-kaurene synthase, in plants (Aach et al., 1997; Helliwell et al., 2001). Then ent-kaurene is converted to GA12 by two cytochrome P450 enzymes, ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO). KO locates in the outer membrane of the plastid, and KAO locates in the endoplasmic reticulum (Helliwell et al., 2001).
The enzymes in this stage are each encoded by a single-copy gene, and loss-of-function mutations at these loci result in a severe dwarf phenotype in Arabidopsis thaliana. GA12 is converted to GA4 via GA15, GA24, and GA9 precursors and is also converted to GA1 through the precursors GA53, GA44, GA19, and GA20 in the 13-hydroxylated pathway. GA1 and GA4 are thought to function as bioactive forms in plants. Therefore, the rates of GA synthesis and deactivation determine the concentration of bioactive forms in plants. ELONGATED UPPERMOST INTERNODE epoxidizes GA4, GA9, and GA12 as a new GA deactivation mechanism (Zhu et al., 2006). Overexpression of rice YAB1 resulted in a semidwarf phenotype and a decrease in GA1 levels, which might be caused by downregulation of the GA3ox2 gene and upregulation of the GA2ox3 gene (Dai et al., 2007). However, the direct regulation at the transcription level in the GA metabolism pathway is poorly known.
Kinesins are a family of microtubule (MT)-based molecules capable of transporting vesicles containing cytoplasmic cargo to specific destinations in higher eukaryotes (Hirokawa, 1998; Miki et al., 2001; Schuyler et al., 2003). Within the kinesin superfamily, the kinesin-4 subfamily typically plays roles in chromatid motility and chromosome condensation activities associated with mitosis (Wang and Adler, 1995; Kwon et al., 2004; Mazumdar and Misteli, 2005) and vesicle/organelle transport (Hirokawa, 1998) in animals. Structurally, kinesin-4 contains a highly conserved ATPase domain at the N-terminal region and a long coiled-coil domain in the middle (stalk region), followed by a globular domain at the C-terminal tail (Mazumdar and Misteli, 2005). The ATPase domain is the motor providing MT-based mechanochemical activity; the coiled-coil in the stalk region is thought to be important for protein–protein interaction; the C-terminal domain is considered the cargo-docking domain (i.e., the region responsible for capturing cargos such as cytoplasmic vesicles or organelles) (Mazumdar and Misteli, 2005). In animals, kinesin-4 has been studied extensively for its function in chromosome alignment (Murphy and Karpen, 1995; Antonio et al., 2000; Funabiki and Murray, 2000), chromosome orientation, oscillation of chromosome arms (Levesque and Compton, 2001), chromosome positioning, bipolar spindle stabilization in egg extracts (Vernos et al., 1995), and chromatin–MT interactions in vitro (Walczak et al., 1998). In addition, Kif4A in humans (Homo sapiens) was found to have a role in the DNA damage response by modulating the BRCA2/Rad51 pathway (Wu et al., 2008), and the kinesin Costal2/Kif7 was found to have an important function in transcriptional regulation in mouse (Mus musculus) through involvement in the Hedgehog signaling pathway (Cheung et al., 2009). The Arabidopsis kinesin-4 FRA1 is involved in the MT-based effects on cellulose microfibril order and cell elongation (Zhong et al., 2002). Recently, the kinesin-4 protein BRITTLE CULM12 (BC12) was found mainly involved in regulation of cell cycle progression and wall properties in rice (Zhang et al., 2010).
In this study, we characterized a mutant allele of BC12, which we have called gdd1, and cloned the BC12/GDD1 gene by a map-based approach. Our data show that besides binding to MTs in an ATP-dependent manner, the GDD1 kinesin-like protein can act as a transcriptional activator that might regulate KO2 gene expression.
RESULTS
Characterization of the gdd1 Mutant
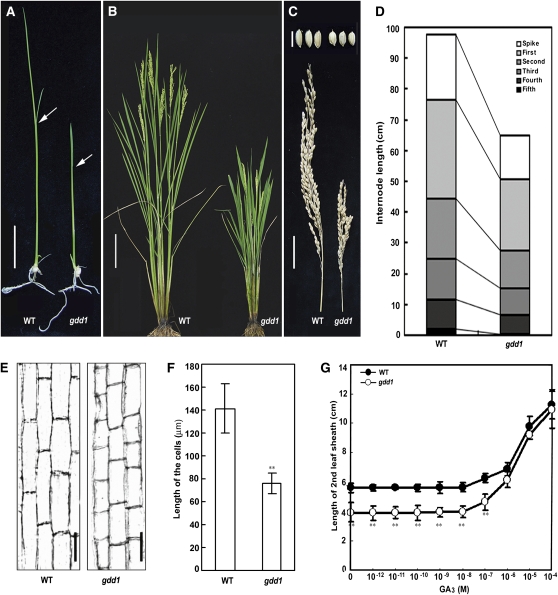
A rice mutant with greatly decreased height was isolated from a transgene line after Agrobacterium tumifaciens–mediated T-DNA insertion. The mutant, designated gibberellin-deficient dwarf1 (gdd1), retained a stable phenotype and showed segregation of the β-glucuronidase (GUS) gene with the T-DNA insertions until T2 progeny (see Supplemental Figure 1 online), suggesting that the phenotype was independent of the T-DNA insertion, the mutant with neither the T-DNA insertion nor GUS fragment from the T2 progeny was used for analysis the function of GDD1. Figure 1 shows the gross morphology of the wild type and mutant plants. The mutant was shorter than the wild type at the three-leaf and heading stages (Figures 1A and 1B). Also, mutant spikes and grains were slightly shorter than those of the wild type (Figure 1C; see Supplemental Figure 2 online). Nearly every internode of the mutant was shorter than that in the wild type (Figure 1D). The stem parenchyma cells were significantly shorter than those of the wild type (P ≤ 0.01) (Figures 1E and 1F), and the cells in the second leaf sheath were also shorter in the mutant (see Supplemental Figure 3 online). Therefore, the mutation of GDD1 inhibits cell elongation in both vegetative and reproductive growth.
Figure 1.
Phenotypic Characterization of the gdd1 Mutant.
(A) Seedling phenotype of the gdd1 mutant and wild type (WT). Arrows indicate the second leaf sheath. Bar = 1 cm.
(B) The gdd1 mutant and wild type at 20 d after heading stage. Bar = 10 cm.
(C) Spike and seeds of the gdd1 mutant and wild type. Bottom bar = 3 cm (spike). Top bar = 5 mm (seeds).
(D) Internode lengths of the gdd1 mutant and wild type. The average values were calculated from measurement of at least 20 plants.
(E) Parenchyma cells in the third internode in the gdd1 mutant and wild type. Bars = 50 μm.
(F) Quantitative measurement of the cell length of gdd1 and the wild type (n = 20). Data are mean ± sd. Asterisks indicate significant difference at P ≤ 0.01 compared with the wild type by Student’s t test.
(G) Elongation of the second leaf sheath in gdd1 and the wild type in response to GA3. Data are mean ± sd (n = 25). Asterisks indicate significant difference at P ≤ 0.01 compared with the wild type by Student’s t test.
To determine whether the dwarf phenotype of mutant plants was caused by GA deficiency or insensitivity, we analyzed the response to exogenous GA3 treatment. The second leaf sheath length of both the mutant and the wild type did not differ with GA3 treatment up to 10−8 M. With GA3 treatment up to 10−6 M, the mutant sheath was almost as long as the wild-type sheath, although the base length of gdd1 was shorter than that of the wild type (Figure 1G). This finding suggests that the mutant gdd1 can respond to exogenous GA3 to rescue the dwarf phenotype, and gdd1 may be deficient in active GA.
Map-Based Cloning of GDD1
To understand the molecular mechanism responsible for the gdd1 phenotype, we used a map-based cloning approach to isolate GDD1. The mutant was crossed with 9311, a wild-type polymorphic indica variety. All F1 progeny showed a height phenotype similar to that of the wild type. Tests of heterozygotes with F2 progeny yielded a segregation of 401 normal and 144 dwarf plants (χ2 [3:1] = 0.59 < χ20.05 = 3.84; P > 0.05), indicating that the dwarf phenotype of the gdd1 mutant is caused by a recessive mutation in a single nuclear gene.
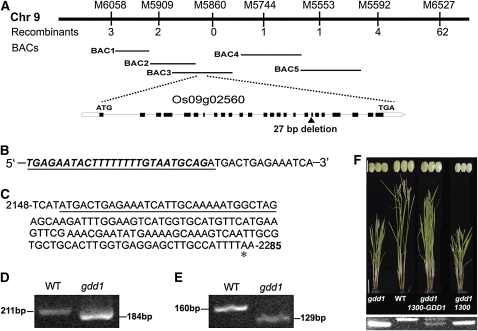
To map GDD1, we used 3300 dwarf plants from F2 populations. The GDD1 locus was initially mapped to the short arm of chromosome 9 between the molecular markers M5909 and M5744 and its location was subsequently localized to BAC3 (Figure 2A). All of the putative open reading frames (ORFs) identified in the 160-kb DNA sequence by annotation analysis were sequenced in the mutant and compared with those in the wild type. A 27-bp deletion was found in one putative gene, Os09g02650, referred to hereafter as GDD1 (Figures 2A, 2B, and 2D). The 27-bp deletion covers the junction of the 19th intron and 20th exon, which suggests that the mutation may disrupt the splicing of the premature RNA. Indeed, an additional 31-bp deletion in the GDD1 transcript was demonstrated by sequencing and matching comparison of the cDNA products of gdd1 and the wild type (Figures 2C and 2E), causing a frame shift that produced a premature protein with normal 717 amino acids.
Figure 2.
Map-Based Cloning of GDD1 and Complementation Test.
(A) Physical mapping of GDD1. GDD1 was localized on BAC3. Accession numbers of the five BAC clones are given at the end of Methods. White boxes, introns; black boxes, exons.
(B) to (E) Mutation site of the gdd1 mutant.
(B) The 27-bp deletion in the gdd1 DNA sequence (underlined) corresponded to 26 bp in the 19th intron (bold italics) and 1 bp in the 20th exon (not italic).
(C) The 31-bp deletion in the cDNA (underlined) and the stop codon (asterisk).
(D) and (E) The different sizes of amplified fragments for the wild type (WT) and gdd1 are shown using genomic DNA (D) and cDNA (E).
(F) Rescue of gdd1 phenotype with the 1300-GDD1 transgene and RT-PCR identification. Bars = 10 cm (stem) and 5 mm (seeds).
Genetic complementation analysis confirmed that the mutant phenotype is due to a lesion of the GDD1 gene. A fragment containing the GDD1 promoter region (~2 kb) and the entire ORF from the wild type was transformed into the gdd1 mutants by Agrobacterium-mediated transformation. Ten independent transgenic lines rescued the mutant phenotypes (Figure 2F; see Supplemental Table 1 online), whereas the control lines with the empty vector failed to complement the gdd1 mutant. Therefore, Os09g02650 is the GDD1 gene.
GDD1 Encodes the KIF4 Family Protein BC12
The GDD1 full-length cDNA is 4309 nucleotides in length with an ORF of 3105 nucleotides that encodes a protein of 1035 amino acids with a predicted molecular mass of 117 kD. A BLAST search of the rice genome revealed only one copy of GDD1. A search of PROSITE (http://www.expasy.org/prosite) revealed that GDD1 has a conserved KIF4 domain at the N terminus (see Supplemental Figure 5 online), which was identified as a kinesin, named BC12 (Zhang et al., 2010). Phylogenetic relationships of BC12/GDD1 and other known KIF4 family members showed that BC12/GDD1 was most similar to At5g47820 (FRA1) of Arabidopsis (see Supplemental Figure 4B and Supplemental Data Set 1 online).
A His fusion protein (His-GDD1-N) containing the GDD1 motor domain (amino acids 1 to 370) was expressed in Escherichia coli and purified (see Supplemental Figure 4 online). This fusion protein was used in MT cosedimentation experiments. In the absence of an exogenous nucleotide, His-GDD1-N was largely cosedimented with the MT pellet. In the presence of ATP, at least 50% of the fusion protein was found in the supernatant rather than the MT pellet. When ATP was replaced by the nonhydrolyzable ATP analog adenylyl imidodiphosphate (AMPPNP), the fusion protein appeared almost exclusively in the MT pellet (see Supplemental Figure 4 online). Thus, GDD1 can bind to MTs in an ATP-dependent manner, consistent with it encoding a kinesin-like protein.
Localization and Expression Pattern of GDD1
The subcellular localization of GDD1 was determined by transient transfection assay. The fluorescent signals of GDD1-green fluorescent protein (GFP) were targeted mainly to nucleus and relatively few to the cytoplasm (see Supplemental Figures 5 and 6 online), which was consistent with the subcellular localization of BC12 (Zhang et al., 2010). Real-time PCR revealed GDD1 mRNA expressed in all organs, with a predominant pattern in the culm and panicle (see Supplemental Figure 5 online). The ubiquitous expression of GDD1 is consistent with the phenotype of the gdd1 mutant, with defects in stem, spike, and seeds, and is similar to that of GA biosynthetic genes (Kaneko et al., 2003).
Genome-Wide mRNA Expression Analysis in the gdd1 Mutant
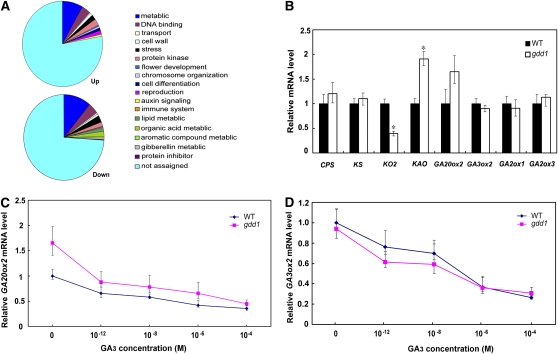
To characterize the molecular events defective in the gdd1 mutant, we compared the mRNA profile of gdd1 and the wild-type seedlings using an Affymetrix whole-genome microarray chip. The reliability of the chip results was confirmed by quantitative RT-PCR (see Supplemental Figure 7 online). We found 116 downregulated and 125 upregulated genes in the gdd1 mutant compared with the wild type (more than fourfold expression change; Figure 3A; see Supplemental Tables 3 and 4 online). Of note, the genes involved in cell wall expansion were significantly altered in expression in the gdd1 mutant. Those encoding xylanase inhibitor protein and cellulose synthase (CESA6) were greatly upregulated. By contrast, the gene for lignin forming anionic peroxidase was downregulated. Therefore, GDD1 might be involved in the regulation of genes associated with cell wall assembly, as has been demonstrated by an alteration in cellulose microfibril orientation and wall composition resulting in brittleness in the bc12 mutant (Zhang et al., 2010). Of note, the expression of the rice KO gene KO2, a key enzyme in early GA synthesis, was greatly decreased in gdd1 compared with the wild type. Plants with mutated KO display the characteristic phenotype of internode stunting and leaf darkening (Helliwell et al., 1998; Davidson et al., 2004; Itoh et al., 2004).
Figure 3.
Mutation of GDD1 Altered GA Metabolic Gene Expression.
(A) Classification of the genes up- or downregulated fourfold or more in gdd1 plants by whole-genome DNA microarray analysis.
(B) Real-time RT-PCR analysis of transcript levels of the eight genes in the GA synthesis pathway. Expression was normalized to that of Actin. Transcript levels from the wild type (WT) were set to 1. Data are means ± sd (n = 3). Asterisk indicates significant difference at P ≤ 0.05 compared with the wild type by Student’s t test.
(C) and (D) Effect of GA3 on the relative expression of GA20ox2 (C) and GA3ox2 (D) in the gdd1 mutant and wild type. Transcript levels from the wild type were set to 1 for real-time PCR. Data are means ± sd (n = 3).
Using quantitative RT-PCR, we further compared the expression of representative GA biosynthesis-related genes, such as ent-copalyl diphosphate synthase (CPS), ent-kaurene synthase (KS), KO2, KAO, GA2ox1, and GA2ox3, as well as GA20ox2/SD1 and GA3ox2/D18 in gdd1 (Figure 3B). Interestingly, the expression of KO2 was downregulated in gdd1, which confirmed the result of the chip assay (Figure 3B; see Supplemental Table 4 online). In gdd1, the transcript level of KAO, encoding a protein that catalyzes the oxidation of ent-kaurenoic acid, was about twofold that of the wild type by an unknown mechanism, which was similar with that in dgl1 (Komorisono et al., 2005). We found no significant differences between the mutant and wild type in expression patterns of CPS, KS, GA2ox1, and GA2ox3. We noted the known negative feedback of GA20ox2/SD1 and GA3ox2/D18 to GA level (Itoh et al., 2001, 2002). As expected, GA20ox2/SD1 showed increased expression in the gdd1 mutant. Also, GA3 treatment produced a downregulated pattern in both the gdd1 mutant and the wild type (Figures 3C and 3D), which may be explained by a GA defect in the mutant. By contrast, the mutant and the wild type did not differ in expression of GA3ox2/D18, although both showed negative induced patterns with GA3 treatment (Figures 3C and 3D). This difference in feedback response between GA20ox2 and GA3ox2 might be due to differences in the underlying mechanisms. For example, YAB1 may directly bind to promoter of GA3ox2 instead of GA20ox2 and suppress its expression to negatively regulate GA homeostasis (Dai et al., 2007).
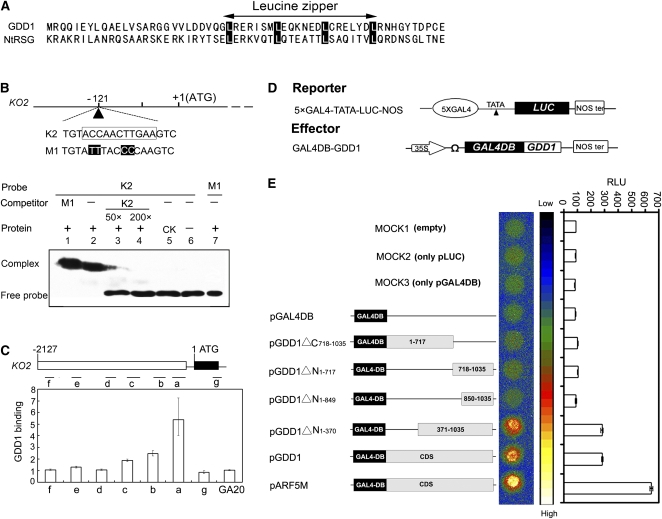
GDD1 Binds to the KO2 Promoter and Activates Transcription
Motif analysis with the bioinformatics software PROSITE (http://www.expasy.org/prosite) revealed a Leu zipper motif, characterized by conserved Leu residues in the bZIP transcription factors, located within the amino acid residues 413 to 434 in the GDD1 protein (Figure 4A). The bZIP proteins were proposed to bind at the site of RF2a, CCA(N)nTGG (Yin et al., 1997). Alignment analysis revealed that the region at −121 nucleotides of the KO2 promoter shared a homologous sequence, ACCAACTTGAA (Figure 4B). To determine whether GDD1 could bind to the KO2 promoter, electrophoretic mobility shift assay (EMSA) was performed using the His fusion protein (His-GDD1ΔC701-1035) containing the Leu zipper domain expressed in E. coli and affinity purified. The nucleotide sequences of the GDD1 binding element of KO2 (K2) and its mutated form (M1) were used as probes. His-GDD1ΔC701-1035 bound to the sequence ACCAACTTGAA (K2) that corresponded to the sequence in the promoter of KO2 but not bind to the mutated version M1, with ACCAACTTGAA changed to ACTTACTCCAA (Figure 4B). Furthermore, the formation of a complex of K2 with His-GDD1ΔC701-1035 was not inhibited in the presence of an excess amount of M1 sequence or unrelated DNA sequences (Figure 4B).
Figure 4.
GDD1 Has DNA Binding and Transactivation Activity.
(A) Alignment of amino acid sequences of the bZIP domain of tobacco RSG and rice GDD1. Highlighted residues indicate the position of Leu residues conserved in the bZIP proteins.
(B) EMSA showing His-GDD1ΔC701-1035 fusion protein binding to the KO2 promoter. Oligonucleotides containing K2, representing the KO2 promoter binding site, or M1, a mutation of K2, were used as the biotin-labeled probes. The K2 sequence is boxed and the mutated bases are highlighted in M1. CK indicates the sonicated extract of E. coli expressing His-GDD1-N without the bZIP domain as a negative control (lane 5). The (+) presence or (−) absence of His-GDD1ΔC701-1035 fusion protein is indicated.
(C) ChIP assays of binding of GDD1 to the promoters of KO2. Open boxes show promoter regions, black lines show untranslated regions and introns, and closed boxes show coding sequences. Regions analyzed by real-time PCR are shown by short lines marked with letters (a to g), and the quantity of binding by GDD1 is shown in the bar graph as fold of enrichment over the control gene GA20ox2. Data are means ± sd (n = 3).
(D) Schematic representation of the reporter and the effector constructs in the transcription activity assay. The reporter consisted of five copies of binding sites for GAL4 in tandem fused upstream of a firefly luciferase gene (LUC). The effector constructs contained the cauliflower mosaic virus 35S promoter, the coding region of the GAL4DB-GDD1 fusion, and the nos polyadenylation signal (Nos ter). A translational enhancer sequence, Ω, from tobacco mosaic virus was located upstream of the sites of translation initiation.
(E) Relative luciferase activities in Arabidopsis mesophyll protoplasts after transfection with reporter plasmids and effectors of various constructs. MOCK1 was a negative control without the reporter and effector. MOCK2 was a negative control with only the pLUC reporter. MOCK3 was a negative control with only pGAL4DB. pARF5M was a positive control. Data are the mean of three independent experiments. RLU, relative light units.
Chromatin immunoprecipitaiton (ChIP) assay was further used to analyze ChIP DNA for promoter regions containing the binding site in KO2. For the ChIP assays, we used an affinity-purified polyclonal anti-BC12/GDD1 antibody that specifically recognizes GDD1 (see Supplemental Figure 8 online). Real-time PCR showed that the fragment “a,” including the binding site in the KO2 promoter was greatly enriched in ChIP. By contrast, the remaining regions from b to g, as well as the negative control, were less amplified in the ChIP assay (Figure 4C). Thus, GDD1 binds directly to the promoter of KO2 in vivo.
We performed a transcriptional activation assay using the GAL4 DB-GDD1 fusion protein in Arabidopsis protoplasts. Luciferase was used as a reporter to check the effector of GDD1 in the system. The activity of luciferase using the pLUC/pGAL4DB-GDD1 construct was much higher than that in the negative control (pLUC/pGAL4DB) and mock treatment (Figures 4D and 4E). The results suggested that GDD1 has transactivation activity in vivo. To understand whether the truncated GDD1 protein in the gdd1 mutant was responsible for the transactivation activity, we further constructed GDD1△C718-1035 to mimic the mutation of GDD1 gene in gdd1. As shown in Figure 4E, deletion of amino acids 718 to 1035 on the C terminus, like the truncated GDD1 in gdd1, had no transactivation activity. Similarly, neither GDD1△N1-717 nor GDD1△N1-849 conferred transactivation activity. By contrast, GDD1△N1-370 still had the same activity as GDD1. Therefore, the C terminus (371 to 1035) could be indispensable for the transactivation activity, and mutation of the GDD1 protein in the gdd1 plant led a deficiency of its transactivation activity.
Endogenous GA Level and Cortical MT Orientation Are Altered in the gdd1 Mutant
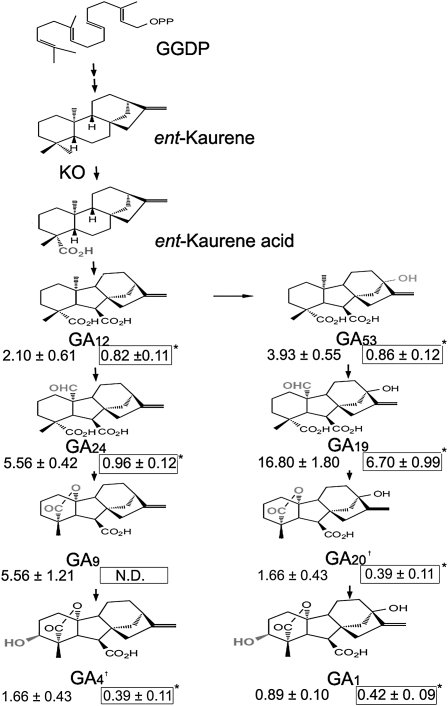
KO converts ent-kaurene to ent-kaurenoic acid, which then goes through a series of processes to produce ramifications of GAs. We determined the endogenous GA level in the gdd1 mutant and the wild type. As shown in the schematic representation of GA biosynthesis (Figure 5), the endogenous level of all GAs tested (i.e., GA9, GA12, GA24, GA53, GA19, GA1, GA20, and GA4) was significantly lower in gdd1 than in the wild type. Therefore, KO2 was suppressed to decrease the GA level.
Figure 5.
Decreased GAs Abundance in gdd1 Plants.
Schematic representation of GA biosynthesis. The numbers are GA endogenous level in the wild type (left) or gdd1 mutant (right, boxed). N.D., not detected due to low abundance; asterisk indicates significant difference at P ≤ 0.05 compared with the wild type by Student’s t test. †The sum content of GA4 and GA20 because chemical structures are too similar to distinguish. Data are means ± sd from three trials (ng/g fresh weight).
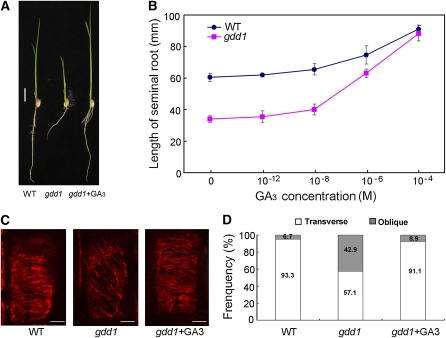
Like the phenotype of reduced height on the stem, the seminal root of gdd1 mutant was shorter than that of the wild type (Figure 6A). GA3 treatment could rescue the root phenotype of the mutant (Figure 6B). To explore whether the reduced GA level affected cortical MT (CMT) orientation in gdd1, we analyzed the CMT arrangement by immuofluorescence assay. As shown in Figures 6C and 6D, CMTs were generally transverse along the long axis, with 93% cells in the elongating zone of root in the wild type. By contrast, oblique CMTs were distributed in >40% of cells of the gdd1 mutant. The oblique CMT abnormal organizations in gdd1 could be restored after exogenous application of GA3 (Figures 6C and 6D). Therefore, mutation of GDD1 resulted in the alteration of GA level, the abnormal organization of CMTs, and abnormalities in cell length. This observation is consistent with known GA-deficient cells that show impaired cell growth and elongation by regulating the orientation of CMTs (Baluška et al., 1993; Wenzel et al., 2000).
Figure 6.
Visualization of CMT Orientation in Cells of the gdd1 Mutant and Wild Type.
(A) Seminal root phenotype of gdd1 mutant can be rescued by GA3. Seeds were germinated in hydroponic culture media for 14 d and then transfered to hydroponic culture media containing 1 μM GA3 for 10 d. Bar = 10 mm. WT, wild type.
(B) Elongation of the seminal root in gdd1 and the wild type in response to GA3. Seeds were germinated in hydroponic culture media for 14 d and then transfered to hydroponic culture media containing different GA3 concentrations. The lengths of seminal roots were measured 10 d after treatment. Data are means ± sd (n = 20).
(C) MT orientation patterns in the elongating root cells of the wild type and gdd1. The wild type shows transverse CMTs along the long axis. The gdd1 mutant shows obliquely oriented CMTs and shows transversely oriented CMTs after 1 μM GA3 treatment. Bars = 5 μm.
(D) Frequency of MT orientation patterns in the elongating root cells of the wild type and gdd1 (n ≥ 30).
DISCUSSION
GDD1 Regulates GA Synthesis
GDD1 has a conserved KIF4 domain at the N terminus and high sequence similarity with other known KIF4 family members, especially with At5g47280 (FRA1) of Arabidopsis (see Supplemental Figure 4 online). Phylogenetic analysis and MT binding assay revealed that GDD1/BC12 belongs to the KIF4 kinesin protein family. The kinesin BC12 mediates cellulose microfibril deposition and wall composition to impact brittleness of culms and to function in cell cycle progression (Zhang et al., 2010). The gdd1 is a new allele of BC12. This allele provides new understanding of how GDD1 relates to the GA biosynthetic pathway. Our mutant shared the phenotypes with bc12 on brittleness of culms and dwarf (Figure 1; see Supplemental Figure 9 online; Zhang et al., 2010). Like its Arabidopsis homolog mutant fra1 (Zhong et al., 2002), gdd1 showed a phenotype of reduced cell length (Figure 1E; see Supplemental Figure 3 online). By contrast, bc12 had an altered cell cycle progression (Zhang et al., 2010). Different mutation sites of the gene may lead a different cytological phenotype with unknown mechanism. Another possibility is that the gene may be different spatio-temporal function characters in various development stages. Therefore, it is possible that GDD1/BC12 may function in both cell elongation and cell cycle progression.
In general, kinesins function as MT-based molecules capable of transporting cytoplasmic cargos to certain destinations in higher eukaryotes (Hirokawa, 1998; Miki et al., 2001; Schuyler et al., 2003). Among the kinesin superfamily, the kinesin-4 subfamily typically plays roles in chromatid motility and chromosome condensation activities associated with mitosis (Wang and Adler, 1995; Kwon et al., 2004; Mazumdar and Misteli, 2005) and vesicle/organelle transport (Hirokawa, 1998) in animals. In Arabidopsis, FRA1 affects the oriented deposition of cellulose microfibrils in secondary walls of fibers, thus resulting in brittleness (Zhong et al., 2002). Unlike At FRA1, which localizes only in cytoplasm (Zhong et al., 2002), and KIF4s of animals, which localize only in the nucleus (Wang and Adler, 1995), GDD1/BC12 is localized in both the cytoplasm and nucleus (see Supplemental Figures 5 and 6 online; Zhang et al., 2010). The localization of cytoplasm supports that function of BC12 in cytoplasm involved in cellulose microfibril deposition and cell wall composition for the brittleness (Zhang et al., 2010). A 17–amino acid fragment (amino acids 971 to 987) of BC12 was identified to be a functional nuclear localization signal (Zhang et al., 2010). Therefore, our data support a possible function of GDD1/BC12 in nucleus.
Unlike the known function of KIF4-like proteins in cell division and brittleness, our data suggest that the KIF4 protein GDD1 is involved in regulation in GA-mediated cell elongation. We found that cell elongation was impaired in the gdd1 mutant (Figure 1), elongation and height were rescued by GA3 treatment (Figure 1), and the expression patterns of genes involved in cell wall formation and GA metabolism were altered (Figure 3).
GDD1 Has DNA Binding and Transactivation Activity
GDD1 possesses the characteristics of kinesin but also shares a motif of the conserved domain of bZIP transcription factors that recognizes the binding site RF2a, CCA(N)nTGG (Yin et al., 1997). Our EMSA and ChIP results and those from transcription activation assay supported that GDD1 could bind to the cis-element sequence of KO2 promoter ACCAACTTGAA at −121 nucleotides (Figure 4C). The five KOL (1 to 5) genes are arranged as tandem repeats in the same direction within a 120-kb sequence. Sequence analysis and complementation experiments demonstrated that KOL2/KO2 corresponds to D35 (Itoh et al., 2004). KOL3 may be a pseudogene, and KOL4 and KOL5 likely take part in phytoalexin biosynthesis (Itoh et al., 2004). Analysis for promoter sequence of KO2 and 3 KO-like genes (see Supplemental Figure 10 online) and ChIP assay revealed that GDD1 could also bind the promoter regions of the KO1 and KOL5 genes with the cis-elements, but the expression pattern of KO1, KOL4, and KOL5 genes was not changed in the gdd1 mutant (see Supplemental Figure 11 online). In the GDD1 complementary rice lines, we observed that the expression of KO2 was also rescued to the wild type level (see Supplemental Figure 12 online). Because some active GAs can still be detected in the mutant (Figure 6), factors other than GDD1 are probably involved in regulating GA biosynthesis. Therefore, KO2 may be one regulated target of GDD1.
The coiled-coil domain (stalk region) was thought to be important for protein–protein interaction (Mazumdar and Misteli, 2005). When the coiled-coil and C-terminal domains were disrupted, transactivation activity of the truncated GDD1 was lost (Figure 4E). In fact, a series of examples that KO2 gene was targeted to lead to alteration of GA level and dwarf plant. The rice dwarf virus P2 protein can interact with KO2, thus causing reduced bioactive GA1 and leading to a dwarf phenotype (Zhu et al., 2005). In addition, tobacco (Nicotiniana tabacum) RSG is a transcriptional activator that binds to the promoter of the Arabidopsis KO gene (Fukazawa et al., 2000). Overexpression of a dominant-negative version in tobacco of REPRESSION OF SHOOT GROWTH (RSG) decreased the expression of Nt KO and prominently decreased plant height (Fukazawa et al., 2000). Our physiological analysis revealed that the gdd1 mutant with decreased GA level could be rescued after GA3 treatment (Figure 1). So, GDD1 is a new member involved in KO2 transcriptional regulation to regulate cell length.
Recently, the kinesin Costal2/Kif7 was found to regulate transcription indirectly but by involving in the signaling pathway in animal (Cheung et al., 2009). Our data support that kinesins with the Leu zipper motif target its cis-element and may regulate gene transcription to mediate development. This finding also suggests the activity of a kinesin in regulating transcription to modulate development.
Possible Roles of GDD1 in the Linkage between MT Orientation and GA Level
The stabilization and maintenance of transverse CMTs by GA3 is well documented (Mita and Katsumi, 1986; Akashi and Shibaoka, 1987; Baluška et al., 1993; Yuan et al., 1994; Shibaoka, 1994; Duckett and Lloyd, 1994; Huang and Lloyd, 1999; Wenzel et al., 2000). For example, GA can induce reorientation of CMTs in living plant cells (Lloyd et al., 1996) and stabilize MTs in maize (Zea mays) suspension cells (Huang and Lloyd, 1999). However, aberrant CMT arrangement in the rice dgl1 mutant causes the upregulation of GA biosynthesis genes such as GA20ox2/SD1, GA20ox1, and GA3ox2/D18, but not KO2 (Komorisono et al., 2005). However, Arabidopsis KSS1 (Bouquin et al., 2003) and rice DGL1 indirectly regulate GA biosynthesis gene expression via an unknown mechanism to integrate MT organization and cell elongation (Foster et al., 2003). Therefore, the GDD1 regulates GA biosynthesis genes expression differently with DGL1, although both dgl1 and gdd1 mutant caused aberrant CMT arrangement.
The mutant gdd1 plants had more cells with oriented CMTs abnormally in the root elongation region (Figure 6). This result is consistent with reports that GA-deficient d5 maize impaired polarity of cell growth in roots through the regulation of the arrangement and orientation of CMTs (Baluška, et al., 1993) and that the GA-deficient M489 dwarf mutant of barley (Hordeum vulgare) could not maintain transverse CMTs of epidermal cells for the elongation zone (Wenzel et al., 2000). The application of exogenous GA can increase the wood tension by enhancing the thickness cell walls and orientating cellulose microfibril parallel or nearly parallel to the longitudinal axis of the fibers (Funada et al., 2008). Our data suggest that mutation of a kinesin-like protein in the rice gdd1 mutant significantly reduces the level of endogenous GAs and cell length by directly downregulating KO2 expression. Changes in GA level may cause the different organization of MTs (Figure 6). Our finding brings new knowledge and a new understanding of the function of the kinesin-4 subfamily.
In conclusion, we demonstrated that GDD1/BC12, a kinesin-like protein, plays a novel role in directly regulating the expression of the KO2 gene in the GA biosynthesis pathway for regulation of MT arrangement and cell elongation to modulate development in rice.
METHODS
Plant Material and Growth Conditions
A recessive mutant, gdd1, was isolated from the transformed line Oryza sativa ssp japonica, cv zhonghua10, which was independent of T-DNA insertion. Surface-sterilized seeds of the wild-type and gdd1 plants were soaked in water for 3 d and then placed in artificial soil for 11 d and grown in the experimental field. Mutant segregants were distinguished from normal segregants by the dwarf phenotype. The mutant was crossed with 9311, a wild-type polymorphic indica variety. Dwarf plants from F2 populations were used for map-based cloning.
GA Induction in Cell Elongation
Shoot elongation was quantified as described (Matsukura et al., 1998) with modification. Seeds of the wild type and gdd1 (n > 25) plants were surface sterilized for 30 min with a 3% NaClO solution, washed three times with sterile distilled water, then placed on a 1% agar plate by adding GA3 of 10−12 ~10−4 M and grown under fluorescent light at 30°C. After 1 week, the lengths of the second leaf sheaths were measured.
Genetic Complementation Test
The GDD1 gene, including the promoter region and the coding region cloned from wild-type genomic DNA and cDNA, was inserted into the vector pCAMBIA1300. This construct and the empty vector pCAMBIA1300 were introduced into gdd1 mutant plants by Agrobacterium tumefaciens–mediated transformation (Wang et al., 2008). The 10 independent transgenic lines were independently isolated and identified. The transgenic plants were detected by RT-PCR.
Affymetrix GeneChip Analysis
Genome-wide expression studies using the rice Affymetrix GeneChip were done with the wild type and gdd1 independently grown under the same growth conditions. Total RNA was isolated from seedlings at the three-leaf stage with the RNA extraction kit (TRIzol reagent; Invitrogen). Statistical analysis of the microarray data was as previously described (Wang et al., 2008). GeneChip Operating Software (GCOS) was used for data collection and normalization. The overall intensity of all probe sets of each array was scaled to 500 to ensure equal hybridization intensity; each probe set was assigned P, A, or M and a P value from the algorithm in GCOS. Information on the GeneChip Rice Genome Array (MAS 5.0) was accessed from the Affymetrix website (http://www.affymetrix.com/products/arrays/specific/rice.affx).
RNA Isolation and Quantitative RT-PCR
Total RNA was extracted from root, stem, mature leaf, sheath, and panicles by use of a RNA extraction kit for analysis of GDD1 mRNA expression (TRIzol reagent). For analysis of transcripts of GAox genes, 2-week-old seedlings sprayed with or without 10−4 M GA3 for 12 h were harvested. For real-time quantitative PCR analysis, 2 μg total RNA was treated with DNase I (Invtrogen) and then transcribed in a total volume of 25 μL with 0.5 μg oligo(dT)15, 0.75 mM deoxynucleotide triphosphate, and 200 units avian myeloblastosis virus reverse transcriptase (Promega). RT-PCR involved use of gene-specific primers (see Supplemental Table 2 online) in a total volume of 15 μL with 3 μL of reverse transcription reactions, 0.25 μM primers, and 7.5 μL SYBR Green Master mix (Applied Biosystems) on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions. The PCR signals were normalized to that of Actin.
DNA Gel Blot Analysis
Genomic DNA was isolated from 10-d-old seedlings and digested with EcoRI. The fractionated DNA was electrophoresed on 0.7% agarose gel and blotted on a nylon membrane. The GUS gene used as a probe was labeled with [α-32P]dCTP. Hybridization was as described (Xu et al., 2005).
Protein Gel Blot Analysis
Protein gel blotting was performed as described (Han et al., 2005). SDS-PAGE and immunoblotting were performed in the mini-gel apparatus and submarine gel transfer systems (Bio-Rad), respectively. Samples separated on 10% acrylaminde were transferred to nitrocellulose membrane at a constant current of 80 mA for 1.5 h in the ice-water bath. The membrane was blocked by 5% BSA at room temperature overnight and then incubated with anti- GDD1 (1:100) and anti-tubulin (Beyotime) at 1:1000 dilution at 4°C overnight, respectively. Goat anti-rabbit IgG horseradish peroxidase conjugate (Sigma-Aldrich) were used as the secondary antibodies for 2 h at 37°C. Proteins were visualized after incubating the membrane with horseradish peroxidase substrate and autoradiography.
Bioinformatic Sequence Analysis
Coiled-coil domains of GDD1 were predicted by the software http://www.ch.embnet.org/software/COILS_form.html. We performed a BLASTsearch of GDD1 homologs using the National Center for Biotechnology Information BLAST server (http://blast.ncbi.nlm.nih.gov/Blast). Amino acid sequences of the KIF4 family members were obtained from the Kinesin webpage (http://proweb.org/kinesin/BE6a_Chromo.html). The obtained sequences were aligned, and the neighbor-joining tree was produced with bootstrap replication using MEGA v4.0.1.
Phylogenetic Analysis
A BLAST search program (http://www.ncbi.nlm.nih.gov/BLAST/) was used to find protein sequences homologous to GDD1. Amino acid sequences of KIF4 family members were obtained from the kinesin Web page (http://proweb.org/kinesin/BE6a_Chromo.html). The obtained protein sequences were aligned using MEGA version 4.0 software (Tamura et al., 2007; see Supplemental Data Set 1 online), and the midpoint-rooted neighbor-joining tree (see Supplemental Figure 4 online) was generated using the same software with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates; random seed).
MT Cosedimentation Assay
This assay was performed as described (Lee and Liu, 2000). In brief, purified His-GDD1-N (1 to 370 amino acids) fusion protein (5 μg) was incubated with 25 μg of MTs (tubulin from the fresh pig brain), 10 μM taxol, and 2 mM ATP or adenylyl imidodiphosphate (AMPPNP) in PEM buffer (80 mM PIPES, 1 mM MgCl2, and 1 mM EGTA, pH 6.8). After incubation at room temperature, the reaction was centrifuged at 55,000g. The supernatant and the MT pellet were analyzed by SDS-PAGE. Proteins of the supernatant and pellet were visualized by Coomassie Brilliant Blue R 250 staining. This assay was repeated at least three times.
Localization of the GDD1-GFP Fusion Protein
The GDD1 coding region was fused in frame to cDNA for GFP, and the fusion DNA was ligated into the 3′ terminus of the cauliflower mosaic virus 35S promoter in the vector pBI221.Transient expression of the GDD1-GFP fusion protein and GFP alone as a control in rice protoplast was as described (Bart et al., 2006). After incubation for 24 h, images were obtained by confocal laser microscope (Zeiss LSM510).
EMSA
This assay was performed essentially as described (Ma et al., 2009). The coding sequence of GDD1 (1 to 2098) was amplified by PCR and cloned into the EcoRI and NotI sites of the expression vector pET-28a. The construct was transformed into Escherichia coli BL21 (DE3). Cells were grown at 30°C and induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 1 mM when the OD600 of the cultured cell was 0.5 to 0.9. The fusion protein was purified with Ni-NTA His·Bind Superflow (Novagen).
Nucleotide sequences of the double-stranded oligonucleotides were for K2 (5′-GTGTCGCTGTACCAACTTGAAGTCT-3′ and 5′-GAGAGACTTCAAGTTGGTACAGCGA-3′) and M1 (5′-GTGTCGCTGTACTTACTCCAAGTCT-3′ and 5′-GAGAGACTTGGAGTAAGTACAGCGA-3′). The oligonucleotides were annealed and then labeled with use of the Biotin 3′ End DNA labeling kit (Pierce). Standard reaction mixtures (20 μL) for EMSA contained 2 μg purified proteins, 2 μL biotin-labeled annealed oligonucleotides, 2 μL 10× binding buffer (100 mM Tris, 500 mM KCl, and 10 mM DTT, pH 7.5), 1 μL 50% glycerol, 1 μL 1% Nonidet P-40, 1 μL 1 M KCl, 1 μL 100 mM MgCl2, 1 μL 200 mM EDTA, 1 μL 1 μg/μL poly(dG-dC), and 8 μL ultrapure water. The reactions were incubated at room temperature (25°C) for 20 min and loaded onto 10% native polyacrylamide gel containing 45 mM Tris, 45 mM boric acid, and 1 mM EDTA, pH 8.3. The gel was sandwiched and transferred to N+ nylon membrane (Millipore) in 0.5× TBE buffer at 380 mA in a 4°C refrigerator for 60 min. Biotin-labeled DNA was detected by use of the LightShift Chemiluminescent EMSA kit (20148; Pierce) following the manufacturer’s instructions.
ChIP
ChIP assay with wild-type seedlings was as described (He et al., 2005). An affinity-purified anti-GDD1 polyclonal antibody was used for immunoprecipitation. Equal amounts of the input DNA and ChIP products were analyzed by quantitative real-time PCR. For the CHIP real-time PCR data analysis as described (Zhang et al., 2009), the threshold cycle (Ct value) of GA20 and different promoter region of KO2 was obtained after quantitative real-time PCR reaction. In brief, the normalizer GA20 Ct value in gdd1 was subtracted from the Ct of GA20 and the different promoter region (a-g) of KO2 in the wild type to produce the dCt. The dCt value of the GA20 was subtracted from the dCt value of every region of KO2 to produce the ddCt value. For every region of KO, we evaluated 2 to the −ddCt power (2−ddCt) as the relative binding levels. The relative binding level of each PCR product was calculated and analyzed from three independent reactions. The ChIP experiments were replicated independently at least three times, with representative experimental data presented in the manuscript.
Transient Assay for Transactivational Activity in Vivo
Reporter plasmids and effector plasmids were constructed as described (Ohta et al., 2000). For construction of effector plasmids encoding GAL4DB-GDD1, site-directed mutagenesis was used to eliminate the SalI site in the ORF of GDD1 in the T-easy vector (Promega), then the coding region of GDD1 was amplified by PCR and inserted, in frame, in the 35S-GAL4 DB at SmaI and SalI sites.
Protoplasts were prepared from mesophyll tissues of Arabidopsis of the ecotype Columbia, and the constructs were transformed into the protoplasts with polyethylene glycol as described (Yoo et al., 2007). Luciferase assay was as described (Fujikawa and Kato, 2007), except d-Luciferin (for firefly luciferase; Gold Bio Technology) replaced the ViviRen Live Cell Substrate. Luminescence images were obtained using a charge-coupled device camera (DU434-BV; Andor Technology). Counts of luminescence were quantified with a 20/20n Luminometer (Turner BioSystems).
Quantification of Endogenous GAs
Rice leaves (3 g) were frozen in liquid nitrogen, ground to fine powder, and extracted with 15 mL 80% (v/v) methanol at 4°C for 12 h. [2H2] GA1 (1.00 ng/g), [2H2] GA4 (2.00 ng/g), [2H2] GA12 (2.00 ng/g), [2H2] GA24 (6.00 ng/g), and [2H2] GA53 (4.00 ng/g) were added to plant samples before grinding as internal standards. After centrifugation (10,000g, 4°C, 20 min), the supernatant was collected and passed through a C-18 SPE-cartridge (12 mL, 1.5 g) preconditioned with 8 mL water, 8 mL methanol, and 8 mL 80% (v/v) methanol. The elution was pooled and evaporated under a nitrogen gas stream and reconstituted in 3 mL water. The solution was acidified with 360 μL 0.1 mol/L hydrochloric acid and extracted with ethyl ether (10 × 0.5 mL). The ether phases were combined, dried under nitrogen gas, and reconstituted with 112 μL acetonitrile. Then, 180 μL Et3N (20 μmol/mL) and 108 μL 3-bromoacetonyltrimethylammonium bromide (20 μmol/mL) were added. The reaction solution was vortex mixed for 10 min and evaporated under a stream of nitrogen gas to dryness, and the residue was dissolved in 30 μL water. The resulting sample solution was injected by 25 kV × 1 min and separated by 100-cm amino groups, coated capillary electrophoresis coupled with electrospray ionization quadrupole–time-of-flight mass spectrometry for analysis.
Immunolocalizaion of CMTs
MTs in root cells were visualized by immunolabeling α-tubulins as described (Baluška et al., 1992; Lloyd et al., 1996) with modification. Briefly, segments (5 to 8 mm) were fixed in 4% paraformaldehyde (Sigma-Aldrich) in PEM buffer for 2 h and embedded in Steedman’s wax. The sections were rehydrated and blocked in PBS containing 1% (w/v) BSA for 0.5 h. After a rinsing three times (10 min/time) with PBS, MTs were probed with the anti-α-tubulin antibody (Beyotime). Secondary antibodies were Cy3 conjugated with goat anti-mouse IgG (Beyotime). Images were obtained by confocal laser microscopy (Zeiss LSM510).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL database under the following accession numbers: FRA1, At5g47820; At F11C1.80, At3g50240; At MSL3.5, At5g60930; GDD1, Os09g02560; KO2, Os06g37300; xylanase inhibitor protein genes, Os07g23640, Os07g23660; cellulose synthase gene, Os07g14850; lignin forming anionic peroxidase gene, Os06g32990. Accession numbers for BAC clones on chromosome 9 can be found in GenBank/EMBL: BAC1, AP006058.3; BAC2, AP005909.2; BAC3, AP005860.2; BAC4, AP005744.2; and BAC5, AP005592.2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DNA Gel Blot Analysis of gdd1 Plants with 32P -Labeled GUS.
Supplemental Figure 2. Comparison of Agronomic Traits in the Wild Type (ZH10) and gdd1 Mutant.
Supplemental Figure 3. Comparison of Cell Length in Leaf Sheath or Cell Number in Root in the Wild Type (ZH10) and gdd1 Mutant.
Supplemental Figure 4. In Vitro Binding of Microtubules by the GDD1 Motor Domain.
Supplemental Figure 5. Subcellular Localization and Expression Pattern of GDD1.
Supplemental Figure 6. The Effects of GA and PAC on the Immunolocalization of GDD1 in Cytoplasm and Nucleus in Mature Regions of Rice Root.
Supplemental Figure 7. Quantitative RT-PCR Analysis of Selective Genes in Rice Whole-Genome DNA Microarray.
Supplemental Figure 8. Protein Gel Blot Analysis of GDD1 Protein in the Wild Type and gdd1 Seedlings Showing the Specificity of the Antibody Used in ChIP Assays (Figure 4C) and Immunolocalization (Supplemental Figure 6).
Supplemental Figure 9. The Culm Brittle Phenotype of gdd1.
Supplemental Figure 10. Analysis GDD1 Binding Sites in the 2-kb Promoters of KO1, KO2, KOL4, and KOL5.
Supplemental Figure 11. ChIP Assays of Binding of GDD1 to the Promoters of KO(L).
Supplemental Figure 12. The Expression of KO2 Was Rescued in the GDD1 Transgenic Lines.
Supplemental Table 1. Culm Length and Seed Size of the GDD1 Complement Lines and Their Progenitor.
Supplemental Table 2. Primers Used in This Work.
Supplemental Table 3. Microarray Analysis of gdd1 Mutant Plants.
Supplemental Table 4. Microarray Analysis of gdd1 Mutant Plants.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 4B Online.
Acknowledgments
We thank Hui Chen, Yuan Zhao, and Rongxi Jiang (Institute of Botany, Chinese Academy of Sciences) for their assistance in rice gene transformation and mutant cultivation; Yihua Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the gift of the BC12/GDD1 antibody; and Jiang Hu (China National Rice Research Institute) for help in rice field management. This work was supported by a grant from the National Nature Science Foundation of China for Innovative Research Groups (No. 30821007) and the state Hi-Tech Research and Development program of China (No. 2006AA10A101).
References
- Aach H., Bode H., Robinson D.G., Graebe J.E. (1997). ent-Kaurene synthase is located in proplastids of meristematic shoot tissues. Planta 202: 211–219 [Google Scholar]
- Akashi T., Shibaoka H. (1987). Effects of gibberellin on the arrangement and the cold stability of cortical microtubules in epidermal cells of pea internodes. Plant Cell Physiol. 28: 339–348 [Google Scholar]
- Antonio C., Ferby I., Wilhelm H., Jones M., Karsenti E., Nebreda A.R., Vernos I. (2000). Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell 102: 425–435 [DOI] [PubMed] [Google Scholar]
- Baluška F., Parker J.S., Barlow P.W. (1992). Specific patterns of cortical and endoplasmic microtubules associated with cell growth and tissue differentiation in roots of maize (Zea mays L.). J. Cell Sci. 103: 191–200 [Google Scholar]
- Baluška F., Parker J.S., Barlow P.W. (1993). A role for gibberellic acid in orienting microtubules and regulating cell growth polarity in the maize root cortex. Planta 191: 149–157 [Google Scholar]
- Bart R., Chern M., Park C.J., Bartley L., Ronald P.C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin T., Mattsson O., Naested H., Foster R., Mundy J. (2003). The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J. Cell Sci. 116: 791–801 [DOI] [PubMed] [Google Scholar]
- Cheung H.O., Zhang X., Ribeiro A., Mo R., Makino S., Puviindran V., Law K.K., Briscoe J., Hui C.C. (2009). The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci. Signal. 2: ra29. [DOI] [PubMed] [Google Scholar]
- Dai M., Zhao Y., Ma Q., Hu Y., Hedden P., Zhang Q., Zhou D.X. (2007). The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 144: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S.E., Smith J.J., Helliwell C.A., Poole A.T., Reid J.B. (2004). The pea gene LH encodes ent-kaurene oxidase. Plant Physiol. 134: 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett C.M., Lloyd C.W. (1994). Gibberellic acid-induced microtubule reorientation in dwarf peas is accompanied by rapid modification of an α-tubulin isotype. Plant J. 5: 363–372 [Google Scholar]
- Foster R., Mattsson O., Mundy J. (2003). Plants flex their skeletons. Trends Plant Sci. 8: 202–204 [DOI] [PubMed] [Google Scholar]
- Fujikawa Y., Kato N. (2007). Split luciferase complementation assay to study protein-protein interactions in Arabidopsis protoplasts. Plant J. 52: 185–195 [DOI] [PubMed] [Google Scholar]
- Fukazawa J., Sakai T., Ishida S., Yamaguchi I., Kamiya Y., Takahashi Y. (2000). Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12: 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Murray A.W. (2000). The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102: 411–424 [DOI] [PubMed] [Google Scholar]
- Funada R., Miura T., Shimizu Y., Kinase T., Nakaba S., Kubo T., Sano Y. (2008). Gibberellin-induced formation of tension wood in angiosperm trees. Planta 227: 1409–1414 [DOI] [PubMed] [Google Scholar]
- Han Y., Jiang J.F., Liu H.L., Ma Q.B., Xu W.Z., Xu Y.Y., Xu Z.H., Chong K. (2005). Overexpression of OsSIN, encoding a novel small protein, causes short internodes in Oryza sativa. Plant Sci. 169: 487–495 [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Phillips A.L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Helliwell C.A., Chandler P.M., Poole A., Dennis E.S., Peacock W.J. (2001). The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 98: 2065–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C.A., Sheldon C.C., Olive M.R., Walker A.R., Zeevaart J.A., Peacock W.J., Dennis E.S. (1998). Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc. Natl. Acad. Sci. USA 95: 9019–9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. (1998). Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279: 519–526 [DOI] [PubMed] [Google Scholar]
- Huang R.F., Lloyd C.W. (1999). Gibberellic acid stabilises microtubules in maize suspension cells to cold and stimulates acetylation of alpha-tubulin. FEBS Lett. 443: 317–320 [DOI] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sato Y., Ashikari M., Matsuoka M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sentoku N., Kitano H., Matsuoka M., Kobayashi M. (2001). Cloning and functional analysis of two gibberellin 3 beta -hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 98: 8909–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Tatsumi T., Sakamoto T., Otomo K., Toyomasu T., Kitano H., Ashikari M., Ichihara S., Matsuoka M. (2004). A rice semi-dwarf gene, Tan-Ginbozu (D35), encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Mol. Biol. 54: 533–547 [DOI] [PubMed] [Google Scholar]
- Kaneko M., Itoh H., Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Ashikari M., Matsuoka M. (2003). Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 35: 104–115 [DOI] [PubMed] [Google Scholar]
- Komorisono M., Ueguchi-Tanaka M., Aichi I., Hasegawa Y., Ashikari M., Kitano H., Matsuoka M., Sazuka T. (2005). Analysis of the rice mutant dwarf and gladius leaf 1. Aberrant katanin-mediated microtubule organization causes up-regulation of gibberellin biosynthetic genes independently of gibberellin signaling. Plant Physiol. 138: 1982–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M., Morales-Mulia S., Brust-Mascher I., Rogers G.C., Sharp D.J., Scholey J.M. (2004). The chromokinesin, KLP3A, dives mitotic spindle pole separation during prometaphase and anaphase and facilitates chromatid motility. Mol. Biol. Cell 15: 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.R., Liu B. (2000). Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr. Biol. 10: 797–800 [DOI] [PubMed] [Google Scholar]
- Levesque A.A., Compton D.A. (2001). The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154: 1135–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C.W., Shaw P.J., Warn R.M., Yuan M. (1996). Gibberellic acid-induced reorientation of cortical microtubules in living plant cells. J. Microsc. 181: 140–144 [Google Scholar]
- Ma Q., Dai X., Xu Y., Guo J., Liu Y., Chen N., Xiao J., Zhang D., Xu Z., Zhang X., Chong K. (2009). Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 150: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura C., Itoh S., Nemoto K., Tanimoto E., Yamaguchi J. (1998). Promotion of leaf sheath growth by gibberellic acid in a dwarf mutant of rice. Planta 205: 145–152 [Google Scholar]
- Matsuo T., Futsuhara Y., Kikuchi F., Yamaguchi H. (1997). Science of the Rice Plant (Tokyo: Nobunkyo; ). [Google Scholar]
- Mazumdar M., Misteli T. (2005). Chromokinesins: Multitalented players in mitosis. Trends Cell Biol. 15: 349–355 [DOI] [PubMed] [Google Scholar]
- Miki H., Setou M., Kaneshiro K., Hirokawa N. (2001). All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. USA 98: 7004–7011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita T., Katsumi M. (1986). Gibberellin control of microtubule arrangement in the mesocotyl epidermal cells of the d5 mutant of Zea mays L. Plant Cell Physiol. 27: 651–659 [Google Scholar]
- Murphy T.D., Karpen G.H. (1995). Interactions between the nod+ kinesin-like gene and extracentromeric sequences are required for transmission of a Drosophila minichromosome. Cell 81: 139–148 [DOI] [PubMed] [Google Scholar]
- Ohta M., Ohme-Takagi M., Shinshi H. (2000). Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J. 22: 29–38 [DOI] [PubMed] [Google Scholar]
- Olszewski N., Sun T.P., Gubler F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14(Suppl): S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Ashikari M., Ueguchi-Tanaka M., Itoh H., Nishimura A., Swapan D., Ishiyama K., Saito T., Kobayashi M., Khush G.S., Kitano H., Matsuoka M. (2002). Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Jeong D.H., An G., Kitano H., Ashikari M., Matsuoka M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Schuyler S.C., Liu J.Y., Pellman D. (2003). The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 160: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibaoka H. (1994). Plant hormone-induced changes in the orientation of cortical microtubules: Alterations in the cross-linking between microtubules and the plasma membrane. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45: 527–544 [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T.Y., Hsing Y.I., Kitano H., Yamaguchi I., Matsuoka M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Vernos I., Raats J., Hirano T., Heasman J., Karsenti E., Wylie C. (1995). Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell 81: 117–127 [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Vernos I., Mitchison T.J., Karsenti E., Heald R. (1998). A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8: 903–913 [DOI] [PubMed] [Google Scholar]
- Wang L., Xu Y., Zhang C., Ma Q., Joo S.H., Kim S.K., Xu Z., Chong K. (2008). OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 3: e3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.Z., Adler R. (1995). Chromokinesin: a DNA-binding, kinesin-like nuclear protein. J. Cell Biol. 128: 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel C.L., Williamson R.E., Wasteneys G.O. (2000). Gibberellin-induced changes in growth anisotropy precede gibberellin-dependent changes in cortical microtubule orientation in developing epidermal cells of barley leaves. Kinematic and cytological studies on a gibberellin-responsive dwarf mutant, M489. Plant Physiol. 124: 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Zhou L., Khidr L., Guo X.E., Kim W., Lee Y.M., Krasieva T., Chen P.L. (2008). A novel role of the chromokinesin Kif4A in DNA damage response. Cell Cycle 7: 2013–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M.L., Jiang J.F., Ge L., Xu Y.Y., Chen H., Zhao Y., Bi Y.R., Wen J.Q., Chong K. (2005). FPF1 transgene leads to altered flowering time and root development in rice. Plant Cell Rep. 24: 79–85 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Kamiya Y. (2000). Gibberellin biosynthesis: Its regulation by endogenous and environmental signals. Plant Cell Physiol. 41: 251–257 [DOI] [PubMed] [Google Scholar]
- Yin Y., Zhu Q., Dai S., Lamb C., Beachy R.N. (1997). RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J. 16: 5247–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yuan M., Shaw P.J., Warn R.M., Lloyd C.W. (1994). Dynamic reorientation of cortical microtubules, from transverse to longitudinal, in living plant cells. Proc. Natl. Acad. Sci. USA 91: 6050–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.Y., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhang B., Qian Q., Yu Y., Li R., Zhang J., Liu X., Zeng D., Li J., Zhou Y. (2010). Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J. 63: 312–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Burk D.H., Morrison W.H., III, Ye Z.H. (2002). A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14: 3101–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Gao F., Cao X., Chen M., Ye G., Wei C., Li Y. (2005). The rice dwarf virus P2 protein interacts with ent-kaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol. 139: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., et al. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]